Abstract
Background
The clinical relevance of infections with the novel human coronavirus NL63 (HCoV-NL63) has not been investigated systematically. We therefore determined its association with disease in young children with lower respiratory tract infection (LRTI).
Methods and Findings
Nine hundred forty-nine samples of nasopharyngeal secretions from children under 3 y of age with LRTIs were analysed by a quantitative HCoV-NL63-specific real-time PCR. The samples had been collected from hospitalised patients and outpatients from December 1999 to October 2001 in four different regions in Germany as part of the prospective population-based PRI.DE study and analysed for RNA from respiratory viruses. Forty-nine samples (5.2%), mainly derived from the winter season, were positive for HCoV-NL63 RNA. The viral RNA was more prevalent in samples from outpatients (7.9%) than from hospitalised patients (3.2%, p = 0.003), and co-infection with either respiratory syncytial virus or parainfluenza virus 3 was observed frequently. Samples in which only HCoV-NL63 RNA could be detected had a significantly higher viral load than samples containing additional respiratory viruses (median 2.1 × 106 versus 2.7 × 102 copies/ml, p = 0.0006). A strong association with croup was apparent: 43% of the HCoV-NL63-positive patients with high HCoV-NL63 load and absence of co-infection suffered from croup, compared to 6% in the HCoV-NL63-negative group, p < 0.0001. A significantly higher fraction (17.4%) of samples from croup patients than from non-croup patients (4.2%) contained HCoV-NL63 RNA.
Conclusion
HCoV-NL63 infections occur frequently in young children with LRTI and show a strong association with croup, suggesting a causal relationship.
The novel human coronavirus HCoV-NL63 shows a strong association with croup.
Introduction
Respiratory tract infections are among the most frequent diseases in the first years of life. Although there is a large number of viruses that are known to be involved in symptomatic respiratory tract infections, including respiratory syncytial virus (RSV), influenza virus (INF), parainfluenza virus (PIV), and human metapneumovirus, none of the known pathogens is detected in a substantial number of cases. Recently we identified a novel coronavirus in a child with bronchiolitis: human coronavirus NL63 (HCoV-NL63) [1,2]. This virus, together with SARS-CoV, is one of the new members of the Coronaviridae family [3–6].
Screening of respiratory samples in Amsterdam and Rotterdam confirmed that HCoV-NL63 is circulating among humans with respiratory disease in the Netherlands [1,7]. To investigate the prevalence of HCoV-NL63 and its involvement in respiratory diseases, we now analysed 949 samples from the Paediatric Respiratory Infection in Germany (PRI.DE) study, a prospective population-based study on lower respiratory tract infections (LRTIs) in children under 3 y of age in Germany [8,9]. The PRI.DE study represents the German population by (i) including multicentre sampling (one city each in the north, east, south, and west of the country) and by (ii) recruiting children in paediatric practices and in referral children's hospitals. We were particularly interested in the presence of HCoV-NL63 in respiratory disease for which no other viral pathogen could be detected, in order to identify clinical symptoms associated with HCoV-NL63 infection. Nasopharyngeal secretion (NPS) of the patients had already been tested for RSV, INF, and PIV, the principal viruses responsible for LRTI in young children [8]. However, RNA of these viruses could not be detected in 58% of samples for outpatients and 51% of samples for hospitalised patients. A second study that examined a subset of these negative samples for human metapneumovirus RNA showed that this virus could be detected in only 0.3% of the patients [9]. To explore the potential contribution of HCoV-NL63 to LRTI and to define clinical symptoms associated with HCoV-NL63 infection, a subset of the PRI.DE samples were analysed in this study by a HCoV-NL63-specific quantitative real-time RT-PCR.
Methods
Participants and Materials
The PRI.DE study is a population-based prospective German multicentre study. Patients from Hamburg, Bochum, Freiburg, and Dresden are been included from paediatric practices (outpatients) or hospitals (inpatients) [8]. Children were recruited from November 1999 to October 2001 and included in the study if they showed clinical signs of laryngotracheitis (croup), bronchitis, bronchiolitis, pneumonia, or apnoea, the last only in infants less than 6 mo of age. Signs and symptoms of clinical diagnoses were defined according to criteria of Denny and Clyde [10]. The definition of croup in the inclusion criteria was hoarseness of voice, barking cough, and inspiratory stridor due to laryngeal obstruction. Before the start of the study this definition had been consented to by the heads of the participating hospitals and had been communicated in writing (study protocol) as well as verbally (investigators meetings in the different cities) to the participating physicians. NPS was taken in a standardised manner by introducing a wetted catheter through the lower nasal meatus into the epipharynx and retracting it with a suction of 200 Pa. Liquid nitrogen freezers had been situated at every practice/hospital to snap freeze the specimen immediately after collection. The samples were transported to the central testing laboratory on dry ice. RNA was extracted from 220 μl of NPS using the Qiamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and eluted in 50 μl of preheated RNase free water (80 °C) for 5 min. The Hexaplex PCR test kit (Prodesse, Milwaukee, Wisconsin, United States) was used to test for the presence of genomic material of seven respiratory viruses: RSV-A, RSV-B, PIV1, PIV2, PIV3, INF A, and INF B [8,11]. The remaining RNA was stored in a −70 °C freezer.
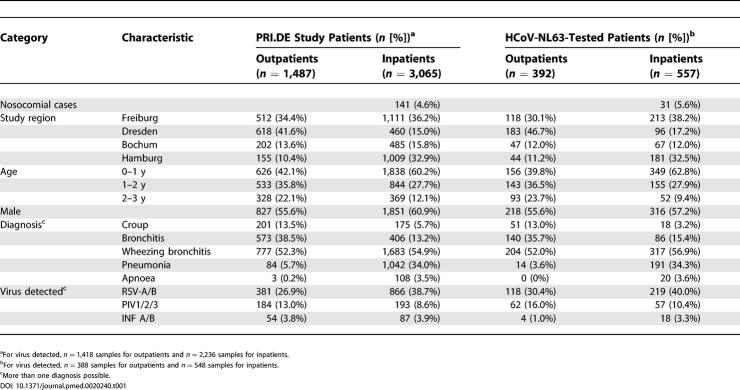
Of the total of 3,654 NPSs that were collected within the PRI.DE study, half were randomly assigned to this study. Randomisation was performed via a program generating random numbers in order to distribute the samples equally between two different laboratories. Randomisation was performed separately for each study centre. A subgroup of 949 samples, for which sufficient amounts of RNA were available, were tested for HCoV-NL63 RNA without knowledge of the result of the Hexaplex assay. For a number of important parameters the 949 samples were a proper representation of the complete PRI.DE study (shown in Table 1). Samples from all seasons were roughly equally represented (20.0% of all samples were collected in the winter months December/January/February, 32.9% of all samples were from March/April/May, 38.9% of all samples were from June/July/August, and 28.0 % of all samples were collected in September/October/November). The only period from which only a few samples were analysed was December 1999 to March 2000 (see Figure 1). The study protocol was approved by the Ethics Committee of the University Hospital Freiburg, and by the ethics committee of each participating centre. Written informed consent was obtained from the parents of all participants.
Table 1. Demographic, Clinical, and Virological Characteristics of PRI.DE Study Population and the Patients included in HCoV-NL63 Analysis.
Figure 1. Seasonal Distribution of HCoV-NL63.
Bars represent the percentage of HCoV-NL63-positive samples per month. Numbers above the columns for each month give the number of HCoV-NL63-positive samples over the number of samples tested.
Laboratory Testing
For real-time HCoV-NL63 PCR, the primers repSZ-1 (5′- GTGATGCATATGCTAATTTG) and repSZ-3 (5′- CTCTTGCAGGTATAATCCTA) [1] were used with a commercially available RT-PCR kit (QuantiTect Probe RT-PCR Kit with separate addition of SYBR Green, Qiagen, Hilden, Germany). RNA (5 μl) was reverse-transcribed in a final volume of 20 μl for 20 min at 50 °C followed by denaturation at 95 °C for 15 min. Forty-nine cycles of PCR amplification were performed in a Rotor-Gene 3000 (Corbett-Research, Mortlake, Australia) at 95 °C for 10 s, followed by an annealing step for 1 min at 55 °C. Elongation was performed at 72 °C for 30 s. Accumulation of PCR products was monitored by measuring fluorescence of intercalated SYBR Green at 72 °C and 81 °C for 15 s. The specificity of the PCR products of samples showing a relevant increase in SYBR Green fluorescence was confirmed by melting point analysis (peak at approximately 86 °C), by determining the size of the PCR product (237 bp) through agarose gel electrophoresis, and by hybridisation to an HCoV-NL63-specific internal probe (5′- AGGGTCCTCCTGGTAGTGGTAAGTCACATTGTTCC). In addition, 17 of the positive PCR products were sequenced to further confirm the identity of the PCR product. Standard precautions were taken to prevent PCR cross-contamination during specimen extraction and PCR amplification. Negative controls included in each run did not show a relevant increase of SYBR Green intercalation.
For quantitative evaluation, a PCR product of HCoV-NL63 was cloned in plasmid TOPO PCR II using the TA-cloning kit of Invitrogen (Carlsbad, California, United States). RNA was in vitro transcribed using the AmpliScribe T7 High Yield Transcription Kit (Epicentre Technologies, Madison, Wisconsin, United States) following the manufacturer's instructions. The synthesised RNA was analysed by agarose gel electrophoresis for intactness of the transcript, and the concentration was determined by the Ribo-Green method. Two independently transcribed RNA samples were adjusted to 109 copies/ml, and RT-PCR analysis of serial dilutions of both control RNAs revealed that five RNA copies per reaction could be detected. This corresponds to a detection limit of ≤225 copies/ml NPS. Both in vitro transcribed control RNAs were also used to quantify the RNA copy number in the supernatant of a HCoV-NL63-infected culture. Serial dilutions of the RNA preparation from the virus culture were used as standards in each run of the real-time RT-PCR. The inter-assay variability of the crossing points of RT-PCR samples containing between 260 and 260,000 copies was below 2%, and the inter-assay variability of their copy numbers was below 35%. Since 260 RNA copies per RT-PCR reaction corresponds to a viral load of approximately 104 RNA copies/ml NPS, 104 copies/ml was taken as the lower limit for accurate quantification.
Statistical Methods
Calculations were performed using the SAS system, version 8.2 (SAS Institute, Cary, North Carolina, United States). Proportions were compared using the χ2 test, and comparisons of viral load were performed by means of Wilcoxon's two-sample test. To determine the association of the investigated viruses with disease (croup or not), odds ratios and corresponding 95% confidence intervals were derived in separate bivariate calculations first. The odds ratios present the probability of croup in children infected with HCoV-NL63 in relation to the probability of croup in patients not infected with HCoV-NL63 within this population of patients with LRTI. Results were confirmed by performing a logistic regression with occurrence of croup as the dependent variable, and detection of RSV, PIV, or HCoV-NL63 as independent variables. Double infections were thus accounted for. INF could not be considered in the model, because no INF-positive croup patients were found.
Results
HCoV-NL63 Infections
Of the 949 PRI.DE samples tested, 392 were from outpatients at the four study sites, and the remaining 557 samples were from hospitalised patients. In total, 49 of the 949 samples (5.2%) were positive for HCoV-NL63. More HCoV-NL63 infections were found in the outpatients (31 patients, 7.9%) than in hospitalised patients (18 patients, 3.2%, p = 0.003). Various clinical diagnoses of lower respiratory tract disease were given for the HCoV-NL63-positive patients, including croup, bronchitis, bronchiolitis, and pneumonia (Table 2). The ages of the HCoV-NL63-infected children ranged from 0 to 2.9 y, with a median age of 0.7 y for the hospitalised patients and 1.5 y for the outpatients. As may be expected based on knowledge of other human coronaviruses, there is a strong seasonal distribution of HCoV-NL63, with preferential detection in the period between November and March (Figure 1). Peaks were observed in December 2000 (14% of patients positive) and February 2001 (12% of patients positive). We found no positive samples in the winter months of 1999 and 2000, but the analysed PRI.DE samples were unequally distributed and only 17 samples were analysed from the period December 1999 to March 2000.
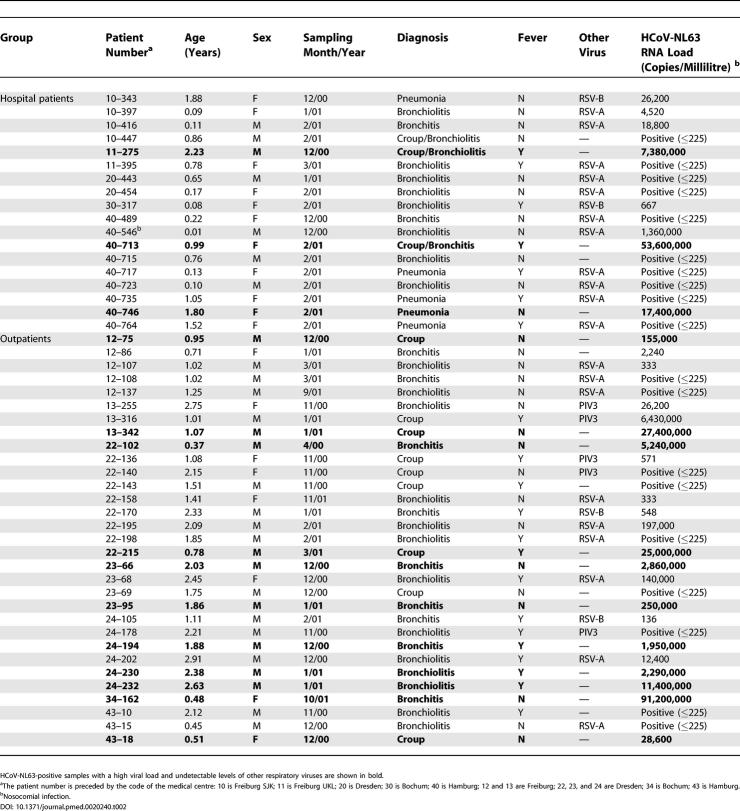
Table 2. HCoV-NL63-Positive Patients.
HCoV-NL63 Co-Infections with RSV-A and PIV3
Since the same samples had been tested previously for the presence of RSV, PIV, and INF RNA [8], the HCoV-NL63-positive samples were analysed for co-infections with these viruses. Co-infections were apparent in 29 of the 49 HCoV-NL63-positive samples: 20 patients were co-infected with RSV-A, four with RSV-B, and five with PIV3. Double infections were observed in the hospitalised patients with HCoV-NL63 (72%) but also in the outpatients (52%). HCoV-NL63 co-infection with RSV-A occurred predominantly in the hospitalised patients (61%) rather than the outpatient group (29%). In contrast, HCoV-NL63 co-infections with PIV3 were exclusively present in the outpatient group (16%). Similar trends were also observed when looking at the overall prevalence of the viruses: RSV-A occurred in 32% of hospitalised patients versus 21% of outpatients, and PIV3 occurred in 5% of hospitalised patients and 8% of outpatients [8]. The RNA load of HCoV-NL63 differed considerably from less than 225 copies/ml (but detectable) to 9 × 107 copies/ml aspirate. Interestingly, the HCoV-NL63 load was significantly higher in samples with undetectable levels of the other viral RNAs (median viral load 2.1 × 106 copies/ml) than in samples that had co-infections with RSV or PIV3 (2.7 × 102 copies/ml, p = 0.0006; Figure 2).
Figure 2. HCoV-NL63 Viral Load in Single and Double Infections.
The median viral load in the two groups is indicated (in red) together with the p-value, showing a significant difference between the singly HCoV-NL63-infected group and the group with a co-infection of either RSV or PIV3.
The HCoV-NL63 Load as a Function of Time after Infection
As mentioned above, the HCoV-NL63 load in the singly infected group (n = 20) was on average high, although there were a few samples with a load below 10,000 copies/ml (n = 6). The HCoV-NL63 load may be related to the date of sampling relative to the onset of disease. We therefore compared the viral load of these patients with the number of days after the onset of disease that the sample was taken. For the group with a high viral load (>10,000 copies/ml), the median time of sampling after onset of disease was 2 d, whereas the median interval was 6 d for the group with the lower viral load. However, the variation in number of days from onset of disease to sampling was high, and the difference between the high and low viral load group was not statistically significant (p = 0.11).
Association of HCoV-NL63 Infection with Clinical Symptoms
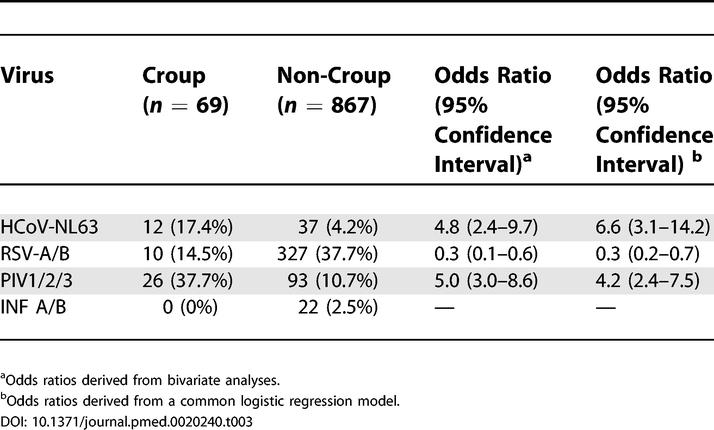
The high frequency of co-infections in HCoV-NL63-positive samples makes it difficult to define HCoV-NL63-induced symptoms. However, in 20 of the 49 HCoV-NL63-positive samples no other virus (RSV, PIV, or INF) could be detected. At least 14 of these samples also had a high viral RNA load (>10,000 copies/ml aspirate), cases that may be best suited to study the clinical symptoms associated with HCoV-NL63 infection. Six of the 14 children (43%) of this group had croup compared to only 54 of 900 HCoV-NL63-negative children (6%, p < 0.0001). A similar high frequency of croup (45%) was also observed for the 20 samples in which only HCoV-NL63 RNA was detected, independent of the viral load. The association of HCoV-NL63 with croup also held for all analysed samples: 24% in the HCoV-NL63-positive group had croup compared to 6% of the 900 HCoV-NL63-negative patients (p < 0.0001). HCoV-NL63 was detectable in 17.4% of all samples from croup patients. The chance of croup is estimated to be 6.6 times higher in HCoV-NL63-positive LRTI patients than in HCoV-NL63-negative LRTI patients (95% confidence interval 3.1–14.2), similar to that found for PIV1, PIV2, and PIV3 (Table 3).
Table 3. Respiratory Viruses Associated with Croup.
In addition to croup, we also observed bronchitis (n = 6, of which one also had croup), bronchiolitis (n = 3, of which one also had croup), and pneumonia (n = 1) in the 14 patients with a high HCoV-NL63 load. None of these diseases was significantly associated with single infection with HCoV-NL63.
Discussion
The newly discovered coronavirus HCoV-NL63 was detected in a considerable number of nasal aspirates of children under the age of 3 y with LRTIs. With an overall occurrence of 5.2%, it is the third most frequently detected pathogen in this patient group, in which RSV is detected in 31.4%, PIV3 in 9.6%, PIV1 in 2.5%, INF A or INF B in 2.4%, and PIV2 in 0.6%. These viruses were detected with similar frequency in the PRI.DE study [8], arguing against a bias during selection of the analysed samples. HCoV-NL63 is more frequently found in the outpatient group with LRTI (7.9%) than among hospitalised patients (3.2%). PIV3 follows the same pattern (8% and 5%, respectively), but the reverse pattern is observed for RSV (21% and 32%, respectively; [8]). Thus, HCoV-NL63 infection seems to be less pathogenic than RSV infection. Hospitalised HCoV-NL63-positive patients are frequently co-infected with RSV. Nevertheless, several severe disease cases that required uptake in the intensive care unit were linked exclusively to HCoV-NL63 infection in this and our previous survey [1].
Croup is a common manifestation of LRTI in children. The cause is generally assumed to be a respiratory virus and PIV1 has frequently been implicated [12]. Among the 69 samples of patients analysed with croup, croup was indeed frequently linked to PIV1 (14.5%), but PIV3 (15.9%), RSV-A (13.0%), PIV2 (7.2%), and RSV-B (1.4%) were also detected in a considerable percentage of samples. HCoV-NL63 could be detected in 17.4% of these croup patients and was therefore the most frequently identified respiratory virus for croup. Since most of the samples tested were derived from the year 2000–2001, we cannot exclude that the high percentage of HCoV-NL63-positive samples is due to a strong viral activity in this particular year, and long-term studies are needed to determine whether HCoV-NL63 infections occur in cycles peaking every two to three years as observed for other respiratory viruses.
Croup has been reported to occur mostly in boys, and it shows a peak occurrence in the second year of life and predominantly in the late fall or early winter season [12]. HCoV-NL63 infection seems to follow these trends: the ratio of boys infected to girls infected is 10:4, the median age in the outpatient group with HCoV-NL63 is 1.55 y, and this virus is circulating mainly in the winter months. Thus, it will be of interest to study the underlying biological reasons for the increased susceptibility of young boys to HCoV-NL63, as this may also explain the higher number of male patients with croup. A preferential occurrence in boys has been described for other respiratory diseases including asthma [13], and human coronavirus infections have previously been associated with exacerbations of asthma [14]. It will therefore also be of interest to study this link for HCoV-NL63.
Results from the PRI.DE study indicated that co-infections with multiple respiratory viruses are rare (1.5% of all samples tested; [8]). In contrast, we report detection of more than one viral RNA in 59% of HCoV-NL63-positive samples, which represents 3% of the 949 samples tested. Most co-infections are with RSV-A, probably because of the high percentage of RSV-A infections and an overlap in seasonality. In the winter season the percentage of RSV-positive samples per month can exceed 50% and even the overall difference in percentage of RSV-positive samples among HCoV-NL63-negative and -positive samples was not significant (Fisher's exact test, two-tailed). This contrasts with results obtained for PIV1- and PIV3-positive patients, for whom co-infection with RSV is rarely observed (data not shown). Therefore, infection with RSV seems to reduce infection by PIV1 and PIV3 (or vice versa), but not infection by HCoV-NL63 (or vice versa).
Quantitative PCR analysis for HCoV-NL63 revealed a significantly lower HCoV-NL63 viral load in patients co-infected with RSV or PIV3 than in patients infected with HCoV-NL63 alone. This interference effect might be explained by direct competition for the same target cell in the respiratory organs or an elevated activation status of innate immune responses. Prolonged persistence of HCoV-NL63 at low levels is another explanation. The HCoV-NL63 load was found to vary with respect to the time of sampling relative to the time of disease onset, with the higher viral loads in early samples (day 1 or 2 after disease onset). This most likely reflects viral clearance by the immune system. This timing effect may also relate to the differences in HCoV-NL63 load in single versus double infections. For instance, an initial HCoV-NL63 infection may set the stage for a subsequent RSV infection. At the time that this second virus is causing symptoms and NPS samples are collected, the HCoV-NL63 infection may already be under control by the immune system.
The initial study on HCoV-NL63 in patients from Amsterdam with respiratory tract illness listed 20% co-infections, which is significantly lower that the 59% that we report in the present study. Differences in handling of the clinical samples may in part explain this. NPS samples were taken in a standardised manner and immediately frozen in liquid nitrogen in the PRI.DE study. Nucleic acid extraction was performed immediately after defrosting. Such optimal sampling and subsequent handling may allow the detection of lower HCoV-NL63 loads (as observed mainly in co-infections). In contrast, the Amsterdam samples were frozen and defrosted repeatedly before nucleic acid isolation, which may have rendered the low load samples negative. This may also explain the higher proportion of HCoV-NL63 positives measured in the current study (5.2% versus 1.4%). Alternatively, HCoV-NL63 might cycle with periods of increased activity like other respiratory viruses and peak every 2–3 y, and the 5.2% incidence in 2000–2001 may represent such a peak year.
In conclusion, our study revealed that HCoV-NL63 belongs to the group of most frequently detected viruses in children under 3 y of age with LRTI and that this virus is strongly associated with croup. Recent articles on HCoV-NL63 show that this virus is spread worldwide [15–19]. The virus has been found in different parts of the world: Australia, Canada, Japan, Belgium, and the United States. Thus, HCoV-NL63 is a human respiratory virus that should be added to the list of pathogens that can cause numerous LRTIs in young children.
Patient Summary
Background
Lower respiratory infections (those that are not just coughs and colds) are very frequent in young children and can be serious. Most infections are caused by viruses, the best known of which are respiratory syncytial virus (RSV) and influenza virus (INF).
Why Was This Study Done?
Recently, a new virus known as HCoV-NL63 has been discovered, which is a human coronavirus (SARS is another coronavirus). Previous work has shown that this virus was present in a child with bronchiolitis. The researchers wanted to know whether the virus was associated with other respiratory diseases.
What Did the Researchers Do and Find?
They looked at 949 samples taken from children with lower respiratory infections in Germany between 1999 and 2001. They found that the virus HCoV-NL63 was more common in outpatients than inpatients, and that more than 40% of children infected with only this virus had croup compared with around 6% of children who did not carry the virus.
What Do These Findings Mean?
Although the numbers of all the children infected with this new virus was not high, these results do suggest that this virus is a cause of croup in some children and may be the third most common cause of lower respiratory infections in this group. More work needs to be done to see whether this is true in other years and other countries.
Where Can I Get More Information Online?
MedlinePlus has Web pages on croup:
http://www.nlm.nih.gov/medlineplus/ency/article/000959.htm
The Mayo Clinic has information about croup for parents:
Acknowledgments
Financial support was received from Wyeth Pharma, Münster, Germany. The Center for Clinical Trials receives funding from the German Federal Ministry of Education and Research (BMBF).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- HCoV-NL63
human coronavirus NL63
- INF
influenza virus
- LRTI
lower respiratory tract infection
- NPS
nasopharyngeal secretion
- PIV
parainfluenza virus
- PRI.DE
Paediatric Respiratory Infection in Germany
- RSV
respiratory syncytial virus
Footnotes
Citation: van der Hoek L, Sure K, Ihorst G, Stang A, Pyrc K, et al. (2005) Croup is associated with the novel coronavirus NL63. PLoS Med 2(8): e240.
References
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K, Jebbink MF, Berkhout B, van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Osterhaus AD, Fouchier RA, Kuiken T. The aetiology of SARS: Koch's postulates fulfilled. Philos Trans R Soc Lond B Biol Sci. 2004;359:1081–1082. doi: 10.1098/rstb.2004.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster J, Ihorst G, Rieger CH, Stephan V, Frank HD. Prospective population-based study of viral lower respiratory tract infections in children under 3 years of age (the PRI.DE study) Eur J Pediatr. 2004;163:709–716. doi: 10.1007/s00431-004-1523-9. [DOI] [PubMed] [Google Scholar]
- Konig B, Konig W, Arnold R, Werchau H, Ihorst G. Prospective study of human metapneumovirus infection in children less than 3 years of age. J Clin Microbiol. 2004;42:4632–4635. doi: 10.1128/JCM.42.10.4632-4635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny FW, Clyde WA. Acute lower respiratory tract infections in nonhospitalized children. J Pediatr. 1986;108:635–646. doi: 10.1016/s0022-3476(86)81034-4. [DOI] [PubMed] [Google Scholar]
- Kehl SC, Henrickson KJ, Hua W, Fan J. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J Clin Microbiol. 2001;39:1696–1701. doi: 10.1128/JCM.39.5.1696-1701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny FW, Murphy TF, Clyde WA, Collier AM, Henderson FW. Croup: An 11-year study in a pediatric practice. Pediatrics. 1983;71:871–876. [PubMed] [Google Scholar]
- Ellis EF. Asthma in childhood. J Allergy Clin Immunol. 1983;72:526–539. doi: 10.1016/0091-6749(83)90479-7. [DOI] [PubMed] [Google Scholar]
- McIntosh K, Ellis EF, Hoffman LS, Lybass TG, Eller JJ. The association of viral and bacterial respiratory infections with exacerbations of wheezing in young asthmatic children. J Pediatr. 1973;82:578–590. doi: 10.1016/S0022-3476(73)80582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden KE, Nissen MD, Sloots TP, Mackay IM. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005;75:455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T, Endo R, Ma X, Ishiguro N, Kikuta H. Detection of human coronavirus NL63 in young children with bronchiolitis. J Med Virol. 2005;75:463–465. doi: 10.1002/jmv.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, Anderson K, Hart L, Van Caeseele P, Brandt K. Human coronavirus NL63 infection in Canada. J Infect Dis. 2005;191:503–506. doi: 10.1086/426869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis. 2005;191:492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes E, Vijgen L, Keyaerts E, Zlateva K, Li S. A novel pancoronavirus RT-PCR assay: Frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]