Abstract
CPS (capsular polysaccharide) is a major virulence factor in Streptococcus pneumoniae. Biosynthesis of CPS RU (repeat unit) proceeds by sequential transfer of sugar residues from the appropriate sugar donor to an activated lipid carrier by committed GTs (glycosyltransferases). While the nucleotide sequence of many cps loci is already known, the real substrate specificity of the hypothetical GTs, as well as the sequence of sugar addition is unclear. In the present paper, we report the biochemical characterization of one α-galactosyltransferase, WciS (Cap8H), a member of GT family 4. This enzyme is implicated in the tetrasaccharide RU biosynthetic pathway of Strep. pneumoniae CPS 8 ([→4)-α-D-Glcp-(1→4)-α-D-Galp-(1→4)-β-D-GlcAp-(1→4)-β-D-Glcp-(1→]n). Expression of WciS–His6 in Escherichia coli BL21 (DE3) strains or BL21 (DE3)/ΔgalU strain resulted in synthesis of a 39 kDa membrane-associated protein identified by N-terminal sequencing and recognized by anti-His6-tag antibody. This protein was capable of adding a galactose residue cellobiuronic acid [β-D-GlcAp-(1→4)-D-Glcp]-pyrophosphate-polyprenol from UDP-Gal. The newly added galactose residue is removed by α-galactosidase, indicating that WciS is a retaining GT. Our results suggest that WciS catalyses the addition of the third sugar residue of the CPS 8 RU. The recombinant WciS–His6 was solubilized and purified as a soluble multimer, opening the way for structural studies.
Keywords: capsular polysaccharide, galactosyltransferase, glycosyltransferase, Streptococcus pneumoniae, virulence factor, WciS
Abbreviations: CPS, capsular polysaccharide; GT, glycosyltransferase; IMAC, immobilized metal-affinity chromatography; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; LPS, lipopolysaccharide; ORF, open reading frame; PPL, pyrophosphate-polyprenol; RU, repeating unit; SEC, size-exclusion chromatography
INTRODUCTION
Streptococcus pneumoniae is an important bacterial pathogen causing invasive diseases such as pneumonia, otitis media and meningitis. Control of this Gram-positive pathogen is based on two completely different ways: the curative action of β-lactam agents, which is becoming less efficient because of the rise in antibiotic multi-resistant strains, and vaccination with a multivalent vaccine based on CPS (capsular polysaccharide) vaccine.
The extracellular CPS of Strep. pneumoniae is the major virulence factor, because it prevents phagocytosis by the host immune system [1]. The CPS also induces T-cell-independent antibodies and this immunoprotection is serotype-specific [1], providing the basis for vaccine development. The current pneumococcal vaccine consists of purified CPS from 23 serotypes out of the 90 different known CPSs [2,3]. Although this vaccine protects healthy adults against pneumococcal infections, its efficiency decreases in high-risk human groups such as young children, elderly people or immunocompromised patients [1], and certain capsular types are poorly immunogenic [4]. A better understanding of CPS biosynthesis pathway may help to elaborate new anti-infectious agents, or even provide biotechnological tools for the synthesis of more efficient conjugated vaccines [5–7].
The first committed step for CPS biosynthesis is the activation of monosaccharides by their conversion into nucleotide derivatives. With the exception of two serotypes (3 and 37) that make use of a processive biosynthetic system [8,9], it has been proposed that a membrane-associated GT (glycosyltransferase) complex catalyses the assembly of the lipid-linked RU (repeating unit) by successive transfer of monosaccharides. This first step is followed by polymerization of the RU, and subsequent export and attachment of the complete CPS to the cell surface [10]. Despite the large number of cps loci and chemical structures of CPS available [11], only the biosynthetic pathway of Cps14 has been reported [12–14].
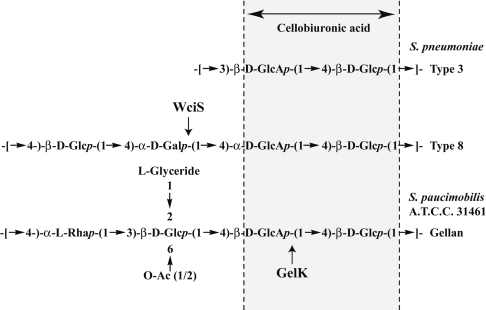
We have studied the serotype 8 CPS, which is included in the 23 CPS-valent vaccine [2]. The chemical structure of the RU of this serotype [15] includes a cellobiuronic acid [β-D-GlcAp-(1→4)-D-Glcp] that is also present in pneumococcal serotype 3 [8] and other bacterial exopolysaccharides, such as gellan (Figure 1) [16,17]. The cps8 locus was initially identified by Muñoz et al. [18] in a DNA fragment between the dexB and aliA loci [19], and was capable of transforming the non-capsulated Strep. pneumoniae M24 to a capsulated serotype 8 transformant [18]. Analysis of the DNA sequence of the serotype 8 CPS gene cluster [11,18] revealed the presence of 12 hypothetical ORFs (open reading frames). In the present paper, we adopt the nomenclature of Jiang et al. [11] who later renamed the genes according to the Bacterial Polysaccharide Gene Database (BPGD) nomenclature system [20] (see http://www.microbio.usyd.edu.au/BPGD/default.htm). The biosynthesis of the cellobiuronic moiety differs with the species. Strep. pneumoniae serotype 3 CPS uses a single processive GT [9] (Figure 1). In Sphingomonas paucimobilis or Rhizobium leguminosarum, the synthesis of cellobiouronic-glycolipid requires two enzymatic activities [21–23], namely a phosphoglucosyltransferase (SpsB, PssA) and a glucuronosyltransferase (GelK, SpsK, PssDE). In the case of CPS 8, which is similar to that of Sphing. paucimobilis or R. leguminosarum, five ORFs deduced from the cps8 locus sequences code for hypothetical proteins that share sequence similarity with enzymes involved in RU assembly of Gram-negative bacterial CPS [11,18]. These proteins are WchA (Cap8E, a glucose-1-phosphate transferase [24]), WciQR {Cap8FG, a putative inverting heterodimeric GT from the Carbohydrate Active enZYme's (CAZy) family 1 (http://afmb.cnrs-mrs.fr/CAZY/index.html) [25]}, WciS and WciT (Cap8H and Cap8J, two putative retaining GTs classified in CAZy's family 4 and 32 respectively). Since there are only two sugars with α-linkages in the serotype 8 CPS RU [15], the putative GTs, WciS and WciT, are predicted to catalyse the addition of the α-D-Glcp or the α-D-Galp moieties.
Figure 1. Structure of the RU of the serotype 8 pneumococcal CPS.
The corresponding hypothetical RU of serotype 3 CPS and gellan from Sphing. paucimobilis A.T.C.C. 31461 are shown for comparison. The glucuronosyltransferase GelK using D-Glcp-PPL as the expected acceptor is indicated by an arrow. Our results suggest that the α-galactosyl-transferase WciS catalyses the transfer (see the arrow) of a galactose residue to the cellobiuronic acid moiety common to the pneumococcal serotypes and gellan exopolysaccharides.
In the present paper, we report the molecular and biochemical characterization of one of the GTs, WciS (Cap8H). Our results show that the purified recombinant GT (WciS–His6) is an α-galactosyltransferase. This work also suggests that WciS adds the third sugar residue of the RU deduced from the serotype 8 CPS structure.
MATERIALS AND METHODS
Materials, culture conditions and bacterial strains
Escherichia coli strains BL21 (DE3) from Novagen were grown in LB (Luria–Bertani) broth at 37 °C with orbital agitation. The E. coli BL21 (DE3)/ΔgalU strain was constructed by generalized P1 transduction [26] using E. coli MC4100 galU95::Tn10 (TetR) as the donor [27]. This strain was kindly provided by Dr R. Hengge-Aronis and Dr E. Klauck (Institüt für Biologie, Pflanzenphysiologie/Mikrobiologie, Berlin, Germany). Sphing. paucimobilis A.T.C.C. 31461 (American Type Culture Collection), kindly provided by Professor I. Correia (Instituto Superior Técnico, Lisboa, Portugal), was grown at 30 °C in S medium as described previously [28]. When required, antibiotics were used at the following concentrations: ampicillin (0.1 mg/l), chloramphenicol (0.03 mg/l) and kanamycin (0.1 mg/l). Strep. pneumoniae serotype 8 chromosomal DNA was from the A.T.C.C. strain. The pET29b and pET23a+ expression vectors were from Novagen. The pET29b expression vector carrying the gelK gene (pET-gelK) was a gift from Professor I. Correia. Oligonucleotide primers for PCR were purchased from Eurogentec (Seraing, Belgium). UDP-[U-14C]Gal (267 mCi/mmol) and UDP-[U-14C]-Glc (303 mCi/mmol) were from Amersham Biosciences and UDP-[U-14C]GlcA (313 mCi/mmol) was from ICI Biochemicals.
Cloning of the wciS gene
The wciS gene was amplified from Strep. pneumoniae type 8 chromosomal DNA by PCR using the Expand High Fidelity PCR system (Roche) with the synthetic oligonucleotides wciS-for (5′-GGAAATTCATATGACAAAAGTTTTGCAAATTGG-3′) as a 5′ primer (the engineered NdeI site is underlined) and wciS-rev (5′-TAGCTCGAGTTTTTTCGTTTCTAATAGAGTATT-3′) as a 3′ primer (the engineered XhoI site is underlined). The PCR was optimized and carried out in 100 μl of a mixture containing 100 ng of Strep. pneumoniae type 8 chromosomal DNA, 200 μM each dNTP (Promega), 1 μM each primer and 5 units Taq DNA polymerase according to the recommendations of the manufacturer (Roche). The predicted 1064 bp PCR product obtained was cloned into the NdeI/XhoI-digested pET23a+ expression vector (Novagen) and sequenced (Genome Express, Meylan, France). The new plasmid was called pET-wciS.
Expression and purification of recombinant protein WciS–His6
E. coli BL21(DE3) (Novagen) and BL21 (DE3)/ΔgalU cells harbouring the plasmid pET-wciS or the cloning vector pET23a+ were grown at 37 °C in 500 ml of LB. At a D600 of 0.6, protein expression was induced by addition of IPTG (isopropyl β-D-thiogalactoside) to 0.4 mM. The bacterial culture was incubated at 37 °C for an additional 2 h. Cells were harvested by centrifugation (5000 g), washed once with 70 mM Tris/HCl, pH 8.2, resuspended in 70 mM Tris/HCl, pH 8.2, 300 mM NaCl, 10 mM EDTA and 1% Tween 20 at 40 D600 equivalents/ml. For subcellular localization of the recombinant protein in E. coli, Tween 20 in the extraction buffer was omitted. The bacterial cells were broken by two passages through a French press (SLM-Amico Instruments, Thermo Spectronic, Rochester, NY, U.S.A.) at 80 MPa and submitted to continuous stirring for 30 min at 4 °C. This last step was performed only with extraction buffer containing detergent. Cell debris and inclusion bodies were then pelletted by centrifugation at 12000 g (12K) for 10 min at 4 °C (Sigma 6K15, rotor Nr 12166) and the supernatant was submitted to a further centrifugation at 50000 g (50K) (Beckman Avanti J-30I, rotor JA-30.50) for 1 h at 4 °C. The recovered insoluble and soluble fractions were stored at −20 °C or were used directly for the GT assay.
For WciS–His6 purification, the extraction buffer was supplemented with 15 mM imidazole. The 50K supernatant fractionated through IMAC (immobilized metal-affinity chromatography) were prepared in 70 mM Tris/HCl, pH 8.2, 300 mM NaCl and 1% Tween 20. The sample was loaded on to a nickel-nitrilotriacetic acid Superflow agarose column (Amersham Biosciences) pre-equilibrated in the same buffer. After a washing step with 70 mM Tris/HCl, pH 8.2, 300 mM NaCl, 0.05% Tween 20 and 50 mM imidazole, the elution was carried out with an imidazole step gradient (50–500 mM) in the same buffer. The fractions containing the recombinant protein were pooled, desalted (Sephadex G-25M; Amersham Biosciences), concentrated and subjected to SEC (size-exclusion chromatography).
SEC
Samples of 500 μl containing 200 μg of IMAC-purified recombinant protein were loaded on to a Superdex 200 HR 10/30 column (Amersham Biosciences) equilibrated with 70 mM Tris/HCl, pH 8.2, 0.2 mM EDTA, 500 mM NaCl, 10% glycerol and 0.05% Tween 20. The chromatographic run was carried out in the same buffer at a flow rate of 0.4 ml/min at 4 °C using the FPLC BioLogic (Bio-Rad). Fractions of 2.5 ml were collected. Fractions containing WciS–His6 were pooled and concentrated by centrifugation at 5000 g using a Centricon YM-10 filter (Amicon Bioseparations) according to the manufacturer's recommendations. Protein samples (10 μl) containing 0.25–12 mg/ml of WciS–His6 were analysed directly by native or denaturing PAGE, or were used for the GT assay. The column was calibrated by running a set of gel-filtration markers (Amersham Biosciences).
Protein quantification, SDS/PAGE, Western blot and amino acid sequencing
The protein concentrations were determined using a Bradford protein assay kit (Bio-Rad) and BSA as a standard. The purification procedure of WciS–His6 was monitored by SDS/PAGE [12.5% acrylamide/bisacrylamide (29:1) staining with Coomassie Brilliant Blue R250]. Pre-stained protein standards (Amersham Biosciences) were used for molecular mass estimation. Proteins were electrotransferred from SDS/PAGE gels on to PVDF membranes (Amersham Biosciences) as described previously [29]. Immunoblot analysis was performed with the Promega His6-tag Western-blotting kit, using alkaline phosphatase His6–protein conjugate, according to the manufacturer's instructions. The blots were developed with NBT (Nitro Blue Tetrazolium)/BCIP (5-bromo-4-chloroindol-3-yl phosphate) (Sigma–Aldrich) systems. N-terminal sequencing was performed with 20 μg of purified recombinant protein as described in [29].
Electrophoresis of proteins under non-denaturing conditions
Protein samples containing WciS–His6 were analysed in native PAGE using a gradient gel [4–20% acrylamide/bisacrylamide (29:1)] in TBE buffer (90 mM Tris/borate, pH 8.4, and 2 mM EDTA). Electrophoresis was at 2 mA/gel using a Bio-Rad Mini-Protean II system at 4 °C, and the gel was stained with Coomassie Brilliant Blue R250. For localization of radiolabelled compounds, the gels were incubated directly for 30 min in ethanol/ethanoic (acetic) acid/water (5:1:4, by vol.) mixture, dried under vacuum and analysed with a phosphoimager (Fujifilm FLA-8000) after 72 h of exposure. The C-terminal His6-tag epitope fusion was detected in the native gels as described above.
Preparation of Sphing. paucimobilis and E. coli glycolipid acceptors
Cells of Sphing. paucimobilis were prepared as described previously [22] from 100 ml cultures of E. coli BL21 (DE3) cells carrying either pET-gelK, pET23a+ or pET29b. EDTA-permeabilized bacterial cells were obtained using the method described by Tomalsky et al. [30], modified slightly as described in [22]. Radiolabelled and non-radiolabelled glycolipid acceptors were synthesized in vitro by incubating 150 μl of permeabilized cells in the presence of the relevant sugar donor (2 mM UDP-Glc, 2 mM UDP-GlcA, 0.25 μCi of UDP-[U-14C]Glc or 0.25 μCi of UDP-[U-14C]GlcA), as described previously [22]. The glycolipids were then recovered as described previously [31]. Permeabilized cell suspensions of BL21 (DE3) carrying pET23a+ (150 μl) were used directly to prepare endogenous glycolipid acceptors. Extracts were finally dried in a SpeedVac to a final volume of 10 μl and were stored at −80 °C until use.
Galactosyltransferase assay
Galactosyltransferase assays were performed in 100 μl volumes containing 10 μl of the glycolipid acceptor, 70 mM Tris/HCl, pH 8.2, 1% Triton X-100, 12.5 mM MgCl2 and 0.10 μCi of UDP-[U-14C]Gal (267 mCi/mmol) (or 2 mM UDP-Gal), the reaction being started by addition of WciS–His6 (approx. 30–300 μg of total protein). Assays were performed in triplicate. After 1 h at 37 °C, the reaction was stopped by the addition of 100 μl of water and 200 μl of chloroform/methanol (1:1, v/v) or analysed directly in native PAGE. Glycolipids were isolated (see below), and the radioactivity was determined by liquid-scintillation counting. The oligosaccharide moiety was released as described previously [22,31] and submitted to analytical TLC [31]. Glucose, galactose, glucuronic acid and oligomaltose (dimer to pentamer) were used as standards. Unlabelled sugar standards on the TLC plate were revealed by charring (100 °C for 10 min) after treatment with 5% H2SO4 in ethanol. The remaining portion of the plate containing the radiolabelled oligosaccharides was analysed with a phosphoimager Fujifilm FLA-8000.
Galactosidase digestion
To determine the linkage configuration (α or β) of the galactose residue transferred in vitro, the oligosaccharides co-migrating with WciS–His6 in the gel or released directly after mild acid hydrolysis were incubated overnight either in 10 mM Mops, pH 6.5, containing 0.6 units of α-galactosidase from green coffee beans (G8507, Sigma–Aldrich), or in 100 mM Tris/HCl, pH 7.3, containing 11 units of β-galactosidase (G3153, Sigma–Aldrich) from E. coli at 25 and 37 °C respectively. Controls were performed with oligosaccharides incubated in the respective buffers without glycosylhydrolases.
RESULTS
WciS–His6 is expressed mainly as a membrane-associated protein in E. coli
The Strep. pneumoniae wciS gene was cloned into the expression vector pET23a+ by PCR amplification based on available nucleotide sequences [10,18]. The resulting recombinant protein, WciS–His6, is predicted to have 354 amino acids with a C-terminal His6-tag. The production of WciS–His6 by E. coli BL21 (DE3) harbouring pET-wciS was confirmed by SDS/PAGE and His6-tag Western blotting. A 39 kDa protein was detected after 1 h of IPTG-induction (results not shown). The amount of protein increased for up to 3 h after induction and remained constant after overnight culture (results not shown). In all subsequent experiments, we prepared protein samples from cultures following 2 h of induction. The 39 kDa protein was identified as WciS–His6 by Western blotting and immunodetection using an antibody against the His6-tag epitope (see below) and N-terminal amino acid sequencing.
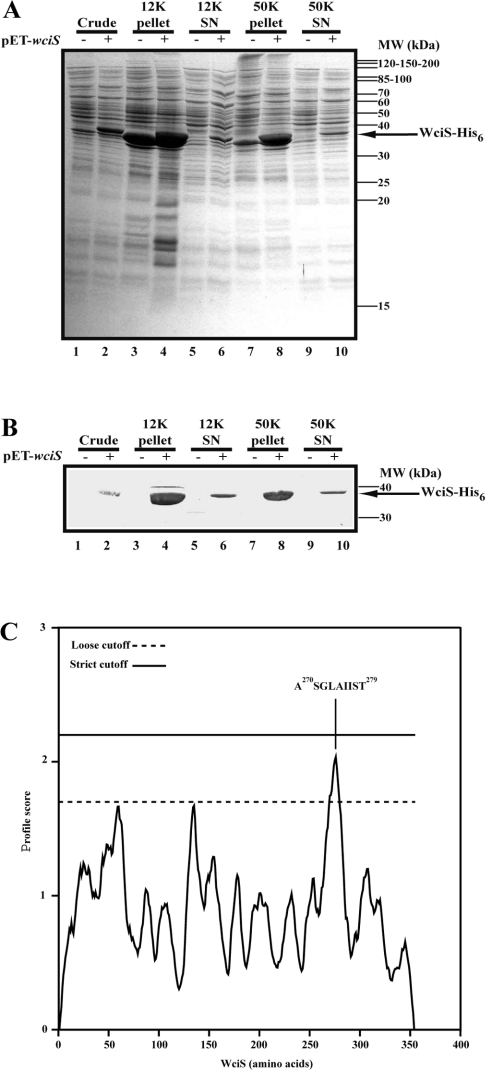
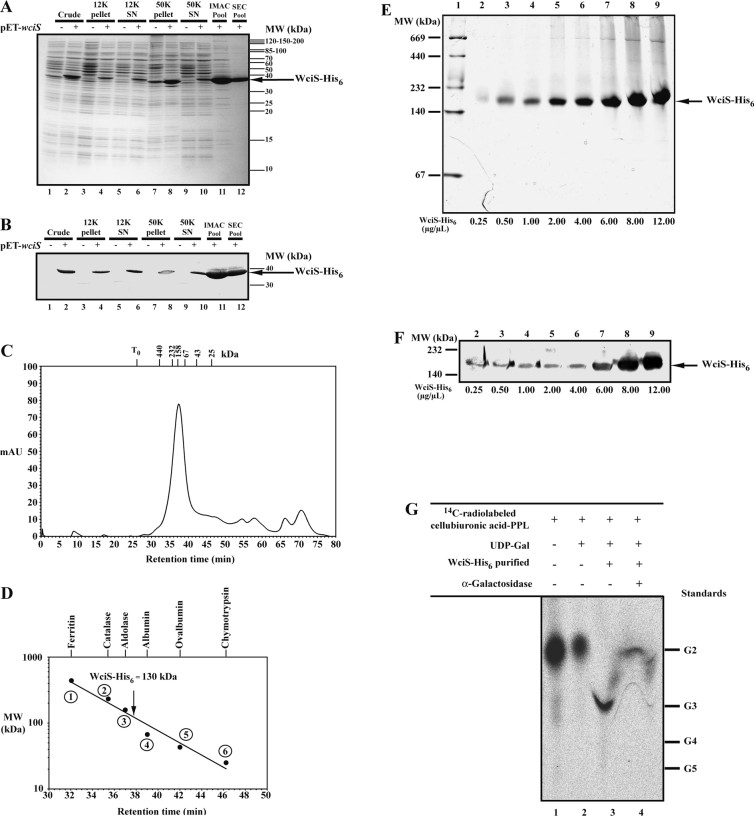
To investigate the subcellular localization of WciS–His6 in E. coli, crude French press extracts were prepared and submitted to differential centrifugation using the strain harbouring pET23a+ without the wciS insert as a control. The protein content of each fraction was analysed by SDS/PAGE (Figure 2A) and Western blot (Figure 2B). WciS–His6 polypeptide was the only significant protein expressed and recognized by anti-His6-tag antibody. A significant amount of the recombinant protein was recovered in the 12K pellet, suggesting that a portion of WciS–His6 formed inclusion bodies (Figures 2A and 2B, lanes 4). Recovery of WciS–His6 in the 50K pellet was almost quantitative (Figures 2A and 2B, lanes 8). Resuspension of the 50K pellet and centrifugation at 50000 g did not result in release of WciS–His6 (results not shown). Furthermore, when membranes were purified by high-speed centrifugation on a 30% (w/v) glycerol cushion, the recombinant protein remained associated to the membrane (L. Pelosi, unpublished work). These data suggest that the recombinant WciS is associated with E. coli membrane (Figures 2A and 2B, lanes 8). The presence of a hypothetical transmembrane helix was not predicted by only the DAS (dense alignment surface) method [32] at strict cut-off, a peptide (A270SGLAIIST279) within the loose cut-off was detected (Figure 2C). Thus it is possible that the association of WciS–His6 with the membrane is not related to the presence of a clear transmembrane domain, as it is also known for MurG [33] or AceA [34]. Interestingly, a fraction of recombinant WciS–His6 was also present in the 50K supernatant (Figures 2A and 2B, lanes 10), even after 3 h of centrifugation (results not shown). However, the ‘soluble’ protein precipitated after 12 h at 4 °C. The 12K and 50K supernatants were frozen at −20 °C, to preclude precipitation.
Figure 2. Membrane localization of WciS–His6.
(A) and (B) E. coli BL21 (DE3) strain harbouring pET23a+ with (+) or without (−) the insert was grown, induced for 2 h and disrupted. The crude fractions obtained (lanes 1 and 2) were subjected to sequential 12000 g (12K; lanes 3–6) and 50000 g (50K; lanes 7–10) centrifugation. Electrophoretic profiles of the pellets and the corresponding supernatants (SN) are presented in lanes 3, 4, 7 and 8, and lanes 5, 6, 9 and 10 respectively. (A) Analysis of each aliquot by Coomassie Blue-staining SDS/12.5% PAGE. Approx. 20 μg of proteins were loaded in each lane. Pre-stained protein standards were used for molecular mass (MW) estimation. (B) His6-tag Western blotting with alkaline phosphatase His6–protein conjugate of each aliquot. The recombinant polypeptide, WciS–His6, expressed in the present is indicated by an arrow. (C) Transmembrane segment prediction profile obtained from WciS sequence using the DAS (dense alignment surface) method. The sequence of the main peptide which could interact with the membrane is indicated.
WciS–His6 displays α-galactosyltransferase activity in vitro on to an endogenous acceptor
In order to understand the function of WciS–His6 in the biosynthetic pathway of the CPS 8 RU and to rule out the possibility of the presence of a hypothetical endogenous acceptor in E. coli, we incubated EDTA-permeabilized BL21 (DE3) cells expressing WciS–His6 with sugar precursors (UDP-[U-14C]Gal, UDP-[U-14C]Glc or UDP-[U-14C]GlcA) under conditions described previously for other GTs [22,31,34]. The radioactivity incorporated into the glycolipid fraction was measured as described in the Materials and methods section. We did not detect any significant incorporation into the glycolipid fraction when E. coli BL21 (DE3) cells harbouring pET23a+ extracts were incubated with each radiolabelled UDP-sugar, showing the absence of active endogenous GTs in the assay (Table 1). Incubation in the absence of added acceptor resulted in the incorporation of 458 pmol/h of [U-14C]Gal moiety into the organic phase (Table 1). This result indicates the presence of an endogenous acceptor. In previous functional characterization of recombinant GT [22,31,34], we have not detected any endogenous acceptor. Neither [U-14C]Glc nor [U-14C]GlcA residue was incorporated into the glycolipid fraction in a WciS–His6-dependent way (Table 1), suggesting that WciS is a galactosyltransferase. This galactosyltransferase activity was also found both in the 12K and 50K supernatants (results not shown), suggesting that the endogenous acceptor co-purifies with the recombinant GT during differential centrifugation.
Table 1. Incorporation of [U-14C]sugar mediated by WciS–His6.
Activity corresponding to 150 μl of EDTA-permeabilized cells of E. coli BL21 (DE3)/pET-wciS or BL21 (DE3)/pET23a+ was expressed as pmol of [U-14C]sugar (Glc, GlcA or Gal) incorporated per hour from corresponding UDP-[U-14C]sugar onto the E. coli BL21 (DE3) endogenous glycolipids.
| Sugar donor | ||||
|---|---|---|---|---|
| Plasmid expressed in BL21 (DE3) | UDP-[U-14C]Glc | UDP-[U-14C]GlcA | UDP-[U-14C]Gal | [14C]sugar incorporated into the E. coli glycolipid fraction (pmol/h) |
| pET23a+ | + | − | − | <3 |
| pET-wciS | + | − | − | <3 |
| pET23a+ | − | + | − | <3 |
| pET-wciS | − | + | − | <3 |
| pET23a+ | − | − | + | <3 |
| pET-wciS | − | − | + | 458±76 |
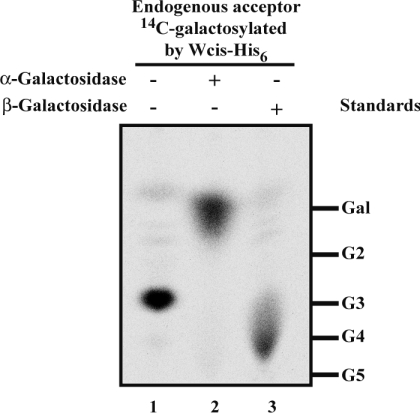
The 14C-galactosylated moiety of the E. coli endogenous acceptor was released from the organic phase by mild acid hydrolysis followed by treatment with bovine alkaline phosphatase, suggesting a pyrophosphate linkage to the lipid moiety (results not shown). Upon delipidation and dephosphorylation, a significant amount of the radiolabelled galactoside was bound to the ion-exchange resin (results not shown), suggesting the presence of a charged sugar. The size of the [U-14C]Gal residue-containing moiety was assessed by TLC (Figure 3, lane 1). A major radioactive spot was detected by phosphoimaging at the same position as the maltotriose marker. The glycosyl moiety of the endogenous acceptor was therefore probably a disaccharide, compatible with the putative disaccharide moiety of the native acceptor of WciS: cellobiuronic acid-PPL (pyrophosphate-polyprenol). The linkage of the added [U-14C]Gal was assessed by digestion with α- and β-galactosidases. The α-galactosidase treatment resulted in the release of Gal (Figure 3, lane 2). Thus WciS–His6 is an α-galactosyltransferase. The streaky migration of the spot on the silica gel corresponding to radiolabelled trisaccharide treated by the β-galactosidase (Figure 3, lane 3) was observed only with this hydrolase, and probably due to the commercial formulation of this enzyme.
Figure 3. Characterization of the 14C-galactosylated endogenous acceptor in E. coli BL21 (DE3).
Phosphoimaging of 14C-galactosylated oligosaccharides moiety. EDTA-permeabilized E. coli BL21 (DE3) cells expressing WciS–His6 were incubated in the presence of UDP-[U-14C]Gal. The radiolabelled acceptor was incubated overnight without galactosylhydrolases (lane 1), in the presence of α-galactosidase (lane 2) or β-galactosidase (lane 3). Galactose (Gal) and oligomaltoses (G2, maltobiose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose) were used as standards.
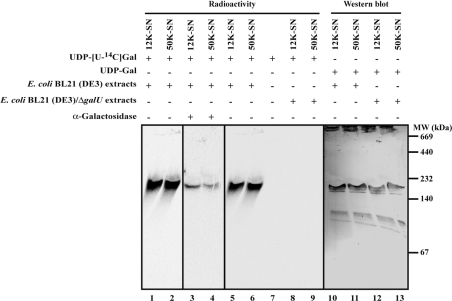
The interactions between WciS–His6 and its glycolipid reaction product were investigated using native PAGE analysis of the 12K and 50K supernatants containing WciS–His6. Incubation in the presence of radiolabelled UDP-Gal resulted in the labelling of a protein with an apparent molecular mass of approx. 180 kDa (Figure 4, lanes 1–4), that co-migrated with WciS–His6 as revealed by Western blotting (Figure 4, lanes 10 and 11). Incubation of this gel in buffer containing α-galactosidase (Figure 4, lanes 3 and 4) resulted in a significant reduction of the radioactive signals, indicating that the galactosylated glycolipid co-migrates with native WciS–His6. These results suggest that the recombinant WciS–His6 behaves as a multimer, and that the reaction product is associated with this complex.
Figure 4. Detection of 14C-galactosylated endogenous acceptor co-migrating with WciS–His6.
Recombinant proteins (12K and 50K supernatants) were prepared from E. coli BL21 (DE3) expressing WciS–His6 (lanes 1–6, 10 and 11) or E. coli BL21 (DE3)/ΔgalU expressing WciS–His6 (lanes 8, 9, 12 and 13) and incubated in the presence of radiolabelled (lanes 1–6, 8 and 9) or not (lanes 10–13) UDP-Gal. Native gradient PAGE (4–20% acrylamide/bisacrylamide in Tris/borate/EDTA) was performed for each sample. The migration of the 14C-galactosylated acceptor and WciS–His6 expressed in the two strains was monitored in the gel by phosphoimaging and His6-tag Western blotting respectively. A control was realized in the same assay conditions with only UDP-[U-14C]Gal added to the reaction mixture (lane 7). Identity of radiolabelled product (lanes 1 and 2) was confirmed by α-galactosidase gel treatment (lanes 3 and 4). SN, supernatant; MW, molecular-mass markers (sizes given in kDa).
The WciS–His6-dependent galactosylation is abolished in the E. coli BL21 (DE3)/ΔgalU strain
The presence of an endogenous acceptor precluded the use of better characterized exogenous acceptors. To overcome this problem, we constructed the E. coli BL21 (DE3)/ΔgalU strain (see the Materials and methods section). The galU gene encodes the UTP:α-D-glucose-1-phosphate uridylyltransferase (EC 2.7.7.9) implicated directly in the UDP-Glc biosynthetic pathway [35]. If glucose, galactose or glucuronic acid (derived from UDP-Glc) are present in the endogenous acceptor, the latter will not be synthesized in the ΔgalU strain. Neither the 12K (Table 2) nor the 50K supernatant (results not shown) prepared from E. coli BL21 (DE3)/ΔgalU extracts expressing WciS–His6 incorporated any significant radioactivity into the glycolipid fraction (Table 2, compare with the 458 pmol/h in Table 1). Moreover, using the ΔgalU extracts, we did not detect any radioactive spot corresponding to the 14C-galactosylated oligosaccharide (Figure 5, lanes 1 and 2), nor any radioactivity co-migrating with WciS–His6 in native electrophoresis (Figure 4, lanes 8, 9, 12 and 13). However, the WciS–His6 overexpressed in the ΔgalU strain was active (154 pmol/h of [14C]Gal) towards the lipid acceptor prepared from E. coli BL21 (DE3) harbouring only a control vector (Table 2). Taken together, these results indicate that the mutant strain lacks the endogenous glycolipid acceptor. In addition, these data point to the presence of glucose, galactose, glucuronic acid and/or galacturonic acid in the endogenous acceptor, as expected for the hypothetical physiological substrate of WciS: the cellobiuronic acid-PPL. The E. coli BL21 (DE3)/ΔgalU harbouring pET-wciS was therefore used for the rest of the present study.
Table 2. Incorporation of [U-14C]Gal mediated by WciS–His6 into the GelK product.
The 12K supernatants were prepared from E. coli BL21 (DE3)/ΔgalU cells expressing WciS–His6 (pET-wciS) or not (control vector, pET23a+). Activity corresponding to 50 μl of GT preparation (approx. 300 μg of total proteins) was expressed as pmol of [U-14C]Glc incorporated per h from UDP-[U-14C]Glc on to the glycolipid fraction prepared from a E. coli BL21 (DE3) strain expressing GelK (pET-gelK) or harbouring the corresponding control vector (pET29b, see the Materials and methods section for further details).
| Plasmid expressed | pET29b glycolipid | pET-gelK glycoloipid | UDP-[U-14C]Gal | [14C]Gal incorporated into the acceptor (pmol/h) |
|---|---|---|---|---|
| pET23a+ | − | − | + | <3 |
| pET-wciS | − | − | + | <3 |
| pET23a+ | + | − | + | <3 |
| pET-wciS | + | − | + | 154±31 |
| pET23a+ | − | + | + | <3 |
| pET-wciS | − | + | + | 301±54 |
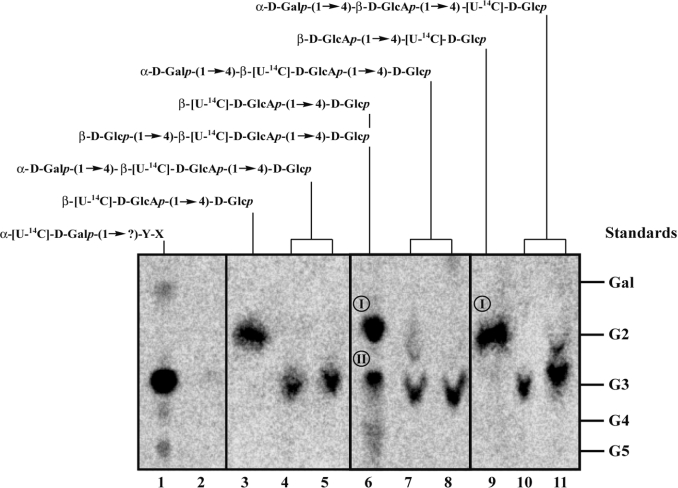
Figure 5. Analytical TLC of oligosaccharides released after WciS–His6-mediated transfer of galactose residues on to endogenous and exogenous acceptors.
Lanes 1 and 2: oligosaccharides released from the lipid fraction by mild acid hydrolysis after incubation of 12K supernatants prepared from E. coli BL21 (DE3) (lane 1) or BL21 (DE3)/ΔgalU (lane 2) expressing WciS–His6 with UDP-[U-14C]Gal. Lanes 3–5: the glycolipid β-[U-14C]D-GlcAp-(1→4)-D-Glcp-PPL was prepared by incubating E. coli BL21 (DE3) expressing GelK extracts with UDP-[U-14C]GlcA (see the Materials and methods section). Lanes 6–8 and lanes 9–11: the glycolipid β-[U-14C]D-GlcAp-(1→4)-D-Glcp-PPL and β-D-GlcAp-(1→4)-[U-14C]D-Glcp-PPL were prepared by incubating Sphing. paucimobilis A.T.C.C. 31461 extracts with the relevant UDP-sugar radiolabelled or not (see the Materials and methods section for further details). The oligosaccharides released by mild acid hydrolysis were analysed by TLC before (lanes 3, 6 and 9) or after incubation with WciS–His6 prepared from E. coli BL21 (DE3) 12K supernatants (lanes 4, 7 and 10) or 50K supernatants (lanes 5, 8 and 11). Glucose and oligomaltoses (G2, maltobiose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose) were used as standards. The predicted chemical structure of each oligosaccharide moieties synthesized in vitro is indicated. X and Y are unknown sugar residues.
The cellobiuronic acid-PPL is an acceptor of WciS–His6
The expected in vivo acceptor of WciS is the cellobiuronic acid-PPL [18,24]. Since this product is not commercially available, we decided to synthesize it in vitro using two kinds of enzymatic extracts [22]. The first one involved the β-(1→4)-D-glucuronosyltransferase, GelK, from Sphing. paucimobilis A.T.C.C. 31461 expressed in E. coli BL21 (DE3) strain harbouring pET-gelK [22]. The glycolipidic acceptors were partially purified (see the Materials and methods section) and added to the reaction mixture containing WciS–His6 (12K supernatant) and UDP-[U-14C]Gal (Table 2). A control was performed in parallel with the glycolipid fraction prepared from a E. coli BL21 (DE3) strain harbouring the corresponding control vector (pET29b) to assess the contribution of the endogenous acceptor. Under these conditions, a higher incorporation of [14C]Gal residues (301 pmol/h) in the glycolipid fraction was found in the presence of exogenous GelK-product, WciS–His6 and UDP-[U-14C]Gal (Table 2). Similar results were obtained with the WciS–His6 50K supernatant (results not shown). The converse experiment, using non-labelled UDP-Gal and β-[U-14C]D-GlcAp-(1→4)-D-Glcp-PPL was also performed (Figure 5, lanes 3–5). The strong decrease in intensity of the radioactive spot, which was the expected β-[U-14C]D-GlcAp-(1→4)-D-Glcp-PPL product (Figure 5, lane 3), was accompanied by the appearance of a radiolabelled trisaccharide after incubation with UDP-Gal and WciS–His6 12K or WciS–His6 50K supernatant (Figure 5, lanes 4 and 5). Incubation with α-galactosidase released the disaccharide (results not shown). These results, showing that the GelK product (cellobiuronic acid-PPL) is a substrate for WciS in vitro are concordant with the proposed biosynthetic RU.
These results were verified by a different approach using radiolabelled lipid-linked oligosaccharides from Sphing. paucimobilis A.T.C.C. 31461. To synthesize the appropriated acceptor, EDTA-permeabilized cells from Sphing. paucimobilis were incubated in the presence of UDP-Glc/UDP-[U-14C]GlcA or UDP-[U-14C]Glc/UDP-GlcA. The oligosaccharides synthesized in vitro by endogenous GTs were released from the glycolipid fraction and were analysed by TLC. Two radioactive spots (I and II) were detected by phosphoimaging after incubation with UDP-Glc and UDP-[U-14C]GlcA. The major oligosaccharide formed (Spot I, Figure 5, lane 6) co-migrated with the maltobiose marker and was assigned to be β-[U-14C]D-GlcAp-(1→4)-D-Glcp, based on data of Videira et al. [22]. Spot II represents probably the trisaccharide β-D-Glcp-(1→4)-β-[U-14C]D-GlcAp-(1→4)-D-Glcp (Figure 5, lane 6), which might be generated by addition of another glucose residue to the previous radiolabelled disaccharide [22]. When permeabilized Sphing. paucimobilis cells were incubated with UDP-[U-14C]Glc and UDP-GlcA, spot I was mainly obtained. After incubation of the radioactive glycolipids in the presence of WciS–His6 from the 12K or 50K supernatant (Figure 5, lanes 7, 8, 10 and 11) and UDP-Gal, a new spot appeared that co-migrated with the maltotriose marker. The corresponding radiolabelled trisaccharide was probably the expected α-D-Galp-(1→4)-β-[U-14C]D-GlcAp-(1→4)-D-Glcp (Figure 5, lanes 7 and 8) and α-D-Galp-(1→4)-β-D-GlcAp-(1→4)-[U-14C]-D-Glcp (Figure 5, lanes 10 and 11). The minor spots revealed in Figure 5, lanes 7 and 11 probably represented the unmodified disaccharide. A similar product {radiolabelled cellobiosyl (β-D-Glcp-(1→4)-D-Glcp)-PPL prepared from Xanthomonas campestris [34]} was tested in parallel. No WciS–His6-dependent incorporation on to this acceptor was found, showing the important role of the GlcA (results not shown). These results support our working hypothesis that WciS catalyses the transfer in vivo of galactose residues from UDP-Gal into the cellobiuronic acid moiety of the Strep. pneumoniae CPS 8 RU.
The SEC-purified WciS–His6 presents an active multimeric structure
We have purified WicS–His6 from the E. coli BL21 (DE3)/ΔgalU strain to exclude the possibilities that the galactosyltransferase activity depends on other endogenous proteins or small molecules present in the cell extracts. To avoid the precipitation of WciS–His6 and to increase the solubility of the protein, E. coli protein extracts were prepared using 1% Tween 20. Under these conditions, most WciS–His6 was found in the 50K supernatant (Figures 6A and 6B, lanes 1–10). Additionally, this treatment prevented the precipitation of the recombinant protein (see above). The WciS–His6 purification, performed in the presence of Tween 20, was monitored using SDS/PAGE and Western blot (results not shown). The 50K supernatant (Figures 6A and 6B, lane 10) was purified by IMAC on a Ni2+-chelate column, as described in the Materials and methods section. A 39 kDa protein that was eluted as a major peak at 50 mM imidazole was identified by Western blot analysis as WciS–His6 (Figures 6A and 6B, lane 11). Both to purify further and to assess the size of WciS–His6, we carried out SEC experiments (Figure 6C). The major protein fraction eluted at 37.5 min (Figure 6D, N-terminus: TKVLQIGPS) with an apparent molecular mass of 130 kDa. After denaturation and SDS/PAGE of this sample, only the monomer of 39 kDa was present (Figures 6A and 6B, lane 12), suggesting that WciS is in multimeric.
Figure 6. Purification of active WciS–His6 in the presence of Tween 20.
(A) Coomassie-Blue-stained SDS/PAGE gel of the WciS–His6 protein samples taken during the purification procedure. (B) His6-tag Western blotting of each aliquot. Cells of the BL21 (DE3)/ΔgalU strain harbouring pET23a+ with (+) or without (−) the insert were disrupted in the extraction buffer containing 1% Tween 20 and subjected to a differential centrifugation (see legend to Figure 1, lanes 1–10). The recombinant protein from the 50K supernatant was subjected to IMAC purification using a nickel-containing column (lane 11) followed by analytical FPLC gel filtration (SEC) (lane 12). The apparent molecular mass of the native WciS–His6 was estimated by filtration on a Superose 6 column. (C) Retention profile of the protein purified from the chromatographic column (the position of the retention times of the protein markers used to calibrate the column is indicated by crossbars; T0, retention time of the void volume of the column, determined using Dextran Blue). (D) Calibration curve of the gel filtration column; the following protein standards (Amersham Biosciences) were used: (1) ferritin (440 kDa), (2) catalase (232 kDa), (3) aldolase (158 kDa), (4) albumin (67 kDa), (5) ovalbumin (43 kDa) and (6) chymotrypsin (25 kDa). (E) SEC-purified WciS–His6 was analysed by native gradient PAGE (4–20% acrylamide/bisacrylamide in Tris/borate/EDTA). Lane 1, molecular-mass protein standards (Amersham Biosciences). Lanes 2–9, monitoring of the concentration of WciS–His6 prepared by centrifugal filtration (from 0.25 to 12.00 μg/μl). The expected WciS–His6 multimer is indicated by an arrow. Each sample (10 μl) was stained with Coomassie Blue. (F) His6-tag Western blotting of the corresponding samples in (E). (G) Analytical TLC of oligosaccharides released after WciS–His6-mediated transfer of galactose residue into radiolabelled cellobiuronic acid-PPL acceptor. The glycolipid β-D-GlcAp-(1→4)-[U-14C]D-Glcp-PPL was prepared as described in Figure 5 (lane 9) and incubated in the presence of UDP-Gal with (lane 3) or without (lane 2) SEC-purified WciS–His6 (30 μg of total protein). The resulting trisaccharide was then incubated overnight in the presence of α-galactosidase (lane 4). Galactose and oligomaltoses (G2, maltobiose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose) were used as standards. WciS–His6 polypeptide was indicated by an arrow. SN, supernatant; MW, molecular-mass markers (sizes in kDa).
Since protein crystallization requires concentrated protein, we studied the behaviour of WciS–His6 in concentrated solutions. Native PAGE was performed with concentrations ranging from 0.25 to 12 μg/μl (Figures 6E and 6F). The apparent average molecular mass of WciS–His6 (approx. 180 kDa) did not change with the concentrations tested. All these results indicate that the multimeric form of WciS–His6 is stable through all the purification procedures (see results above and Figure 4). The discrepancies observed in the native apparent molecular mass of WciS–His6 estimated by SEC and native PAGE (130 and 180 kDa respectively) rely on the fact that the two experimental approaches separate proteins on the basis of two different properties.
The α-galactosyltransferase activity of the SEC-purified WciS–His6 was assayed in the presence of radiolabelled glycolipid acceptor (β-D-GlcAp-(1→4)-[U-14C]D-Glcp-PPL) (Figure 6G, lane 1). The TLC analysis showed that the resulting radiolabelled galactosylated oligosaccharide co-migrated with the maltotriose marker (Figure 6G, lane 3) and was hydrolysed in the presence of α-galactosidases (Figure 6G, lane 4), indicating that the purified galactosyltransferase is active. It is worthy of mention that in purifications performed using the wild-type E. coli strain, the endogenous acceptor co-purified all the way with WciS–His6.
DISCUSSION
In the present study, we report on the functional characterization of WciS as an α-galactosyltransferase that uses cellobiuronic acid-PPL as an acceptor. This function is in agreement with the sugar sequence of the CPS type 8 and previous proposals based on sequence analyses [11,18], as well as its classification in the CAZy family 4 of retaining GTs [25]. WciS is therefore the first of the three putative GTs, deduced from the cps8 gene cluster sequence, to be characterized using a biochemical approach. The recombinant WciS co-precipitates on centrifugation with membranes; however, the nature of the association remains unclear. Solubilization of WciS was achieved in the presence of detergents (the present study, and L. Pelosi, unpublished work). We have also found that WciS behaves as a multimeric protein.
As proposed for other GTs that are involved in LPS (lipopolysaccharide) biosynthesis, such as AceA from Acetobacter xylinum [34], ExoM from Sinorhizobium meliloti [31] or GelK from Sphing. paucimobilis [22], features of PPL moiety are required for WciS–His6 activity (results not shown). We have also found that WciS–His6 galactosylates an endogenous glycolipid acceptor. The endogenous acceptor co-purifies with WciS. Moreover, the reaction product co-migrates in native electrophoresis with the multimeric protein. The strong interaction between WciS–His6 and the reaction product can be understood either as a non-specific hydrophobic interaction or as a specific interaction with the active site of the protein. Although our data cannot distinguish between these two hypotheses, we can speculate that the specific interaction leads to channelling of RU biosynthesis.
Although a chemical characterization of the endogenous acceptor and the WciS products are beyond the purpose of the present paper, our data supports that the endogenous acceptor is a cellobiouronic glycolipid. Indeed, the properties of the product formed with the endogenous acceptor, as well as the absence of it in the ΔgalU strain, are compatible with a cellobiuronic acid. Unfortunately, there are no data available on extracellular polysaccharides produced by E. coli B strains, precluding any hypothesis on the origin of such a disaccharide. Further structural studies will be necessary to gain further information about the actual chemical structure of the endogenous acceptor. The E. coli BL21 (DE3)/ΔgalU strain may represent a new biological tool for both expression and biochemical characterization of recombinant GTs in decreasing the probability to have endogenous acceptors.
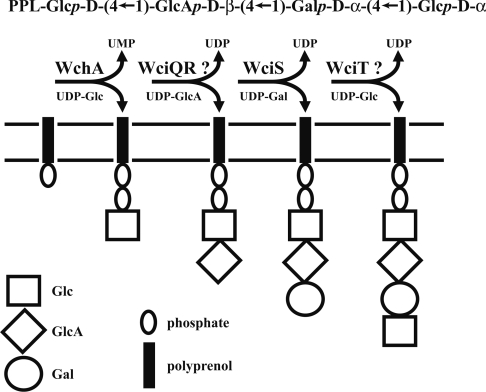
The present study, in addition to the CAZy classifications, allows us to provide a model for the biosynthesis of the RU of serotype 8 CPS (Figure 7). In a previous study, we have shown that WchA primes the biosynthesis of the serotype 8 CPS by transfer of a phosphoglucose residue to a lipid [24]. Given the presence of the cellobiuronic acid in CPS type 8 and the similarity of WciQR with SpsK, GelK and PssDE, it is possible that the second step is the transfer of a glucuronic acid residue catalysed by WciQR. Thus this is the acceptor of WciS that transfers the galactose residue to form the trisaccharide. Finally, WciT may add the remaining glucose. The synthesis in vitro of the complete RU may be useful for the study of translocation and the polymerization events.
Figure 7. Model of the assignment of biochemical function to GTs involved in RU assembly of Strep. pneumoniae CPS 8.
Like the Streptococcal gene products, such as Cps14E, Cps9vE and CpsIIIE characterized previously [12,36,37], we propose that WchA is a UDP-Glc: PPL glucosyl-1-phosphate transferase [24]. WciS catalyses the transfer of galactose residue, from UDP-Gal, on to a cellobiuronic acid-PPL through an α-linkage. The functions of WciQR and WciT are unknown. However, based on bioinformatic analysis and assuming a sequential expression of the presumed GT genes identified in the CPS 8 cluster, we propose that WciQR and WciT catalyse the addition of the second (GlcA) and fourth (glucose) residues of the CPS 8 RU.
Acknowledgments
We thank R. Hengge-Aronis and E. Klauck for providing E. coli strains, D. Schneider for help with P1 transduction, P. Videira for her critical reading of the manuscript, and N. Potier and E. Coursange for technical assistance. The N-terminal amino acid sequence of recombinant protein was determined by J.-P. Andrieu, Institut de Biologie Structurale (IBS), Grenoble, France. L.P. and N.S. are supported by a post-doctoral (CNRS) and a doctoral (CNRSL) fellowship respectively. This work was supported by CNRS (France) grants.
References
- 1.Alonso de Velasco E., Verheul A. F. M., Verhoef J., Snippe H. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol. Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbins J. B., Austrian R., Lee C. J., Rastogi S. C., Schiffman G., Henrichsen J., Mäkelä P. H., Broome C. V., Fracklam R. R., Tiesjema R. H., Parke J. C. J. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J. Infect. Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 3.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler J. C., Breiman R. F., Campbell J. F., Lipman H. B., Broome C. V., Facklam R. R. Pneumococcal polysaccharide vaccine efficacy: an evaluation of current recommendations. JAMA, J. Am. Med. Assoc. 1993;270:1826–1831. [PubMed] [Google Scholar]
- 5.Makela P. H., Kayhty H. Evolution of conjugate vaccines. Expert Rev. Vaccines. 2002;1:399–410. doi: 10.1586/14760584.1.3.399. [DOI] [PubMed] [Google Scholar]
- 6.Alexander J., Guercio M. F., Frame B., Maewal A., Sette A., Nahm M. H., Newman M. J. Development of experimental carbohydrate-conjugate vaccine composed of Streptococcus pneumoniae capsular polysaccharides and the universal helper T-lymphocyte epitope (PADRE®) Vaccine. 2004;22:2362–2367. doi: 10.1016/j.vaccine.2003.11.061. [DOI] [PubMed] [Google Scholar]
- 7.Lee C. J., Lee L. H., Frasch C. E. Protective immunity of pneumococcal glycoconjugates. Crit. Rev. Microbiol. 2003;29:333–349. doi: 10.1080/713608018. [DOI] [PubMed] [Google Scholar]
- 8.Cartee R. T., Forsee W. T., Jensen J. W., Yother J. Expression of the Streptococcus pneumoniae type 3 synthase in Escherichia coli: assembly of the type 3 polysaccharide on a lipid carrier. J. Biol. Chem. 2001;276:48831–48839. doi: 10.1074/jbc.M106481200. [DOI] [PubMed] [Google Scholar]
- 9.Llull D., Garcia E., Lopez R. Tts, a processive β-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in Pneumococcus and other Gram-positive species. J. Biol. Chem. 2001;276:21053–21061. doi: 10.1074/jbc.M010287200. [DOI] [PubMed] [Google Scholar]
- 10.Whitfield C. Biosynthesis of lipopolysaccharide O-antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 11.Jiang S. M., Wang L., Reeves P. R. Molecular characterization of Streptococcus pneumoniae Type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect. Immun. 2001;69:1244–1255. doi: 10.1128/IAI.69.3.1244-1255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolkman M. A. B., Morrison D. A., van der Zeijst B. A. M., Nuijten P. J. M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyltransferase gene cps14E. J. Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolkman M. A., van der Zeijst B. A. M., Nuijten P. J. M. Functional analysis of glycosyltransferase encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J. Biol. Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 14.Kolkman M. A. B., Wakarchuk W., Nuijten P. J. M., van der Zeijst B. A. M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for biosynthesis of the tetrasaccharide subunit. Mol. Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones J. K. N., Perry M. B. The structure of the type VIII Pneumococcus specific polysaccharide. J. Am. Chem. Soc. 1957;79:2787–2793. [Google Scholar]
- 16.O'Neil M. A., Selvendran R. R., Morris V. J. Structure of the acidic extracellular gelling polysaccharide produced by Pseudomonas elodea. Carbohydr. Res. 1993;124:123–133. doi: 10.1016/0008-6215(86)80037-4. [DOI] [PubMed] [Google Scholar]
- 17.Kuo M. S., Mort A. J., Dell A. Identification and location of L-glycerate, an unusual substituent in gellan gum. Carbohydr. Res. 1986;56:173–183. [Google Scholar]
- 18.Muñoz R., Mollerach M., Lopez R., Garcia E. Characterization of type 8 capsular gene cluster of Streptococcus pneumoniae. J. Bacteriol. 1999;181:6214–6219. doi: 10.1128/jb.181.19.6214-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia E., Lopez R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol. Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 20.Reeves P. R., Hobbs M., Valvano M., Skurnik M., Whitfield C., Coplin D., Kido N., Klena J., Maskell D., Raetz C., Rick P. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 21.Pollock T. J., van Workum W. A., Thorne L., Mikolajczak M. J., Yamazaki M., Kijne J. W., Armentrout R. W. Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J. Bacteriol. 1998;180:586–593. doi: 10.1128/jb.180.3.586-593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Videira P., Fialho A., Geremia R. A., Breton C., Correia I. Biochemical characterization of the (1→4)-β-glucuronosyltransferase GelK in the gellan gum-producing strain Sphingomonas paucimobilis A.T.C.C. 31461. Biochem. J. 2001;358:457–464. doi: 10.1042/0264-6021:3580457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki M., Thorne L., Mikolajczak M., Armentrout R. W., Pollock T. J. Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J. Bacteriol. 1996;178:2676–2687. doi: 10.1128/jb.178.9.2676-2687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelosi L., Boumedienne M., Saksouk N., Geiselmann J., Geremia R. A. The glucosyl-1-phosphate transferase WchA (Cap8E) primes the capsular polysaccharide repeat unit biosynthesis of Streptococcus pneumoniae serotype 8. Biochem. Biophys. Res. Commun. 2004;327:857–865. doi: 10.1016/j.bbrc.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 25.Campbell J. A., Davies G. J., Bulone V., Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J. H. Molecular Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1972. Experiments. [Google Scholar]
- 27.Bohringer J., Fischer D., Mosler G., Hengge-Aronis R. UDP-glucose is a potential intracellular signal molecule in the control of expression of σS and σS-dependent genes in Escherichia coli. J. Bacteriol. 1995;177:413–422. doi: 10.1128/jb.177.2.413-422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Videira P. A., Cortes L. L., Fialho A. M., Correia I. Identification of the pgmG gene, encoding a bifunctional protein with phosphoglucomutase and phosphomannomutase activities, in the gellan gum-producing strain Sphingomonas paucimobilis A.T.C.C. 31461. Appl. Environ. Microbiol. 2000;66:2252–2258. doi: 10.1128/aem.66.5.2252-2258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 30.Tolmasky M. E., Staneloni R. J., Leloir L. F. Lipid-bound saccharides in Rhizobium meliloti. J. Biol. Chem. 1982;257:6751–6757. [PubMed] [Google Scholar]
- 31.Lellouch A. C., Geremia R. A. Expression and study of recombinant ExoM, a (1→4)-β-D-glucosyltransferase involved in succinoglycan biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 1999;181:1141–1148. doi: 10.1128/jb.181.4.1141-1148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cserzo M., Wallin E., Simon I., von Heijne G., Elofsson A. Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 33.Mengin-Lecreulx D., Texier L., Rousseau M., van Heijenoort J. The murG gene of Escherichia coli code for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geremia R. A., Roux M., Ferreiro D. U., Dauphin-Dubois R., Lellouch A. C., Ielpi L. Expression and biochemical characterisation of recombinant AceA, a bacterial α-mannosyltransferase. Mol. Gen. Genet. 1999;261:933–940. doi: 10.1007/s004380051040. [DOI] [PubMed] [Google Scholar]
- 35.Frey P. A. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 36.van Selm S., Kolkman M. A. B., van der Zeijst B. A. M., Zwaagstra K. A., Gaastra W., van Putten J. P. M. Organization and characterization of the capsule biosynthesis locus of Streptococcus pneumoniae serotype 9V. Microbiology. 2002;148:1747–1755. doi: 10.1099/00221287-148-6-1747. [DOI] [PubMed] [Google Scholar]
- 37.Chaffin D. O., Beres S. B., Yim H. H., Rubens C. E. The serotype of type Ia and II group B Streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 2000;182:4466–4477. doi: 10.1128/jb.182.16.4466-4477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]