Abstract
HS (heparan sulphate) proteoglycans bind secreted signalling proteins, including FGFs (fibroblast growth factors) through their HS side chains. Such chains contain a wealth of differentially sulphated saccharide epitopes. Whereas specific HS structures are commonly believed to modulate FGF-binding and activity, selective binding of defined HS epitopes to FGFs has generally not been demonstrated. In the present paper, we have identified a series of sulphated HS octasaccharide epitopes, derived from authentic HS or from biosynthetic libraries that bind with graded affinities to FGF4, FGF7 and FGF8b. These HS species, along with previously identified oligosaccharides that interact with FGF1 and FGF2, constitute the first comprehensive survey of FGF-binding HS epitopes based on carbohydrate sequence analysis. Unexpectedly, our results demonstrate that selective modulation of FGF activity cannot be explained in terms of binding of individual FGFs to specific HS target epitopes. Instead, different FGFs bind to identical HS epitopes with similar relative affinities and low selectivity, such that the strength of these interactions increases with increasing saccharide charge density. We conclude that FGFs show extensive sharing of binding sites in HS. This conclusion challenges the current notion of specificity in HS–FGF interactions, and instead suggests that a set of common HS motifs mediates cellular targeting of different FGFs.
Keywords: co-receptor, epitope, fibroblast growth factor, glycosaminoglycan, heparan sulphate proteoglycan
Abbreviations: [1-3H]aManR, 2,5-anhydro-D-[1-3H]mannitol; FGF, fibroblast growth factor; FGFR, FGF receptor; GAG, glycosaminoglycan; GlcA, D-glucuronic acid; GlcNAc, N-acetylglucosamine; GlcNS, N-sulphated glucosamine; HS, heparan sulphate; IdoA, L-iduronic acid; PG, proteoglycan
INTRODUCTION
The functional significance of the structural variability exhibited by GAG (glycosaminoglycan)-type polysaccharides remains a fundamental question in cell and developmental biology. GAGs are abundant on cell surfaces and in the extracellular matrix, and have been shown to regulate cell behaviour in the broadest sense. GAGs dynamically influence cell adhesion, migration, proliferation and differentiation, either through direct binding to secreted signalling proteins, thereby modulating their activities, or through interactions with molecules of the extracellular matrix [1]. HS (heparan sulphate) GAGs, abundantly implicated with regulation of growth factor activity, are ubiquitous in animal organisms. HS chains occur covalently attached to PG (proteoglycan) core proteins, either as membrane-intercalated HSPGs (syndecans and glypicans) at cell surfaces, or deposited in the extracellular matrix (perlecan, agrin and collagen XVIII). Genetic studies in Drosophila and other model systems have shown that HS is essential for normal FGF (fibroblast growth factor), Wingless/Wnt, Dpp (decapentaplegic)/BMP (bone morphogenetic protein) and Hedgehog activity. Mutants that are defective in enzymes associated with HS biosynthesis show anomalous distribution and signalling of these growth factors/morphogens [2–4].
HS is synthesized as a linear polymer of up to several hundred units of alternating GlcA (D-glucuronic acid) and GlcNAc (N-acetylglucosamine) units. This precursor is partially modified through a series of reactions, including N-deacetylation and N-sulphation of GlcNAc units, C5-epimerization of GlcA into IdoA (L-iduronic acid), sulphation at 3-O- and 6-O-positions of glucosamine and at 2-O-position of GlcA and IdoA units. Owing to the variable regulation of these reactions in different cells and tissues, and at different developmental stages, the mature HS products display sulphated domains of variable length and composition [5,6]. These domains provide binding sites for protein ligands. The structural diversity is strictly regulated, as revealed by compositional analysis of HS from various organs and by immunohistochemical application of antibodies recognizing different HS epitopes [7].
FGFs (22 distinct protein species are currently recognized) are of fundamental importance in embryogenesis. The various FGFs generally require HS for biological activity, presumably to generate ternary signalling complexes with FGF receptors [8–12]. The marked temporal and topological selectivity of FGF action during embryonic organogenesis has been attributed to modulations of HS fine structure [13,14]. To test this hypothesis, we have compared a series of HS epitopes with regard to saccharide sequence and FGF binding. The results demonstrate extensive sharing of growth-factor-binding HS epitopes between different FGF species, and thus challenge the notion of specificity in these interactions.
EXPERIMENTAL
Materials
FGF8b was produced in Escherichia coli and purified through its His6-tag under denaturing conditions. After refolding by dialysis, FGF8b was purified on a heparin–Sepharose CL-6B column, and the bound protein was eluted with NaCl [15]. FGF7 was produced in bacteria and was purified as described previously [16]. The mature secreted form of human FGF4, spanning amino acids Ala-31 to Leu-206, was expressed from the PET-15b bacterial expression vector after transformation into E. coli BL21 and induction with isopropyl β-D-thiogalactoside. FGF4 protein was purified as described in [17].
Fully N-sulphated oligosaccharides were prepared from pig intestinal mucosa HS as described in [18]. Briefly, the polysaccharide was N-deacetylated by hydrazinolysis, followed by deamination with nitrous acid (pH 3.9). The products were reduced with NaB3H4 (64 Ci/mmol) to create terminal [1-3H]aManR (2,5-anhydro-D-[1-3H]mannitol) residues. The specific radioactivity for octamers was typically 5×103 d.p.m./pmol. The labelled saccharides were separated by size-exclusion chromatography, and the octasaccharide fraction was isolated [19]. In-vitro-generated oligosaccharide libraries were obtained as described in [20]. Briefly, fully N-sulphated partially O-desulphated heparin octasaccharides, or N-sulphated octasaccharides isolated from E. coli K5 capsular polysaccharide, were subjected to enzymatic O-sulphation by incubation with various O-sulphotransferases.
Affinity chromatography
Recombinant histidine-tagged FGF8b (2.3 mg) dissolved in 17 ml of 1 M NaCl was circulated twice through 0.5 ml of Ni-NTA (Ni2+-nitrilotriacetate) Superflow nickel-chelating Sepharose (Qiagen). The gel was rinsed with 20 mM imidazole buffer according to the manufacturer's instructions before further use. The degree of coupling was estimated to be 70% by measurement of the absorbance of the protein solution before and after circulation through the Ni-NTA resin. Approx. 1 mg of recombinant FGF4 or FGF7 was immobilized on to CH Sepharose-4B as described in [19]. A control column of CH Sepharose lacking FGF protein did not bind highly sulphated heparin oligosaccharides. The FGF columns (generally a total volume of 1 ml, containing 60–180 nmol of total protein, or 1–3 mg of protein/ml of gel) were equilibrated in 0.14 M NaCl, 50 mM Tris/HCl, pH 7.4, before sample addition. Oligosaccharide samples for affinity chromatography were derived from authentic HS, as well as from biosynthetic libraries. For preparative runs, 50 nmol of saccharide (250×106 d.p.m.) was applied to the FGF columns. Octamers of already known sequence [19–21], typically 1–5 pmol, were also applied individually to the FGF columns for analytical runs. Samples were generally applied in 200 μl of the equilibration buffer, retained for 10 min at 4 °C and eluted with a stepwise gradient (total volume 35 ml) ranging from 0.2 to 1.0 M NaCl, buffered as above. Fractions of 1 ml were collected and analysed for radioactivity.
Ion-exchange chromatography
Oligosaccharide fractions eluted from the FGF affinity matrices were desalted on PD-10 columns (Amersham Biosciences) run in water, and further resolved by anion-exchange chromatography on a 2 mm×250 mm PropacPA-1 column (Dionex, Camberley, Surrey, U.K.). Samples were eluted with a linear gradient (total volume 100 ml) extending from 1 to 1.5 M NaCl, adjusted with HCl to pH 3. Saccharides corresponding to peak fractions were pooled, desalted and dried in a centrifugal evaporator.
Sequence analysis
Sequence analysis was performed as described previously [19,22]. Briefly, 20×103 d.p.m. (∼4 pmol) of N-sulphated [1-3H]aManR end-labelled octasaccharides were subjected to partial deaminative cleavage (pH 1.5 procedure) in 20 μl of nitrous acid (2 mM NaNO2 and 20 mM HCl) on ice. The reaction was terminated at various time points (15, 30, 45, 60 and 75 min) by transferring 4-μl aliquots to 10 μl of 200 mM sodium acetate, pH 6. The resultant pool was split into four equal portions that were variously subjected to enzyme digestion. Enzymes used were iduronate-2-sulphatase, α-L-iduronidase and glucosamine-6-sulphatase (Glyko, Novato, CA, U.S.A.). The samples were incubated as indicated in the legend to Figure 1, at 37 °C for 16 h according to the manufacturer's instructions. The sequence data were assembled by comparing the PropacPA-1 anion-exchange patterns of samples before and after enzyme treatments. The PropacPA-1 column was run in gradients (0.01 ml/min) of 0–1 M NaCl (total volume 100 ml) or 0–1.5 M NaCl (total volume 150 ml), adjusted to pH 3 with HCl, and analysed online using a radioactivity flow detector (Radiomatic 500 TR, Packard, Meriden, CT, U.S.A.).
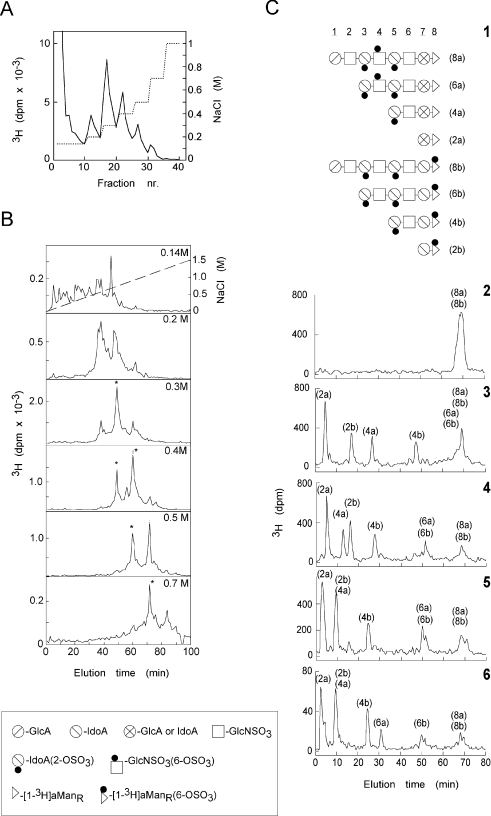
Figure 1. Sequence analysis of HS octasaccharides with different affinities for FGF8b.
(A) Affinity chromatography of [1-3H]aManR-labelled HS octasaccharides isolated from pig intestinal mucosa on immobilized FGF8b. Bound saccharides were eluted with a stepwise gradient of NaCl (dotted line). (B) Fractions recovered at different NaCl concentrations (indicated in the upper right corner of each panel) were separated further with respect to charge by anion-exchange HPLC. Peak fractions indicated by asterisks were subjected to sequence analysis. (C) Sequence analysis (see the Experimental section) of octasaccharides eluted from the FGF8b matrix at 0.3 M NaCl. Panel 1: the elucidated structures of the two different octasaccharides, denoted 8a and 8b (corresponding to structures 2/2-1/6-a and 2/2-1/6-c in Table 1) found in this affinity pool, along with subfragments generated by chemical scission of the two octasaccharides. Symbols used to depict non-sulphated and sulphated HS monosaccharide units (and in Table 1) are shown in the box beneath (B). Panels 2–6 show analytical ion-exchange chromatograms of intact octasaccharides (Panel 2), and of fragments (di-, tetra- and hexa-saccharides) obtained by partial HNO2 degradation, either with no subsequent enzyme treatment (Panel 3) or followed by digestion with iduronate 2-sulphatase (Panel 4), iduronate 2-sulphatase and α-L-iduronidase (Panel 5), or iduronate 2-sulphatase, α-L-iduronidase and glucosamine 6-sulphatase (Panel 6). The enzyme digestions (releasing sulphate groups or IdoA units) induce shifts in the elution positions of individual subfragments. The shifts were related to the elution positions of a panel of previously characterized oligosaccharide standards [19]. The end label (at the [1-3H]aManR residue, designated by a triangle in Panel 1), provides a common reference point that enables sequence readout.
RESULTS
Data from crystallography and affinity chromatography have shown that FGF-binding domains may be accommodated within octasaccharides generated by deaminative cleavage of HS. Such octamers, derived from N-sulphated domains of authentic HS, were isolated after selective chemical degradation of the polysaccharide. In addition, octamer libraries were generated by enzymatic O-sulphation of 6-O-desulphated and partially 2-O-desulphated (re-N-sulphated) heparin octasaccharides. The biosynthetic libraries enable assessment of rare epitopes that are present at low concentration in tissues, and provide a valuable complement to tissue-derived saccharides in assessing the binding specificities of protein ligands. Both types of fragments were previously affinity-fractionated on immobilized FGF1, and selected species were sequence analysed using a technique based on combined chemical and enzymatic degradation of terminally radiolabelled octamers [19,20]. While these analyses showed an overall correlation between FGF1 affinity and O-sulphate content, sequences containing an internal -IdoA2S-GlcNS6S-IdoA2S- (where GlcNS is N-sulphated glucosamine) structure were in particular implicated with FGF1 binding. In the present study, we took a similar approach to probe the saccharide-binding requirements of other FGF species that are known to be selectively recruited in embryo/organo-genesis, primarily FGF4 and FGF8b.
Structural analysis of FGF8b-binding octasaccharides
HS-octasaccharides isolated from N-sulphated domains of pig intestinal mucosa, and end-labelled by reduction with NaB3H4, were shown previously to encompass a range of epitopes with graded affinities for FGF1 and FGF2 [19]. Following application of such an octasaccharide mixture to an FGF8b–Sepharose column, approx. 50% of the 3H-labelled fragments were retained by the immobilized growth factor at physiological salt conditions, and could be eluted at stepwise increasing salt concentrations, from 0.2 to 1 M NaCl (Figure 1A). Each affinity fraction was separated further by anion-exchange HPLC (Figure 1B). The flow-through fraction (Figure 1B, top panel) contained several components with no or only few O-sulphate groups, as judged by their early elution positions in the chromatogram. The FGF8b-binding fractions contained limited numbers of distinct subfractions with different O-sulphate contents, and with apparent correlation between charge density and affinity. Charge-homogeneous octasaccharides, recovered from fractions indicated by asterisks in Figure 1(B), were subjected to sequence analysis based on exoenzyme digestion of fragments obtained by partial deaminative cleavage [19]. Figure 1(C) illustrates the sequence identification of octasaccharides eluted from the FGF8b column with 0.3 M NaCl, and purified by anion-exchange HPLC (fraction emerging after ∼49 min; panel marked 0.3 M in Figure 1B). Sequence readout revealed a mixture of two distinct octasaccharides (designated 8a and 8b in Figure 1C; 2/2-1/6-a and 2/2-1/6-c respectively in Table 1), both carrying two 2-O-sulphate groups and one 6-O-sulphate group.
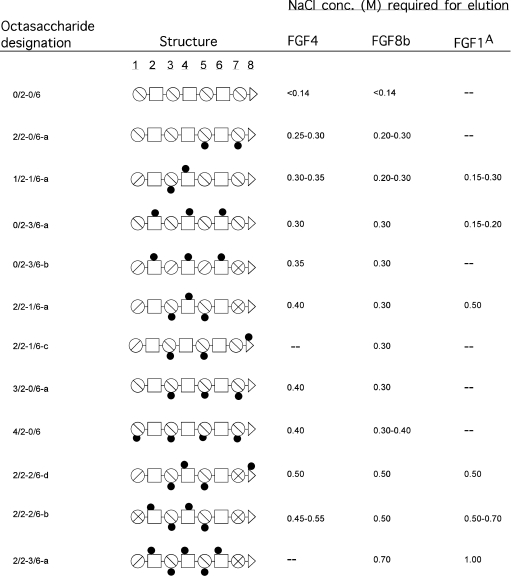
Table 1. Selection of HS octasaccharides that interact with FGF4, FGF8b and FGF1.
AData from Kreuger et al. [19] and Jemth et al. [20]. A complete listing of all HS sequences probed for interaction with these proteins, and with FGF7, has been compiled in Supplementary Table 1 (see http://www.BiochemJ.org/bj/389/bj3890145add.htm). A key to symbols can be found in Figure 1.
The sequence information for all octasaccharides separated with regard to affinity for FGF8b is compiled in Supplementary Table 1 (http://www.BiochemJ.org/bj/389/bj3890145add.htm). The selection of FGF8b-binding HS epitopes in Table 1 illustrates our key findings, relating saccharide sequence information to growth factor affinity (expressed in terms of the salt concentration required for elution from immobilized FGF8b). The exclusively N-sulphated octamer, 0/2-0/6, did not bind FGF8b at physiological salt concentration, whereas octamers with two or more O-sulphate groups bound with appreciable affinity. Evidently, HS saccharides need to carry O-sulphate groups in order to bind FGFs. Surprisingly, a general lack of preference for one or the other type of O-sulphate group was observed for interactions between FGF8b and HS. For example, an octamer with three 2-O-sulphate groups and no 6-O-sulphate (3/2-0/6-a) bound FGF8b as tightly as an octamer carrying three 6-O-sulphate groups and no 2-O-sulphate (0/2-3/6-a). Another unexpected finding was that the C5 configuration of the hexuronic acid residues within the HS epitopes seems to have little importance for FGF8b binding. A 6-O-sulphated fragment with predominantly GlcA units (0/2-3/6-b) thus binds FGF8b as avidly as an identically sulphated fragment containing only IdoA units (0/2-3/6-a). Thus the affinity for FGF8b generally increases along with the charge density of the HS octamer epitopes (see also [23]). Motifs with clusters of both 6-O- and 2-O-sulphate groups tend to provide slightly stronger binding to FGF8b, compared with motifs that contain the same number of more evenly spaced O-sulphate groups (compare, for example, 2/2-2/6-b with 4/2-0/6).
Binding of sequence-defined octasaccharides to FGF4 and FGF7
Interactions with FGF4 were probed by applying a panel of already sequence-defined octasaccharides (from authentic HS, as well as from biosynthetic libraries; see Supplementary Table 1 at http://www.BiochemJ.org/bj/389/bj3890145add.htm) to affinity chromatography on the immobilized growth factor. FGF4 was able to bind essentially all HS epitopes recognized by FGF8b, the relative affinities toward the two proteins varying only slightly (Table 1). Analogous to FGF8b, FGF4 binds with similar affinity to tri-6-O-sulphated octasaccharides containing either all GlcA or IdoA residues. Also, the most efficient FGF4 ligands tend to exhibit charge clusters made up of both 2-O- and 6-O-sulphate groups. Altogether, the similarities in HS-epitope recognition between FGF8b and FGF4 are remarkable (Table 1).
Analogous experiments with FGF7 revealed overall weaker binding of octamers to this species than to FGF4 and FGF8b (shown in Supplementary Table 1 at http://www.BiochemJ.org/bj/389/bj3890145add.htm). However, all motifs that bind FGF7 also interact with FGF4 and FGF8b. Notably, FGF7 seems to favour saccharide motifs with spread-out, rather than clustered, O-sulphate groups.
DISCUSSION
Several developmental processes are known to depend on selective FGF involvement in temporally controlled mode [13]. For example, limb and brain development involve selective FGF activation [24]. We demonstrated previously differential structural requirements for HS binding to FGF1 and FGF2, such that FGF2 interacts strongly with an epitope containing an -IdoA2S-GlcNS-IdoA2S-trisaccharide sequence, whereas FGF1 binding requires, in addition, 6-O-sulphation of the internal GlcNS unit for substantial binding [19]. This finding, and other data on differential O-sulphate requirements in HS–FGF interactions [25], suggested that selective binding of different FGF species to HS motifs could be of primary importance in regulation of FGF action. Our present study, the first survey of HS–FGF interactions based on actual saccharide sequence information, does not support this notion. In fact, an octamer (fragment 2/2-1/6-a in Table 1) containing the tri-O-sulphated trisaccharide sequence implicated with FGF1 binding was found to interact with similar affinity also with FGF4 and FGF8b. Moreover, the survey reveals alternative binding structures that may be shared by several different FGFs at physiological, or higher, ionic strength (Table 1). This conclusion applies primarily to FGF1, FGF4 and FGF8b, but also to FGF7, although binding of octasaccharides to FGF7 was generally weaker (Supplementary Table 1 at http://www.BiochemJ.org/bj/389/bj3890145add.htm).
Highly sulphated sequences with both 2-O- and 6-O-sulphate groups generally bind strongly to FGFs. FGF2 alone appears capable of strong binding to sequences containing only one type of O-sulphates (i.e. 2-O-sulphate groups [19,20,26]). Epitopes that bind more weakly at close to physiological ionic strength could conceivably be more discriminative, hence functionally important. Again, our results argue against such selectivity (Table 1, and Supplementary Table 1 at http://www.BiochemJ.org/bj/389/bj3890145add.htm) [19,20]. In fact, FGF1, FGF4, FGF7 and FGF8b all bind N-sulphated octasaccharides with three 6-O-sulphate groups, but no 2-O-sulphate, at physiological ionic strength. The same growth factors (FGF1 was not tested) also bind octamers with three or four 2-O-sulphate groups, but no 6-O-sulphate. These findings agree with studies on FGF7 by Ostrovsky et al. [27], but are at some variance with those of Ashikari-Hada et al. [25], who proposed that FGF4 and FGF7 bind only to HS epitopes containing both 2-O- and 6-O-sulphate groups. The lack of interaction between FGF8 and the sulphated oligosaccharides tested by Ashikari-Hada et al. [25] may be due to the selection of the target FGF8 isoform (FGF8b, a heparin-binding isoform, was used in the present study).
It may be argued that salt elution of oligosaccharides from the various immobilized FGFs would affect mainly the electrostatic component of these interactions, and not assess the substantial contribution to the binding affinity (and specificity) from van der Waals forces and hydrogen bonds [28,29]. Our results do not exclude some uncertainty in this regard. On the other hand, the data of Thompson et al. [28] show a linear correlation between affinity (as measured by isothermal titration calorimetry) and the NaCl concentration needed to elute various FGF2 mutants from a heparin column. In this case, saccharides were immobilized and not the proteins, but the experiment is energetically equivalent to those performed in the present study. Furthermore, we have determined the dissociation constants for several different HS octasaccharides with FGF1 using a filter-binding assay [30] and again found a good correlation between affinity and critical NaCl concentration (J. Kreuger, P. Jemth and U. Lindahl, unpublished work). We therefore assume that our data reflect the true affinity of these interactions, and not only the strength of the electrostatic bonds.
Our findings indicate that the regulation of FGF action in vivo cannot be explained in terms of the selectivity of the primary interactions between FGF species and HS chains. Yet other studies point to the importance of HS fine structure in such regulation [13]. It has been proposed that formation of the FGF–HS–FGFR (FGF receptor) ternary complex is initiated by FGF–HS interaction, followed by transition of the growth factor to a HS domain interacting with both proteins [14,31]. Selectivity achieved through formation of a HS-binding pocket by an FGF–FGFR pair would not necessarily reflect the binding properties of either protein alone [13,14,31]. Apart from the distribution of sulphate groups, also the organization of protein-binding domains along the HS chain may need to be considered [32]. Previous findings implicating HS-binding sites on both FGF and its receptor [8] have been amply verified through crystallography of ternary FGF–oligosaccharide–FGFR complexes [9,11].
On the other hand, the comparatively restricted phenotype derangements resulting from targeted disruption of genes encoding certain enzymes (2-O-sulphotransferase, glucuronyl C5-epimerase) in murine HS biosynthesis clearly argue against a critical role for specific HS epitopes in FGF regulation [2,33,34]. In fact, the only functionally implicated selective HS epitope identified to date is the antithrombin-binding pentasaccharide sequence in heparin and HS [5]. Notably, this sequence, like the (yet to be fully characterized) HS epitope interacting with the Herpes simplex gD glycoprotein [35], contains a rare, and functionally essential, glucosamine 3-O-sulphate unit.
The present study reveals that selective modulation of FGF activity cannot be explained by binding of individual FGFs to specific HS target epitopes. No HS epitope has yet been shown to selectively interact with any given protein owing to its unique disposition of the commonly expressed N-, (IdoA-)2-O-, and 6-O-sulphate groups. Pending the disclosure of such specific interactions between HS epitopes and protein ligands, we suggest that the binding of proteins to commonly sulphated HS domains is dictated primarily by charge density rather than by the precise disposition of various sulphate substituents. This suggestion does not detract from the functional significance of the HS–protein encounters [36].
Online data
Acknowledgments
The FGF8b expression vector was a gift from C. MacArthur. J.K. is supported by the Wenner–Gren Foundations. Supported by grants from the Swedish Research Council (15023), the Swedish Cancer Society (4708), the Swedish Foundation for Strategic Research (A303:156e), and Polysackaridforskning AB (Uppsala, Sweden).
References
- 1.Bernfield M., Gotte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 2.Bullock S. L., Fletcher J. M., Beddington R. S., Wilson V. A. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998;12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellaiche Y., The I., Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature (London) 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 4.Takei Y., Ozawa Y., Sato M., Watanabe A., Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2004;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- 5.Esko J. D., Lindahl U. Molecular diversity of heparan sulfate. J. Clin. Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher J. T. Heparan sulfate: growth control with a restricted sequence menu. J. Clin. Invest. 2001;108:357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology: further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- 8.Guimond S., Maccarana M., Olwin B. B., Lindahl U., Rapraeger A. C. Activating and inhibitory heparin sequences for FGF-2 (basic FGF): distinct requirements for FGF-1, FGF-2, and FGF-4. J. Biol. Chem. 1993;268:23906–23914. [PubMed] [Google Scholar]
- 9.Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature (London) 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 10.Rapraeger A. C., Krufka A., Olwin B. B. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 11.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 12.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 13.Allen B. L., Rapraeger A. C. Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly. J. Cell Biol. 2003;163:637–648. doi: 10.1083/jcb.200307053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z. L., Zhang L., Yabe T., Kuberan B., Beeler D. L., Love A., Rosenberg R. D. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J. Biol. Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 15.MacArthur C. A., Lawshe A., Xu J., Santos-Ocampo S., Heikinheimo M., Chellaiah A. T., Ornitz D. M. FGF-8 isoforms activate receptor splice forms that are expressed in mesenchymal regions of mouse development. Development. 1995;121:3603–3613. doi: 10.1242/dev.121.11.3603. [DOI] [PubMed] [Google Scholar]
- 16.Sher I., Weizman A., Lubinsky-Mink S., Lang T., Adir N., Schomburg D., Ron D. Mutations uncouple human fibroblast growth factor (FGF)-7 biological activity and receptor binding and support broad specificity in the secondary receptor binding site of FGFs. J. Biol. Chem. 1999;274:35016–35022. doi: 10.1074/jbc.274.49.35016. [DOI] [PubMed] [Google Scholar]
- 17.Bellosta P., Iwahori A., Plotnikov A. N., Eliseenkova A. V., Basilico C., Mohammadi M. Identification of receptor and heparin binding sites in fibroblast growth factor 4 by structure-based mutagenesis. Mol. Cell. Biol. 2001;21:5946–5957. doi: 10.1128/MCB.21.17.5946-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreuger J., Prydz K., Pettersson R. F., Lindahl U., Salmivirta M. Characterization of fibroblast growth factor 1 binding heparan sulfate domain. Glycobiology. 1999;9:723–729. doi: 10.1093/glycob/9.7.723. [DOI] [PubMed] [Google Scholar]
- 19.Kreuger J., Salmivirta M., Sturiale L., Gimenez-Gallego G., Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J. Biol. Chem. 2001;276:30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 20.Jemth P., Kreuger J., Kusche-Gullberg M., Sturiale L., Gimenez-Gallego G., Lindahl U. Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs: exploring the structural diversity by screening for fibroblast growth factor (FGF)1 and FGF2 binding. J. Biol. Chem. 2002;277:30567–30573. doi: 10.1074/jbc.M203404200. [DOI] [PubMed] [Google Scholar]
- 21.Jemth P., Smeds E., Do A. T., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. Oligosaccharide library-based assessment of heparan sulfate 6-O-sulfotransferase substrate specificity. J. Biol. Chem. 2003;278:24371–24376. doi: 10.1074/jbc.M212155200. [DOI] [PubMed] [Google Scholar]
- 22.Vives R. R., Pye D. A., Salmivirta M., Hopwood J. J., Lindahl U., Gallagher J. T. Sequence analysis of heparan sulphate and heparin oligosaccharides. Biochem. J. 1999;339:767–773. [PMC free article] [PubMed] [Google Scholar]
- 23.Loo B. M., Salmivirta M. Heparin/heparan sulfate domains in binding and signaling of fibroblast growth factor 8b. J. Biol. Chem. 2002;277:32616–32623. doi: 10.1074/jbc.M204961200. [DOI] [PubMed] [Google Scholar]
- 24.Inatani M., Irie F., Plump A. S., Tessier-Lavigne M., Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 25.Ashikari-Hada S., Habuchi H., Kariya Y., Itoh N., Reddi A. H., Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 26.Turnbull J. E., Fernig D. G., Ke Y., Wilkinson M. C., Gallagher J. T. Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. J. Biol. Chem. 1992;267:10337–10341. [PubMed] [Google Scholar]
- 27.Ostrovsky O., Berman B., Gallagher J., Mulloy B., Fernig D. G., Delehedde M., Ron D. Differential effects of heparin saccharides on the formation of specific fibroblast growth factor (FGF) and FGF receptor complexes. J. Biol. Chem. 2002;277:2444–2453. doi: 10.1074/jbc.M108540200. [DOI] [PubMed] [Google Scholar]
- 28.Thompson L. D., Pantoliano M. W., Springer B. A. Energetic characterization of the basic fibroblast growth factor–heparin interaction: identification of the heparin binding domain. Biochemistry. 1994;33:3831–3840. doi: 10.1021/bi00179a006. [DOI] [PubMed] [Google Scholar]
- 29.Raman R., Venkataraman G., Ernst S., Sasisekharan V., Sasisekharan R. Structural specificity of heparin binding in the fibroblast growth factor family of proteins. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreuger J., Lindahl U., Jemth P. Nitrocellulose filter binding to assess binding of glycosaminoglycans to proteins. Methods Enzymol. 2003;363:327–339. doi: 10.1016/S0076-6879(03)01062-0. [DOI] [PubMed] [Google Scholar]
- 31.Ibrahimi O. A., Zhang F., Lang Hrstka S. C., Mohammadi M., Linhardt R. J. Kinetic model for FGF, FGFR, and proteoglycan signal transduction complex assembly. Biochemistry. 2004;43:4724–4730. doi: 10.1021/bi0352320. [DOI] [PubMed] [Google Scholar]
- 32.Kreuger J., Matsumoto T., Vanwildemeersch M., Sasaki T., Timpl R., Claesson-Welsh L., Spillmann D., Lindahl U. Role of heparan sulfate domain organization in endostatin inhibition of endothelial cell function. EMBO J. 2002;21:6303–6311. doi: 10.1093/emboj/cdf638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J. P., Gong F., Hagner-McWhirter A., Forsberg E., Abrink M., Kisilevsky R., Zhang X., Lindahl U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J. Biol. Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- 34.Merry C. L., Bullock S. L., Swan D. C., Backen A. C., Lyon M., Beddington R. S., Wilson V. A., Gallagher J. T. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse. J. Biol. Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- 35.Shukla D., Liu J., Blaiklock P., Shworak N. W., Bai X., Esko J. D., Cohen G. H., Eisenberg R. J., Rosenberg R. D., Spear P. G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 36.Lander A. D., Selleck S. B. The elusive functions of proteoglycans: in vivo veritas. J. Cell Biol. 2000;148:227–232. doi: 10.1083/jcb.148.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.