Abstract
The endophytic Gram-negative bacterium Gluconacetobacter diazotrophicus SRT4 secretes a constitutively expressed levansucrase (LsdA, EC 2.4.1.10), which converts sucrose into fructooligosaccharides and levan. The enzyme is included in GH (glycoside hydrolase) family 68 of the sequence-based classification of glycosidases. The three-dimensional structure of LsdA has been determined by X-ray crystallography at a resolution of 2.5 Å (1 Å=0.1 nm). The structure was solved by molecular replacement using the homologous Bacillus subtilis (Bs) levansucrase (Protein Data Bank accession code 1OYG) as a search model. LsdA displays a five-bladed β-propeller architecture, where the catalytic residues that are responsible for sucrose hydrolysis are perfectly superimposable with the equivalent residues of the Bs homologue. The comparison of both structures, the mutagenesis data and the analysis of GH68 family multiple sequences alignment show a strong conservation of the sucrose hydrolytic machinery among levansucrases and also a structural equivalence of the Bs levansucrase Ca2+-binding site to the LsdA Cys339–Cys395 disulphide bridge, suggesting similar fold-stabilizing roles. Despite the strong conservation of the sucrose-recognition site observed in LsdA, Bs levansucrase and GH32 family Thermotoga maritima invertase, structural differences appear around residues involved in the transfructosylation reaction.
Keywords: five-bladed β-propeller, fructosyltransferase, Gluconacetobacter diazotrophicus, glycoside hydrolase, glycoside hydrolase 68 family (GH68 family), levansucrase
Abbreviations: Bs, Bacillus subtilis; FTF, fructosyltransferase; Gd, Gluconacetobacter diazotrophicus; GH, glycoside hydrolase; NCS, non-crystallographic symmetry; R.M.S.D., root mean square deviation
INTRODUCTION
Fructans (β-D-fructofuranosyl polymers with a terminal D-glucosyl moiety) constitute the primary carbohydrate reserve in 15% of higher plants comprising approx. 40000 species and appear in a wide range of bacterial and fungal species [1]. In plants, fructans are known to be involved in the stabilization of cellular membranes conferring protection from water stress caused by drought or low temperatures [2]. The physiological roles of fructans in bacteria have been less described, and it has been postulated that fructan biosynthesis may be involved in a variety of processes, such as survival of bacteria in soil, phytopathogenesis and symbiosis [3]. Of emerging importance is the use of fructo-oligosaccharides as health-promoting ‘pre-biotics’, which act as food sources for beneficial bacteria, such as bifidobacteria.
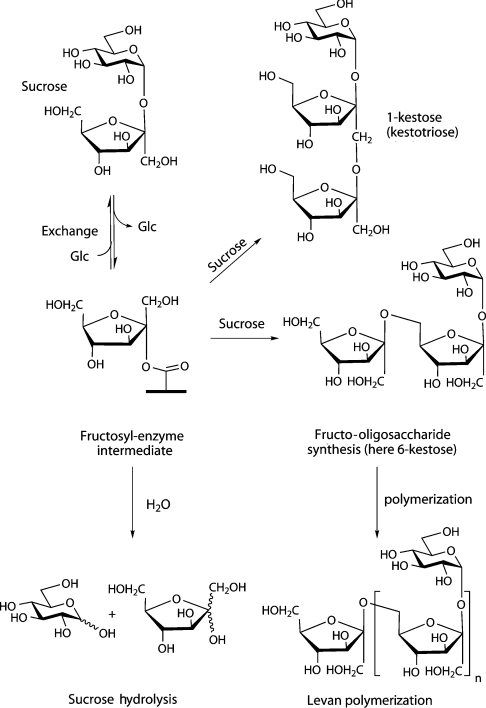
The synthesis of fructans starts with a transfructosylation reaction in which a sucrose molecule plays the role of fructosyl donor and with a second molecule of sucrose the initial acceptor of the fructosyl moiety (Scheme 1). Following initiation, the extension, the type of linkage and the branching of the fructan chain varies according to the enzyme and organism. Fructan synthesis in plants is carried out by the action of two or more FTFs (fructosyltransferases) that are able to produce complex mixtures of fructans with a great variety of linkages of the fructosides [2]. In bacteria, fructan biosynthesis is somewhat simpler because only one multifunctional enzyme is involved. This enzyme is named levansucrase (EC 2.4.1.10) when the product is a fructan of the type levan (β-2,6-linked polyfructan) or inulosucrase (EC 2.4.1.9) when the fructan is of the type inulin (β-2,1-linked polyfructan). Levansucrase catalyses the transfer of the fructosyl residue, from sucrose, to a variety of acceptors including water (sucrose hydrolysis), glucose (exchange), sucrose (oligofructoside synthesis) and fructan (polymerase reaction). The attack of sucrose can occur via O1 to form 1-kestose (1-kestotriose, isokestose; the basis of β-2,1-linked inulins) or via O6 to form the β-2,6-linked fructo-oligosaccharide 6-kestose. These reactions all occur via a Ping Pong mechanism involving the formation of a covalent fructosyl–enzyme intermediate [4–6] (Scheme 1).
Scheme 1. Various reactions catalysed by levansucrase.
Within the sequence-derived classification of GHs (glycoside hydrolases) and transglycosidases [7,8], bacterial FTFs are classified in family GH68 (http://afmb.cnrs-mrs.fr/CAZY) and, recently, the first three-dimensional structure of an FTF was published [9]. FTFs have also been shown to be structurally similar to sucrose-6-phosphate hydrolases, invertases, fructanases and eukaryotic FTFs found in family GH32. Together GH68 and GH32 thus comprise the β-fructofuranosidase clan GH-J.
The recent crystal structure of Gram-positive bacterium Bacillus subtilis (hereafter Bs) levansucrase and its complex with sucrose revealed the structural determinants for substrate recognition and the spatial disposition of three key catalytic acidic residues at the active site. The structure displays a five-bladed β-propeller topology that is shared with the elucidated crystal structures of Thermotoga maritima invertase (GH32) [10], Cellvibrio japonicus α-L-arabinanase (GH43) [11] and the Cichorium intybus fructan 1-exohydrolase IIa (GH32) [12]. Despite their high degree of structural similarities in the disposition of catalytic residues at the active site, the GH43 α-L-arabinanase operates with inversion of the anomeric configuration of the substrate, while levansucrases and invertases are ‘retaining’ enzymes.
The endophytic Gram-negative bacterium Gluconacetobacter diazotrophicus (hereafter Gd) secretes a constitutively expressed levansucrase (LsdA), which is responsible for utilization of the natural substrate sucrose [5,13]. The kinetic parameters of the levansucrases from the Bs and Gd are of the same order of magnitude in the initial step of the reaction, the formation of the fructosyl–enzyme intermediate, but there are remarkable differences in the affinity of the intermediates for the various fructosyl acceptors [5,14]. Bs levansucrase catalyses the formation of high-molecular-mass levan without accumulation of transient oligofructans, whereas Gd levansucrase synthesizes and accumulates a large amount of β-2,1-linked tri- and tetra-saccharides, with a much lower yield of levan under the same reaction conditions [5]. The structural determinants for the polymerization reaction of FTFs remain at present poorly understood, not least because a single structure makes it hard to mine the 48 (as of 25 April 2005) GH68 family members. The combined analysis of protein sequences, mutagenesis data and three-dimensional structures of levansucrases with different catalytic performances should thus contribute to a deeper insight into the polymerization mechanism of FTFs. In the present paper, as part of this process, we report the crystal structure of Gd levansucrase at a resolution of 2.5 Å (1 Å=0.1 nm) in its ligand-free state and limited biochemical analyses to support a detailed comparison with its Bs homologue.
MATERIALS AND METHODS
Bacterial strains and media
Escherichia coli strains DH5α [Φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169] and S17-1 {thi pro hsdR recA [RP4-2(Tc::Mu) (Km::Tn7)]} [15] were used as a cloning host and as a conjugative donor host respectively. E. coli strains were grown at 37 °C in Luria–Bertani broth supplemented with ampicillin (50 μg/ml) and tetracycline (12.5 μg/ml) when needed. Gd strains SRT4 (wildtype, Lev+) and AD5 (SRT4 lsdA::nptII-ble cassette, Lev−) [16] were grown at 28 °C in LGIE medium containing 5% (w/v) sucrose and 1% (v/v) glycerol [17]. Chloramphenicol (30 μg/ml) and tetracycline (20 μg/ml) were added to LGIE agar medium to select the transconjugants.
Site-directed mutagenesis
Plasmid pALS137 carries the 3.2-kb HindIII/NruI fragment from plasmid pALS5 [17] inserted into pUC8 digested with HindIII/SmaI. Plasmid pALS137 containing the levansucrase gene (lsdA) under its own promoter was used as template for mutagenesis using the GeneEditor Kit (Promega). The oligonucleotides 5′-GATGTCTGGGTCTGGAACACGTGGACCCTGATCGAC-3′, 5′-CCAGCGTGGCGTCGCGAACAGCACCGAGGCCGATC-3′ and 5′-CTGATTTCGGCCAACAGCGTCAATGATCAGACCGAACGGCCG-3′ were used to generate the mutations D135N, C339S and C395S respectively (amino acid substitutions are in bold, restriction sites created by silent mutations are underlined). Plasmids from E. coli transformants resistant to the GeneEditor Antibiotic Selection Mix were screened by digestion with PmlI (D135N), NruI (C339S) and BclI (C395S), and mutations were confirmed by DNA sequencing. The plasmids carrying the mutations D135N, C339S and C395S in the lsdA gene were named pALS191, pALS201 and pALS203 respectively.
Enzyme expression in lsdA::nptII-ble Gd mutant
Plasmids pALS137, pALS190, pALS201 and pALS203 were HindIII-linearized and inserted into the broad-range cloning vector pRK293 [18]. The resulting plasmids were named pALS148, pALS192, pALS205 and pALS207 respectively and were used to transform E. coli S17-1 (tra+). The conjugal mating procedure as described by Arrieta et al. [17] was used to mobilize plasmids pALS148, pALS192, pALS205 and pALS207 into the Gd mutant AD5, in which the lsdA gene is replaced by the nptII-ble cassette.
Crystallization, data collection and refinement
Details of protein expression, purification and crystallization were as reported previously [19]. Crystals grew over 2 weeks using ammonium sulphate in the range 0.35–0.6 M in 0.1 M sodium citrate, pH 5.6, with 30–60% ethanol and the detergents SDS (1.5–5.0 mM) and β-octylglucoside (10–40 mM) present. Handling crystals grown in ∼50% ethanol was extremely difficult and approx. 50 crystals had to be screened in order to gain a single ordered specimen for data collection. A 2.5 Å resolution dataset was collected from a single crystal at 100 K on beamline ID14-EH1 [ESRF (European Synchrotron Radiation Facility), Grenoble, France] using an ADSC Quantum-4 detector. Diffraction images were integrated with MOSFLM and scaled and reduced with SCALA from the CCP4 suite of programs [20]. Data collection statistics are summarized in Table 1. The structure was solved by molecular replacement using the co-ordinates of Bs levansucrase as a search model (Protein Data Bank accession code 1OYG) with all residues (except alanine and glycine) truncated to serine. The program AMoRe [21] was used with data in the resolution range 20–4 Å and an outer radius of Patterson integration of 37 Å.
Table 1. Summary of data collection and refinement statistics.
Values in parentheses refer to the highest resolution shell, 2.64–2.5 Å, for the final dataset. Rsym=Σ(I−〈I〉)2/ΣI2.
| Parameter | Value |
|---|---|
| Data collection | |
| High resolution (Å) | 2.5 |
| Completeness (%) | 99.2 (98.5) |
| Multiplicity | 3.3 (3.2) |
| Rsym | 0.078 (0.318) |
| 〈I/σI〉 | 11.6 (4.6) |
| Refinement | |
| Resolution used in refinement (Å) | 105.4–2.5 |
| Total number of non-hydrogen atoms | 7848 |
| Number of water molecules | 194 |
| Number of sulphate ions | 4 |
| Rcryst (%) | 19.6 |
| Rfree (%)* | 24.9 |
| Average B-factor | |
| Protein atoms (Å2) | 25.7 |
| Sulphate ions atoms (Å2) | 74.3 |
| Solvent atoms (Å2) | 34.1 |
| R.M.S.D. from ideal values† | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.51 |
| B-factor for bonded atoms | |
| Main chain bonds (Å2) | 0.612 |
| Main chain angles (Å2) | 0.786 |
| Side chain bonds (Å2) | 1.232 |
| Side chain angles (Å2) | 1.920 |
| Protein Data Bank accession code | 1W18 |
* Rfree calculated using 5% of total reflections omitted from refinement.
† Data from [45].
For refinement, 5% of the observations were set aside for cross-validation analysis [22] and were used to monitor various refinement strategies. Simulated annealing by slow cooling and positional refinements were initially performed on the molecular replacement solution with CNS [23] using NCS (non-crystallographic symmetry) restraints. CNS density modification procedures and 2-fold map averaging were applied to the calculated 2Fo-Fc maps, and this allowed the placement of the structurally most conserved residues. After several cycles of NCS refinement with NCSref [20] interspersed with cycles of manual building with Xfit [24], it was possible to place approx. 70% of the residues. Using phases calculated from this incomplete model, ARP/wARP [20] built approx. 90% of the residues as serine, valine, alanine and glycine that were later mutated to the correct side chains. The rest of the residues, mainly in loop regions, were placed manually with Xfit [24]. Final refinement cycles were performed with REFMAC [25] using NCS restraints and TLS (translation, libration and screw-rotation) corrections [26]. Water molecules and four sulphate ions were placed at peak positions of σa-weighted [27] Fo-Fc maps and 2Fo-Fc maps greater than 3σ and 1σ respectively and were checked at the graphic display. A summary of refinement statistics is shown in Table 1. The final model was validated against composite annealed omit maps from CNS. Ramachandran [28] statistics produced by PROCHECK [29] indicate that 82.9% of the residues are located in the most favoured regions of the plot and 16.1% in additional allowed regions. Eight residues were found in generous allowed regions with no residues located in the disallowed regions. Co-ordinates have been deposited at the Protein Data Bank [30] under accession code 1W18.
RESULTS AND DISCUSSION
Structure of Gd levansucrase LsdA
The open reading frame of Gd levansucrase (LsdA) encodes a 584-residue protein with a 30-residue N-terminal signal peptide that is cleaved off during secretion. The mature protein thus comprises a 553-residue polypeptide (numbering 31–584) with an N-terminal pyroglutamate. LsdA contains three cysteine residues (Cys157, Cys339 and Cys395), with Cys339 and Cys395 engaged in a disulphide bridge [31].
The crystal structure of LsdA (residues 62–554) was solved by molecular replacement using the homologous Bs levansucrase [9] as the search model at a resolution of 2.5 Å and was subsequently refined to crystallographic R-factors of 19.6% (Rcryst) and 24.9% (Rfree) (Table 1). The asymmetric unit contains two copies of the polypeptide chain, and, in addition, 194 water molecules and four sulphate ions have been modelled. Tight NCS restraints were maintained throughout the refinement and model-building harnessed electron density maps averaged according to the co-ordinate-derived NCS operators.
LsdA displays the five-bladed β-propeller fold (Figure 1), as first reported for the N-acetylglucosamine-binding tachylectin [32] and as first observed as an enzyme fold for the GH43 arabinanase [11] and then subsequently for the invertase from T. maritima [10] and the fructan 1-exohydrolase from Ci. intybus [12]. The five β-sheet modules are arranged in consecutive order with 5-fold pseudosymmetry around a central cavity. As in other β-propellers described so far, the four β-strands composing each β-sheet display a strong twist with an average of 90° between the first and the last strand. Similarly to Bs levansucrase and arabinanase, LsdA does not possess ‘molecular Velcro’, a term colloquially used to describe the closure of the propeller fold in which the N-terminal first strand forms the outermost β-strand of the C-terminal blade [33,34]. Fold stabilization is, however, provided by the packing of the N- and C-termini of the polypeptide. As in Bs levansucrase, the LsdA N-terminus runs along the perimeter of blade IV forming a clamp-like loop that adds a fifth β-strand to blade III (numbered as βIIIE in Figure 2). Additionally in LsdA, the N-terminus (residues 76–78) forms a short two-stranded antiparallel β-sheet that extends to a β-strand (residues 442–444) located in the insertion between βIVC and βIVD (β-strands numbered as β1 and β4). Different from Bs levansucrase, the C-terminus in LsdA does engage in hydrogen-bonding interactions to the central core through the formation of a two-stranded parallel β-sheet between β-strand 2 (residues 147–150) and β-strand 5 (residues 542–545).
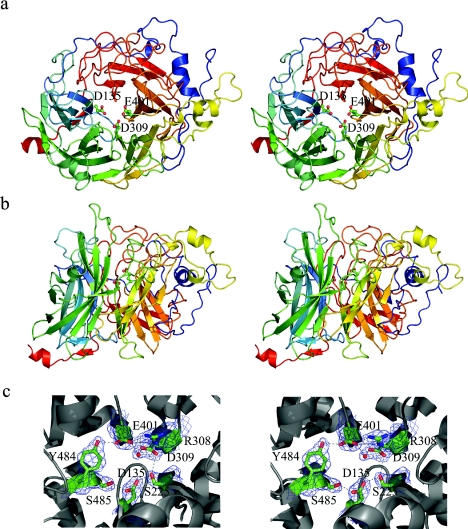
Figure 1. Three-dimensional structure of LsdA.
Superior (a) and lateral (b) stereo views of the five-bladed β-propeller fold. The colour is ‘ramped’ from N- (blue) to C- (red) terminus. Catalytic residues Asp135, Asp309 and Glu401 are shown in ball-and-stick representation. (c) Stereo view of the electron density map (contoured at 1σ level) ‘carved’ around catalytic residues and other residues involved in the hydrogen-bond (broken lines) network at the active site. These Figures were prepared with PYMOL [47].
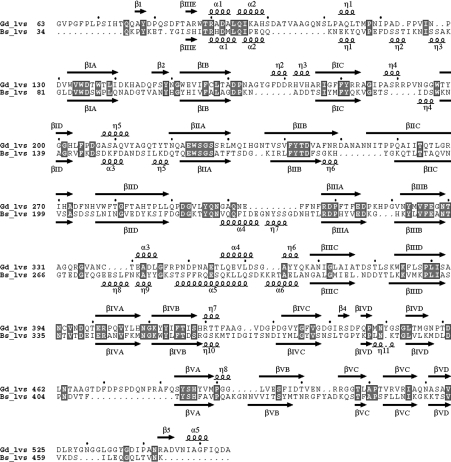
Figure 2. LsdA (Gd_lvs) and Bs levansucrase (Bs_lvs) sequence alignment based on structural superimposition generated by TOP [20].
Secondary-structural elements were assigned using DSSP [48] and are indicated by arrows (β-strands) and squiggles (helices). These elements were labelled following the Bs levansucrase numbering in [9]. A roman numeral (I–V) is assigned to each β-sheet module, and, in each module, the β-strands are numbered with a capital letter (A–D). α- and 310-helices (labelled α and η respectively) are numbered consecutively. Strictly conserved residues are boxed. The Figure was produced with ESPript [49].
Comparison with the Bs levansucrase
LsdA presents an overall sequence identity with its Bs homologue of 26%, but, in the 283 structurally equivalent residues, the sequences share 31% identity. The secondary-structure elements superimpose well, with an R.M.S.D. (root mean square deviation) of 1.24 Å over 283 equivalent Cα atoms. Not surprisingly, greater variation is observed in insertions into surface loops, a feature commonly found in the β-propeller fold [35]. Several helices, including short stretches of 310-helix, are conserved along the sequence alignment (Figure 2). In both structures, the longest insertion is found between β-strands IIIB and IIIC, where three helical stretches are interspersed.
LsdA contains a disulphide bridge (Cys339–Cys395) that links the extended loop between β-strands IIIB and IIIC with the insertion located between blades III and IV. The replacement of either Cys339 or Cys395 by serine reduced the kcat for sucrose hydrolysis approx. 60-fold (Table 2). Although equivalent cysteine residues appear in the levansucrases from the high-GC content Gram-positive species Arthrobacter sp. K-1 (GenBank® accession number BAB72022) and Actinomyces naeslundii (GenBank® accession number AAG09737) and the Gram-negative species Burkholderia cepacia R1808 (GenBank® accession number ZP_00218900) and Burkholderia pseudomallei K96243 (GenBank® accession number YP_110564), there is no general conservation of this disulphide bridge among GH68 family members. Indeed, a pair of cysteine residues is absent in most Gram-positive FTFs, including Bs levansucrase. Levansucrases from Gram-negative bacteria Rahnella aquatilis, Pseudomonas syringae and Pseudomonas aurantica display two highly conserved cysteine residues elsewhere, which are not equivalent to those found in LsdA and that might also be implicated in a disulphide bridge. It has been reported that the incubation with dithiothreitol or 2-mercaptoethanol decreased the enzyme activity of Ps. syringae levansucrase [36]. In Zymomonas mobilis levansucrase the substitution of serine for cysteine residues 121, 151 and 244 abolished the levan-forming activity and halved the sucrose hydrolysis activity [37]. In contrast, Bs levansucrase contains a Ca2+ ion connecting residues Asp241, Gln272, Leu308 and Asn310 located in the longest insert between β-strands IIIB and IIIC, with Asp339 placed in the insertion between blades III and IV. Leu308 is not found strictly conserved in Gram-positive levansucrases, possibly due to the fact that the Ca2+ interaction with this residue is through the backbone carbonyl oxygen. The residues involved in Ca2+ binding are strictly conserved among FTFs from Gram-positive bacteria, except the high-GC content species Ac. naeslundii and Arthrobacter sp., but variable in the levansucrases of Gram-negative bacteria (see partial alignment in Figure 3). Thus the activity of LsdA and other levansucrases from Gram-negative bacteria is not affected in the presence of metal ions, such as Ca2+, or the chelating agent EDTA [5,37,38]. In contrast, FTFs from the Gram-positive species Streptococcus mutans [39], Streptococcus salivarius [6] and Lactobacillus reuteri [40,41] show maximal activity only upon addition of 1 mM Ca2+. The fact that both the disulphide bridge in LsdA and the Ca2+-binding site in Bs levansucrase map in structurally equivalent regions in their respective molecular architectures suggests that these features play a similar role in structural maintenance of the catalytic glutamate residue (Glu401 in LsdA and Glu342 in Bs levansucrase), found in the highly conserved ‘D(E/Q)(T/I/V)ER’ motif, poised for catalysis [9].
Table 2. Kinetic parameters of wild-type and site-directed mutant enzymes.
The native and mutant enzymes were purified from culture supernatants as described by Arrieta et al. [17] and quantified by the Bradford procedure using BSA as a standard. The reaction mixtures (50 μl) containing 200 mM sucrose in 0.1 M sodium acetate, pH 5.0, were incubated with 0.3 unit of purified enzyme at 30 °C. After 15 min, the reaction was stopped by heating in a boiling-water bath for 5 min. Glucose released from sucrose hydrolysis was measured by a glucose oxidase/peroxidase-coupled colorimetric reaction. The kcat was calculated using 60000 as the Mr of a functional unit of levansucrase. The Km for sucrose was determined within a substrate concentration range 1–150 mM at 30 °C and pH 5.0. The kinetic parameters were determined by non-linear regression analysis of substrate–velocity plots using the LEONORA program [46]. One unit of enzyme activity is defined as the amount of enzyme releasing 1 μmol of glucose/min based on initial-velocity measurements under the following conditions: 0.2 M sucrose in 0.1 M sodium acetate buffer, pH 5.0, at 30 °C. Values are means±S.D. (n=6).
| Strain | Mutation | kcat (min−1) | Km (mM) | kcat/Km (M−1·min−1) |
|---|---|---|---|---|
| AD5 (pALS148) | Wild-type | (3.9±0.2)×103 | 11.9±0.4 | 3.3×105 |
| AD5 (pALS192) | D135N | 1.7±0.2 | 12.3±0.9 | 1.4×102 |
| AD5 (pALS205) | C339S | 66.9±2.3 | 13.8±1.2 | 4.8×103 |
| AD5 (pALS207) | C395S | 71.8±6.4 | 13.1±1.4 | 5.5×103 |
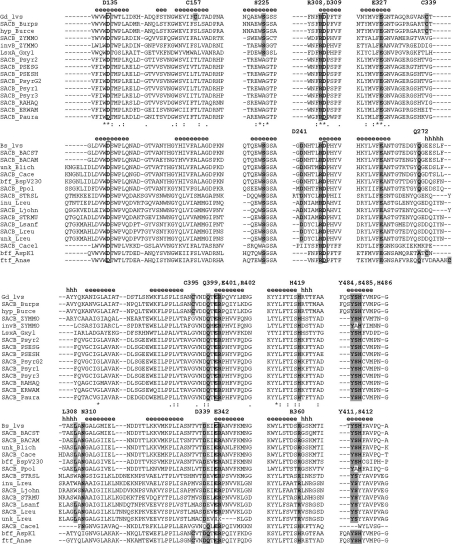
Figure 3. Multiple sequence alignment of selected fragments of GH68 family members.
The sequences are divided into two groups, Gram-negative and Gram-positive levansucrases headed by LsdA (Gd_lvs) and Bs levansucrase (Bs_lvs) respectively. Protein identifiers and access codes for each sequence in the SwissProt/TrEMBL and GenBank® databases are as follows: Gd LsdA (Gd_lvs, Q43998); B. pseudomallei K96243 levansucrase (SACB_Burps, YP_110564); B. cepacia R1808 hypothetical protein (hyp_Burce, ZP_00218900); Z. mobilis levansucrase (SACB_ZYMMO, Q60114, Q06487, Q60116), and invertase (invB_ZYMMO, AAA61488); Gluconacetobacter xylinus levansucrase (LsxA_Gxyl, BAA93720); Ps. syringae pv. tomato levansucrase (SACB_Psyr1, AAO54974; SACB_Psyr2, AAO55819; SACB_Psyr3, AAO59056); Ps. syringae pv. glycinea levansucrase (SACB_PSESG, O52408; SACB_PsyrG2, AAK49952); Ps. syringae pv. phaseolicola levansucrase (SACB_PSESH, O68609); R. aquatilis levansucrase (SACB_RAHAQ, O54435); Erwinia amylovora levansucrase (SACB_ERWAM, Q46654); Ps. aurantiaca levansucrase (SACB_Paura, AAL09386); Bs levansucrase (Bs_lvs, P05655, P70984); Bacillus stearothermophilus levansucrase (SACB_BACST, P94468); Bacillus amyloliquefaciens levansucrase (SACB_BACAM, P21130); Bacillus licheniformis unnamed protein product (unk_Blich, CAF05486); Clostridium acetobutylicum levansucrase (SACB_Cace, CAC1772; SACB_Cace1, CAC1774); Bacillus sp. V230 β-fructofuranosidase (bff_BspV230, BAA32083); Pa. polymyxa levansucrase (SACB_Ppol, CAB39327); Streptococcus salivarius levansucrase (SACB_STRSL, Q55242); L. reuteri inulosucrase (inu_Lreu, AAN05575), levansucrase (SACB_Lreu, AAO14618) and unnamed protein product (unk_Lreu, CAD19335); Lactobacillus johnsonii levansucrase (SACB_Ljohn, NP_964768); Streptococcus mutans levansucrase (SACB_STRMU, P11701); Lactobacillus sanfranciscensis levansucrase (SACB_Lsanf, CAD48195); Arthrobacter sp. K-1 β-fructofuranosidase (bff_AspK1, BAB72022); Ac. naeslundii FTF (ftf_Anae, AAG09737). Secondary-structural elements were assigned using DSSP [48] and are indicated by ‘e’ (β-strands) and ‘h’ (helices). Catalytic residues Asp135, Asp309 and Glu401 are boxed.
The active site of LsdA
Three strictly conserved acid residues have been identified in alignments of protein sequences corresponding to GH families 32, 43, 62 and 68 [42] (boxed residues in Figure 3). Site-directed mutagenesis of these carboxy residues has been performed for various representatives of GH68. On the basis of the mutagenesis and the structural information, it has been proposed that acidic residues Asp135, Asp309 and Glu401 of LsdA act as the catalytic nucleophile, the ‘transition-state stabilizer’ and the general acid–base catalyst respectively. Substitution of asparagine for Asp135 resulted in an enzyme with substantially impaired activity, with a 2300-fold decrease in kcat (Table 2). It has also been reported that mutation of Asp309 to asparagine in the RPD (Arg-Pro-Asp) motif reduced the kcat 75-fold, again without change in the Km [38].
In the crystal structure of LsdA, the catalytic constellation of carboxy residues is located at the bottom of a funnel-shaped cavity at the centre of the β-propeller (Figure 1). Each one of these residues, namely Asp135, Asp309 and Glu401, belong to the first and innermost β-strand of blades I, III and IV respectively. This feature is commonly found in this type of fold, where the surface of the central cavity is mostly lined by residues that belong to the first strand of each modular sheet [35]. The three carboxylic residues in LsdA are perfectly superimposable on to the equivalent residues in Bs levansucrase and are spaced some 4.8–7.4 Å from each other. The proposed catalytic nucleophile, Asp135, side chain forms two hydrogen bonds with side chains of Ser225 and Ser485 (Figure 1c); the same pattern of interactions is present in Bs levansucrase for the equivalent residue. Both serine residues are found highly conserved in the GH68 family multiple sequence alignment, except that the equivalent to Ser225 in some of the Gram-negative bacteria, including Pseudomonas, Rhanella and Erwinia species, have an alanine residue in this position (Figure 3). Interestingly, the LsdA motif Tyr484-Ser485-(His/Tyr/Trp)486 is strictly conserved in GH68 family, except in Z. mobilis invertase and Paenibacillus polymyxa levansucrase, where Ser485 is found replaced by alanine. The structurally equivalent residues of LsdA Ser485 and Bs levansucrase Ser412 in T. maritima invertase and Ci. intybus fructan 1-exohydrolase are Ala241 and Ala275 respectively, and, in the multiple sequence alignment of the GH32 family, an alanine residue is found highly conserved in this position (this motif was described as motif F in [43]). The high conservation of serine in this position in GH68 levansucrases and its replacement by alanine in GH32 invertases, levanases or sucrases suggests that this H-bond interaction could play an important role in the orientation of the nucleophilic residue during transfructosylation catalysis.
The side chain of Asp309 forms hydrogen bonds to Ser225, the Arg402 backbone amide nitrogen and a water molecule that, at the same time, forms a hydrogen bond with the catalytic residue Glu401. The Glu401 side chain in turn establishes hydrogen bonds with side chains of Glu327 and Tyr484 and a salt bridge with Arg308 (Figure 1c). The equivalent positions of LsdA Arg308 and Tyr484 are found strictly conserved in GH68 family and are also conserved in GH32 family members (described as motifs D and F in [43]). The LsdA equivalent Glu327 position is strictly conserved in GH68 family, but is highly variable in representatives of GH32 family.
Analysis of the sucrose complex of the inactive mutant of Bs levansucrase (Protein Data Bank accession code 1PT2) [9] revealed the residues involved in substrate recognition. All of these residues, except for Gln399/Glu340 and His419/Arg360 (LsdA/Bs levansucrase numbering), are present in LsdA. Indeed, structural overlap of this subset of co-ordinates gives a R.M.S.D. of 0.75 Å for the Cα atoms of residues that are responsible for sucrose recognition and catalysis. This close similarity allows the modelling and analysis of contacts of the sucrose molecule at the active site in LsdA. Figure 4 shows the LsdA sucrose-binding site and the potential hydrogen-bonding interactions of the sucrose molecule with LsdA based on the actual interactions with Bs levansucrase inactive mutant (Protein Data Bank accession number 1PT2). Most of the hydrogen-bonding interactions observed in the Bs levansucrase sucrose complex appear to be conserved in the modelled interaction of sucrose and LsdA. This high degree of structural conservation at the sucrose-binding site is in agreement with the enzymatic data that report kinetic constants of the same order of magnitude for the initial step of the transfructosylation reaction [5], i.e. the formation of the fructosyl–enzyme intermediate, and the overall sucrose hydrolysis reaction (Table 3). In addition, the residues involved in the hydrogen-bond network of interactions at the sucrose hydrolysis active site are found strictly conserved in the GH68 sequence alignment (Figure 3).
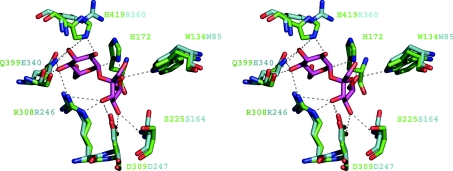
Figure 4. Sucrose-binding site of LsdA.
The Figure shows an overlap of the co-ordinates of the Bs levansucrase E342A-sucrose complex (Protein Data Bank accession code 1PT2) with the Gv levansucrase, LsdA. Potential hydrogen-bonding interactions are represented by broken lines. LsdA residues are bond-coloured green and Bs levansucrase are turquoise. Labels of depicted residues follow the same colour pattern.
Table 3. Apparent kinetic constants for the Bs and Gd levansucrases (adapted from [5]).
Values are given for the (forward) rate of intermediate formation (k+2) and subsequent hydrolysis, levan (lev) production and inulin (inu) production. Under conditions of 1 M sucrose, the Gd enzyme transfers approx. one-third of the fructosyl–enzyme intermediate to sucrose to form 1-kestose. Such a reaction was not observed for the Bs enzyme.
| Origin | k+2 (s−1) | khydrolysis (s−1) | klev (M−1·s−1) | kinu (M−1·s−1) |
|---|---|---|---|---|
| Gd | 550 | 51 | 1×104 | Not detected |
| Bs | 260 | 35 | 4×104 | 7×103 |
The sucrose hydrolytic machinery is located at the bottom of the catalytic pocket, and the residues that line this zone are located in the first β-strand of each blade. Immediately above this zone and surrounding the modelled glucosyl moiety of the sucrose molecule, it is possible to describe a fringe of residues that belong to the inserts that link β-strand B with C in each blade (see Figure 4). His419 of LsdA is located in this region and is the structural equivalent of Bs levansucrase Arg360 and the sequence equivalent of Z. mobilis levansucrase His296. Indeed, in Gram-negative levansucrases, a histidine residue is found strictly conserved in the equivalent position of LsdA His419, while in Gram-positive representatives an arginine residue is present (see the alignment in Figure 3). In both LsdA and Bs levansucrase, a tyrosine residue, whose aromatic ring is placed parallel to the plane of the histidine ring or the guanidinium group in LsdA His419 or Bs levansucrase Arg360 (Tyr484 in LsdA and Tyr411 in Bs levansucrase) is found strictly conserved in all levansucrases. Both Bs levansucrase Arg360 and Z. mobilis levansucrase His296 are reported to be crucial for the catalysis of the transfructosylation reaction [14,44], suggesting that a positively charged and electron delocalizing environment is required around this position. This suggestion is supported further by the fact that T. maritima invertase [10] does not have a structural equivalent residue to LsdA His419 or Bs levansucrase Arg360, and Ci. intybus fructan 1-exohydrolase shows a glutamate residue (Glu234) pointing in the opposite direction to Bs levansucrase Arg360 [12]. The presence of residual transfructosylation activity in Bs levansucrase Arg331→Ser/Leu mutants [14] also indicates that the structural determinants for the transfructosylation catalysis are more complex and at present not fully understood. Indeed, the absence of structural data for complexes between fructosyl acceptors and levansucrases makes it difficult to reach conclusions about what determines the outcome of the transfructosylation reaction in which LsdA releases large amount of 1-kestotriose (Glc- and kestotetraose in the initial phase of the reaction [5]), while Bs levansucrase synthesizes long-chain fructans under similar reaction conditions (Table 3). The crystallographic structures currently available of members of GH68 and GH32 families, together with the sequences and biochemical data, strongly support the hypothesis that FTFs evolved from invertases through comparatively small mutational events [2]. The LsdA structure presented in the present paper sheds some light on some of these changes and should help inform subsequent mutagenesis experiments in order to explore and harness the structural determinants that are responsible for transfructosylation.
Acknowledgments
We thank Professor Guy Dodson from the National Institute for Medical Research in London and the Chemistry Department, University of York for his support on this project. Collaboration between C.M.-F. and the University of York was made possible through funding from the Royal Society. G.J.D. is a Royal Society University Research Fellow.
References
- 1.Cairns A. J. Fructan biosynthesis in transgenic plants. J. Exp. Bot. 2003;54:549–567. doi: 10.1093/jxb/erg056. [DOI] [PubMed] [Google Scholar]
- 2.Vijn I., Smeekens S. Fructan: more than a reserve carbohydrate? Plant Physiol. 1999;120:351–360. doi: 10.1104/pp.120.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cote G. L., Ahlgren J. A. Metabolism in microorganisms, part I: levan and levansucrase. In: Suzuki M., Chatterton N. J., editors. Science and Technology of Fructans. Boca Raton: CRC Press; 1993. pp. 141–168. [Google Scholar]
- 4.Chambert R., Treboul G., Dedonder R. Kinetic studies of levansucrase of Bacillus subtilis. Eur. J. Biochem. 1974;41:285–300. doi: 10.1111/j.1432-1033.1974.tb03269.x. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez L., Arrieta J., Menendez C., Vazquez R., Coego A., Suarez V., Selman G., Petit-Glatron M. F., Chambert R. Isolation and enzymic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem. J. 1995;309:113–118. doi: 10.1042/bj3090113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song D. D., Jacques N. A. Purification and enzymic properties of the fructosyltransferase of Streptococcus salivarius ATCC 25975. Biochem. J. 1999;341:285–291. [PMC free article] [PubMed] [Google Scholar]
- 7.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrissat B., Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 9.Meng G., Futterer K. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 2003;10:935–941. doi: 10.1038/nsb974. [DOI] [PubMed] [Google Scholar]
- 10.Alberto F., Bignon C., Sulzenbacher G., Henrissat B., Czjzek M. The three-dimensional structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J. Biol. Chem. 2004;279:18903–18910. doi: 10.1074/jbc.M313911200. [DOI] [PubMed] [Google Scholar]
- 11.Nurizzo D., Turkenburg J. P., Charnock S. J., Roberts S. M., Dodson E. J., McKie V. A., Taylor E. J., Gilbert H. J., Davies G. J. Cellvibrio japonicus α-L-arabinanase 43A has a novel five-blade β-propeller fold. Nat. Struct. Biol. 2002;9:665–668. doi: 10.1038/nsb835. [DOI] [PubMed] [Google Scholar]
- 12.Verhaest M., Ende W. V., Roy K. L., De Ranter C. J., Laere A. V., Rabijns A. X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J. 2005;41:400–411. doi: 10.1111/j.1365-313X.2004.02304.x. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez L., Sotolongo M., Rosabal Y., Menendez C., Ramirez R., Caballero-Mellado J., Arrieta J. Structural levansucrase gene (lsdA) constitutes a functional locus conserved in the species Gluconacetobacter diazotrophicus. Arch. Microbiol. 2000;174:120–124. doi: 10.1007/s002030000173. [DOI] [PubMed] [Google Scholar]
- 14.Chambert R., Petit-Glatron M. F. Polymerase and hydrolase activities of Bacillus subtilis levansucrase can be separately modulated by site-directed mutagenesis. Biochem. J. 1991;279:35–41. doi: 10.1042/bj2790035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon R., Priefer U., Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 16.Hernandez L., Arrieta J., Betancourt L., Falcon V., Madrazo J., Coego A., Menendez C. Levansucrase from Acetobacter diazotrophicus SRT4 is secreted via periplasm by a signal-peptide-dependent pathway. Curr. Microbiol. 1999;39:146–152. doi: 10.1007/s002849900436. [DOI] [PubMed] [Google Scholar]
- 17.Arrieta J., Hernandez L., Coego A., Suarez V., Balmori E., Menendez C., Petit-Glatron M. F., Chambert R., Selman-Housein G. Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiology. 1996;142:1077–1085. doi: 10.1099/13500872-142-5-1077. [DOI] [PubMed] [Google Scholar]
- 18.Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Fleites C., Tarbouriech N., Ortiz-Lombardia M., Taylor E., Rodriguez A., Ramirez R., Hernandez L., Davies G. J. Crystallization and preliminary X-ray diffraction analysis of levansucrase (LsdA) from Gluconacetobacter diazotrophicus SRT4. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:181–183. doi: 10.1107/s0907444903025514. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A. T. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 23.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.McRee D. E. XtalView/Xfit – a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov G. N. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Winn M. D., Isupov M. N., Murshudov G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 27.Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. Sect. A Found. Crystallogr. 1986;42:140–149. [Google Scholar]
- 28.Ramachandran G. N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski R. A., Moss D. S., Thornton J. M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 30.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betancourt L., Takao T., Hernandez L., Padron G., Shimonishi Y. Structural characterization of Acetobacter diazotropicus levansucrase by matrix-assisted laser desorption/ionization mass spectrometry: identification of an N-terminal blocking group and a free-thiol cysteine residue. J. Mass Spectrom. 1999;34:169–174. doi: 10.1002/(SICI)1096-9888(199903)34:3<169::AID-JMS780>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Beisel H. G., Kawabata S., Iwanaga S., Huber R., Bode W. Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker S. C., Saunders N. F., Willis A. C., Ferguson S. J., Hajdu J., Fulop V. Cytochrome cd1 structure: unusual haem environments in a nitrite reductase and analysis of factors contributing to β-propeller folds. J. Mol. Biol. 1997;269:440–455. doi: 10.1006/jmbi.1997.1070. [DOI] [PubMed] [Google Scholar]
- 34.Neer E. J., Smith T. F. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 35.Paoli M. Protein folds propelled by diversity. Prog. Biophys. Mol. Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 36.Hettwer U., Gross M., Rudolph K. Purification and characterization of an extracellular levansucrase from Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 1995;177:2834–2839. doi: 10.1128/jb.177.10.2834-2839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senthilkumar V., Busby S. J., Gunasekaran P., Senthikumar V., Bushby S. J. Serine substitution for cysteine residues in levansucrase selectively abolishes levan forming activity. Biotechnol. Lett. 2003;25:1653–1656. doi: 10.1023/a:1025650928313. [DOI] [PubMed] [Google Scholar]
- 38.Ohtsuka K., Hino S., Fukushima T., Ozawa O., Kanematsu T., Uchida T. Characterization of levansucrase from Rahnella aquatilis Jcm-1683. Biosci. Biotechnol. Biochem. 1992;56:1373–1377. [Google Scholar]
- 39.Scales W. R., Long L. W., Edwards J. R. Purification and characterization of a glycosyltransferase complex from the culture broth of Streptococcus mutans FA1. Carbohydr. Res. 1975;42:325–338. doi: 10.1016/s0008-6215(00)84274-3. [DOI] [PubMed] [Google Scholar]
- 40.van Hijum S. A., Szalowska E., van der Maarel M. J., Dijkhuizen L. Biochemical and molecular characterization of a levansucrase from Lactobacillus reuteri. Microbiology. 2004;150:621–630. doi: 10.1099/mic.0.26671-0. [DOI] [PubMed] [Google Scholar]
- 41.van Hijum S. A., van der Maarel M. J., Dijkhuizen L. Kinetic properties of an inulosucrase from Lactobacillus reuteri 121. FEBS Lett. 2003;534:207–210. doi: 10.1016/s0014-5793(02)03841-3. [DOI] [PubMed] [Google Scholar]
- 42.Pons T., Naumoff D. G., Martinez-Fleites C., Hernandez L. Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins. 2004;54:424–432. doi: 10.1002/prot.10604. [DOI] [PubMed] [Google Scholar]
- 43.Pons T., Olmea O., Chinea G., Beldarrain A., Marquez G., Acosta N., Rodriguez L., Valencia A. Structural model for family 32 of glycosyl-hydrolase enzymes. Proteins. 1998;33:383–395. doi: 10.1002/(sici)1097-0134(19981115)33:3<383::aid-prot7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 44.Yanase H., Maeda M., Hagiwara E., Yagi H., Taniguchi K., Okamoto K. Identification of functionally important amino acid residues in Zymomonas mobilis levansucrase. J. Biochem. (Tokyo) 2002;132:565–572. doi: 10.1093/oxfordjournals.jbchem.a003258. [DOI] [PubMed] [Google Scholar]
- 45.Engh R. A., Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:392–400. [Google Scholar]
- 46.Cornish-Bowden A. Oxford: Oxford University Press; 1995. Analysis of Enzyme Kinetic Data. [Google Scholar]
- 47.DeLano W. L. San Carlos, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 48.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 49.Gouet P., Courcelle E., Stuart D. I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]