Abstract
AurA (Aurora-A) is a ubiquitous protein kinase regulating entry into mitosis and shown to promote transformation upon overexpression. In order to gain information on the structural features determining its substrate specificity, we assayed human recombinant AurA on a variety of phosphoacceptor peptide substrates including a series of properly modified derivatives of the Kemptide (ALRRASLGAA). The data presented here show that AurA is a basophilic Ser/Thr protein kinase recognizing the consensus R/K/N-R-X-S/T-B, where B denotes any hydrophobic residue with the exception of Pro. We show that the presence of a Pro at position n+1 fully abrogates phosphorylation of the peptide substrate. Although the consensus for AurA is reminiscent of that of PKA (protein kinase A), it significantly differs from the latter for a much more stringent dependence on the hydrophobic residue at n+1 and for its tolerance of residues other than Arg at position n−3. Based on the finding that the peptide ALKRASLGAA is not a substrate of PKA while still providing a sensitive assay of AurA activity, we suggest that this peptide may be used for differential screening of the two kinases. We have further validated the AurA consensus by generating a peptide (APSSRRTT288LCGT) that comprises the main AurA autophosphorylation site and by showing that AurA phosphorylated this peptide exclusively at one site fulfilling its consensus (Thr288). Moreover, we show that AurA could autophosphorylate at Thr288 through an intermolecular mechanism of reaction and that, in vivo, PKA was not involved with Thr288 phosphorylation. The evidence obtained in the present study provides a rational tool for predicting AurA sites in potential substrates of physiological significance.
Keywords: Aurora-A, consensus sequence, peptide substrate, phosphorylation, protein kinase, site specificity
Abbreviations: AurA, Aurora-A; CDC2, cell division cycle 2 kinase; CK1, casein kinase 1; DTT, dithiothreitol; HEK-293T cells, human embryonic kidney 293T cells; kd-AurA, kinase-dead AurA; PKA, protein kinase A; wt-AurA, wild-type AurA
INTRODUCTION
Aurora protein kinases were initially identified in yeast and Drosophila for their role in the control of chromosome segregation and cytokinesis through the regulation of microtubule activity [1]. Budding yeast mutants for Ipl1, the single form of the Aurora kinase present in this organism [2], are characterized by abnormal ploidy [3]. Aurora mutants in Drosophila typically display monopolar spindles that result from defective centrosome separation [4]. Of the three human homologues so far described, AurA (Aurora-A) and AurB were isolated in a screening for kinases overexpressed in colon carcinoma [5]. AurA is expressed at low-levels in most tissues but is particularly abundant in tumour cell lines [6,7] and accordingly, the gene coding for AurA maps to a region frequently amplified in human tumours and cancer cell lines [5,6,8,9]. AurA, which is localized at the centrosomes during interphase and at the spindle throughout mitosis [10], displays a peak of kinase activity at G2 phase (growth 2 phase), before CDC2 (cell division cycle 2 kinase) and AurB [5]. AurA kinase activity results from the balance of positive and negative phosphorylation: whereas autophosphorylation [11] and phosphorylation by as yet unknown upstream kinases [12] contribute to confer full AurA activation, phosphorylation by GSK3 (glycogen synthase kinase 3) at two sites close to the T-loop apparently triggers AurA autophosphorylation at an inhibitory site [13]. A further layer of complexity in the control of AurA activity is conferred by interaction with protein phosphatase 1, which results in mutual regulation of the two enzymatic activities [14]. Degradation of AurA occurs at mitotic exit and is mediated by the anaphase-promoting complex/cyclosome (APC/C) [15–17]. Depletion of AurA by siRNA (small interfering RNA) interference resulted in impaired activation of cyclin B/CDK1 (cyclin-dependent kinase 1) and was followed by an almost complete block of entry into mitosis [18]. On the other hand, overexpression of AurA was shown to cause transformation of Rat1 and NIH3T3 cells, which in turn could grow as tumours in nude mice [5]. Among the physiological targets of AurA so far identified are histone H3, BRCA1 (breast-cancer susceptibility gene 1) and p53 [19–21]. Although in all cases phosphorylation by AurA was shown to affect the substrate's biological properties, the sites phosphorylated in these proteins could not be demonstrated to be exclusive targets of AurA.
To gain information on physiologically relevant AurA targets, we set out to define the consensus sequence of this kinase. Using human recombinant AurA, expressed in Escherichia coli and purified to near homogeneity, we screened a set of synthetic peptides derived from the Kemptide as well as peptides modelled on other phosphoacceptor sites. This has led to the definition of the consensus sequence recognized by AurA and the identification of a number of local features acting as positive or negative specificity determinants.
MATERIALS AND METHODS
AurA cloning, expression and purification
Full-length human AurA was obtained by means of PCR using Pfu polymerase (Stratagene, Cambridge, U.K.). The first-strand cDNA template was synthesized on polyadenylated mRNA purified from mitotic HeLa cells using M-MLV reverse transcriptase (Promega, Chilworth, Southampton, U.K.). The gene-specific forward and reverse primers used in the PCR were: 5′-CGCGGATCCATGGACCGATCTAAAGAAAACTGCATTTC-3′ and 5′-GGCGAGCTCCTAAGACTGTTTGCTAGCTGATTCTTTG-3′ respectively. The 1.2 kb PCR product was purified using the QIAquick PCR purification kit (Qiagen, Chatsworth, CA, U.S.A.), subcloned into pBluescript SK+ (Stratagene) via BamHI/XhoI sites and controlled by sequencing. The wt (wild-type) and kd (kinase-dead) (Asp274>Asn) AurA open reading frames were subcloned in pTXB3 (New England Biolabs, Hitchin, Herts., U.K.) and recombinant AurA was expressed as Intein–AurA fusion protein in the BL21 E. coli strain. In order to purify AurA, cells were lysed in prechilled buffer A (20 mM Tris, pH 8.0, 500 mM NaCl, 0.1% Triton X-100, 1 mM EDTA and 10% glycerol, containing Roche complete protease inhibitor cocktail) and extracts were sonicated before centrifugation for 30 min at 38724 g in SS-34 rotor (Sorvall Centrifuges, Kendro Laboratory Products, Bishop's Stortford, Herts., U.K.) at 4 °C. The soluble fraction was filtered and loaded on to a 10 ml chitin column (New England Biolabs), pre-equilibrated in buffer A at 4 °C. After washing with 200 ml of buffer A, flow was stopped and the column was left overnight in buffer A containing 100 mM DTT (dithiothreitol) to facilitate intein self-splicing. On the following day, proteins were step-eluted using buffer A and aliquots of each fraction were examined by SDS/PAGE.
Cell culture
HeLa and HEK-293T (human embryonic kidney-293T) cells were maintained in Dulbecco's modified Eagle's medium (OmniLab, Mettmenstetten, Switzerland) supplemented with 10% (v/v) fetal calf serum (Life Technologies, Basel, Switzerland), penicillin (100 units/ml) and streptomycin (100 μg/ml) (complete medium). For synchronization experiments, cells were seeded at 1×106 in 10-cm-diameter dishes and treated after 24 h with 2 mM thymidine (SynGen, Cambridge, U.K.) for 16 h, released for 8 h in complete medium and thymidine (2 mM) was added for a second period (15 h). The extent of synchronization was controlled by flow cytometric analysis of DNA. Transient transfections were performed using FuGene6 (Roche, Lewes, East Sussex, U.K.). Treatment of cells with the PKA (protein kinase A) inhibitor H89 (Calbiochem, Bad Soden, Germany) or the PKA activators forskolin and 8Br-cAMP (Calbiochem) are described in the Figure legends.
Western blotting
A polyclonal serum to AurA (AurA-Pab36) was generated in two rabbits (Clonestar, Brno, Czech Republic) using full-length AurA. IgGs were purified by FPLC on a Protein A–Sepharose column (Amersham Biosciences). The antibody to AurA phospho-Thr288 and CDC2 phospho-Tyr15 were purchased from Cell Signaling Technology (Beverly, MA, U.S.A.). The antibodies to CDC2 and histone H3 phospho-Ser10 were from Upstate Biotechnology (Lake Placid, NY, U.S.A.). Western-blot analysis was performed after separation of proteins on polyacrylamide gels under denaturing conditions. Proteins were transferred to PVDF membrane (Amersham Biosciences), the membrane was probed with specific antibodies and bands were revealed with the ECL® system (Amersham Biosciences).
MS analysis
Aliquots corresponding to 10 μg of purified wt- and kd-AurA were precipitated by the addition of 125 μg/ml sodium deoxycholate and 10% (w/v) trichloroacetic acid [22]. Proteins were dissolved in electrophoresis sample buffer (100 mM Tris/HCl, pH 6.8, 100 mM DTT, 2%, w/v, SDS, 30% glycerol and 0.2% Bromophenol Blue) [23] and heated for 5 min at 95 °C. Samples were alkylated with idoacetamide (55 mM) for 45 min in the dark before subjecting them to SDS/PAGE. The gel was stained with Coomassie Blue and bands corresponding to wt- or kd-AurA were excised and digested with 1 μg of trypsin (Promega) in 50 mM ammonium bicarbonate (pH 8.0) at 37 °C for 16 h [24]. The resulting peptides were analysed by capillary liquid chromatography tandem MS (LC-MS/MS) using a Magic C18 100 μm×10 cm HPLC column (Spectronex, Basel, Switzerland) connected on line to an ion-trap Finnigan DecaXP (ThermoFinnigan, San Jose, CA, U.S.A.). A linear gradient from 5 to 50% B [0.1% formic acid and 80% (v/v) acetonitrile in water] in A (0.1% formic acid and 2% acetonitrile in water) in 60 min was delivered with a Rheos 2000 HPLC system (Flux, Basel, Switzerland) at 100 μl/min. A precolumn flow splitter reduced the flow to approx. 300 nl/min and the peptides were manually loaded with a 10 μl Hamilton syringe on a peptide cap-trap (Michrom BioResources, Auburn, CA, U.S.A.) mounted in the injection loop of the MS. The eluting peptides were ionized by electrospray ionization, detected and the peptide ions were automatically selected and fragmented by collision-induced dissociation (MS/MS) in the ion-trap. Individual MS/MS spectra were compared against the known protein sequence using TurboSequest software [25]. Phosphorylated peptides were sequenced more than once.
Peptide synthesis
Solvents, resin and coupling reagents for peptide synthesis were from Applied Biosystems (Foster City, CA, U.S.A.). All resins and protected amino acids were purchased from Novabiochem (Laufelfingen, Switzerland). HPLC grade solvents were obtained from Merck (Darmstadt, Germany). Synthetic peptides were prepared by solid-phase peptide synthesis using an automated peptide synthesizer (model 431-A, Applied Biosystems). The fluoren-9-ylmethoxycarbonyl (Fmoc) strategy was used throughout the peptide chain assembly [26]. Wang resin (loading of 0.96 mmol/g) was used as solid support to obtain 0.1 mmol of each peptide. A parallel synthesis of the peptide APSSRRTTLCGT was also performed on the amino PEGA resin (Novabiochem) using the same procedure described above in order to obtain a permanently anchored peptide to be employed for direct Edman sequencing analysis. Side-chain-protecting groups were tert-butyloxycarbonyl for Lys and Trp, trytyl for Gln, Asn and Cys; tert-butyl ester for Asp and Glu; tert-butyl ether for Ser, Tyr and Thr; 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulphonyl for Arg. Coupling was performed with a single reaction for 40–50 min by a 0.45 M solution in N,N-dimethylformamide (DMF) of 2-(1-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and N-hydroxybenzotriazole (HOBt) in the presence of N-ethyldi-isopropylamine (DIEA), following the manufacturer's instructions. Peptide cleavage was usually performed by reacting the peptidyl-resins with trifluoroacetic acid/water/thioanisole/ethane dithiol/phenol (10 ml/0.5 ml/0.5 ml/0.250 ml/750 mg) for 2–2.5 h. The peptides were precipitated with ice-cold ethyl ether and isolated by centrifugation. Pellets were washed several times with ether, dissolved in abundant water and freeze-dried. Purity was checked by analytical reversed-phase HPLC on a 5 μm C18 Symmetry300 column, 4.6×250 mm (Waters, Milford, MA, U.S.A.) using a linear gradient of 5–40% acetonitrile in 0.1% trifluoroacetic acid at 1 ml/min. The purity of the peptides was in the range of 85–95%. The peptide AcFNRTSLPWQGLKAATKKQKY was purified by a preparative reversed-phase HPLC column (Prep Nova-Pak HR C18, 250 mm×10 cm, 6 μm bead size; Waters) at 12 ml/min using a linear gradient as described above. The molecular mass of the peptide was confirmed by MS with direct infusion on a Micromass ZMD-4000 mass spectrometer (Waters-Micromass, Milford, MA, U.S.A.).
Edman sequencing
The peptide APSSRRTTLCGT, which was covalently bound to the beads of the solid support employed during synthesis, was phosphorylated under conditions described below and exhaustively washed to eliminate excess ATP and other reagents. Subsequently, few beads bearing the radiolabelled peptide were loaded on a Procise HT 491 protein sequencer (Applied Biosystems). Modified cartridge chemistry cycle was used to isolate ATZ amino acids, according to the manufacturer's instructions. No-flask cycle or HPLC gradient cycles were loaded. At every cycle the removed ATZ amino acids were quantitatively transferred to an external fraction collector connected to the ATZ port, using 90% (v/v) methanol and 10% water as solvent (S1). The collected fractions (600 μl) were supplemented with 2 ml scintillation cocktail and counted for 1 min in a liquid-scintillation counter. The recovered/measured 33P radioactivity at every cycle was plotted against the primary sequence of the peptide substrate.
Phosphorylation assay
Phosphorylation reactions were performed by incubating the phosphorylatable protein or peptide substrate in 25 μl of a medium containing 50 mM Tris/HCl (pH 8), 10 mM MgCl2, 1 mM DTT, 50 μM [γ-33P]ATP (specific radioactivity, 1–2 μCi/nmol) and 6 ng of recombinant protein kinase AurA for 10 min at 37 °C. PKA activity was similarly assayed by incubating peptide substrates in a solution containing 50 mM Tris/HCl (pH 7.5), 10 mM MgCl2, 10−6 M cAMP and 10–20 ng of PKA holoenzyme. All activity assays were linear with respect to time and enzyme concentration. The in vitro reactions shown in Figure 1 were performed in the conditions described above with the exception that [γ-32P]ATP (specific activity, 5–10 μCi/nmol) was used. The phosphate incorporated into protein substrates was evaluated by subjecting samples to SDS/PAGE, staining and autoradiography. The radiolabelled peptides were isolated and quantified by a procedure using phosphocellulose filters [27]. Km values were calculated from Lineweaver–Burk double-reciprocal plots of the data. The values obtained represent the mean of at least three independent experiments.
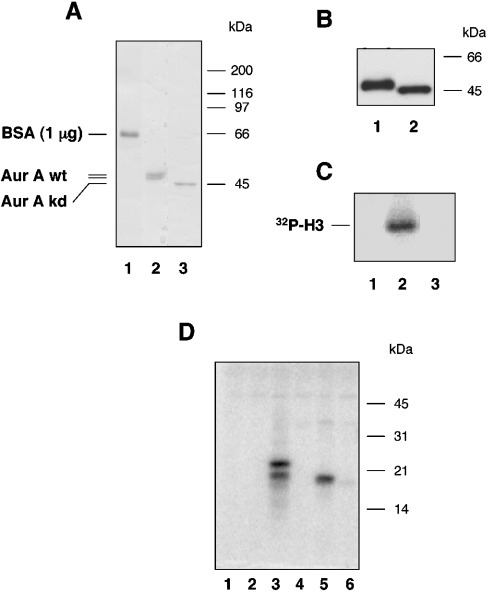
Figure 1. Analysis and characterization of recombinant AurA.
(A) The extent of AurA purification was examined by resolving an aliquot of the first chitin-column fraction on an SDS/polyacrylamide gel. Aliquots derived from preparation of wt-AurA (lane 2) or kd-AurA (lane 3) are shown. To appreciate the protein amount, the Coomassie Blue signal given by 1 μg of BSA is shown in lane 1. (B) Aliquots of the same material were examined by Western-blot analysis using purified antibody AurA-Pab36. Lane 1, wt-AurA; and lane 2, kd-AurA. (C) Kinase activity of the purified fractions shown in (A) was assayed on histone H3, as indicated in the Materials and methods section. Lane 1, blank; lane 2, wt-AurA; and lane 3, kd-AurA. (D) Purified AurA was tested on a set of artificial substrates used at 0.1 mg/ml final concentration and resolved on 15% (w/v) Laemmli gel. Lane 1, blank; lane 2, PCNA; lane 3, MBP; lane 4, histone H1; lane 5, histone H3; and lane 6, histone HIII-S (Lys-rich).
Phosphoamino acid analysis
Aliquots of radiolabelled peptide substrates were subjected to partial acid hydrolysis in 6 M HCl at 105 °C for 4 h followed by high-voltage paper electrophoresis and corrections for hydrolytic loss of pSer (48%) and pThr (14%) as described previously [28]. The hydrolytic loss was not significantly influenced by the structure of the peptide substrates.
RESULTS
Purification and characterization of human recombinant AurA
BL-21 cells were transformed with plasmids encoding either wt- or kd-AurA as C-terminal fusion to the self-splicing protein intein. AurA was purified to near-homogeneity in one single chromatographic step (Figure 1A). Coomassie Blue stained bands corresponding to wt- and kd-AurA clearly displayed distinct electrophoretic mobility. Western-blot analysis performed with a purified AurA-Pab36 (Figure 1B), confirmed the identity of the protein bands observed in the Coomassie Blue-stained gel. Wt-AurA was capable of phosphorylating histone H3, whereas the kd-form was completely inactive (Figure 1C). The specific activity of the purified kinase was found to be 0.42×104 pmol·min−1·mg−1 using histone H3 as substrate. Wt-AurA could also phosphorylate MBP (myelin basic protein) with comparable efficiency to histone H3, but was incapable of phosphorylating histone H1, a mixture of Lys-rich histones or PCNA (proliferating-cell nuclear antigen; Figure 1D).
Specificity determinants of AurA
Preliminary screening of AurA activity was performed towards a wide spectrum of peptide substrates previously developed to characterize the consensus of a variety of protein kinases, including PKA, PKC, p90RSK, CDC2, DYRK1α (dual-specificity tyrosine-phosphorylated and regulated kinase 1α), CK1 (casein kinase 1), CK2 and Golgi casein kinase [27]. The results obtained disclosed the basophilic nature of AurA, which proved inactive towards peptide substrates of acidophilic and proline-directed kinases (results not shown). Among the basic peptides readily phosphorylated by AurA the best one was a slightly extended version of the so-called Kemptide, ALRRASLGAA, a typical substrate of PKA as well as of several other basophilic protein kinases. This peptide was in fact phosphorylated by AurA with kinetic constants comparable with those of PKA (Table 1, peptide 1). Phosphorylation of Kemptide by PKA, as well as by a number of other basophilic protein kinases, is specified by the two arginine residues at positions n−2 and n−3. This was also the case of AurA, as shown by the detrimental effect of replacing both arginine residues with lysines (compare peptide 1 with peptide 5) and of transposing them to positions n−4/n−5 (peptide 4). Although both these modifications also abrogated phosphorylation by PKA, other substitutions disclosed significant differences between the sequence specificities of AurA and PKA. Of special interest in this respect is the effect of replacing the hydrophobic residue at position n+1 (Leu): substitution of this single residue with Ala (peptide 12) caused an approx. 30-fold reduction in phosphorylation efficiency by AurA. This was mostly accounted for by a significant decrease in Kcat, whereas its effect on PKA-catalysed phosphorylation was modest, entirely accounted for by just a 2-fold decrease in Kcat. This suggested that, in the case of AurA, a hydrophobic residue at position n+1 is a specificity determinant almost as powerful as the Arg-Arg doublet at positions n−2/n−3. To corroborate this conclusion, we introduced additional substitutions of Leu at n+1. The data showed that Ile and Phe, unlike Ala, were still compatible with high phosphorylation efficiency. However, whereas the n+1 substitution to Arg was relatively deleterious on both Km and Kcat in the case of AurA, it almost tripled the Kcat with PKA. The latter was also insensitive to the replacement of the n+1 Leu with the artificial bulky apolar residue of tert-butyl-glycine, which in contrast hampered phosphorylation by AurA. It is worth noting that a Pro residue at position n+1 (peptide 13) completely abrogated phosphorylation by AurA as well as by PKA. The special relevance of a hydrophobic determinant at position n+1 in AurA peptide substrates may also explain why peptide 4, in which n−2 and n−3 are occupied by Ala while Leu is still present at n+1, was no more a substrate for PKA but was still significantly phosphorylated by AurA. The efficiency of phosphorylation of peptide 4 by AurA was comparable with that of peptide 12: in the latter, the Arg-Arg doublet was still at its optimal position but the Leu at n+1 was replaced by Ala.
Table 1. Kinetic constants for AurA and PKA determined on a set of peptide substrates.
Assays were performed in triplicate as indicated in the Materials and methods section; n.d., not determined due to undetectable phosphorylation. Residues whose displacement and/or substitution are discussed are shown in boldface.
| AurA | PKA | ||||||
|---|---|---|---|---|---|---|---|
| Substrate | Km (μM) | Kcat (s−1) | Efficiency (Kcat/Km)×102 | Km (μM) | Kcat (s−1) | Efficiency (Kcat/Km)×102 | |
| 1 | ALRRASLGAA | 263 | 5.50 | 2.09 | 60 | 2.3 | 3.94 |
| 2 | ALARRSLGAA | 200 | 0.39 | 0.19 | 1000 | 0.1 | 0.01 |
| 3 | ARRAASLGAA | 222 | 0.39 | 0.19 | 500 | 0.4 | 0.08 |
| 4 | RRAAASLGAA | 500 | 0.29 | 0.05 | n.d. | n.d. | – |
| 5 | ALKKASLGAA | n.d. | n.d. | – | n.d. | n.d. | – |
| 6 | ALKRASLGAA | 357 | 1.11 | 0.31 | n.d. | n.d. | – |
| 7 | ALRKASLGAA | 1000 | 0.27 | 0.02 | 294 | 1.3 | 0.44 |
| 8 | ALRRASIGAA | 370 | 5.36 | 1.45 | 44 | 2.1 | 4.77 |
| 9 | ALRRASFGAA | 555 | 5.56 | 1.00 | 76 | 2.7 | 3.55 |
| 10 | ALRRASRGAA | 1084 | 3.38 | 0.31 | 322 | 6.9 | 2.14 |
| 11 | ALRRAStBuGlyGAA | 714 | 3.82 | 0.53 | 48 | 2.0 | 4.16 |
| 12 | ALRRASAGAA | 430 | 0.32 | 0.07 | 53 | 1.3 | 2.45 |
| 13 | ALRRASPGAA | n.d. | n.d. | – | n.d. | n.d. | – |
| 14 | ALRRATLGAA | 5000 | 5.18 | 0.10 | 526 | 1.4 | 0.26 |
| 15 | ALRRAYLGAA | n.d. | n.d. | – | n.d. | n.d. | – |
| 16 | AcFNRTSLPWQGLKAATKKQKY | 868 | 1.23 | 0.14 | n.d. | n.d. | – |
| 17 | NKRRRSVTPPE | 1010 | 1.91 | 0.18 | 41 | 1.22 | 2.97 |
| 18 | NKRRRSPTPPE | n.d. | n.d. | – | n.d. | n.d. | – |
| 19 | Histone H3 | 10 | 0.35 | 3.50 | n.d. | n.d. | – |
Also remarkable was the different response of AurA and PKA to the individual substitution of the two crucial arginine residues with lysines: AurA tolerated much better the substitution at position n−3, the one at position n−2 being hardly compatible with detectable phosphorylation (compare peptides 6 and 7). The opposite applied to PKA, which still appreciably phosphorylated the Arg-Lys peptide while failing to phosphorylate the Lys-Arg peptide (peptide 6). Consequently, the latter may represent a promising lead for the development of specific substrates to selectively monitor AurA. In the same vein, it is worth remarking that AurA phosphorylated fairly efficiently a peptide encompassing the 213–232 sequence of CK1 (AcFNRTSLPWQGLKAATKKQKY) (peptide 16). This peptide, which was phosphorylated by AurA at a single Ser residue flanked by an Arg at n−2 and an Asn at n−3 (motif: NRxSL), was instead unaffected by PKA.
To sum up, it can be concluded that the consensus sequence of AurA conforms to the motif R/K/N-R-X-S/T-B, where B stands for any hydrophobic residue except Pro. The relevance of the hydrophobic in n+1 is comparable with that of the Arg at position n−2. Not surprisingly therefore, a peptide reproducing the segment around CDC25B Ser353, recently identified as an AurA target [29], in which both crucial determinants are present (peptide 17 in Table 1) was readily phosphorylated by AurA with favourable kinetic constants. Interestingly, also in this case phosphorylation was abrogated if the Val at position n+1 is replaced by Pro (see peptides 17 and 18 in Table 1).
Identification of the residues phosphorylated in bacterially expressed AurA
Activation of AurA is associated with phosphorylation at the T-loop Thr288 [30]. In order to examine whether the human recombinant AurA that we expressed in E. coli was phosphorylated, we submitted wt- and kd-AurA to LC-MS/MS analysis. Among the approx. 100 tryptic peptides that were analysed, resulting in coverage of 70–80% of the protein sequence, we could identify 14 phosphopeptides in wt-AurA, accounting for a total of 15 phosphorylation sites (Table 2). No phosphopeptide could be detected in kd-AurA, consistent with the view that all the phosphates were incorporated through an autocatalytic mechanism. Figure 2 shows the MS/MS spectra of two representative phosphopeptides with the assignment of the respective sites of phosphorylation. Somewhat surprisingly, only one among all sites identified (i.e. Thr 288 in the activation loop) fully conforms to the consensus outlined above, whereas another residue, Ser342, partially matches it for displaying the K-R-X-S motif. Particularly striking was the identification of Tyr148 among the phosphorylation sites present in the recombinant kinase, which flatly contradicts the failure of AurA to phosphorylate its optimal peptide substrate with Ser replaced by Tyr (Table 1, peptide 15).
Table 2. Identification of the sites of phosphorylation in AurA.
Tryptic peptides derived from digestion of human recombinant wt-AurA were analysed by MS/MS. The sites of phosphorylation identified in phosphopeptides present among the peptides detected are indicated.
| Sequence | Residues | Phosphorylated residue |
|---|---|---|
| SKENCISGPVK | 4–14 | Ser10 |
| SKENCISGPVKATAPVGGPK | 4–23 | Thr16 |
| RVLVTQQFPCQNPLPVNSGQAQR | 24–46 | Ser41 |
| VLCPSNSSQR | 47–56 | Ser53 or Ser54 |
| LVSSHKPVQNQK | 64–75 | Ser67 |
| QLQATSVPHPVSR | 78–90 | Ser83 |
| SKQPLPSAPENNPEEELASK | 98–117 | Ser98 |
| SKQPLPSAPENNPEEELASK | 98–117 | Ser104 |
| GKFGNVYLAR | 142–151 | Tyr148 |
| ELQKLSKFDEQR | 221–232 | Ser226 |
| DIKPENLLLGSAGELK | 256–271 | Ser266 |
| IADFGWSVHAPSSR | 272–285 | Ser278 |
| RTTLCGTLDYLPPEMIEGR | 286–304 | Thr287 |
| RTTLCGTLDYLPPEMIEGR | 286–304 | Thr288 |
| RISRVEFTFPDFVTEGAR | 340–357 | Ser342 |
| EVLEHPWITANSSKPSNCQNK | 376–396 | Ser391 |
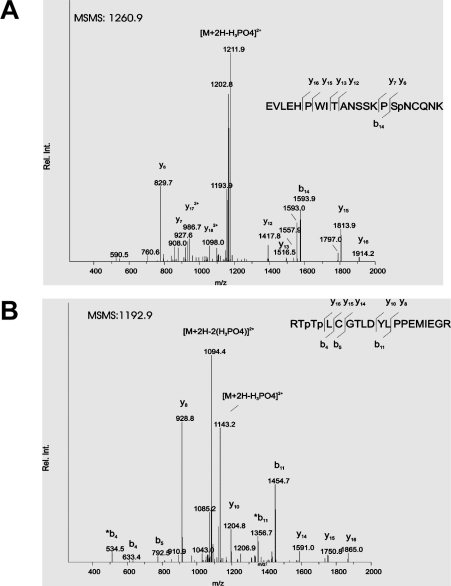
Figure 2. MS/MS spectra of two tryptic phosphopeptides identified by TurboSequest.
(A) MS/MS spectrum of m/z 1260.9. The major fragment generated by neutral loss of H3PO4, indicative of phosphorylation, is marked [M+2H-H3PO4]2+. The y- ( ) and b-fragments (
) and b-fragments ( ) detected upon collision-induced fragmentation are indicated in the sequence. The ions y6, y7 and b14 show that phosphorylation is located in the C-terminal part of the peptide allowing assignment of phosphorylation to the indicated Ser (Sp). (B) MS/MS spectrum of m/z 1192.9. The neutral losses of 1×H3PO4 and 2×H3PO4, marked as [M+2H−H3PO4]2+ and [M+2H−2(H3PO4)]2+ respectively indicate two phosphorylation sites in this peptide. The ions y14–y16 and b4 and b5 allowed location of the sites of phosphorylation to the N-terminus of the peptide, with the two indicated Thr (Tp) assigned as phosphorylation sites.
) detected upon collision-induced fragmentation are indicated in the sequence. The ions y6, y7 and b14 show that phosphorylation is located in the C-terminal part of the peptide allowing assignment of phosphorylation to the indicated Ser (Sp). (B) MS/MS spectrum of m/z 1192.9. The neutral losses of 1×H3PO4 and 2×H3PO4, marked as [M+2H−H3PO4]2+ and [M+2H−2(H3PO4)]2+ respectively indicate two phosphorylation sites in this peptide. The ions y14–y16 and b4 and b5 allowed location of the sites of phosphorylation to the N-terminus of the peptide, with the two indicated Thr (Tp) assigned as phosphorylation sites.
AurA specifically phosphorylated Thr288 within a synthetic peptide derived from its activation loop
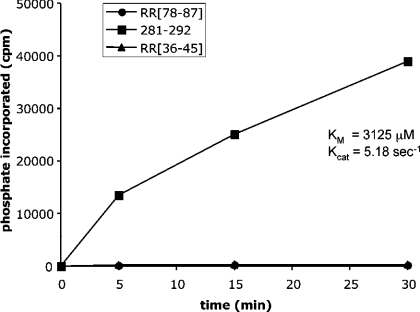
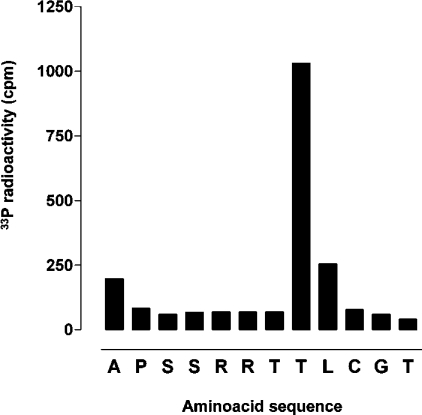
MS analysis disclosed up to 15 residues phosphorylated in bacterially expressed AurA with only a couple of them displaying the motif R/K-R-X-S/T. This finding prompted us to synthesize peptides reproducing some of these sites to assess the significance of their phosphorylation. One such peptide encompassed the 281–292 sequence. This included the only phosphorylated residue (Thr288) conforming to the optimal R-R-X-S/T-B consensus, along with another site found phosphorylated in recombinant AurA (Thr287) and three potential phosphoacceptor residues. Two additional peptides were synthesized in order to reproduce the segments 36–45 and 78–87. These included the phosphorylated residues Ser41 and Ser83, neither of which conformed to the AurA consensus, although the latter displayed the hydrophobic determinant at position n+1. As shown in Figure 3, only the 281–292 peptide was phosphorylated by AurA, whereas the other two peptides were totally unaffected. Upon phosphorylation by AurA, the 281–292 peptide was subjected to automated-Edman degradation to localize the phosphorylated residue(s). The data shown in Figure 4 provide the unambiguous demonstration that the only residue phosphorylated in this peptide was Thr288, while no detectable amount of radiolabelled phosphate was released from Ser283, Ser284, Thr287 or Thr292. The Thr288 homologue in Xenopus AurA (Thr295) was found to be almost 100% phosphorylated upon expression of the recombinant kinase in E. coli [11]. Interestingly, Thr288 is the structural homologue of PKA Thr297, a crucial residue the phosphorylation of which is a prerequisite for the activation of PKA as well as a number of other ‘Arg-Asp’ protein kinases [31,32]. As for AurA (Figure 1A), autophosphorylation of bacterially expressed PKA at Thr297 was reported to cause retarded migration of the protein in SDS/PAGE [33]. To confirm our findings on the mechanism of AurA autophosphorylation, we examined the ability of active AurA to phosphorylate Thr288 in full-length kd-AurA in vitro. Incubation of catalytic amounts of wt-AurA with kd-AurA resulted in evident phosphorylation of the latter as shown by incorporation of 32P (Figure 5B) as well as by reactivity to an antibody directed to phospho-Thr288 (Figure 5A).
Figure 3. Phosphorylation of peptides reproducing AurA autophosphorylation sites.
Synthetic peptides spanning residues 36–45, 78–87 and 281–292 of human AurA were subjected to in vitro phosphorylation with purified recombinant AurA. The Km and Kcat values calculated for the optimal peptide substrate are indicated.
Figure 4. Identification of the phosphoacceptor residue within the peptide 281–292 of AurA.
The synthetic peptide spanning residues 281–292 was subjected to in vitro phosphorylation with purified recombinant AurA followed by purification of the peptide and automated Edman degradation. Release of phosphate at each cycle of Edman degradation was quantified by liquid scintillation. The signal obtained at cycle 1 was probably due to the fact that phosphorylation was performed using the peptide substrate covalently bound to the beads of the solid support employed during synthesis (see the Materials and methods section). Release of non-specifically bound phosphate may have occurred at the first cycle of Edman degradation.
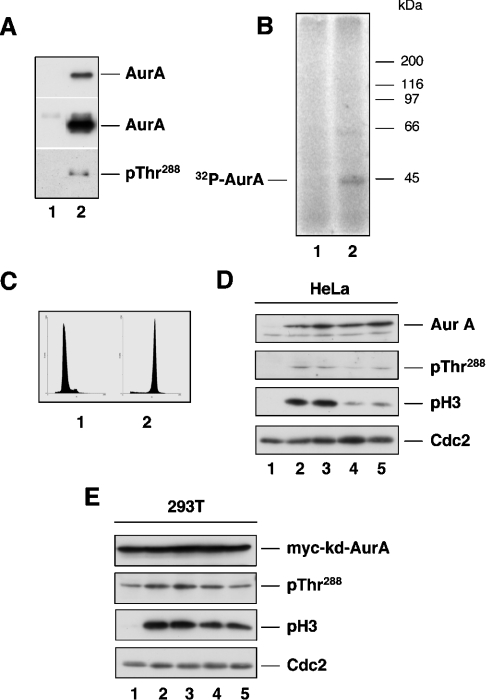
Figure 5. Effect of PKA inhibitors or activators on the in vivo phosphorylation of AurA Thr288.
In vitro phosphorylation reactions were performed using human recombinant wt-AurA (24 ng) incubated alone (lane 1) or in the presence of kd-AurA (750 ng, lane 2) under conditions described in the Materials and methods section. Reactions were performed using non-labelled ATP (A) or [γ-32P]ATP (B). Aliquots of the reactions were resolved by SDS/PAGE and bands were either directly detected by autoradiography (B) or by Western-blot analysis using antibodies specific to AurA and to phosphorylated Thr288 (A). The upper and middle insets in (A) are different exposures of the same PVDF membrane that allow to appreciate the ratio of wt- and kd-AurA employed in the assay. (C) Flow cytometric analysis of double-thymidine synchronized (G1/S, panel 1) and nocodazole-treated (G2/M, panel 2) HeLa cells. (D) G1/S synchronized HeLa cells (lane 1) were treated with nocodazole (0.4 ng/ml) 1 h after release from the second thymidine block (lanes 2–5). H89 (0.5 μM, lane 3), forskolin (20 μM, lane 4) or 8Br-cAMP (1 mM, lane 5) was added at 6 h and cells were harvested at 10 h post-release. AurA expression and phosphorylation at Thr288 as well as histone H3 phosphorylation were detected using antibodies described in the Materials and methods section. CDC2 was used as loading control. (E) G1/S synchronized HEK-293T cells (lane 1) transfected with Myc-tagged kd-AurA were treated with nocodazole (0.4 ng/ml) 1 h after release (lanes 2–5). H89 (0.5 μM, lane 3), forskolin (20 μM, lane 4) or 8Br-cAMP were added at 5 h, cells were harvested at 8 h post-release and analysed as described in (D).
In vivo phosphorylation of AurA at Thr288 occurs in a PKA-independent manner
Given the results described above, we set out to assess the claim that PKA might be responsible for AurA phosphorylation at Thr288 in vivo [30]. To this end, HeLa cells were synchronized by double-thymidine block-release and treated with nocodazole to prevent cells to move beyond mitosis. The degree of cell synchronization was controlled by flow cytometric analysis of DNA (Figure 5C) and phosphorylation of the mitotic marker histone H3 (Figure 5D). The PKA inhibitor H89 or either one of the two PKA activators forskolin and 8Br-cAMP was added at 8 h post-release and cells were analysed at 10 h. In synchronized cells, AurA protein level resulted to be low at G1/S and high at G2/M, as described in the literature [5]. Mitotic phosphorylation of AurA at Thr288 could be detected in control cells as well as in cells treated with the PKA inhibitor H89 (Figure 5D, lanes 2 and 3). The slightly lower level of Thr288 phosphorylation observed in cells that were treated with PKA activators (Figure 5D, lanes 4 and 5) was paralleled by a decrease rate of mitotic entry in this setting. This was visually estimated from the lower number of rounded-up cells (∼30% in treated versus ∼70% in control cells) and by the lower extent of histone H3 phosphorylation in cells treated with PKA activators (Figure 5D, lanes 4 and 5). To extend this observation we employed HEK-293T cells, which are amenable to transient expression studies and display a slightly shorter cell cycle than HeLa. Double-thymidine synchronized HEK-293T cells were transfected with kd-AurA between the two blocks and treated with nocodazole after release from the second block. H89, forskolin or 8Br-cAMP was added 5 h post-release and cells were examined at 8 h. The extent of Thr288 phosphorylation in Myc-tagged kd-AurA appeared to increase with progression to mitosis (Figure 5E). This was not surprising, as it probably resulted from an intermolecular autophosphorylation reaction catalysed by endogenous AurA, similar to the one observed in vitro (Figures 5A and 5B). As for HeLa, HEK-293T cells showed no change in the pattern of Thr288 phosphorylation upon treatment with the PKA inhibitor H89 (Figure 5E, lane 3). The PKA activators forskolin or 8Br-cAMP resulted in a pattern of delayed entry into mitosis, although less pronounced compared with HeLa cells, and a proportional decrease of Thr288 phosphorylation (Figure 5E, lanes 4 and 5).
DISCUSSION
Mitosis is a highly regulated process during which sister chromatids are segregated into two newly formed cells [34]. Errors in this process may lead to aneuploidy and facilitate the onset of cancer [35]. Central to the correct execution of mitosis is the protein kinase CDC2, although a number of studies have pointed to the essential auxiliary role played by Polo-like, NIMA (never in mitosis in Aspergillus nidulans)-related and Aurora-related kinases [34,36]. Despite the growing evidence on the importance of AurA in mitosis, clarification of its exact role requires the identification of physiological substrates and, in turn, of downstream pathways triggered by the kinase. One way to address this issue is to define the specificity determinants for recognition and phosphorylation of substrates by the kinase. In the present study, we have taken such an approach by testing human recombinant AurA on a set of peptide substrates appropriately designed to elucidate its site specificity.
A library of peptide substrates previously developed to assay a wide variety of protein kinases was used for a preliminary inspection of the site specificity of AurA. Once ascertained that only basic peptides were appreciably phosphorylated by AurA, we set out to precisely define the local structural determinants recognized by AurA. To this end, we synthesized a number of derivatives of an extended version of the ‘Kemptide’ (ALRRRASLGAA), which was taken as representative of the best substrate for AurA. From this analysis, it appeared that an Arg at position n−2 and a hydrophobic residue at position n+1, relative to the phosphorylatable residue, are of crucial importance to determine site recognition by AurA. The resulting consensus sequence was R/K/N-R-X-S/T-B, where B stands for any hydrophobic residue, with the notable exception of Pro (see below). The sequence recognized by human AurA appeared to be similar to the putative consensus reported for yeast Ipl1, which was deduced on the basis of the sequence flanking sites phosphorylated by Ipl1 in a set of kinetochore proteins [37]. Although mammalian AurB is believed to be the functional homologue of yeast Ipl1, it is likely that, given the high conservation of the catalytic domains, the specificity of Aurora family members be similar. In the present study, we found that AurA shared with PKA, as well as with many other basophilic Ser/Thr protein kinases, the absolute requirement for an Arg at position n−2. However, whereas AurA displayed a stringent requirement for a hydrophobic residue at n+1, this seemed to be much less important for PKA. Additionally, AurA showed a wider tolerance of the nature of the n−3 residue, with a Lys or even an Asn, albeit in a peptide unrelated to the Kemptide (peptide 16, Table 1), fulfilling the criteria for phosphorylation. In contrast, PKA showed a stringent requirement for Arg at n−3. The inspection of PKA and AurA structures may provide a partial explanation for their different site specificity. Whereas in fact all three acidic residues interacting with the Arg n−2 in the PKA/pseudosubstrate peptide complex [38] are conserved in AurA (Table 3), the corresponding three acidic residues in PKA that recognize the n−3 Arg are either absent from AurA, due to its shorter C-terminal domain, or replaced by a Thr (Thr217 instead of Glu127). Moreover, the hydrophobic pocket of the p+1 loop of PKA, which is formed by Leu198, Pro202 and Leu205 and functions by accommodating the side chain of the n+1 residue of the substrate, is conserved but more hydrophobic in AurA, with Pro202 replaced by a third Leu. This may account for the more stringent requirement of AurA for a hydrophobic determinant at n+1. Apart from the rationale underlying the different consensus of AurA and PKA, our results indicated that the peculiar requirement of AurA could be exploited for the development of specific peptide substrates to be used for monitoring AurA kinase activity. Such peptides will be unaffected or marginally affected by PKA and possibly other basophilic protein kinases. Two representative examples are peptides 6 and 16 of Table 1: neither of them was phosphorylated by PKA, while they proved to be fairly good substrates for AurA by virtue of their K/N-R-X-S-L motif. The performance of these peptides could be further improved by modifications aimed at optimizing their phosphoacceptor properties without loosing specificity. Useful hints may come from screening a peptide library where the Z-K-R-X-S-L-Z′ scaffold is maintained invariant while the X, Z and Z′ residues are randomly modified.
Table 3. Residues of AurA corresponding to the amino acids implicated in p+1, p−2 and p−3 recognition sites of PKA [38].
| Recognition site | AurA | PKA |
|---|---|---|
| p+1 | Leu289 | Leu198 |
| Leu293 | Pro202 | |
| Leu296 | Leu205 | |
| p−2 | Glu260 | Glu170 |
| Asp294 | Glu203 | |
| Glu321 | Glu230 | |
| p−3 | Thr217 | Glu127 |
| – | Asp329 | |
| – | Glu331 |
The AurA consensus, which was deduced from the analysis of Kemptide derivatives, was further validated by a parallel approach based on peptides encompassing sites that we found phosphorylated in the bacterially expressed kinase. MS analysis showed that the 15 sites present in human recombinant AurA (see Table 2) largely coincided with the 10 phosphorylated residues identified in the Xenopus kinase [11]. Especially relevant was Thr288, the homologue of Thr197 in PKA. In both kinases the latter is located in the activation loop, also defined as the T-loop after PKA Thr197. Phosphorylation of a residue in the activation loop has been reported to occur in a number of kinases and was shown to cause conformational changes that facilitate enzymatic activation [32]. A mechanistic explanation of the role of T-loop phosphorylation was provided by structural studies on the so-called ‘Arg-Asp’ kinases, in which an Arg adjacent to the catalytic Asp is believed to hamper catalysis unless it is neutralized by interaction with the phosphorylated residue of the activation loop [39]. AurA also contains an ‘Arg-Asp’ motif (residues 255–256) in the catalytic loop and the T-loop Thr288 is extensively phosphorylated in the bacterially expressed protein (the present study and [11]). This is consistent with an autocatalytic reaction and it has a rational explanation in the AurA consensus defined in the present study. The definite demonstration that Thr288 phosphorylation by AurA is driven by its consensus was provided by analysis of a peptide encompassing the 281–292 segment of AurA activation loop. This peptide was readily phosphorylated by AurA (Figure 3), but, more importantly, phosphorylation occurred at the only site (Thr288) fulfilling the consensus for AurA among five potential phosphoacceptor residues present in the peptide (Figure 4). The ability of AurA to autophosphorylate at Thr288 was further confirmed using full-length kd-AurA as substrate for catalytically active AurA (Figures 5A and 5B). The finding that AurA could efficiently autophosphorylate at Thr288 questions the claim that PKA might be responsible for the phosphorylation of this site [30], particularly in view of the fact that cAMP level and, in turn, PKA activity decrease at the onset of mitosis [40]. Our results showing that in vivo phosphorylation at Thr288 was not affected by the PKA inhibitor H89 or the PKA activators forskolin and 8Br-cAMP, confirmed the doubts on the involvement of PKA in the process of AurA activation.
Another site displaying AurA consensus is Ser342. Phosphorylation at Ser349 in Xenopus AurA (Ser342 in the human protein) was reported to be an autocatalytic event, which was apparently triggered by GSK3-mediated phosphorylation of two serine residues located at n−4 with respect to the T-loop residues Thr290 and Thr291 (Thr287 and Thr288 in human AurA). Autophosphorylation at Ser349 was claimed to result in the inhibition of Xenopus AurA activity [13]. Nonetheless, both human (the present study) and Xenopus AurA [11] were found to be fully phosphorylated at Ser342 and Ser349 respectively and, despite this, to be highly active kinases. It is also interesting to note that Thr287 and several other residues that do not fulfil the consensus for AurA were found to be phosphorylated in bacterially expressed AurA, albeit to an extent lower than what was observed for Thr288 ([11] and the present study, results not shown). Phosphorylation at these sites was probably an artifact due to the conditions of bacterial expression, where protein kinases reach abnormally high concentrations that are never observed under physiological conditions. This, in turn, could favour the reversible formation of aggregates where mis-phosphorylation of exposed residues would occur irrespective of their location in consensus sequences. A situation similar to what we observed with AurA has been reported for other protein kinases. Notably, bacterial expression of the recombinant catalytic subunit of PKA resulted in phosphorylation at Thr197 and at several other residues that, contrary to Thr197, do not conform to the PKA consensus sequence [33]. The fact that some of the phosphorylation sites identified in bacterially expressed AurA may be the result of unspecific events is also supported by two observations: first, synthetic peptides reproducing two of the sites found in the bacterially expressed kinase and lacking consensus for AurA proved totally refractory to phosphorylation by AurA (Figure 3); secondly, one of the phosphosites identified in both human (the present study) and Xenopus AurA [11] was Tyr148. Nonetheless, replacement of Ser with Tyr in the synthetic peptide displaying the optimal consensus for AurA resulted in undetectable phosphorylation of this peptide (Table 1).
Another interesting point emerging from our screening is the importance of the nature of the n+1 residue in the peptide substrate: a Pro at this position entirely abrogated phosphorylation by AurA. Such an absolute lack of recognition of the S/T-P motif, which is common to other Ser/Thr protein kinases [41], may reflect a device that is in place in the cell to control the onset of mitosis: by allowing synchronization of activity without overlapping of function with proline-directed kinases, AurA and CDC2 can regulate independent processes during the progression to and the execution of mitosis. Incidentally, the intolerance for a Pro at n+1, in conjunction with lack of any residual consensus for AurA around Ser315 in p53 (TSSSS315PQP), would argue against the possibility that this site might be efficiently phosphorylated by AurA. A synthetic peptide including Ser315 (residues 309–321) was entirely unaffected by AurA in vitro (results not shown). On the other hand, the finding that Ser315 is phosphorylated in full-length p53, although less readily than Ser215 [42], supports the view that sometimes higher-order structural features may enable AurA to phosphorylate residues that do not display the consensus sequence. In sharp contrast, Ser353 in CDC25B, which is embedded in the sequence RRRS353V and which was recently reported to be a target of AurA [29], perfectly fits in with the consensus outlined with our peptide substrates.
It should be borne in mind in this connection that, although the substrate's primary sequence around target sites is of key importance to determine phosphorylation, this is likely to be one of the requirements for substrate recognition by protein kinases, which is also contributed by higher-order structures, supra-molecular association, compartmentalization etc. Pertinent to this remark is the case of histone H3, a putative target of AurA [19]: although histone H3 was phosphorylated much more slowly than several peptide substrates, it displayed a Km value (10 μM) that was 26-fold lower than that of the optimal peptide substrate (Table 1). Despite this, however, the use of peptide substrates remains an excellent choice to determine the optimal consensus for protein kinases, and definition of such a consensus, in turn, facilitates the prediction of potential sites of phosphorylation in physiological substrates.
Acknowledgments
We are indebted to P. Sassone-Corsi, IGBMC (Institute of Genetics and Molecular and Cellular Biology), Strasbourg, France, for providing the AurA kinase-dead cDNA construct and to C. Koenig for invaluable technical assistance. This study was supported by the European grant LSHB-CT-2004-503467 and a grant from CIB (Consorzio Interuniversitario Biotecnologie) to L.A.P. and by a Zurich-Cancer-League grant and the Swiss National Science Foundation grant 31-100090/1 to S.F.
References
- 1.Giet R., Prigent C. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 1999;112:3591–3601. doi: 10.1242/jcs.112.21.3591. [DOI] [PubMed] [Google Scholar]
- 2.Chan C. S., Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., Murray A. W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover D. M., Leibowitz M. H., McLean D. A., Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell (Cambridge, Mass.) 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff J. R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B., Schryver B., Flanagan P., Clairvoyant F., Ginther C., et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Kuang J., Zhong L., Kuo W. L., Gray J. W., Sahin A., Brinkley B. R., Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff J. R., Plowman G. D. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- 8.Sen S., Zhou H., White R. A. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 9.Shindo M., Nakano H., Kuroyanagi H., Shirasawa T., Mihara M., Gilbert D. J., Jenkins N. A., Copeland N. G., Yagita H., Okumura K. cDNA cloning, expression, subcellular localization, and chromosomal assignment of mammalian aurora homologues, aurora-related kinase (ARK) 1 and 2. Biochem. Biophys. Res. Commun. 1998;244:285–292. doi: 10.1006/bbrc.1998.8250. [DOI] [PubMed] [Google Scholar]
- 10.Stenoien D. L., Sen S., Mancini M. A., Brinkley B. R. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil. Cytoskeleton. 2003;55:134–146. doi: 10.1002/cm.10120. [DOI] [PubMed] [Google Scholar]
- 11.Haydon C. E., Eyers P. A., Aveline-Wolf L. D., Resing K. A., Maller J. L., Ahn N. G. Identification of novel phosphorylation sites on Xenopus laevis Aurora A and analysis of phosphopeptide enrichment by immobilized metal-affinity chromatography. Mol. Cell. Proteom. 2003;2:1055–1067. doi: 10.1074/mcp.M300054-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Littlepage L. E., Wu H., Andresson T., Deanehan J. K., Amundadottir L. T., Ruderman J. V. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkissian M., Mendez R., Richter J. D. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev. 2004;18:48–61. doi: 10.1101/gad.1136004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama H., Zhou H., Li Q., Tatsuka M., Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J. Biol. Chem. 2001;276:46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- 15.Honda K., Mihara H., Kato Y., Yamaguchi A., Tanaka H., Yasuda H., Furukawa K., Urano T. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex–ubiquitin–proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- 16.Castro A., Arlot-Bonnemains Y., Vigneron S., Labbe J. C., Prigent C., Lorca T. APC/Fizzy-related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littlepage L. E., Ruderman J. V. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell (Cambridge, Mass.) 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 19.Crosio C., Fimia G. M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C. D., Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouchi M., Fujiuchi N., Sasai K., Katayama H., Minamishima Y. A., Ongusaha P. P., Deng C., Sen S., Lee S. W., Ouchi T. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J. Biol. Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- 21.Katayama H., Sasai K., Kawai H., Yuan Z. M., Bondaruk J., Suzuki F., Fujii S., Arlinghaus R. B., Czerniak B. A., Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat. Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 22.Bensadoun A., Weinstein D. Assay of proteins in the presence of interfering materials. Anal. Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 25.Gatlin C. L., Eng J. K., Cross S. T., Detter J. C., Yates J. R., III Automated identification of amino acid sequence variations in proteins by HPLC/microspray tandem mass spectrometry. Anal. Chem. 2000;72:757–763. doi: 10.1021/ac991025n. [DOI] [PubMed] [Google Scholar]
- 26.Fields G. B., Noble R. L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 27.Ruzzene M., Pinna L. A. Assay of protein kinases and phosphatases using specific peptide substrates. In: Hardie D. G., editor. Protein Phosphorylation – A Practical Approach. Oxford: Oxford University Press; 1999. pp. 221–253. [Google Scholar]
- 28.Perich J. W., Meggio F., Reynolds E. C., Marin O., Pinna L. A. Role of phosphorylated aminoacyl residues in generating atypical consensus sequences which are recognized by casein kinase-2 but not by casein kinase-1. Biochemistry. 1992;31:5893–5897. doi: 10.1021/bi00140a027. [DOI] [PubMed] [Google Scholar]
- 29.Dutertre S., Cazales M., Quaranta M., Froment C., Trabut V., Dozier C., Mirey G., Bouche J. P., Theis-Febvre N., Schmitt E., et al. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2–M transition. J. Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 30.Walter A. O., Seghezzi W., Korver W., Sheung J., Lees E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene. 2000;19:4906–4916. doi: 10.1038/sj.onc.1203847. [DOI] [PubMed] [Google Scholar]
- 31.Johnson L. N., Noble M. E., Owen D. J. Active and inactive protein kinases: structural basis for regulation. Cell (Cambridge, Mass.) 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 32.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases: controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Yonemoto W., Garrod S. M., Bell S. M., Taylor S. S. Identification of phosphorylation sites in the recombinant catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 1993;268:18626–18632. [PubMed] [Google Scholar]
- 34.Nigg E. A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 35.Gisselsson D. Chromosome instability in cancer: how, when, and why? Adv. Cancer Res. 2003;87:1–29. doi: 10.1016/s0065-230x(03)87164-6. [DOI] [PubMed] [Google Scholar]
- 36.Carmena M., Earnshaw W. C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 37.Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., III, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell (Cambridge, Mass.) 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 38.Knighton D. R., Zheng J. H., Ten Eyck L. F., Xuong N. H., Taylor S. S., Sowadski J. M. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 39.Johnson L. N., Lewis R. J. Structural basis for control by phosphorylation. Chem. Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 40.Grieco D., Porcellini A., Avvedimento E. V., Gottesman M. E. Requirement for cAMP-PKA pathway activation by M phase-promoting factor in the transition from mitosis to interphase. Science. 1996;271:1718–1723. doi: 10.1126/science.271.5256.1718. [DOI] [PubMed] [Google Scholar]
- 41.Pinna L. A., Ruzzene M. How do protein kinases recognize their substrates? Biochim. Biophys. Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 42.Liu Q., Kaneko S., Yang L., Feldman R. I., Nicosia S. V., Chen J., Cheng J. Q. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J. Biol. Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]