Abstract
The rapid growth in the number of newly identified DNA and RNA viral sequences underscores the need for an accurate and comprehensive classification system for all viral realms at different taxonomic levels. Here, we establish the Viral Taxonomic Assignment Pipeline (VITAP), which addresses classification challenges by integrating alignment-based techniques with graphs, offering high precision in classifying both DNA and RNA viral sequences and providing confidence level for each taxonomic unit. This tool automatically updates its database in sync with the latest references from the International Committee on Taxonomy of Viruses (ICTV), efficiently classifying viral sequences as short as 1,000 base pairs to genus level. VITAP possesses good generalization capabilities, maintaining accuracy comparable to other pipelines while achieving higher annotation rates across most DNA and RNA viral phyla. Its application in deep-sea viromes has led to significant taxonomic updates, providing comprehensive diversity information of viruses from deep-sea. VITAP is available at https://github.com/DrKaiyangZheng/VITAP.
Subject terms: Software, Classification and taxonomy, Water microbiology
High-throughput sequencing retrieves viral sequences from diverse environments, but these sequences can be difficult to classify. The here developed VITAP is a taxonomic assignment pipeline for DNA and RNA viruses that efficiently classifies incomplete viral sequences (as short as 1000-bp) down to genus level.
Introduction
Viruses possess a range of replication and mobility strategies and play a vital role in ecology1–6. Cross-species infection by viruses allows them to transfer genes horizontally, driving the formation of the “web of life”, which replaces the traditional view of the “tree of life”7–12. Viruses can also hijack host metabolic pathways during infection periods, altering their physiological state and possibly bringing metabolic capability to their hosts1,13–16. Biological community structure can be shaped through the cell lysis that has resulted from viral infection1,17,18. This allows viruses to indirectly influence global biogeochemical cycling through the interference of cellular metabolisms and lifespans1,5,13,19. Although viruses are passive opportunists, they can produce essential ecological effects by manipulating three life domains.
The development of high-throughput sequencing for viruses provides the opportunity to deeply profile human-associated and environmental DNA and RNA viral communities. In-depth sequencing of the gut viral metagenomes has led to the construction of a human intestinal DNA virome database20–22, which has revealed the dominant viral lineages in the human gut23,24. The large-scale metatranscriptome sampling of different animal tissues has expanded the RNA viral host spectrum, revealing many RNA viruses infecting both invertebrates and vertebrates25,26. The investigation of animal-derived metatranscriptomes is capable of tracing pathogen migration and mutation and provides a potential contribution to public health27. In the environment, the recovery of viromes from diverse habitats has revealed their effects on local ecosystems. For example, DNA viruses have been shown to influence microbial hydrocarbon biodegradation at cold seeps and utilize refractory organic matter in the deep sea4,28. The large-scale metatranscriptome sampling from a range of ecosystems has revealed a cryptic RNA virosphere that was little known before29–33. These culture-independent investigations of DNA and RNA viruses highlight critical ecological effects that were unknown before.
Sound taxonomy, which underpins all ecological and evolutionary investigations, depends on the identification of biological marker genes. An understanding of virus dynamics is impeded by the absence of universally applicable marker genes, such as rRNA genes encoded by cellular organisms34–36. Consequently, methodologies analogous to those used in cellular organisms are inadequate for viral phylogenetics. The rapid growth in the number of viral genomes has prompted the International Committee on Taxonomy of Viruses (ICTV) to propose special taxonomic criteria for viruses, such as virion morphology and single-/multiple-gene phylogenies37. Currently, five primary methods of automated viral classification have enabled taxonomic research on a large number of uncultured viral genomes. The first, an openly available online tool (VICTOR), utilizes a prokaryotic classifying algorithm (genome blast distance phylogeny, GBDP) to categorize prokaryotic viral genomes through phylogenetic-based clustering38. VICTOR helped to establish nine new viral families associated with Flavobacteriia, which have been accepted by ICTV in 202039. The second, the alignment-based gene-sharing clustering (vConTACT2 and ViPTree) and genome-wide nucleic acids similarity (VIRIDIC), have been widely adopted by ICTV to classify viruses40–42. These methods are specifically efficient for taxonomic assignments of dsDNA viruses of prokaryotes, leading to the establishment of 63 novel families of head-tail viruses43. The third, the present/absent patterns of protein families for genomes, has been used in VPF-class and geNomad to classify viruses44,45. These methods utilized a voting strategy to determine the best-fit taxonomic units of target genomes and are effective for incomplete viral genomes. The fourth, CAT/BAT combined with protein sequence alignment and the last common ancestry approach (LCA), has been used to classify viral genomes46. The fifth, PhaGCN/PhaGCN2, was the first method to introduce deep-learning methods into automatic viral taxonomic assignments and expand the viral taxonomic units of the global ocean virome database (GOV). The aforementioned pipelines have made significant contributions to the field. While some tools can be applied to a wide range of viral taxa, many of them demonstrate optimized performance primarily for specific viral lineages, such as prokaryotic viruses. The adaptability of these pipelines to classify diverse viral taxa comprehensively remains a challenge. Additionally, most pipelines are not designed to allow non-expert users to update taxonomic criteria in real-time, which could align with ICTV’s annual proposals.
Here, we present VITAP (viral taxonomic assignment pipeline), which has a redesigned taxonomic algorithm and provides the confidence level of each taxonomic unit. VITAP is capable of automatically updating its reference database based on the latest viral reference release from the ICTV and can effectively perform taxonomic assignments for viral sequences as short as 1000 base pairs (bp) to genus level. In addition, while maintaining accuracy, precision, and recall comparable to those of other pipelines, VITAP exhibits a high annotation rate for nearly all RNA and DNA viral phyla, not merely confined to prokaryotic dsDNA viruses. With increasing systematic research on metagenomes and metatranscriptomes, this method is expected to provide a more comprehensive, automated viral taxonomic assignment pipeline.
Results
VITAP overview and workflow
The VITAP workflow includes two main sections: generation of a taxonomic-specific database, and taxonomic assignments for target genomes (Fig. 1). The first step enables users to generate a VITAP-specific database based on each release of an ICTV proposal. The genomes included in the viral metadata resource master species list (VMR-MSL) are automatically retrieved and downloaded from GenBank and are used to generate a viral reference protein database. The protein alignment scores (bitscores) are used to calculate the taxonomic units’ thresholds. Hence, the VITAP-specific database includes a viral reference protein database, taxonomic units’ thresholds, and VMR-MSL information. The second step is the taxonomic assignments of target genomes utilizing the VITAP-specific database (Supplementary Note 1). The proteins of target genomes are first aligned to viral reference proteins. Different proteins are assigned different weights for taxonomic signals from the protein alignment between target genomes and viral references; these are used to calculate the taxonomic scores. These taxonomic scores are used in cumulative average calculations to determine the best taxonomic paths, which represent the most likely taxonomic hierarchies and units. Based on their taxonomic scores compared to related thresholds, this result is defined as low-/high-/medium-confidence results. The unique framework of VITAP offers more features compared to other pipelines, providing a more comprehensive resource for taxonomic studies (Table 1).
Fig. 1. Flowchart of VITAP workflow and methodology.
The VITAP contains two main parts: the update part enables to generate the VITAP-specific database based on current viral taxonomic information from ICTV; the prediction part utilizes previously built database to perform taxonomic assignment. The VITAP utilized an algorithm to determine the best-fit taxonomic lineage, and provides result with various confidence level.
Table 1.
Features of viral taxonomic assignment pipelines
| vConTACT2 | CAT | PhaGCN2 | geNomad | VPF-Class | VITAP | |
|---|---|---|---|---|---|---|
| Taxonomy rationale | GB | PB | GB | PB | PB | GB |
| ICTV database adaptation | √ | √ | √ | √ | ||
| Custom database adaptation | √ | √ | √ | |||
| Updates by non-specialist users | √ | √ | √ | |||
| Family-level classification recommendations | √ | √ | √ | √ | √ | √ |
| Genus-level classification recommendations | √ | √ | √ | √ | ||
| Genome-content-based network | √ | √ | ||||
| Short sequence (as low as 1-kb) analysis | √ | √ | √ | √ | √ | |
| Proposals for new taxonomic units | √ | |||||
| Taxonomic assignments across all viral realms | √ | √ | √ | √ |
GB genome-based, PB protein-based.
These features reflect the capabilities of these pipelines but do not pertain to their performance strengths or weaknesses.
Benchmarking VITAP against simulated viromes and new viruses
Based on the taxonomic assignments of viral reference genomic sequences in VMR-MSL38, VITAP demonstrates acceptable generalization performance for most viral phyla. Through tenfold cross-validation compared to vConTACT2 (Fig. 2a, b), VITAP demonstrates comparable accuracy, precision, and recall (over 0.9, on average and median) for family- and genus-level taxonomic assignments (Fig. 2c, d and Supplementary Data 1). Nevertheless, VITAP achieves a significantly higher annotation rate than vConTACT2. Specifically, VITAP’s family-level average annotation rates exceed those of vConTACT2 by 0.53 (at 1-kb) to 0.43 (at 30-kb), whereas VITAP’s genus-level average annotation rates surpass those of vConTACT2 by 0.56 (at 1-kb) to 0.38 (at 30-kb). For different viral phyla, VITAP’s principal advantage over vConTACT2 lies in its annotation rate. For sequences as short as 1 kb, VITAP’s family-level annotation rate exceeds that of vConTACT2 by 0.13 (Cossaviricota) to 0.87 (Phixviricota) (Fig. 2e), while its genus-level annotation rate is higher by 0.13 (Cossaviricota) to 0.94 (Cressdnaviricota) (Fig. 2f). For 30-kb sequences, VITAP’s genus-level annotation rates for Cossaviricota and Preplasmiviricota are 0.20 and 0.05 lower than those of vConTACT2, respectively. VITAP’s family-level annotation rates for Cossaviricota and Saleviricota are 0.07 and 0.04 lower, respectively. Apart from these three phyla, for 30-kb sequences, VITAP’s family-level annotation rate surpasses that of vConTACT2 by 0.27 (Taleaviricota) to 0.85 (Kitrinoviricota) (Fig. 2e); its genus-level annotation rate is higher by 0.06 (Artverviricota) to 0.86 (Kitrinoviricota) (Fig. 2f). Overall, in terms of generalizability, vConTACT2’s strength lies in its very high F1 score, albeit at the cost of severely diminished annotation rates. In contrast, VITAP maintains an acceptable F1 score (over 0.9 on average) while preserving relatively high annotation rates. Specifically, VITAP’s annotation rates for short sequences exceed those of vConTACT2 across all viral phyla, and for nearly complete genomes, VITAP also achieves higher annotation rates for all RNA viral phyla and most DNA viral phyla compared to vConTACT2.
Fig. 2. The generalization ability of VITAP compared to vConTACT2 based on the VMR-MSL38 database.
The VMR-MSL38 database was divided into a training set comprising 70% of the data and a test set comprising 30%. Sequences in the test set were sliced into genome fragments of varying lengths (1-, 5-, 10-, 20-, and 30-kb), and taxonomic assignments were performed using VITAP and vConTACT2, which was built on the 70% training set. The dataset splitting, training, and taxonomic assignment steps were independently repeated ten times. The accuracy, precision, recall, F1 score, and annotation rate were employed to characterize the tenfold cross-validation. The center lines of the boxes indicate the median values of taxonomic assignment matrices on 17 viral phyla. The bounds of the box represent the interquartile range, with the lower and upper bounds, respectively, corresponding to the first and third quartiles. The whiskers denote the lowest and highest values within 1.5 times the interquartile range. a The F1 scores of VITAP and vConTACT2 across ten independent family and genus level taxonomic assignments; b The annotation rate of VITAP and vConTACT2 across ten independent family and genus level taxonomic assignments; c Taxonomic assignment performances of VITAP and vConTACT2 on family levels were evaluated by accuracy, precision, recall F1 score, and annotation rate. The boxplots represent the distribution of averages of five classification matrices for 17 different viral phyla produced by two pipelines; d Taxonomic assignment performances of VITAP and vConTACT2 on genus levels were evaluated by accuracy, precision, recall F1 score, and annotation rate. The boxplots represent the distribution of averages of five classification matrices for 17 different viral phyla produced by two pipelines; e Taxonomic assignment performances of VITAP and vConTACT2 on family-level cross 17 viral phyla were evaluated by F1 score and annotation rate; f Taxonomic assignment performances of VITAP and vConTACT2 on genus level cross 17 viral phyla were evaluated by F1 score and annotation rate.
By employing these pipelines to perform taxonomic assignments on their database-derived sequences, we evaluated each pipeline’s efficiency and performance in utilizing taxonomic databases. For family-level taxonomic assignments on sequences of varying lengths, VITAP and the other four pipelines all achieve average accuracy, precision, and recall values exceeding 0.9 (Fig. 3a). PhaGCN2 is unable to perform taxonomic assignments on sequences as short as 1-kb, but demonstrates comparable classification performance to other pipelines for longer sequences 5-kb. VITAP’s family-level annotation rate exceeds that of most other pipelines, remaining comparable to VPF-Class (both exceeding 0.9). For genus-level taxonomic assignments, VITAP exhibits accuracy approaching 1, comparable to vConTACT2 (Fig. 3b). Although its precision and recall are slightly lower than those of vConTACT2 and CAT, they remain acceptable (above 0.9), remaining comparable to VPF-Class. In terms of genus-level annotation rate, VITAP surpasses vConTACT2 and CAT, remaining comparable to VPF-Class. This result further underscores VITAP’s ability to maintain high taxonomic assignment matrices while achieving a higher annotation rate than other pipelines.
Fig. 3. Database utilization efficiency evaluation of VITAP compared to vConTACT2, CAT, VPF-Class, PhaGCN2, and geNomad.
Boxplots represent the performance metrics (accuracy, precision, recall, F1 score, annotation rate) of five fragment subsets with different lengths (1-, 5-, 10-, 20-, and 30-kb), which were generated from their corresponding database The boxplots represent the distribution of averages of five classification matrices for 17 different viral phyla produced by two pipelines. The center lines of the boxes indicate the median values of taxonomic assignment matrices on 17 viral phyla. The bounds of the box represent the interquartile range, with the lower and upper bounds, respectively, corresponding to the first and third quartiles. The whiskers denote the lowest and highest values within 1.5 times the interquartile range. a The evaluation of taxonomic assignments on family-level. For each box plot, the boxes from left to right represent the statistical data of vConTACT2, CAT, VPF-Class, PhaGCN2, geNomad, and VITAP, respectively; b The evaluation of taxonomic assignments on genus level. The PhaGCN2 and geNomad lack genus-level taxonomic assignment capabilities, hence their related results are not displayed. For each box plot, the boxes from left to right represent the statistical data of vConTACT2, CAT, VPF-Class, and VITAP, respectively.
Next, a taxonomic assignment comparison on newly released viral genomes between VITAP and other pipelines was conducted to validate its potential advantages in viral taxonomy applications. The viral reference genomes collected in previous ICTV releases and NCBI viral RefSeqs (VMR-MSL35, VMR-MSL37, and NCBI RefSeq209) were used to build VITAP databases. These VITAP database versions are consistent with those used by other pipelines to avoid the impact of database version differences on the comparison. Genomes released in NCBI Viruses after January 2020 (for VMR-MSL35) and January 2022 (for VMR-MSL37 and NCBI RefSeq209) were respectively designated as ‘new genomes’ and fragmented to generate simulated viromes47. The simulated viromes were used to perform taxonomic assignments using five state-of-the-art pipelines (VPF-Class: genomes released after January 2020, others: genomes released after January 2022) and VITAP, respectively40,44,46,48,49. Based on the results of VMR-MSL37-based family-level taxonomic assignments, vConTACT2, CAT, PhaGCN2 and VITAP had relatively high accuracy, precision, and recall, resulting in similar F1 scores (Fig. 4a); based on the results of VMR-MSL35-based family-level taxonomic assignments, VITAP had higher accuracy, precision, recall, and annotation rate than VPF-Class (Supplementary Fig. 1a). Hence, the performance differences of family-level taxonomic assignments between most pipelines are mainly reflected by the differences of annotation rate (Fig. 4a and Supplementary Data 2, 3). VITAP’s average annotation rate is higher than other pipelines (family-level: 0.95, genus level: 0.86), and its standard deviation is lower than other pipelines (family-level: 0.15, genus level: 0.2). For genus-level taxonomic assignments, VITAP and CAT exhibit similar performance, with vConTACT2 slightly outperforming both. All three pipelines achieve an average F1 score above 0.9. Nevertheless, CAT and VITAP clearly outperform vConTACT2 in terms of annotation rate, especially for sequences as short as 1 and 5 kb. Even for 20-kb and 30-kb sequences, CAT and VITAP remain superior to vConTACT2. In addition, VITAP achieves a lower standard deviation in genus-level taxonomic assignments across all sequence lengths (vConTACT2: 0.37, CAT: 0.29, VITAP: 0.15) (Fig. 4b). Similar evaluations for the VMR-MSL35-based taxonomic assignments also indicate that VITAP outperforms VPF-Class on accuracy, precision, recall, F1 score and annotation rate (Supplementary Fig. 1a, b).
Fig. 4. Benchmarking performance metrics (accuracy, precision, recall, F1 score, annotation rate) of VITAP compared to vConTACT2, CAT, PhaGCN2, and geNomad based on newly released genomes (3705 viral reference genomes, released after 2022.01).
All of these pipelines are based on the VMR-MSL37/NCBI RefSeq209 database. a The taxonomic assignment performance evaluation on family-level. Boxplots represent the five performance metrics of five fragment subsets with different lengths (1-, 5-, 10-, 20-, and 30-kb), which were generated from the VMR-MSL37/NCBI RefSeq209 database, including 191,596, 40,425, 21,857, 13,622, and 8844 fragments, respectively. These boxplots represent the distribution of averages of five classification matrices for 16 different viral phyla produced by two pipelines. The center lines of the boxes indicate the median values of taxonomic assignment matrices on 16 viral phyla. The bounds of the box represent the interquartile range, with the lower and upper bounds, respectively, corresponding to the first and third quartiles. The whiskers denote the lowest and highest values within 1.5 times the interquartile range. For each box plot, the boxes from left to right represent the statistical data of vConTACT2, CAT, PhaGCN2, geNomad, and VITAP, respectively; b The taxonomic assignment performance evaluation on genus level. For each box plot, the boxes from left to right represent the statistical data of vConTACT2, CAT, and VITAP, respectively; c On the aspect of family-level taxonomic assignments, the F1 score and annotation rate of VITAP and the other four pipelines were compared spanning various sequence lengths; d On the aspect of genus-level taxonomic assignments, the F1 score and annotation rate of VITAP and the other four pipelines were compared spanning various sequence lengths.
To provide more detail on the effectiveness of VITAP taxonomic assignments, the taxonomic assignment performance through six pipelines was compared in terms of viral phyla. Different viral taxonomic ranks within the Baltimore framework have a range of taxonomic criteria, which create difficulties and challenges for automated taxonomic assignments. For the VMR-MSR37-based family-level taxonomic assignments, VITAP consistently achieves F1 scores exceeding 0.9, outperforming or equaling other pipelines for taxonomic assignments on sequences of all lengths (Fig. 4c and Supplementary Data 2). Meanwhile, VITAP attains annotation rates above 0.8 across 13 viral phyla on sequences of all lengths (except for Nucleocytoviricota, Artverviricota and Lenarviricota at 1 kb/5 kb). By contrast, CAT and geNomad are able to maintain annotation rates above 0.8 for all sequence lengths in only five (Peploviricota, Cressdnaviricota, Preplasmiviricota, Duplornaviricota, Kitrinoviricota) and two (Peploviricota, Uroviricota) viral phyla, respectively. PhaGCN2 achieves annotation rates of at least 0.8 for sequences longer than 5 kb from four viral phyla (Taleaviricota, Uroviricota, Saleviricota, Preplasmiviricota), whereas vConTACT2 fails to reach annotation rates of 0.8 for most viral phyla. For the VMR-MSR37-based genus-level taxonomic assignments, VITAP achieves an F1 score exceeding 0.9 for sequences of various lengths from 14 viral phyla (except for Preplasmiviricota and Lenarviricota) (Fig. 4d and Supplementary Data 3). Moreover, VITAP and CAT maintain annotation rates above 0.7 for sequences of various lengths from nine and eight viral phyla, respectively. vConTACT2 only maintains an annotation rate of 0.88 for Nucleocytoviricota, and when the sequence length reaches the 30-kb cutoff, it achieves annotation rates exceeding 0.7 for seven viral phyla. These results further demonstrate that although vConTACT2 exhibits high accuracy, precision, and recall, its annotation rate is suboptimal and is significantly influenced by sequence length. For VMR-MSR35-based taxonomic assignments, VITAP generally achieves higher F1 scores and annotation rates than VPF-Class across all viral phyla (Supplementary Fig. 1c, d and Supplementary Data 3). Notably, for RNA viruses, VPF-Class failed to perform taxonomic assignments for any of the five RNA viral phyla. In sum, these results demonstrate VITAP’s superior performance in genus-/family-level taxonomic assignments compared to other pipelines. The advantage of VITAP lies in its ability to maintain a high F1 score while achieving relatively high annotation rates across multiple viral phyla. In addition, VITAP is particularly suitable for taxonomic assignments on highly incomplete DNA and RNA viral sequences, which constitute the main components of metagenomes, metatranscriptomes, and metaviromes.
High accuracy and robustness of VITAP in taxonomic assignments of short sequences from different viral phyla
Based on the results from tenfold cross-validation, the F1 scores of assigned viral families/genera for short sequences were further analyzed. These results focus solely on those sequences with assigned taxonomic units by VITAP, demonstrating its capability for effective family and genus level taxonomic assignments for sequences as short as 1-kb and 5-kb. For taxonomic assignment of 1-kb sequences at the family-level, VITAP yielded reliable taxonomic assignments (average F1 score >0.9) for 13 viral phyla, and acceptable taxonomic assignments for Cossaviricota (average F1 score = 0.83), Nucleocytoviricota (average F1 score = 0.89), Negarnaviricota (average F1 score = 0.88), and Pisuviricota (average F1 score = 0.88) (Fig. 5a and Supplementary Data 4). At least 80% of families in each viral phylum had an F1 score of higher than 0.9. For taxonomic assignment of 1-kb sequences at the genus level, VITAP yielded similar average F1 scores for most viral phyla except Cressdnaviricota, which had a notably reduced value (decreased from 0.92 to 0.67). In nine viral phyla (Peploviricota, Uroviricota, Cressdnaviricota, Hofneiviricota, Phixviricota, Preplasmiviricota, Kitrinoviricota, Negarnaviricota, and Pisuviricota), less than 80% of genera had F1 scores of higher than 0.9 (Fig. 5a and Supplementary Data 4). For taxonomic assignment of 5-kb sequences at the genus and family levels, VITAP is capable of performing more stable and effective taxonomic assignments than for 1-kb sequences. The family-level average F1 scores for the 17 viral phyla range from 0.91 (Nucleocytoviricota) to 1 (for eight viral phyla); the genus-level average F1 scores range from 0.62 (Pisuviricota) to 1 (for six viral phyla). Nearly 90% of families in each viral phylum had an F1 score higher than 0.9 (Fig. 5a and Supplementary Data 4). In Peploviricota, Cressdnaviricota, Hofneiviricota, Phixviricota, Kitrinoviricota, and Negarnaviricota, 80% of genera have average F1 scores higher than 0.9 when the sequence length reached at least 5-kb. In summary, for short sequences from all 17 viral phyla, VITAP is able to generate reliable family-level taxonomic assignments. For short sequences from the majority of viral phyla (10 out of 17 for 1-kb sequences and 14 out of 17 for 5-kb sequences), VITAP is able to generate acceptable genus-level taxonomic assignments.
Fig. 5. The robustness of VITAP’s taxonomic assignments for short sequences within different viral phyla, and across whole viral genomes.
This result focuses on the taxonomic assignment efficiency of short sequences that were successfully assigned taxonomic units by VITAP, without considering sequences that were not assigned, as the annotation rate of VITAP has already been thoroughly evaluated in previous results. a The comparison of the taxonomic assignment efficiency at the family and genus level for 1- and 5-kb short sequences, which derived from different viral phyla. For 1-kb sequences, VITAP can produce acceptable family-level taxonomic assignment results; For 5-kb sequences, VITAP can produce reliable results on both genus and family levels. Different viral phyla are represented by distinct symbols; DNA viruses and RNA viruses are denoted by different background colors in the scatter plots. b VITAP is capable of capturing taxonomic signals for 1-kb short sequences from different regions of viral genomes. Four viruses (Tequatrovirus T4, human alphaherpesvirus 1, monkeypox virus, and severe acute respiratory syndrome coronavirus 2) were used to demonstrate VITAP’s classification confidence for their different genomic sequences. In the genome organization diagrams, heatmaps below indicate the taxonomic assignment confidence levels at the family and genus levels for the sequences; line graphs represent the alignment scores of these sequences among members within their own taxonomic unit, as well as with members of other taxonomic units.
The relationship between alignment scores and confidence levels of taxonomic assignments were assessed. Four representative genomes from four viral realms were selected to show the taxonomic assignment performances of 1-kb sequences from along the genomes (Fig. 5b). Viral sequences generally have higher alignment scores within their own taxonomic units compared to those from other taxonomic units. The greater the difference between the alignment scores within their taxonomic units and those with other units, the stronger the taxonomic distinctiveness of these sequences, corresponding to higher confidence in their taxonomic assignments. For Human herpesvirus 1 strain 17 (Simplexvirus humanalpha1) and Monkeypox virus strain Zaire-96-I-16 (Monkeypox virus), some sequences located in longer intergenic regions could also be effectively classified, with a high-confidence of taxonomic assignment (ranging from 0.78 to 1). Owing to the end-to-end translation strategy for short non-coding sequences, VITAP is also capable of effectively classifying potential non-coding viral sequences in metagenomes.
Based on the results of the tenfold cross-validation, the accuracy of genus-/family-level taxonomic assignments for short sequences across various viral phyla was further evaluated at different confidence levels. For most phyla, high-/medium-confidence assignments showed an accuracy above 0.7, indicating acceptable performance (Fig. 6 and Supplementary Note 2). Low-confidence assignments varied, with some phyla, such as Taleaviricota and Peploviricota achieving an acceptable accuracy for 5-kb sequences. Notably, the evaluation highlights VITAP’s stable performance in RNA viral taxonomic assignments. For Duplornaviricota, Lenarviricota, and Negarnaviricota, all genus and family-level taxonomic assignments, regardless of confidence level, demonstrated an accuracy exceeding 0.7, indicating consistent reliability (Fig. 6 and Supplementary Note 2). Taxonomic assignments with high-/medium-confidence of Kitrinoviricota and Pisuviricota achieved accuracies above 0.7 across different sequence lengths (Fig. 6 and Supplementary Note 2). This result also confirmed VITAP’s effectiveness in accurately performing taxonomic assignments for RNA viral sequences.
Fig. 6. The VITAP taxonomic assignments’ accuracy of different confidence levels for family and genus level unit assignments across viral phyla.
The pie charts indicate the true positive and false positive results from family and genus levels unit assignments for viral phyla, with the accuracy (and lineage scores calculated by VITAP). If a taxonomic hierarchy in a viral phylum has no results at a certain confidence level, it will be shown as an empty pie chart. The results demonstrate that VITAP can provide accurate taxonomic assignments at a high-confidence level (lineage score ≥0.9) for all viral phyla, and offer acceptable classification for most viral phyla at a medium-confidence level (0.1 ≤ lineage score <0.9).
The re-assessment of viral diversity in deep-sea environments
To update insights into the diversity of deep-sea viruses under a viral taxonomic framework, viruses derived from four deep-sea viromes were re-performed taxonomic assignments using VITAP4,28,50,51. For the original taxonomic profiling from the literature, 93.5% of deep-sea-derived vOTUs were unclassified at the class level4,50. These studies were all based on the taxonomic framework proposed by ICTV prior to 2021 for viral taxonomic criteria, which included morphological-based taxonomic units (Sipoviridae, Myoviridae, and Podoviridae) that are now abolished in the taxonomy of head-tail viruses. Morphology-based taxonomy of head-tail viruses is a low-resolution classification, that fails to adequately reflect the genomic characteristics of these viruses. Nonetheless, these studies filled gaps in the understanding of the ecological functions of deep-sea viruses, but the information they provided on the taxonomy and systematics of these viruses is still quite limited.
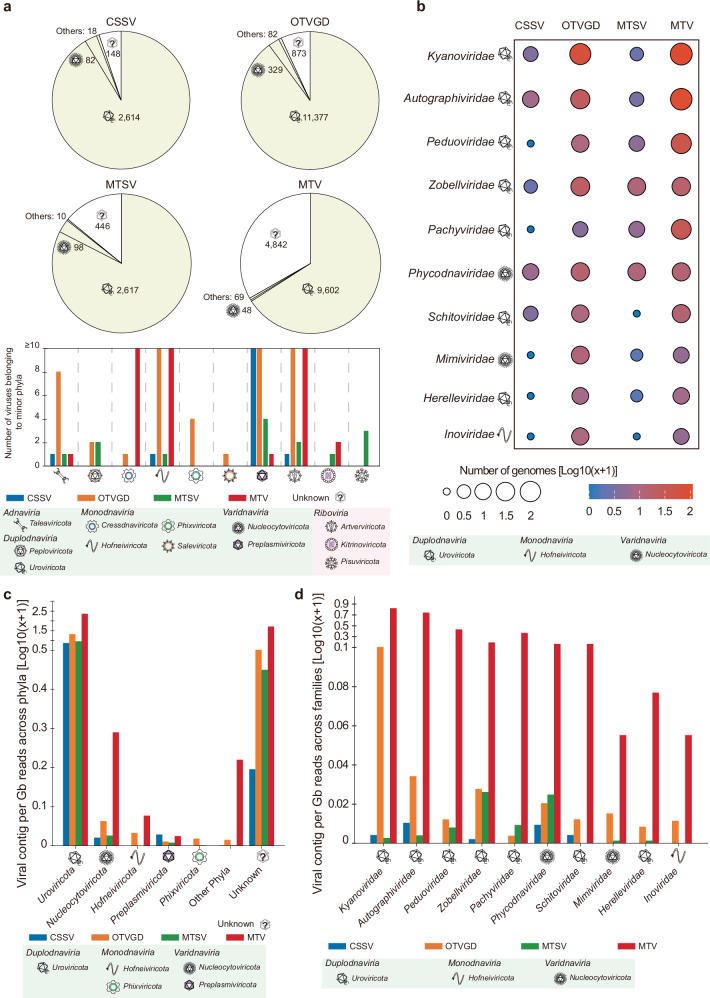
Leveraging the advantages of VITAP in viral taxonomic assignments, the taxonomic profile of deep-sea viruses has been re-established. To achieve more comprehensive results, we performed taxonomic assignments for deep-sea viruses using an expanded database combining IMG/VR and NCBI RefSeqs. Based on the expanded database, VITAP achieved a slightly higher lineage-level annotation rate compared to using the VMR-MSL37 database alone, while significantly improving family- and genus-level annotation rates, with increases of approximately 1.5-fold and 1.7-fold, respectively (Supplementary Fig. 2a, b). With regard to whole lineage annotation, VITAP and geNomad have similar annotation rates on these datasets, which are significantly higher than those of other pipelines (Supplementary Fig. 2a and Supplementary Data 5). They both assigned taxonomic lineages to over 27,000 deep-sea-derived viruses. However, with regard to family and genus level unit assignments, the annotation rate of VITAP is higher than geNomad and other pipelines, further confirming the good generalization ability of VITAP. Although VPF-Class and PhaGCN2 achieved higher genus and family-level unit assignments compared to VITAP, their results were not considered to be highly reliable and exhaustive due to their outdated databases, suboptimal taxonomic assignment efficiency or low lineage annotation (Supplementary Figs. 1 and 2). At the phylum-to-class level, 27,064 vOTUs out of 33,373 could be assigned to established viral phylum or classes (Fig. 7a and Supplementary Data 5). Hence, VITAP increased the proportion of classifiable viruses from the original 6.5 to 81.1%. Most deep-sea viruses belonging to Uroviricota-Caudoviricetes, comprise 77.9, 80.3, 75.0, and 77.9% vOTUs of CSSV (cold seep sediment virome), OTGVD (oceanic trench viral genome dataset), MTSV (Mariana Trench sediment virome), and MTV (Mariana Trench water column virome), respectively. The Varidnaviria is the second largest viral realm, comprising 0.9% vOTUs of MTV to 3.3% vOTUs of MTSV. Notably, 11 vOTUs were assigned to Adnaviria (4 from OTVGD, 3 from the CSSV, and 1 from CSSV), which contained all filamentous dsDNA viruses infecting Archaea52,53. At the family-level, only 2339 vOTUs were assigned to family-level units, indicating that there were a substantial number of undiscovered viral families in deep-sea viruses, especially for head-tail viruses (Supplementary Fig. 2a and Supplementary Data 5). For established viral families, the Kyanoviridae (T4-like viruses infecting cyanobacteria) are the most classified viruses within deep-sea viromes (n = 459). This is followed by Autographiviridae (T7-like viruses) and Peduoviridae (P2-like viruses), which contain 292 and 110 vOTUs, respectively. In addition, 79 and 28 vOTUs (mainly from the OTVGD and MTV) were classified as Phycodnaviridae and Mimiviridae, respectively. The family features of these viral lineages were also confirmed through genome-wide analysis (Supplementary Fig. 3).
Fig. 7. The viral taxonomy of deep-sea viromes.
a The realm-level taxonomy of four deep-sea virome datasets. The pie chart and bar chart indicate the number of viral operated units (vOTUs) contained in each viral phylum, which are represented by distinct viral symbols; b The family-level taxonomy of four deep-sea virome datasets. The bubble chart displays the top ten assigned viral families in each dataset. Viral family names and their respective phylum symbols are annotated on the left side of the heatmap; c The bar graph represents the density of each phylum level taxonomic unit in each dataset based on sequencing depth (giga base, Gb). Different datasets are indicated by bars of varying colors; d The bar graph represents the density of the top ten family-level taxonomic units in each dataset based on sequencing depth (giga base, Gb). Different datasets are indicated by bars of varying colors. Corresponding phylum symbols are annotated beside the viral family names. (CSSV sediment virome of cold seep, OTGVD oceanic trench viral genome dataset, MTSV sediment virome of Mariana Trench, MTV water column virome of Mariana Trench).
Recruitment of a range of viral taxonomic units were normalized based on their metagenome sizes and were used to facilitate comparisons between different deep-sea viromes. At the phylum level, Uroviricota had the highest density across all viromes (from 6.3 to 230.3 vOTUs/GB), followed by Nucleocytoviricota (from 0.05 to 0.95 vOTUs/GB) (Fig. 7c). For vOTUs assigned to established families, Autographiviridae, Kyanoviridae, Zobellviridae and Kyanoviridae had the highest densities in CSSV, OTVGD, MTSV, and MTV, respectively (Fig. 7d). Notably, most viral families from the MTV had higher densities than other three datasets, potentially due to its enrichment process leading to greater efficiency in virion recovery50.
Discussion
Through comparative genomics employing computational techniques, significant progress has been made in mapping out the evolutionary history of key viral groups. This progress has led to the establishment of a systematic viral taxonomic framework, now officially recognized by ICTV54–57. The automated taxonomic assignments of viruses has been a challenge within the field of virology for decades. Particularly, with the rapid development of meta-omics, the vast amount of viral genomic data presents unprecedented challenges for viral taxonomy. Leveraging computational biology techniques, previous research efforts have resulted in the development of a series of high-performance automated classification pipelines, making a significant contribution to advances in the field40–42,46,48. These pipelines are primarily applicable to prokaryotic viruses with dsDNA genomes, providing a crucial reference for the establishment of various taxonomic hierarchies for head-tail viruses as recognized by ICTV58,59. The dsDNA viruses that infect prokaryotes are widely distributed in the environment and have long served as the primary subjects of research in ecological virology4,24,28,60,61. With further advances in metagenomic technologies, an increasing number of studies have been focused on the diversity of viruses beyond head-tail viruses (e.g., ssDNA viruses and RNA viruses), aiming to elucidate their biological and ecological significance12,29–33,62–68. However, no automatic taxonomic assignment pipeline is universally available for all viruses. This was the initial motivation behind the design of VITAP, with the expectation of filling this technical void in the field. With an acceptable time-consuming and memory-consuming requirement (Table 2, Supplementary Fig. 4, and Supplementary Note 3), VITAP is capable of performing efficient taxonomic assignments for viruses from different realms and is continuously updated based on the latest ICTV proposals. VITAP exhibits reliable generalization ability for taxonomic assignments of highly diverse viromes. In the evaluation based on newly released viral RefSeqs, VITAP and other pipelines show comparable accuracy, precision, recall, and F1 score for taxonomic assignments, we demonstrated superior overall annotation rates of VITAP compared to other pipelines, particularly for highly incomplete viral sequences (Fig. 4 and Supplementary Fig. 1). In application, the advantage of VITAP becomes obvious, as reflected in its remarkably higher annotation rate compared to other pipelines for DSV-derived viruses. VITAP can also integrate viral genomes from different databases (e.g., NCBI RefSeqs and IMG/VR) along with their taxonomic information to perform taxonomic assignments. This integration enhances the generalization ability of VITAP’s taxonomic assignment, resulting in a greater number of annotations (Supplementary Figs. 2, 5 and Supplementary Data 5). In addition, it offers a user-friendly interface and provides easily interpretable results.
Table 2.
The time-consuming of different pipelines across viral sequence sets with diverse lengths
| Pipelines | Database version | 1-kb (h) | 5-kb (h) | 10-kb (h) | 20-kb (h) | 30-kb (h) |
|---|---|---|---|---|---|---|
| vConTACT2 | VMR-MSL37 | 2.5 | 1.9 | 1.5 | 1.1 | 1.0 |
| CAT | NCBI RefSeq209 | 1.3 | 1.1 | 1.0 | 0.9 | 0.7 |
| PhaGCN2 | VMR-MSL37 | - | 9.1 | 6.1 | 2.4 | 1.8 |
| geNomad | VMR-MSL37 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| VITAP | VMR-MSL37/NCBI RefSeq209 | 2.7 | 1.8 | 1.4 | 1.3 | 0.9 |
| VPF-Class | VMR-MSL35 | 141.7 | 30.1 | 15.6 | 8.8 | 5.1 |
| VITAP | VMR-MSL35 | 5.3 | 3.3 | 2.8 | 2.3 | 1.8 |
These were recorded by wall-clock time under the programs running with 12 CPUs and 96 GB memory.
VITAP is capable of performing taxonomic assignments for viral sequences as short as 1 kb. However, the taxonomic assignments of these sequences require careful interpretation (Supplementary Note 4). Firstly, researchers need to rigorously quality control the short sequences used for taxonomic assignments to exclude any potential contamination from cellular genomic sequences. Such contamination may arise from artificial concatemer/circular sequences introduced during the reads assembly process45,69, or natural viral genomes with cellular origins due to frequent HGTs between viruses and cellular organisms70,71. For nearly complete genomes, this cellular contamination can be effectively identified through comparison with reference genomes and analysis of G + C skewing72. However, this process becomes challenging for shorter sequences, as much of the information is lost or cannot yield statistically meaningful conclusions73. Secondly, there are extensive HGTs between viruses and cellular organisms, as well as among different viruses63,70,71,74. These exogenous sequences do not reflect the bona fide taxonomic information of the viruses and can lead to abnormal results when present in short sequences. For instance, some genetic elements of head-tail viral ORFs and cellular gene transfer agents (GTAs) share a common origin75–77; there are widespread HGTs between giant viruses78. Thirdly, the boundaries between viral lineages are not always clear. Different viral lineages may share genes26,63,74,79, and smaller viral elements can even integrate into larger viral genomes80,81, which complicates the taxonomic assignments of short sequences. Considering these issues, VITAP attempts to provide local optimum taxonomic hierarchies by making comparisons with reference genomes and calculating characteristic thresholds, although these results are not guaranteed to be correct (Supplementary Note 4). Therefore, the taxonomic assignments of short (≤1-kb) viral sequences remain a challenge for VITAP as well as other current pipelines, albeit VITAP has made some improvements in this aspect.
Unlike genome-content-based taxonomic assignment pipelines, VITAP performs taxonomic assignments on relevant viral contigs in accordance with an existing taxonomic framework. The former approach is based on clustering algorithms, performing taxonomic assignments based on signature sequences present within each group40,41. Initially, adopting a similar methodology for VITAP was considered, but subsequent evaluations revealed the limitations of clustering algorithms for viral taxonomic assignments. Specifically, it is challenging to generate results consistent with the current taxonomic framework for viral genomes from different taxonomic levels/units under the same clustering parameters (e.g., inflation in the Markov clustering algorithm, linkage criteria in Hierarchical Clustering). Therefore, viral taxonomic assignments based on clustering methods require careful consideration of these clustering parameters and the design of specific classification approaches for viruses from different taxonomic hierarchies or units31. However, this introduces a potential problem: often, it is not clear in advance to which taxonomic hierarchy or unit the virus to be classified belongs, creating an obstacle to the effective use of these taxonomic assignment pipelines. Undoubtedly, while genome-content-based taxonomic assignment pipelines centered on clustering algorithms have apparent limitations, they provided an important method for the establishment of novel viral taxonomic units (especially for head-tail viruses)56,58,82,83.
A hybrid annotation strategy can make more comprehensive annotation results than only using a single one. For instance, the hybrid annotation strategy of IMG/VR (v.4) is based on several databases, including the viral RefSeqs database, geNomad’s marker-based assignments, alignment to GenBank NR database, and vOTU consensus45. Based on the benchmarking and DSV-based annotation analysis, the results indicate that, the performance of stand-alone VITAP surpassed or was equal to other stand-alone pipelines on most viral phyla (Supplementary Fig. 2a and Supplementary Data 5). VITAP largely replicates IMG/VR’s taxonomic assignments for DNA viruses, with fewer inconsistencies and higher accuracy in some cases, but shows limited consistency for RNA viruses at the family-level (Supplementary Fig. 6, Supplementary Data 6, and Supplementary Note 5). For genomes with inconsistencies, the IMG/VR hybrid taxonomic annotation pipeline outperforms VITAP by over five-fold, suggesting potential limitation of VITAP due to the lack of reference sequences in ICTV. However, its performance is expected to be improved as the diversity of viral databases increases (Supplementary Fig. 5). In addition, VITAP assigned new family-level taxonomic units to 41,469 previously unclassified vOTUs from IMG/VR (v.4), highlighting the complementary advantages of VITAP compared to other pipelines in taxonomic assignments (Supplementary Fig. 7, Supplementary Data 7, and Supplementary Note 6). Currently, no single pipeline can achieve performance comparable to that of a hybrid annotation strategy. The combining results from multiple pipelines is expected to obtain more comprehensive annotations in meta-omic analysis. Overall, VITAP currently offers a viral taxonomic assignment algorithm framework, including a series of steps to determine the optimal taxonomic hierarchies and units for viral sequences. In future work, we will explore whether the weights generated during the VITAP taxonomic assignment process can be utilized as measures of distance or similarity in clustering algorithms, giving the capacity to guide the generation of novel taxonomic units.
Methods
Database of viral reference genomic sequences used by VITAP
Published viral reference genomes were used to build the VITAP-specific database and perform benchmarking. As the high priority of ICTV in viral taxonomy, the taxonomic assignments by VITAP are highly based on viral genomes accepted by ICTV. After carefully evaluating the differences in taxonomic information between various databases, viral genomes from other databases can also be integrated into the VITAP database to enhance the sensitivity of taxonomic assignments. Genomes collected in ICTV VMRs and NCBI RefSeqs (VMR-MSL35, VMR-MSL37, VMR-MSL38, and NCBI RefSeq209) were retrieved and downloaded from GenBank. These included 10,345, 15,230, 16,238, and 13,650 viral genomes/segments from 6388, 10,245, 11,095, and 10,377 viral species respectively. Based on the viral region information provided by ICTV, viral regions that were integrated into host genomes were extracted from corresponding host chromosomes.
The gene calling of viral genomes/segments was performed using two strategies, open-reading-frame (ORF)-based and end-to-end translation. Prodigal (v.2.6) was used to predict putative viral ORFs with default parameters in “meta” mode84. For those short sequences that could not be processed by Prodigal, the end-to-end translation strategy was applied to generate six possible reading frames using Seqkit (v.2.5)85. The ORFs and end-to-end translated reading frames were merged into a viral reference protein set.
Determination of taxonomic thresholds
Taxon-specific alignment thresholds were calculated for each taxonomic unit based on all-to-all alignment of reference genomes. Unlike the method designed by Lavigne et al., the proportion of shared homologous ORFs within a taxonomic unit was not presupposed86. Instead, the alignment-based sequence similarity was quantified and used to determine the taxon thresholds. This is a more flexible approach, as each taxonomic unit is assigned a specific threshold, thus avoiding the previous rigid taxonomic assignment process. Viral reference proteins were self-aligned using DIAMOND (v.0.9) with 1e-5 as the E-value and other default parameters, reporting the top 1000 hits. Each ORF was subsequently given a taxonomic lineage composition based on the taxonomic lineage of the assigned reference genomes. Different ORFs within a target genome might have different taxonomic lineage compositions. The hits for each ORF were then categorized into three types: self-hit, top-hit, and others, assigning different weights to each type. A self-hit is an ORF’s self-alignment, with a weight of 1; top-hit is the second-ranking hit, excluding the self-hit, and all other hits sharing the same taxonomic unit, with a weight of 1.2; others, including all remaining hits, were assigned a weight of 0.8. A second type of weight based on a voting strategy was then defined. Dominant taxonomic units (constituting at least 50% of all hits for an ORF, referring to geNomad) were assigned a weight of 1.249, while the remaining taxonomic units were assigned a weight of 1. Based on the two types of weights mentioned above ( and ) and the raw bitscore of each hit of each ORF (), the taxon bitscore () of each hit was calculated:
| 1 |
where describes a modified bitscore considering different hit types for each alignment.
Each taxonomic unit was scored for each genome. Firstly, the ratio of the number of homologous ORFs (with 1e-5 as the E-value cut-off of BLASTp) is defined. Let be the number of ORFs of a genome that fall into a taxonomic unit; let be the total number of ORFs of a genome. Based on the calculated and , a parameter is defined by:
| 2 |
where describes the ratio of the number of homologous ORFs (with 1e-5 as the E-value cut-off of BLASTp) appearing in a genome within a taxonomic unit to its total ORFs. For each unit within each taxonomic hierarchy, a distinct will be calculated. In other words, each realm-, kingdom-, phylum-, class-, order-, family-, genus-, and species-level unit within the viral taxonomy framework will have its own specific . This ratio is squared to amplify differences. Secondly, the taxonomic scores for each genome are determined. Let be the sum of for a genome within a taxonomic unit; let be the number of hits within this taxonomic unit. Then, the taxonomic score () for a genome is defined by:
| 3 |
where describes a quantified parameter that considers two features, including ORF alignment scores, and the proportion of homologous ORFs present in a taxonomic unit relative to the total number of ORFs in the genome.
The taxonomic threshold for a taxonomic unit is determined from the set of taxonomic scores of a reference viral genome. Defining and : belongs to a set that contains a series of taxonomic scores, all of which are associated with taxonomic units identical to those of the query genome. The lowest value within this set is defined as . Similarly, belongs to another set that consists of taxonomic scores associated with taxonomic units different from those of the query genome. The highest value within this set is defined as . Based on the calculated and , two types of taxonomic score threshold are defined by:
| 4 |
where and respectively describe the cases where multiple genomes from the same taxonomic unit are present in the database, and the cases where certain taxonomic units in the database contain only a single genome. These equations considered both different taxonomic units with close relationships and relatively independent taxonomic units. If a genome has taxonomic scores in multiple different taxonomic units, the taxonomic score threshold for its bona fide unit is the minimum value among all members that meet the consistency criteria for that unit. If all genomes in a particular unit only have taxonomic scores within that unit, the threshold is set in the upper quartile of the lowest taxonomic score in that unit, ensuring a degree of diversity tolerance.
The taxonomic thresholds were calculated for each taxonomic unit through the algorithm described above. The thresholds were subsequently used to calculate quantitative parameters for the classification of a target genome into taxonomic units at different taxonomic hierarchies. These steps provide the minimum thresholds for the taxonomic validity of each taxonomic unit. These values vary for different taxonomic units and are influenced by the diversity of the reference sequence database.
Optimal taxonomic lineage determination and confidence level assignment
The taxonomic unit assignment of VITAP independently processes in different taxonomic hierarchies (from realm-level to species-level). For annotations of a certain genome, the best-fit taxonomic units belonging to different hierarchies are not always consistent with its real hierarchies. This confusion might have resulted from horizontal gene transfers between different taxonomic units within different taxonomic ranks. Based on a range of taxon scores of a genome, an algorithm to detect all possible taxonomic lineages for that genome and determine one optimal taxonomic lineage was developed. Firstly, based on the genome-taxonomy bipartite graph and taxonomic hierarchy, taxonomic units of a genome were aligned at a range of hierarchies. The genome-taxonomy bipartite graph consists of genome IDs, taxonomic units, and taxonomic scores (), describing the taxonomic score () of each genome for each taxonomic unit. A genome may have multiple taxonomic unit associations, but the mapping between each genome and each taxonomic unit is unique. This step produced all possible taxonomic lineages for a genome. Secondly, based on pre-built taxonomic thresholds ( and ) and taxonomic score for a genome (), the ratio of each taxonomic unit’s taxon score to its corresponding threshold was calculated:
| 5 |
where the was used as the taxon weight (species to realm-level). For a genome, described a quantitative parameter for the likelihood of a genome being attributed to a specific taxonomic unit. Thirdly, the lineage score was calculated based on the array of of a genome. Let and be two types of lineage scores; let be the lowest taxonomic rank at which exceeds 0.3; is the final terminating taxonomic hierarchy, for which the calculated lineage score is the maximum among all lineage scores; let be the number of taxon levels included in a lineage. Then:
| 6 |
In detail, for lineages where all taxon scores are less than 0.3, the determination of involves a series of calculations. Starting from the realm, the taxon scores of lower ranks were progressively summed and the average calculated, resulting in a set of lineage scores . The highest lineage score is designated as ; the corresponding lowest rank is identified as the optimal terminating hierarchy. Based on the current database and taxonomic frameworks, this step determined the lowest classifiable taxonomic hierarchy for a genome. Fourthly, the three confidence levels were assigned based on lineage scores: high-confidence for results with lineage score ≥ 0.6; medium-confidence for results with lineage score ≥ 0.3; low-confidence for the rest of other results. In terms of the significance of varying lineage scores, a lineage score of 1 indicates that a genome meets the threshold of the current taxonomic framework (a high-confidence setting of 0.6 is used to maintain a certain tolerance level). When the lineage score is less than 1, it signifies that a genome does not meet the threshold of the existing taxonomic framework but is, to some extent, close to certain taxonomic lineages (the lineage score describes this degree). Fifthly, the result with the highest lineage score with certain conditions for each genome was selected as the optimal taxonomic lineage (). Let be a set containing all possible taxonomic lineages; let be the number of weights (taxonomic hierarchies) in the set for . Then, for each possible lineage () from the last weight to the first weight, each lineage score :
| 7 |
its ratio to was calculated and was determined:
| 8 |
In summary, this equation aims to find an in the set such that, under the condition , the value of is maximized. This allows VITAP to capture the condition where the optimal taxonomic lineage has the most number of weights (taxonomic hierarchies) and its average weight is at least 0.6 of the maximum weight of all possible taxonomic lineages.
The generalization ability assessment of VITAP
The 16,238 viral genomic sequences collected in VMR-MSL38 were used to characterize the generalization ability of VITAP. 70% of the sequences from the set were selected as the training set, and the remaining sequences as the test set. The sequences in the test set were sliced into fragments with diverse lengths (1-kb, 5-kb, 10-kb, 20-kb, 30-kb). The training set was used to build a database utilized by VITAP. Taxonomic assignments of the test set were performed by VITAP based on the built database in the previous step. The model’s performance on the test set was characterized using four parameters for different taxonomic levels: accuracy, precision, recall, F1 score, and annotation rate. Let , , , be numbers of true positive, true negative, false positive, and false negative, respectively. Then:
| 9 |
| 10 |
| 11 |
| 12 |
| 13 |
where TP refers to the number of viral sequences correctly assigned to the bona fide taxonomic units; TN refers to the number of viral sequences that do not belong to given taxonomic units and were also correctly excluded by the VITAP; FP refers to the number of viral sequences that were incorrectly assigned to a taxonomic unit; FN refers to the number of viral sequences that should have been assigned to a taxonomic unit but were incorrectly excluded by the VITAP and other pipelines. These performance matrices were calculated for family and genus, respectively.
The model is not expected to provide taxonomic units beyond those present in the training set. On the other hand, unlike traditional binary and multi-class supervised learning, viral taxonomic assignment task is a complex hierarchical classification rather than a flat classification. Therefore, applying a strategy to describe the model’s generalization ability objectively was considered: the statistics were performed only on the taxonomic units appearing in the training and test sets. For taxonomic units missing from the training set, if the related sequences were marked by VITAP as “-“ (indicating that they cannot be classified) rather than being assigned to incorrect taxonomic units, these sequences were also considered true positive results. These instances should not be considered failures of VITAP and other pipelines. These steps were repeated ten times independently (tenfold validation) to ensure the stability of the generalization ability assessment.
Benchmarking of VITAP with other pipelines on viral sequences
The performance of VITAP and other state-of-the-art pipelines was evaluated based on a simulated virome generated from ICTV reference genomes. To enable a comparison between different pipelines, three distinct VITAP databases were constructed based on VMR-MSL35 (compared to VPF-Class), VMR-MSL37 (compared to vConTACT2, PhaGCN2, and geNomad), and NCBI RefSeq209 (compared to CAT) to ensure that VITAP and the other pipelines used the same (or contemporaneous) databases. First, we sliced the viral genomes from each database corresponding to each pipeline into 1-kb, 5-kb, 20-kb, and 30-kb sequences and performed taxonomic assignments to evaluate how effectively each pipeline utilized its respective database. Second, two “new” viral datasets (viral RefSeqs released after 2022.01 and after 2020.01, respectively) were established as test datasets based on the release dates of these databases. Sequences of different lengths (1-kb, 5-kb, 10-kb, 20-kb, and 30-kb) from two “new” viruses datasets were generated by slicing genomes, using fragment lengths as the window size and half the fragment length as the step size. The sequences generated in this step exhibited at least 50% overlap in their regions and covered the entire genomes. The database with an older version (VMR-MSL35) corresponds to a larger number of “new” test sequences, while the database with newer versions (VMR-MSL37 and NCBI RefSeqs209) corresponds to fewer “new” test sequences. Consequently, pipelines tested using VMR-MSL35 as the database (e.g., VPF-Class and VITAP) include a greater number of sequences compared to those tested using VMR-MSL37/RefSeqs209 as the database (e.g., vConTACT2, CAT, PhaGCN2, geNomad, and VITAP).
Datasets with diverse lengths were subject to previously published pipelines to perform taxonomic assignments. For vConTACT2, the genomes of VMR-MSL37 were merged with the “new” virus dataset to generate VCs (with “–db none” option and other default parameters). As stated by vConTACT2, the extent to which vConTACT2 can reproduce a situation where one VC and one sub-VC represent one family and genus was monitored, respectively. We strictly followed vConTACT2’s official guideline: “If the user genome is in the same VC but not the same subcluster as a reference, then it’s highly likely the two genomes are related at roughly genus-subfamily level.”40. Therefore, given that a VC may represent a genus or a subfamily, a VC must have purity at the family-level, meaning that all members within the same VC must belong to the same family. Otherwise, all members within that VC were considered false negatives that cannot be efficiently classified. For genus, referring to “If the user genome is within the same VC subcluster as a reference genome, then there is a very high probability that the user genome is part of the same genus,” a sub-VC produced by vConTACT2 represents a genus. All members within the same subVC must belong to the same genus. Otherwise, all members within that subVC were considered false negatives that cannot be efficiently classified. For CAT (with NCBI RefSeq209 database)46, fragments of the “new” viral dataset (released after 2022.01) were used to perform taxonomic assignments. For VPF-Class (with VMR-MSL35-contemporary database)44, fragments of “new” viral datasets (released after 2020.01) were used to perform taxonomic assignments. At that time, Caudovirales had not yet been abolished. For PhaGCN2 (with VMR-MSL37 database)48, fragments of the new viral dataset (released after 2022.01) were used to perform taxonomic assignments. Only fragment sets with lengths over 10-kb can be used in taxonomic assignments of PhaGCN2. 1-kb sequences cannot be recognized by the neural network module of PhaGCN2. For geNomad (with VMR-MSL37 database)45, fragments of “new” viruses set (released after 2022.01) were used to perform taxonomic assignments. The consistency between assigned taxonomic units and bona fide taxonomic units was assessed by accuracy, precision, recall, F1 score, and annotation rate. The calculation approach is the same as the generalization ability assessment.
Comparison of taxonomic annotation on IMG/VR (v.4)
The taxonomic annotation of 2,631,412 vOTUs from the IMG/VR (v.4) database was performed based on the approach described above45. As the GTA-like genomes have been assigned as novel viral families in the current ICTV viral metadata resource (VMR-MSL38, as of September 2023), it was expected that viral genomes belonging to these novel families would be found. The IMG/VR (v.4) genome database was downloaded from the JGI genome portal (https://genome.jgi.doe.gov/portal/IMG_VR/IMG_VR.home.html). For DNA viruses, a vOTU-level cluster was determined if its internal members had 95% average nucleotide identity (ANI) and 85% aligned fraction coverage (AF) across the genome45. For RNA viruses, a vOTU-level cluster was determined if its internal members had 90% ANI and 80% AF for RNA-dependent RNA polymerase (RdRp)67. The annotation of vOTUs by VITAP was compared with their original taxonomic units to assess the consistency.
For viral contigs that showed differences in family-level taxonomic assignments between two pipelines (2,830 inconsistencies and 41,479 newly added) (Supplementary Data 6, 7), their ORFs were aligned with the NR database (as 2023.12.26), using 1e-5 as the E-value cut-off, and reporting the top 1,000 hits. Based on the alignment result and GenBank taxonomy database, let be the number of ORFs of a genome that fell into a family; let be the total number of ORFs of a genome, the participation ratio () of each viral contig at each family-level was calculated:
| 14 |
where is the compositional percentage of a viral sequence at the protein level for a given family. Subsequently, the consistency with the results of VITAP or IMG/VR based on the family with the highest was evaluated. This voting-based evaluation method does not rely on the databases of VITAP and IMG/VR. Although it is influenced by the species diversity present in public databases, and so the results may be relatively objective.
Building the database from diverse genome sources
To enhance classification sensitivity, 4807 and 785 representative genomes from NCBI RefSeq209 and IMG/VR (v. 4) were incorporated into the reference sequence of VITAP, respectively. All 13,650 viral genomes from NCBI RefSeq209 were included. For genomes from IMG/VR (v.4), only viral taxonomic units (vOTUs) labeled as high-confidence and high-quality (completeness >0.9) from IMG/VR workflow, as well as with genus as lowest taxonomic hierarchy, were included (n = 10,335)45. Firstly, viral genomes from NCBI RefSeq209 were aligned to the VMR-MSL37 database using MMSeqs2 with parameters 95 and 100% as the alignment identity and coverage, respectively. This step led to 6840 non-redundant genomes being compared to the VMR-MSL37 database. Secondly, vOTUs from IMG/VR (v.4) were aligned to the VMR-MSL37 genome database and 6,840 non-redundant viral RefSeqs, respectively, leading to 1234 extra non-redundant genomes. Thirdly, taxonomic assignments were performed on these 8074 extra non-redundant genomes by VMR-MSL37-based VITAP. Only genomes where the VITAP-derived taxonomic information is consistent with the original taxonomic information to a certain level were retained: 3627 genomes with consistency down to genus; 909 genomes with consistency down to family (received no genus level units from VITAP); 271 genomes with consistency down to order (received no family and genus level units from VITAP); 501 genomes with consistency down to class (received no order, family and genus level units from VITAP); 284 genomes without any taxonomic assignment by VITAP. The 5592 genomes as final extra reference genomes were combined with the VMR-MSL37 database. These extra genomes and genomes from the VMR-MSL37 database were merged to calculate thresholds of all taxonomic units and build the database utilized by VITAP.
Taxonomic re-annotation of the deep ocean virome
VITAP and five other pipelines (vConTACT2, CAT, VPF-Class, PhaGCN2, and geNomad) were used to perform viral taxonomic assignments of deep ocean viromes datasets derived from four studies. The taxonomic databases used by these pipelines include VMR-MSL37/RefSeq209/IMGVR4-hybrid database (VITAP), VMR-MSL37 (vConTACT2 and VITAP), RefSeq209 (CAT), and pipeline-integrated databases (VPF-Class, PhaGCN2, and geNomad). Four viromes associated with deep sea and corresponding metadata were retrieved and downloaded from public databases. These viromes were derived from oceanic trenches and cold seep sediments4,28,50,51. All viral contigs were performed taxonomic assignments through VITAP using the VMR-MSL37/RefSeq209/IMGVR4-hybrid database and stand-alone VMR-MSL37, respectively. The prokaryotic viral genomes with lengths over 5 kb from the top ten viral families were used to build a genome-content similarity network by vConTACT240 and visualized by Cytoscape87. As the contigs assigned to Herelleviridae have no linkages with Herelleviridae RefSeqs, these contigs were further confirmed by a viral proteome tree using ViPTree (v.4)41. The proteins from two assigned giant viral families (Mimiviridae, and Phycodnaviridae) were subject to Diamond BLASTp against viral proteins from RefSeq223. The average number of hits for each target viral protein was calculated across four different viral groups (Phycodnaviridae in RefSeqs, Mimiviridae in RefSeqs, Head-tail viruses in RefSeqs, and Filamentous phages in RefSeqs).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Jilu Han for his help with cross-platform testing and some bug fixes. We thank the support of the High-Performance Biological Supercomputing Center at the Ocean University of China; the Ocean Negative Carbon Emissions (ONCE); the high-performance servers of Center for High-Performance Computing and System Simulation, Pilot National Laboratory for Marine Science and Technology (Qingdao); the Marine Big Data Center of Institute for Advanced Ocean Study of Ocean University of China, the IEMB-1, a high-performance computing cluster operated by the Institute of Evolution and Marine Biodiversity. This work was supported by the Laoshan Laboratory (LSKJ202203201, Wang M.), 2024 Graduate Self-directed Research Project (2024ZZKY, 202461036, Zheng K.); Natural Science Foundation of China (41976117, 42120104006, 42176111, Wang M. and Liang Y.), and the Fundamental Research Funds for the Central Universities (202172002, 201812002, 202072001, Wang M. and McMinn A.).
Author contributions
Conceptualization: K.Z. and J.S.; Algorithm and code design: K.Z.; Code test: J.S.; Writing manuscript: K.Z., J.S., L.K.; Review and editing: Y.L., D.P.-E., A.M., and L.K.; Computational resource: Y.L. and M.W.; Funding acquisition and supervision: Y.L., A.M., and M.W.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The hybrid database of VITAP and related taxonomic assignments of IMG/VR (v.4) vOTUs generated in this study have been deposited in the Figshare database under data 10.6084/m9.figshare.25426159.v3. The benchmarking and other taxonomic assignment assessment data generated in this study are provided in the Supplementary Information/Source Data file.
Code availability
The open-source Python code of VITAP, auxiliary scripts in analysis, taxonomic assignments of IMG/VR (v.4)-derived vOTUs, are all freely available at Code Ocean, GitHub (https://github.com/DrKaiyangZheng/VITAP), Zenodo (10.5281/zenodo.14873988)88, and Anaconda (https://anaconda.org/bioconda/vitap).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kaiyang Zheng, Jianhua Sun.
Change history
9/22/2025
A Correction to this paper has been published: 10.1038/s41467-025-64370-6
Contributor Information
Yantao Liang, Email: liangyantao@ouc.edu.cn.
Andrew Mcminn, Email: andrew.mcminn@utas.edu.au.
Min Wang, Email: mingwang@ouc.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-57500-7.
References
- 1.Breitbart, M., Bonnain, C., Malki, K. & Sawaya, N. A. Phage puppet masters of the marine microbial realm. Nat. Microbiol.3, 754–766 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Fuhrman, J. A. Marine viruses and their biogeochemical and ecological effects. Nature399, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Nigro, O. D. et al. Viruses in the oceanic basement. mBio8, e02129-16 (2017). [DOI] [PMC free article] [PubMed]
- 4.Li, Z. et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J.15, 2366–2378 (2021). [DOI] [PMC free article] [PubMed]
- 5.Gazitua, M. C. et al. Potential virus-mediated nitrogen cycling in oxygen-depleted oceanic waters. ISME J.15, 981–998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson, L. R. et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl Acad. Sci. USA108, E757–E764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vienne, D. M. Lifemap: exploring the entire tree of life. PLoS Biol.14, e2001624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durzynska, J. & Gozdzicka-Jozefiak, A. Viruses and cells intertwined since the dawn of evolution. Virol. J.12 (2015). [DOI] [PMC free article] [PubMed]
- 9.Soucy, S. M., Huang, J. & Gogarten, J. P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet.16, 472–482 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Liu, H. et al. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol.84, 11876–11887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, H. et al. Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol. Biol.11, 276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz, F. et al. Giant virus diversity and host interactions through global metagenomics. Nature578, 432–436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurwitz, B. L. & U’Ren, J. M. Viral metabolic reprogramming in marine ecosystems. Curr. Opin. Microbiol.31, 161–168 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Yu, Z. C. et al. Filamentous phages prevalent in Pseudoalteromonas spp. confer properties advantageous to host survival in Arctic sea ice. ISME J.9, 871–881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee, A., Willett, J. L. E., Dunny, G. M. & Duerkop, B. A. Phage infection and sub-lethal antibiotic exposure mediate Enterococcus faecalis type VII secretion system dependent inhibition of bystander bacteria. PLoS Genet.17, e1009204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Secor, P. R. et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe18, 549–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitbart, M. Marine viruses: truth or dare. Ann. Rev. Mar. Sci.4, 425–448 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Suttle, C. A. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Trubl, G. et al. Soil viruses are underexplored players in ecosystem carbon processing. mSystems3, e00076-18 (2018). [DOI] [PMC free article] [PubMed]
- 20.Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol.6, 960–970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory, A. C. et al. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe28, 724–740.e728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manor, O. et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun.11, 5206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards, R. A. et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 4, 1727–1736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shkoporov, A. N. et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe26, 527–541.e525 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Shi, M. et al. Redefining the invertebrate RNA virosphere. Nature540, 539–543 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Shi, M. et al. The evolutionary history of vertebrate RNA viruses. Nature556, 197–202 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Wang, J. et al. Individual bat virome analysis reveals co-infection and spillover among bats and virus zoonotic potential. Nat. Commun.14, 4079 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jian, H. et al. Diversity and distribution of viruses inhabiting the deepest ocean on Earth. ISME J. 15, 3094–3110 (2021). [DOI] [PMC free article] [PubMed]
- 29.Wolf, Y. I. et al. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol.5, 1262–1270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starr, E. P., Nuccio, E. E., Pett-Ridge, J., Banfield, J. F. & Firestone, M. K. Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc. Natl Acad. Sci. USA116, 25900–25908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zayed, A. A. et al. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science376, 156–162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez-Huerta, G. et al. Diversity and ecological footprint of Global Ocean RNA viruses. Science376, 1202–1208 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Chen, Y. M. et al. RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat. Microbiol.7, 1312–1323 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Hug, L. A. et al. A new view of the tree of life. Nat. Microbiol.1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Eme, L. et al. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature618, 992–999 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, Y. et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature593, 553–557 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmonds, P. et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol.15, 161–168 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Meier-Kolthoff, J. P. & Goker, M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics33, 3396–3404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartlau, N. et al. Highly diverse flavobacterial phages isolated from North Sea spring blooms. ISME J.16, 555–568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol.37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Nishimura, Y. et al. ViPTree: the viral proteomic tree server. Bioinformatics33, 2379–2380 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Moraru, C., Varsani, A. & Kropinski, A. M. VIRIDIC-A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses12, 1268 (2020). [DOI] [PMC free article] [PubMed]
- 43.Adriaenssens, E. et al. in ICTV Online: International Committee on Taxonomy of Viruses (ICTV) (International Committee on Taxonomy of Viruses (ICTV), 2022).
- 44.Pons, J. C. et al. VPF-Class: Taxonomic assignment and host prediction of uncultivated viruses based on viral protein families. Bioinformatics37, 1805–1813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camargo, A. P. et al. IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res.51, D733–D743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Meijenfeldt, F. A. B., Arkhipova, K., Cambuy, D. D., Coutinho, F. H. & Dutilh, B. E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol.20, 217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brister, J. R., Ako-Adjei, D., Bao, Y. & Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 43, D571–D577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang, J. Z. et al. Virus classification for viral genomic fragments using PhaGCN2. Brief Bioinform24, bbac505 (2023). [DOI] [PubMed]
- 49.Camargo, A. P. et al. Identification of mobile genetic elements with geNomad. Nat. Biotechnol. 42, 1303–1312 (2023). [DOI] [PMC free article] [PubMed]
- 50.Gao, C. et al. Virioplankton assemblages from challenger deep, the deepest place in the oceans. iScience25, 104680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, J. et al. Novel viral communities potentially assisting in carbon, nitrogen, and sulfur metabolism in the upper slope sediments of Mariana Trench. mSystems7, e0135821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krupovic, M. et al. Adnaviria: a new realm for archaeal filamentous viruses with linear A-form double-stranded DNA genomes. J. Virol.95, e0067321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laso-Perez, R. et al. Evolutionary diversification of methanotrophic ANME-1 archaea and their expansive virome. Nat. Microbiol. 8, 231–245 (2023). [DOI] [PMC free article] [PubMed]
- 54.Dutilh, B. E. et al. Perspective on taxonomic classification of uncultivated viruses. Curr. Opin. Virol.51, 207–215 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Koonin, E. V., Kuhn, J. H., Dolja, V. V. & Krupovic, M. Megataxonomy and global ecology of the virosphere. ISME J. 18, wrad042 (2024). [DOI] [PMC free article] [PubMed]
- 56.Zhou, Y., Wang, Y. & Krupovic, M. ICTV virus taxonomy profile: Aoguangviridae 2023. J. Gen. Virol. 104, 001922 (2023). [DOI] [PMC free article] [PubMed]
- 57.Koonin, E. V. & Yutin, N. The crAss-like phage group: how metagenomics reshaped the human virome. Trends Microbiol. 28, 349–359 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Liu, Y. et al. Diversity, taxonomy, and evolution of archaeal viruses of the class Caudoviricetes. PLoS Biol.19, e3001442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krupovic, M. et al. Bacterial viruses subcommittee and archaeal viruses subcommittee of the ICTV: update of taxonomy changes in 2021. Arch. Virol.166, 3239–3244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell177, 1109–1123.e1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuzyakov, Y. & Mason-Jones, K. Viruses in soil: nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol. Biochem.127, 305–317 (2018). [Google Scholar]
- 62.Moniruzzaman, M., Weinheimer, A. R., Martinez-Gutierrez, C. A. & Aylward, F. O. Widespread endogenization of giant viruses shapes genomes of green algae. Nature588, 141–145 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Gaia, M. et al. Mirusviruses link herpesviruses to giant viruses. Nature616, 783–789 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paez-Espino, D. et al. Diversity, evolution, and classification of virophages uncovered through global metagenomics. Microbiome7, 157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roux, S. et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat. Microbiol4, 1895–1906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirchberger, P. C., Martinez, Z. A. & Ochman, H. Organizing the global diversity of microviruses. mBio13, e0058822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neri, U. et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell185, 4023–4037.e18 (2022). [DOI] [PubMed]
- 68.Callanan, J. et al. Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci. Adv.6, eaay5981 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benler, S. & Koonin, E. V. Fishing for phages in metagenomes: what do we catch, what do we miss? Curr. Opin. Virol.49, 142–150 (2021). [DOI] [PubMed] [Google Scholar]
- 70.Irwin, N. A. T., Pittis, A. A., Richards, T. A. & Keeling, P. J. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat. Microbiol. 7, 327–336 (2022). [DOI] [PubMed] [Google Scholar]
- 71.Keen, E. C. et al. Novel “superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio8, e02115-16 (2017). [DOI] [PMC free article] [PubMed]
- 72.Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol.39, 578–585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roux, S. et al. Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol.37, 29–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhan, Y., Huang, S., Voget, S., Simon, M. & Chen, F. A novel roseobacter phage possesses features of podoviruses, siphoviruses, prophages and gene transfer agents. Sci. Rep.6, 30372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuhn, J. H. & Koonin, E. V. Viriforms—a new category of classifiable virus-derived genetic elements. Biomolecules13, 289 (2023). [DOI] [PMC free article] [PubMed]
- 76.Lang, A. S., Zhaxybayeva, O. & Beatty, J. T. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol.10, 472–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhan, Y. & Chen, F. Bacteriophages that infect marine roseobacters: genomics and ecology. Environ. Microbiol. 21, 1885–1895 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Wu, J. et al. Gene transfer among viruses substantially contributes to gene gain of giant viruses. Mol. Biol. Evol.41, msae161 (2024). [DOI] [PMC free article] [PubMed]
- 79.Lawrence, J. G., Hatfull, G. F. & Hendrix, R. W. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol.184, 4891–4905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Filee, J. Giant viruses and their mobile genetic elements: the molecular symbiosis hypothesis. Curr. Opin. Virol.33, 81–88 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Piskurek, O. & Okada, N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl Acad. Sci. USA104, 12046–12051 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wittmann, J. et al. From orphan phage to a proposed new family-the diversity of N4-like viruses. Antibiotics9, 663 (2020). [DOI] [PMC free article] [PubMed]
- 83.Liu, Y. et al. Taxonomy proposal 2021: Create 3 new orders and 14 new families in the class Caudoviricetes (Duplodnaviria, Uroviricota) for classification of tailed archaeal viruses. ICTV Online: International Committee on Taxonomy of Viruses (ICTV) (2022).
- 84.Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE11, e0163962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lavigne, R., Seto, D., Mahadevan, P., Ackermann, H. W. & Kropinski, A. M. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol.159, 406–414 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng, K. et al. VITAP: A High Precision Tool for DNA and RNA Viral Classification Based on Meta-omic Data. Zenodo10.5281/zenodo.14873987 (2025). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The hybrid database of VITAP and related taxonomic assignments of IMG/VR (v.4) vOTUs generated in this study have been deposited in the Figshare database under data 10.6084/m9.figshare.25426159.v3. The benchmarking and other taxonomic assignment assessment data generated in this study are provided in the Supplementary Information/Source Data file.
The open-source Python code of VITAP, auxiliary scripts in analysis, taxonomic assignments of IMG/VR (v.4)-derived vOTUs, are all freely available at Code Ocean, GitHub (https://github.com/DrKaiyangZheng/VITAP), Zenodo (10.5281/zenodo.14873988)88, and Anaconda (https://anaconda.org/bioconda/vitap).