Abstract
Intervertebral disc degeneration (IDD) is a degenerative spinal condition characterized by disc structural damage, narrowing of joint spaces, and nerve root compression, significantly reducing patients’ quality of life. To address this challenge, a novel therapeutic strategy was developed using cellulose supramolecular hydrogel as a carrier to deliver IL4I1-modified MΦ membrane biomimetic nanoparticles (CHG@IL4I1-MNPs) to target tissues. This hydrogel exhibits excellent biocompatibility and mechanical properties while enabling sustained drug release in the degenerative disc microenvironment, enhancing therapeutic outcomes. CHG@IL4I1-MNPs effectively regulate MΦ polarization by promoting M2 MΦ activation, thereby improving immune microenvironment balance. Animal studies demonstrated that CHG@IL4I1-MNPs alleviated symptoms of IDD, reduced inflammation, and supported tissue repair, highlighting its potential to reduce reliance on long-term medication and improve quality of life. The strategy uniquely combines nanoparticle technology with immunomodulation, achieving precise targeting of MΦs. Beyond IDD, this approach offers potential applications in other immune-related diseases, providing a versatile platform for nanomedicine. This study introduces an innovative method to treat IDD and advances the integration of immunotherapy and nanotechnology, offering both clinical benefits and new directions for future research. These findings hold strong potential for improving patient outcomes and expanding treatment options for related diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12951-025-03241-0.
Keywords: Intervertebral disc degeneration, MΦ, IL4I1, Polarization regulation, Nanoparticles, Cellulose hydrogel
Introduction
In the realm of modern medicine, intervertebral disc degeneration (IDD) stands out as a prevalent degenerative disease, with its incidence steadily increasing, profoundly affecting patients’ quality of life and health [1, 2]. The World Health Organization (WHO) estimates that approximately 20% of the global population suffers from chronic low back pain, with a large proportion attributed to pathological changes caused by IDD [3, 4]. IDD places a substantial burden on healthcare resources, with standard treatments including physical therapy, pharmacotherapy, and surgical interventions. These approaches not only incur high medical costs for patients but, due to the chronic nature of the disease, often result in long-term work absences and reduced productivity, further exacerbating socioeconomic burdens [5, 6]. In terms of quality of life, IDD commonly causes varying degrees of functional impairment, including restricted mobility and persistent pain, which, in severe cases, may lead to a loss of independence, greatly impacting daily living [7, 8]. The development of IDD involves diverse cellular, molecular, and tissue-level changes, encompassing intricate biological processes such as cell polarization, inflammatory responses, and tissue repair [9–11]. Prior research underscores the indispensable role of immune cells, particularly MΦs, in IDD progression, with the balance between M1 and M2 MΦs emerging as a pivotal factor in disease advancement [12].
MΦs play crucial roles in the pathophysiological processes of IDD [13, 14]. Overactivated M1 MΦs produce abundant inflammatory mediators, leading to sustained enhancement of the inflammatory response, exacerbating degenerative changes in intervertebral disc (IVD) tissue [15–17]. Conversely, impaired function and reduced numbers of M2 MΦs result in decreased repair and regenerative abilities, further intensifying IDD development [15, 18, 19]. Therefore, regulating the polarization of MΦs to achieve a balanced M1/M2 phenotype may represent an effective strategy for treating IDD, potentially ameliorating clinical symptoms and enhancing the quality of life for patients [20–22].
The key factor IL4I1 plays a significant role in this therapeutic strategy [23, 24]. Studies indicate that IL4I1 can promote the polarization of M2 MΦs, enhancing their anti-inflammatory and reparative functions while inhibiting the excessive inflammatory response of M1 MΦs, thus mitigating inflammatory damage [25, 26]. Consequently, leveraging IL4I1 to regulate the polarization of MΦs emerges as a novel approach to treating IDD [25, 27, 28]. This study employs Cellulose Hydrogel with IL4I1-Modified MΦ Membrane Biomimetic Nanoparticles (CHG@IL4I1-MNPs) to explore the molecular mechanisms of IL4I1 in modulating MΦ polarization and assess its potential application in treating IDD.
Through in-depth research on the IL4I1-regulated MΦ polarization mechanism, our aim is to unveil its significant role in IDD pathogenesis. Functioning as a novel nanocarrier, CHG@IL4I1-MNPs not only efficiently release IL4I1 but also offer robust biological stability and cellular penetration, providing new tools and strategies for treating IDD [29–31]. By evaluating the efficacy of CHG@IL4I1-MNPs in a mouse IDD model, we aim to explore their potential advantages and therapeutic prospects in clinical applications. This study holds significant importance in enhancing the understanding of the critical role of MΦ polarization regulation in IDD pathogenesis while also paving the way for developing new biotherapeutic approaches to improve patient quality of life, with promising scientific and clinical implications.
Through single-cell sequencing (scRNA-seq) and transcriptomic analysis, we delve into the potential mechanisms linking IL4I1 to IDD occurrence and design a series of experiments to validate the impact of IL4I1-MNPs on MΦ polarization. Experimental results demonstrate the successful preparation of CHG@IL4I1-MNPs and their substantial therapeutic effects in a mouse IDD model, unveiling new strategies for IDD treatment. This study aims to achieve effective IDD treatment through the regulation of MΦ polarization and introduce a potential new avenue for biotherapeutic application with clinical significance.
Materials and methods
Downloading and analyzing GEO scRNA-seq data
The scRNA-seq data from dataset GSE244889, obtained through the Gene Expression Omnibus (GEO), comprises 4 samples of mild IDD (MIDD, grades I-II) nucleus pulposus (NP) tissues and 3 samples of severe IDD (SIDD, grades III-V) NP tissues.
The scRNA-seq data was analyzed using the “Seurat” package in R software. Initial steps included a series of quality control measures with set filtering thresholds as follows: nFeature_RNA > 200, nFeature_RNA < 5000, and percent.mt < 10. Batch effects were mitigated using canonical correlation analysis (CCA) followed by data standardization using the LogNormalize function.
To reduce the dimensionality of the scRNA-Seq dataset, principal component analysis (PCA) was performed on the top 2000 variably expressed genes based on variance, selecting the top 30 principal components for UMAP clustering analysis. The FindClusters function in Seurat was employed to identify main cell subgroups with the resolution set to the default (res = 1). Subsequently, the UMAP algorithm was utilized for non-linear dimensionality reduction of scRNA-seq sequencing data. Marker genes for various cell subtypes were identified using the Seurat package, annotations of marker genes for individual cell clusters were further performed using the “SingleR” package, and reference dataset loading was facilitated with the HumanPrimaryCellAtlasData function. Cell annotations were conducted based on known cell lineage-specific marker genes in combination with the CellMarker online resource. Additionally, pseudotime analysis was carried out using the “monocle” package in R software, and cell-cell communication analysis was executed via the “cellchat” package [12, 32].
Downloading and analyzing GEO transcriptome sequencing data
Transcriptome RNA data related to IDD was obtained from the GEO database, specifically datasets GSE167199 and GSE186542. Each dataset comprises 3 normal human NP tissue samples and 3 IDD human NP tissue samples, all obtained through the GPL24676 platform. To ensure an adequate sample size, the GSE167199 and GSE186542 datasets were merged, and batch effects were mitigated using the “sva” package in R [33].
The “limma” package in R was employed to identify DEGs in the merged dataset, with a selection criterion of |log2FC| > 1 and a P-value < 0.05 for M-DEG filtering. Visualization of the M-DEGs was conducted using the “ggplot2” and “pheatmap” packages. Subsequently, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed utilizing the “clusterProfiler”, “org.Hs.eg.db”, “org.Mm.eg.db”, “enrichplot”, “ggplot2”, and “pathview” packages [33, 34].
Following this, machine learning algorithms, including LASSO, SVM-RFE, and Random Forest, were applied to the M-DEGs using the “glmnet”, “e1071”, and “randomForest” packages to identify feature genes significantly associated with IDD. Core genes were extracted by taking the intersection using the “venn” package in R [33].
Cell culture
The human monocytic cell line THP-1 (TCHu 57) was obtained from the Cell Bank of the Chinese Academy of Sciences. Human nucleus pulposus cells (NPCs; #4800) were purchased from ScienCell Research Laboratories. Cells were cultured in DMEM medium (Gibco, 31600034) supplemented with 0.1% non-essential amino acids (Gibco, 11140050), 1% penicillin-streptomycin (Gibco, 15070063), and 10% FBS (Gibco, 10099141) in a cell culture incubator at 37℃ with 5% CO2. NPCs and THP-1 cells in the logarithmic growth phase were dissociated and digested using 0.25% trypsin (Gibco, 25200114). Third-generation cells were utilized for subsequent experiments [35, 36]. THP-1 cells were stimulated with 100 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, P1585) for 24 h to induce macrophage (MΦ) phenotype [10].
Preparation of MΦ cell membranes (MΦCM)
THP-1-derived MΦ were cultured in T-175 flasks until fully confluent, and then cells were harvested by centrifugation at 700 g for 5 min. The cells underwent three washes with 1×PBS (500 g, 10 min each), followed by resuspension of the cell pellet in a homogenous buffer containing 75mM sucrose (Sigma-Aldrich, V900116), 20mM Tris-HCl (pH 7.5, Sigma-Aldrich, T5941), 2mM MgCl2 (Sigma-Aldrich, M4880), 10mM KCl (Sigma-Aldrich, P5405), and a protease/phosphatase inhibitor cocktail (ThermoFisher, A32959). Subsequently, the cell suspension was homogenized using a Dounce homogenizer (Sigma-Aldrich, CLS772215) with 15–25 strokes. After homogenization, the mixture was centrifuged at 800 g for 5 min to collect the supernatant, followed by a second centrifugation at 10,000 g for 25 min to collect the supernatant again. The collected supernatant was then centrifuged at 150,000 g for 50 min to discard the supernatant and obtain cell membrane particles, which were washed once with water. The membrane protein content was quantified using the Pierce BCA assay kit (ThermoFisher, 23227), and the membranes were stored at -80 °C for future experiments [37, 38].
Preparation of IL4I1-NPs and IL4I1-MNPs
IL4I1-NPs were prepared using the double solvent evaporation method. In brief, 1 ml of 2% (w/v) PLGA in ethyl acetate solution was mixed with 10 µg of recombinant human IL4I1 (R&D Systems, 5684-AO) dissolved in 100 µl of sterile DNase/RNase. The solution was then sonicated to form an emulsion. Subsequently, 2% polyvinyl alcohol (PVA; Sigma-Aldrich, 363170) aqueous solution was added, and the organic solvent was evaporated overnight under stirring to obtain a water-oil-water emulsion. Finally, IL4I1-NPs were purified by centrifugation at 13,000 × g for 10 min, followed by resuspension in sterile PBS (pH 7.4). For the preparation of IL4I1-MNPs with MΦ membrane coating, IL4I1-NPs were mixed with MΦCM at a weight ratio of 1:1 for polymer and membrane proteins, subjected to 2–3 min of sonication at 200 W and then extruded through 1000, 400, 200, and 100 nm polycarbonate filters [39, 40].
Characterization of IL4I1-NPs and IL4I1-MNPs
The size and surface zeta potential of IL4I1-NPs and IL4I1-MNPs were measured using dynamic light scattering (DLS) with a Malvern Zetasizer Nano ZS. For nanoparticle morphology assessment, samples were adsorbed onto carbon-coated copper grids (400 mesh, Electron Microscopy Sciences) and stained with 2% uranyl acetate (Electron Microscopy Sciences, 22400-2). Imaging was conducted using a JEOL 1200 EX II transmission electron microscope. To evaluate nanoparticle stability, samples were stored in deionized water (Sigma-Aldrich, 1.01262), 1×PBS, and 50% FBS at 4 °C, and nanoparticle size was measured daily using DLS for 72 h [37, 40].
The concentration of IL4I1 in IL4I1-NPs and IL4I1-MNPs was determined using a human IL4I1 Enzyme-Linked Immunosorbent Assay (ELISA) kit (NOVUS, NBP3-06773) according to the manufacturer’s instructions. Encapsulation efficiency and loading capacity were calculated based on the ratio of loaded IL4I1 to total IL4I1. Specifically, 1 mg of lyophilized NPs was accurately weighed and dissolved in ultrapure water, and the supernatant was used to measure free IL4I1. The remnant was dissolved in dichloromethane for ultrasonic detection of loaded IL4I1. Encapsulation efficiency = loaded IL4I1 / (free IL4I1 + loaded IL4I1) × 100%; loading capacity = loaded IL4I1 / 1 mg. IL4I1 release rate was measured at each time point (0, 1, 4, 8, 24, 48, and 120 h) using the ELISA kit [39].
Characterization of membrane proteins
The protein distribution of MΦ cell lysates, MΦCM, IL4I1-NPs, and IL4I1-MNPs was assessed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Specifically, samples were prepared in sample buffer (Fdbio, FD006) at a protein concentration of 2.0 mg/mL and separated by 10% SDS-PAGE. The resulting gels were stained with Coomassie Brilliant Blue stain (Beyotime, P0017), followed by several washes with distilled water, de-staining, yielding protein bands for MΦ cell lysates, MΦCM, IL4I1-NPs, and IL4I1-MNPs [37, 41].
In vitro cellular uptake experiment
For the experiment, the IL4I1 was replaced with a cell membrane-deep red fluorescent probe (DiR; YEASEN, 40757ES25) to prepare DiR-NPs using the method described previously. The dye was incorporated into PLGA polymer at a concentration of 0.1 wt%. Subsequently, the DiR-NPs were enveloped with cell membranes to form DiR-MNPs. MΦ cells were cultured in a 12-well plate (2 × 105 cells per well) for 24 h. Equal amounts of DiR-NPs or DiR-MNPs (5 µg/mL) were added to each well, and the cells were co-incubated with MΦ for 1, 2, or 4 h. At the specified time points, the cells were washed thrice with PBS. After centrifugation, the fluorescence intensity of DiR within the cells was measured using flow cytometry (Beckman Coulter). Subsequently, the cells were fixed in 4% paraformaldehyde (Biosharp, BL539A) for 20 min, permeabilized in 0.5% Triton X-100 (Sigma-Aldrich, X100) for 20 min, stained with DAPI (Invitrogen, D3571) for 5 minutes, and observed under a confocal laser scanning microscope (CLSM, Leica) to capture images. The fluorescence intensity of three independent fields was quantified using ImageJ software [40, 42].
Synthesis of cellulose supramolecular hydrogels
A solution containing 7% sodium hydroxide and 12% urea was prepared as the solvent for synthesizing the CHG, comprising sodium hydroxide (7 g; Sigma-Aldrich, S8045), urea (12 g; Sigma-Aldrich, U5378), and deionized water (81 ml). The cellulose solution was prepared using a freeze-thaw process, in which 5.0 g of cellulose was dispersed in the sodium hydroxide/urea system and cooled to -12℃. The mixture formed a transparent solution after mechanical stirring for 30 min. Next, 20.0 g of β-cyclodextrin (Sigma-Aldrich, 779334) was added to the clear solution, followed by the addition of 14.7 g of epichlorohydrin (Sigma-Aldrich, 8.03296) to the mixed solution. After stirring at 20℃ for 1 h, the solution was heated to 55℃ and allowed to gel for 5 h. The cellulose supramolecular hydrogel was thoroughly washed with ample deionized water to remove urea/sodium hydroxide completely [43].
Preparation of CHG@IL4I1-MNPs
The CHG@IL4I1-MNPs were prepared using an immersion method. The CHG hydrogel was immersed in the IL4I1-MNPs solution for 24 h, resulting in the formation of CHG@IL4I1-MNPs. The obtained CHG@IL4I1-MNPs were washed with water, freeze-dried, and used for subsequent experiments [43].
Swelling and degradation of hydrogel
Hydrogel (300 µl) was placed in a cell culture dish and soaked in PBS for 24 h. After removing the surface water, the swollen samples were weighed at specific time intervals (0.5, 1, 2, 6, 12, and 24 h). The swelling ratio was calculated using the formula: Swelling ratio = (Wt - Wd) / Wd × 100%, where Wd represents the dry weight of the hydrogel, and Wt represents the weight of the swollen hydrogel. Furthermore, we assessed the degradation of the hydrogel at specific time points (2, 5, 7, 8, 14, 21 days). After removing the surface supernatant, the weight of the hydrogel was measured, and the degradation rate was calculated using the formula: Degradation rate = W1 / W0 × 100%, where W0 is the initial wet weight of the hydrogel, and W1 is the wet weight of the hydrogel at the given time point [44].
Scanning Electron microscope (SEM)
After freeze-drying CHG and CHG@IL4I1-MNPs, their morphology was observed using a scanning electron microscope (JSM-IT200, Hitachi). The samples were sputter-coated with gold and palladium for 60 s on the surface before SEM imaging at an acceleration voltage of 5 kV and a working distance of 13.9 mm. Three samples were selected for electron microscopy scanning at various concentrations [45].
Rheological analysis
The rheological characteristics of the hydrogels CHG and CHG@IL4I1-MNPs were evaluated using a rotational rheometer (Gemini HR Nano 200, Malvern). Parallel plates with a diameter of 20 mm and a gap width of 1 mm were selected for the analysis. Oscillatory frequency sweeps from 0.1 Hz to 10 Hz were conducted to assess the viscoelasticity. Rheological measurements were carried out at a constant strain of 5.0% and a temperature of 25℃. The storage modulus (G’) and loss modulus (G″) were monitored as a function of frequency [46].
In vitro drug release testing of hydrogels
CHG@IL4I1-MNPs hydrogel (10 mg/g) and IL4I1-MNPs (10 mg) were weighed and placed in dialysis bags for the experiment. Each group was immersed in a beaker containing 1 L of PBS solution and shaken at 37 °C (100 rpm). At predetermined time points (1, 2, 3, 4, 5, 7, 9, 11, 13, and 15 days), the dialysis bags were removed from the beaker, and 10 ml of the release medium solution was accurately withdrawn (replacing with blank PBS in the vessel). The collected medium was evaporated, dissolved in 0.5 mL of pure ethanol, and then centrifuged at 10,000 rpm for 30 min after filtration. The supernatant was subjected to IL4I1 ELISA kit analysis to determine the release rate of IL4I1 [44, 46].
Cell transfection
To overexpress FGR, THP-1-derived MΦs were infected with pHBLV-CMV lentivirus carrying the human FGR gene (FGR-OE) or an empty lentivirus (NC-OE) at an Multiplicity of Infection (MOI) of 20 for 6 h. Following this, the cells were cultured in a fresh DMEM complete medium for 48 h before subsequent treatments. Both FGR-OE and NC-OE were obtained from Hanheng Biology [47].
Cell treatment and grouping
THP-1-derived MΦs were induced for 24 h with 100 ng/mL LPS (MCE, HY-D1056) or 50 ng/mL IL-4 (R&D Systems, BT-004) to polarize into M1 or M2 states. Subsequently, to evaluate the impact of IL4I1 on MΦ polarization, 50 ng/mL recombinant human IL4I1 or PBS was co-cultured with MΦs induced by LPS or IL-4 for M1 or M2 polarization for an additional 24 h. Based on the different cytokine additions, the cells were categorized into 7 groups: (1) PBS; (2) LPS; (3) IL-4; (4) LPS + PBS; (5) LPS + IL4I1; (6) IL-4 + PBS; (7) IL-4 + IL4I1 [10, 48, 49].
To validate the role of FGR in the regulation of MΦ polarization by IL4I1-MNPs, NC-OE or FGR-OE-transfected MΦs were treated with IL4I1-MNPs (containing 50 ng/mL IL4I1) and polarized with LPS or IL-4. The cells were divided into 6 groups: (1) LPS + PBS + NC-OE; (2) LPS + IL4I1-MNPs + NC-OE; (3) LPS + IL4I1-MNPs + FGR-OE; (4) IL-4 + PBS + NC-OE; (5) IL-4 + IL4I1-MNPs + NC-OE; (6) IL-4 + IL4I1-MNPs + FGR-OE [10, 48, 49].
Co-culture experiments of MΦs with NPCs: To simulate the verification environment of IDD, NPCs were cultured for 24 h in DEMF complete medium containing 10 ng/mL IL-1β (Sino Biology, 10139-HNAE). Subsequently, different treated MΦs were seeded in the upper chamber of a Transwell membrane with 0.4 μm pores at a density of 1 × 105 cells. IL-1β pre-treated NPCs were seeded in the lower chamber at a density of 1 × 105 cells for co-culture for 48 h. The experiment was divided into 7 groups: (1) NPCs + PBS + NC-OE; (2) NPCs + LPS + PBS + NC-OE; (3) NPCs + LPS + IL4I1-MNPs + NC-OE; (4) NPCs + LPS + IL4I1-MNPs + FGR-OE; (5) NPCs + IL-4 + PBS + NC-OE; (6) NPCs + IL-4 + IL4I1-MNPs + NC-OE; (7) NPCs + IL-4 + IL4I1-MNPs + FGR-OE. The group NPCs + PBS + NC-OE indicates co-culture of NPCs with non-polarized-induced MΦs [50–52].
Immunofluorescence staining
Cell-containing slides were fixed in 4% paraformaldehyde (PFA; Biosharp, BL539A) for 30 min, permeabilized, and blocked with 0.3% Triton X-100 (CS9013, G-clone) combined with 5% BSA (PN4810, G-clone) for 1 h. The slides were then incubated overnight with primary antibodies at 4 °C. The primary antibodies used were as follows: Rabbit anti-human CD80 (1:200; Abcam, ab315832), Rabbit anti-human CD163 (1:50; Abcam, ab316218), Mouse anti-human CD86 (2 µg/mL; Abcam, ab270719), Mouse anti-human CD206 (1 µg/mL; ThermoFisher, MA5-44033), Rabbit anti-human Collagen II (1:2000; Abcam, ab34712), and Rabbit anti-human MMP13 (1:200; Abcam, ab39012). After washing with PBS three times, the slides were incubated at room temperature for 1 h with secondary antibodies: Alexa Fluor™ 488 Goat anti-Rabbit IgG (4 µg/mL, ThermoFisher, A-11008) and Alexa Fluor™ 647 Goat anti-Mouse IgG (2 µg/mL, ThermoFisher, A-21235). Subsequently, cell nuclei were stained with DAPI solution (1:1000; Invitrogen, D1306) for 5 min at room temperature. Sample observation and imaging were conducted using a fluorescence microscope (FV-1000; Olympus). The ImageJ software was utilized to quantify the fluorescence intensity of co-localized CD80 and CD86, CD163 and CD206, as well as Collagen II or MMP13 in 3 independent fields per group [10, 52, 53].
CCK-8
Cell Proliferation: Cell proliferation was assessed using the CCK8 assay kit (Dojindo, CK04) following the manufacturer’s instructions. NPCs co-cultured with MΦ for 24 and 48 h were seeded into 96-well plates. Subsequently, they were incubated at 37 °C in a DMEM medium containing 10% CCK-8 for an additional 3 h. After shaking the culture plates for 10 min, the absorbance was measured at 450 nm [53].
Cell Viability Assessment: The CCK8 assay kit was utilized to measure cell viability. After exposing MΦ to CHG and CHG@IL4I1-MNPs for 1 day, as described previously, the cells were treated with DMEM medium containing 10% CCK-8 and their absorbance at 450 nm was measured [44].
EdU staining
For the EdU assay, after co-culturing NPCs with MΦ, 50 µM EdU (Sigma-Aldrich, BCK-EDU647) was added to each well and incubated for 2 h. Subsequently, cells were fixed with 4% PFA at room temperature for 15 min, followed by permeabilization with 0.3% Triton X-100 at room temperature for 20 min. After washing the cells with PBS three times, 100 µL of 1× Apollo reaction mixture was added to each well and incubated at room temperature for 30 min. Following two PBS washes, cells were stained with DAPI for 5 min. Finally, cells were randomly observed and photographed under a fluorescence microscope (BX63, Olympus), and the percentage of EdU-positive cells was quantified using ImageJ software in three independent fields, calculated as EdU-positive cell count/total cell count ×100%. The total cell count was determined by counting the DAPI-stained cell nuclei [52, 53].
Flow cytometry analysis of cell apoptosis
Cell apoptosis was detected using the Annexin V-FITC/PI apoptosis detection kit (YEASEN, 40302ES20) following the manufacturer’s instructions to identify apoptotic cells (Annexin V+/PI-, lower right quadrant). Specifically, NPCs co-cultured with MΦ were stained with 5 µL Annexin V-FITC and 10 µL PI, after which the cells were incubated in the dark at room temperature for 10–15 min. Subsequently, analysis was performed using the EpicsAltra flow cytometer (Beckman Coulter) [53].
Transwell experiment
The impact of MΦ on the migratory capabilities of NPCs was investigated using 8 μm Transwell chambers (Solelybio, SBM07797). Initially, 200 µL of NPCs suspension was seeded in the upper chamber of the Transwell at a density of 5 × 104 cells per well, while MΦ was placed in the lower chamber. The Transwell system was then incubated in a cell culture incubator at 5% CO2 and 37 °C for 12 and 24 h. Subsequently, the culture medium in both the upper and lower chambers of the Transwell was removed, followed by washing twice with PBS buffer and fixation in 4% PFA at room temperature for 30 min. After fixation, any remaining NPCs on the upper layer of the Transwell were gently wiped off using a cotton swab, followed by staining the lower chamber with 0.1% crystal violet solution (Solarbio, G1063) for 20 min. Finally, observations and photographic documentation were performed under an optical microscope [52].
Animal care and ethical statement
Male SPF C57BL/6J mice aged 12 weeks, weighing 20–25 g, were obtained from Beijing Vitonlihua Experimental Animal Technology Co., Ltd., China. They were individually housed in a controlled environment with a humidity of 60–65% and temperature maintained at 22–25 °C with a 12-hour light-dark cycle in an SPF-grade animal facility. Prior to experimentation, the mice were observed for general health status and fasted for 8–12 h. All animal care procedures and surgical operations followed the guidelines outlined in the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health. Ethical approval for all animal experiments in this study was obtained from the institutional ethics review committee (Approval no. 2024PS1228K), and all procedures were strictly conducted following the regulations for animal welfare and experimental practices. At the end of the experiments, humane euthanasia was performed on all mice using pentobarbital sodium at a dose of 120 mg/kg (Sigma-Aldrich, P3761) [54].
Construction of mouse tail IDD model
A total of 108 male C57BL/6J mice at 12 weeks of age were randomly divided into two groups: Sham (n = 18) and Model (n = 90). The Model group aimed to establish the mouse tail IDD model. Specifically, mice were anesthetized via tail vein injection of pentobarbital sodium (30 mg/kg) and laid prone on the experiment table. A sagittal skin incision was made from Co7 to Co9 to assist in locating the IVD position for needling in the tail region. Subsequently, following the previously described procedure, a needle puncture was made into the Co7-Co8 IVD using a syringe needle. An 24G and 30G syringe needle was inserted percutaneously along the vertical axis into the Co7-Co8 IVD, rotated axially 180°, and held for 30 s. The Co8 and Co9 segments were left undisturbed to serve as control segments. The tail vertebral discs were exposed aseptically during the procedure. In the Sham group, mice received anesthesia and only underwent skin incision [52–55].
For the Sham group, mice were randomly divided into Sham (n = 6), Sham + PBS (n = 6), and Sham + PBS + NC-OE (n = 6); while for the Model group, mice were classified into Mild Model (mild IDD, n = 6), Severe Model (severe IDD, n = 6), PBS (n = 6), DiR-NPs (n = 6), DiR-MNPs (n = 6), Model + PBS (n = 9), Model + IL4I1 (n = 6), Model + IL4I1-NPs (n = 6), Model + IL4I1-MNPs (n = 9), Model + PBS + NC-OE (n = 6), Model + CHG + NC-OE (n = 6), Model + IL4I1-MNPs + NC-OE (n = 6), Model + GHG@IL4I1-MNPs + NC-OE (n = 6), Model + GHG@IL4I1-MNPs + FGR-OE (n = 6).
For the Mild Model and Severe Model groups, needle piercing for IDD model establishment was done with 30G and 24G needles, respectively. The Sham group received skin piercing as a control procedure [52–55].
In the PBS, DiR-NPs, and DiR-MNPs groups, mice received tail vein injections of DiR-labeled NPs DiR-NPs or DiR-MNPs (40 mg/kg, 100 µL) or 100 µL of PBS after 4 weeks of needle piercing for severe Model establishment. This was for subsequent in vivo biodistribution analysis [40, 56].
The Model + PBS, Model + IL4I1, Model + IL4I1-NPs, and Model + IL4I1-MNPs groups underwent tail vein injections of PBS, free IL4I1 (100 ng/10 µL), IL4I1-NPs, or IL4I1-MNPs, respectively, on the 7th, 10th, 13th, 16th, 19th, 22nd, and 25th days after the establishment of the severe Model using a 24G needle for NP puncture in the Co7-Co8 IVD. Imaging and histological analysis of mice were conducted on the 28th day. The Sham + PBS group received a similar volume of PBS after skin piercing. Additionally, three mice, each from the Model + PBS and Model + IL4I1-MNPs groups, underwent transcriptome sequencing [48, 57, 58].
For the Model + PBS + NC-OE, Model + CHG + NC-OE, Model + IL4I1-MNPs + NC-OE, Model + GHG@IL4I1-MNPs + NC-OE, and Model + GHG@IL4I1-MNPs + FGR-OE groups, one week after establishing the Severe Model, 20 µL of PBS, CHG, or GHG@IL4I1-MNPs hydrogel solution was injected into each IVD using a 31G microinjector. Simultaneously, 3 µL of PBS containing 0.5nM lentivirus was injected into the tail vein of each mouse. Lentivirus carrying the mouse FGR-OE for overexpression and NC-OE were obtained from Hanheng Biology. The Sham + PBS + NC-OE group underwent skin piercing and received the same procedures as the Model + PBS + NC-OE group [50, 59].
IVD image analysis
Four weeks after a puncture, mice were anesthetized, and micro-computed tomography (µ-CT, Scanco Medical) scans were performed. The scan parameters were set as follows: voltage = 100 kV, current = 98 µA, and voxel size = 10 μm. NRecon, DataViewer, and CTvox software were utilized for the reconstruction and creation of three-dimensional micro-CT scan images. Image J software was used to measure and calculate the disc height index (DHI). The 3D images represented the gap between two vertebrae, displaying structural changes [60, 61].
Histological examination
The mice were euthanized after the imaging analysis. The Co7-8 IVD was harvested and fixed in 10% neutral formalin for 24 h, followed by decalcification in 10% ethylenediaminetetraacetic acid (EDTA; MCE, HY-Y0682) for 4 weeks. Subsequently, the samples were sectioned into 10 μm slices containing the endplate, annulus fibrosus (AF), and nucleus pulposus (NP). The sections were dewaxed twice in xylene for 5 min each, dehydrated through graded ethanol, and stained according to the manufacturer’s instructions with either hematoxylin and eosin (H&E; Solarbio, G1120) or Safranin-O/Fast green (Solarbio, G1371). Stained slides were observed under a fluorescence microscope (FV-1000, Olympus) and scored based on the Masuda grading system. A higher histological score indicates a higher degree of degeneration [11].
Immunohistochemistry
To observe the specific expression of Collagen II, Aggrecan, ADAMTS5, and MMP13 in tissues, NP tissue sections were subjected to deparaffinization and hydration through a gradient of alcohols, followed by antigen retrieval using a protease. Subsequently, the tissues were blocked in 5% goat serum (Beyotime, C0265) and then incubated overnight at 4 °C with primary antibodies against Collagen II (1:100; Abcam, ab34712), Aggrecan (1:2000; Abcam, ab315486), ADAMTS5 (1:100; ThermoFisher, PA5-27165), and MMP13 (1:2000; Abcam, ab219620). The tissue sections were then incubated for 1 h with an HRP-conjugated Goat anti-Rabbit IgG secondary antibody (1:8000; ThermoFisher, 65-6140). Visualization was achieved by incubating the sections with 3,3’-diaminobenzidine (DAB) tetrahydrochloride (YEASEN, 36201ES03) and counterstaining with hematoxylin (Beyotime, C0107). Three sections from each mouse were examined, with two independent fields of view per section, and the positive staining area percentages of Collagen II, Aggrecan, ADAMTS5, and MMP13 were quantified using ImageJ software [10].
Flow cytometry
After euthanizing mice, the lumbar IVDs were extracted. The collected tissue samples were washed in cold PBS and minced into 1–2 mm fragments. Digestion of the samples into single cells was carried out using Collagenase II (Beyotime, ST2303-100 mg) and Dispase II (Beyotime, ST2303-100 mg). The single cells were filtered through a strainer to remove residual tissue debris and washed three times in cold PBS. Dead cells stained positive for 7-AAD reagent (CST, 72782 S) were excluded. Subsequently, the cells were co-incubated with FITC-labeled CD45 (BD Biosciences, 553079), Alexa Fluor™ 488-labeled CD11b (BD Biosciences, 557672), PE-labeled F4/80 (BD Biosciences, 565410), APC-labeled CD80 (BD Biosciences, 560016), Alexa Fluor™ 700-labeled CD86 (BD Biosciences, 560581), PerCP-eFluor™ 710-labeled CD163 (ThermoFisher, 46-1631-82), and fluorite 450-labeled CD206 (ThermoFisher, 48-2061-82) antibodies. Following antibody addition, cell co-incubation was conducted in the dark at 4 °C for 30 min. Subsequently, the cells were washed twice with PBS and centrifuged at 2000 g for 5 min at 4 °C. Flow cytometric analysis of marker percentages was performed using the BD Accuri C6 flow cytometer (BD Biosciences). All the mentioned antibodies were anti-mouse antibodies. Notably, CD45+CD11b+F4/80+ cells were identified as MΦs [52].
ELISA
Following standardized protocols, mouse tail IVD tissues were homogenized, or cell culture supernatants were collected, then centrifuged at 4 °C and 800 g for 20 min and 10 min, respectively. The resulting supernatants were stored at -80 °C for subsequent analysis. Mouse ELISA kits for TNF-α (ThermoFisher, 88-7324-88), IL-1β (ThermoFisher, 88–7013 A-88), IL-10 (ThermoFisher, BMS614), and TGF-β (ThermoFisher, BMS608-4) were utilized to measure the levels of cytokines in mouse tissue homogenates, while human ELISA kits for TNF-α (ThermoFisher, 88-7346-88), IL-1β (ThermoFisher, BMS224-2), IL-10 (ThermoFisher, 88-7106-88), and TGF-β (ThermoFisher, BMS249-4) were employed to assess cytokine levels in cell culture supernatants [62].
Biocompatibility assessment
Four weeks post-puncture, prior to euthanizing the mice, whole blood was collected via tail vein injection at 24 h post-injection of potassium EDTA into blood collection tubes (Fisher Scientific, NC9980784) for blood cell counting and hematological analysis. The blood cell counts, including White Blood Cell (WBC), Red Blood Cell (RBC), and Platelet (PLT), were performed using the Bright-Line™ Hemocytometer (Z359629). Hematological analysis was conducted using the Indiko Automated Clinical Chemistry Analyzer (Thermo Scientific, 98630000). For histological analysis, post-euthanasia tissue samples of heart, liver, spleen, lung, and kidney were collected, fixed in 4% paraformaldehyde for 24 h, and then embedded in paraffin. Tissue Sect. (5 μm) were stained with H&E (Solarbio, G1120) following the manufacturer’s protocol, visualized, and photographed under an Olympus BX51 microscope to evaluate the potential toxicity of NPs in vivo [58, 63].
In vivo biological distribution, MΦ uptake, and pharmacokinetics
Four weeks post-puncture, mice from the PBS group, DiR-NPs group, and DiR-MNPs group received tail vein injections of PBS, DiR-NPs, or DiR-MNPs, respectively.
Biological distribution assessment: At 4 and 24 h post-tail vein injection, mice were anesthetized, and in vivo imaging was performed using the in vivo Imaging System (IVIS; PerkinElmer). Subsequently, at each designated time point, three mice per group were euthanized, and heart, liver, spleen, lung, kidney, blood, and lumbar IVD samples were collected for ex vivo fluorescence imaging [40, 42, 56].
MΦ uptake in mouse IVDs: Following the aforementioned injections of PBS, DiR-NPs, or DiR-MNPs into the tail vein, lumbar IVDs were collected at 4 and 24 h post-injection for sorting CD45+CD11b+CD68+ MΦs using flow cytometry. Subsequently, the percentage of DiR-positive cells within MΦs was analyzed using the Cytek Aurora Flow Cytometer (Cytek) [40].
In vivo pharmacokinetics: To characterize the systemic pharmacokinetic profiles of DiR-NPs and DiR-MNPs, mouse whole blood was collected via mandibular puncture at 0, 4, 8, 12, and 24 h post-tail vein injection. The serum was separated by centrifugation at 800 × g for 15 min, and fluorescence intensity was measured. The fluorescent signals were quantified using the Tecan Infinite M200 plate reader (Tecan), with fluorescence intensity at 0 h set to 100%. The fluorescence values at subsequent time points were normalized to this baseline [40].
RNA extraction, library construction, and sequencing
Total RNA was isolated from MΦ cells sorted via flow cytometry from the lumbar IVDs of mice in the Model + PBS group and Model + IL4I1-MNPs group using Trizol reagent (15596026, Invitrogen, USA). The concentration and purity of RNA samples were determined using a Nanodrop 2000 spectrophotometer (1011U, Nanodrop, USA). Total RNA samples meeting the criteria for subsequent experiments, including RNA Integrity Number (RIN) ≥ 7.0 and 28 S:18 S ratio ≥ 1.5, were utilized [64].
Sequencing libraries were generated and sequenced by CapitalBio Technology (Beijing, China). A total of 5 µg RNA was used for each sample. In brief, the Ribo-Zero Magnetic Kit (MRZE706, Epicentre Technologies) was employed to deplete ribosomal RNA (rRNA) from total RNA. The NEB Next Ultra RNA Library Prep Kit (#E7775, NEB, USA) was utilized for Illumina library construction and sequencing. Subsequently, RNA was fragmented into approximately 300 bp (bp) fragments in NEB Next First Strand Synthesis Reaction Buffer (5×). The first-strand cDNA was synthesized using reverse transcriptase and random primers, followed by second-strand cDNA synthesis in the presence of dUTP Mix (10×). End repair of cDNA fragments, addition of polyA tails, and ligation of sequencing adapters were performed. After adapter ligation, the second strand of cDNA was digested using USER Enzyme (#M5508, NEB, USA) to create a strand-specific library. The library DNA was amplified, purified, and enriched via PCR. Subsequently, the libraries were assessed using the Agilent 2100 Bioanalyzer, quantified using the KAPA Library Quantification Kit (KK4844, KAPA Biosystems), and finally subjected to paired-end sequencing on the NextSeq 500 (Illumina) sequencing platform [65].
Quality control of sequencing data and alignment to the reference genome
The raw data underwent preprocessing using the Trimmomatic tool to eliminate adapters, and low-quality reads. Throughout the quality control process, counts of low-quality bases at the 30 and 50 ends were conducted, along with calculations of original sequencing amount, effective sequencing volume, Q30 quality scores, GC content, and read counts. Subsequently, the filtered high-quality reads were aligned to the mouse reference genome from the NCBI database using HISAT2 version 2.2.1 software [65].
Bioinformatic analysis of differentially expressed genes (GEGs) from transcriptome sequencing
As described in the GEO transcriptome section, the “limma” package in R language was utilized to identify DEGs from the raw count matrix of MΦ transcriptome sequencing data. DEGs were selected based on a threshold of |log2FC| > 1 and a P value < 0.05. Visualization of the DEGs was carried out using the “ggplot2” and “pheatmap” packages in R. Subsequently, GO and KEGG analyses were performed using packages such as “clusterProfiler” in R. Feature genes mediating IL4I1-induced MΦ polarization were identified through LASSO and SVM-RFE machine learning algorithms, and the Venn diagram was employed to intersect the feature genes [33, 66].
RT-qPCR
The cells were lysed using the Trizol reagent kit (Invitrogen, 10296028CN) to extract total RNA, which was then analyzed for quality and concentration using UV-visible spectrophotometry (ND-1000, Nanodrop, USA).
For mRNA expression analysis, reverse transcription was conducted with the PrimeScript™ RT-qPCR kit (TaKaRa, RR037Q). Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) was carried out using SYBR Premix Ex Taq™ (TaKaRa, RR390A) on the LightCycler 480 system (Roche Diagnostics, Pleasanton, CA, USA). The internal control GAPDH was utilized as the mRNA reference gene. The primers were designed and provided by Shanghai General Bioscience Tech. Co., Ltd. The primer sequences can be found in Table S1. The 2-ΔΔCt method was employed to determine the fold change in target gene expression between the experimental and control groups, as calculated by ΔΔCT = ΔCtExperimental - ΔCtControl, where ΔCt = CtTarget gene – CtReference gene [52].
Western blot (WB)
The cells were lysed using RIPA total protein extraction buffer (AS1004, Aspen Biological Technology Co., Ltd., Wuhan, China), followed by protein quantification using the BCA protein quantification kit (ThermoFisher, 23227).
The proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and then blocked with 5% BSA at room temperature for 1 h. Primary antibodies including Collagen II (1:1000; ThermoFisher, PA1-26206), Aggrecan (1:500; ThermoFisher, MA3-16888), ADAMTS5 (1:500; ThermoFisher, PA5-27165), MMP13 (1:500; ThermoFisher, PA5-27242), IL4I1 (1:500; ThermoFisher, PA5-113266), and GAPDH (1:10000; ThermoFisher, AM4300) were added and incubated overnight at 4 °C.
The membrane was washed three times with TBST (3 × 5 min), followed by incubation with either Anti-Mouse-HRP secondary antibody (1:10000; ThermoFisher, 31430) or Goat anti-Rabbit-HRP secondary antibody (1:10000; ThermoFisher, 31460) at room temperature for 2 h. After another three TBST washes, excess ECL working solution (WBKLS0500, Millipore) was added and incubated for 1 min at room temperature for membrane transfer. Excess ECL reagent was removed, the membrane was sealed with plastic wrap, placed in a dark box, and exposed to X-ray film for 5–10 min for imaging and development. Image J analysis software was utilized for quantitative gray analysis of the band patterns in the WB images, with GAPDH serving as the internal reference [52].
Statistical analysis
All experiments were independently conducted at least three times, and the data were presented as mean ± SD (standard deviation). Differences between groups were analyzed using independent sample t-tests or one-way analysis of variance. If the ANOVA results indicated significant differences, Tukey’s HSD post hoc test was performed to compare differences between individual groups. For non-normally distributed or inhomogeneous variance data, the Mann-Whitney U test or Kruskal-Wallis H test was applied. All statistical analyses were carried out using GraphPad Prism 8.0. A P-value < 0.05 was considered statistically significant [67].
Result
IDD nucleus pulposus tissue comprises 12 cell subtypes
Data from the GEO database (GSE244889) included scRNA-seq data from four mild IDD (MIDD, grades I-II) and three severe IDD (SIDD, grades III-V) Nucleus Pulposus (NP) tissue samples. The R software package “Seurat” was utilized for scRNA-seq data analysis. The cell RNA distribution post quality control and standardization is depicted in Figure S1A. The correlation coefficient between nCount and percent. Mt was r = -0.17, while the correlation coefficient between nCount and nFeature was r = 0.9, indicating that the filtered cell data exhibit favorable quality and are suitable for further analysis (Figure S1B).
Subsequently, the filtered cells underwent analysis, with highly variable genes identified through gene expression variance filtering. The top 2000 variably expressed genes were selected for downstream analysis (Figure S1C). Cell cycle scores were computed using the CellCycleScoring function, and normalization of the data was performed (Figure S1D). PCA was then employed for linear dimensionality reduction based on the chosen highly variable genes. Analysis revealed a total of 50 PCs, with the first 30 PCs effectively capturing the information from the selected highly variable genes and holding significant analytical value (Figure S1E-F). The feature genes of the top two PCs were displayed (Figure S1G) alongside the main gene expression heatmap of the top 6 PCs (Figure S1H).
Furthermore, examining the distribution of cells from different samples in PC 1 and PC 2 unveiled clear batch effects among the samples (Figure S1I). To alleviate inter-sample batch effects and improve the accuracy of cell clustering, batch correction was carried out on the sample data using the harmony package (Figure S1J). The results indicated the successful elimination of sample batch effects, rendering the data ready for further analysis (Figure S1K).
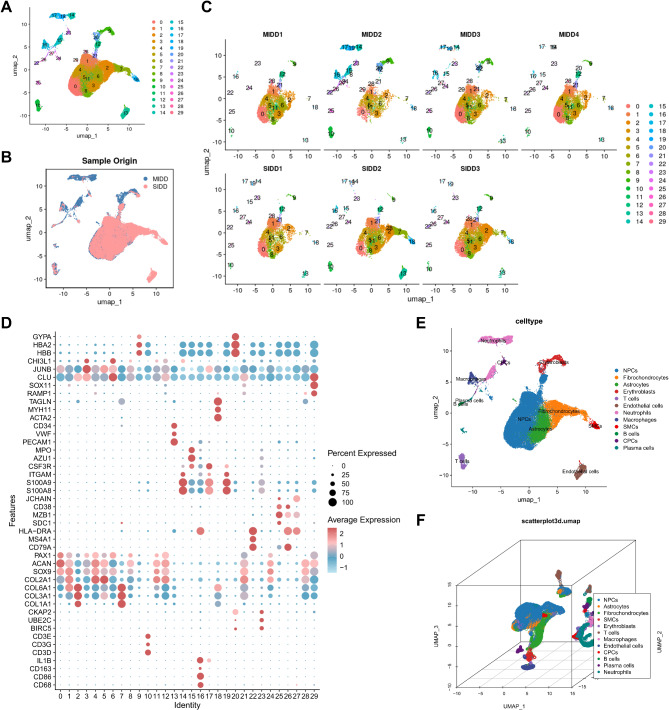
Subsequently, we employed the UMAP algorithm to achieve nonlinear dimensionality reduction on the top 30 PCs, with a resolution set to 1 for cluster analysis (Figure S2). The clustering analysis produced 30 clusters, providing details on the expression profiles of marker genes for each cluster (Fig. 1A-B), along with the distribution of clusters within each sample, as illustrated in Fig. 1C. Cell annotation was then conducted based on the CellMarker database and literature references (Fig. 1D) [61], resulting in the identification of 12 distinct cell categories: NPCs, Fibrochondrocytes, Astrocytes, Erythroblasts, T cells, Endothelial cells, Neutrophils, MΦs, Smooth muscle cells (SMCs), B cells, Cartilage progenitor cells (CPCs), and Plasma cells (Fig. 1E-F).
Fig. 1.
scRNA-seq data cell clustering and annotation. Note: (A) UMAP visualization of cell clustering showing the aggregation and distribution of cells from NP samples in the MIDD group (n = 4) and SIDD group (n = 3), with each color representing a cluster; (B) UMAP visualization of cell clustering in two dimensions, displaying the aggregation and distribution of cells from NP samples in the MIDD group (n = 4) and SIDD group (n = 3), where blue indicates the MIDD group and red indicates the SIDD group; (C) Distribution of cell populations in each sample; (D) Expression levels of known cell lineage-specific marker genes in different clusters, where darker red signifies higher average expression levels and larger circles represent more cells expressing the gene; (E) Three-dimensional visualization of cell annotations based on UMAP clustering, with each color representing a cell subpopulation; (F) Visualization of cell annotations in the MIDD group (n = 4) and SIDD group (n = 3) based on UMAP clustering, where each color denotes a cell subpopulations
These outcomes demonstrate that the NP tissue samples from the MIDD and SIDD groups can be classified into 30 clusters, encompassing 12 cell subtypes. This establishes a foundational understanding for unraveling the characteristic changes in key cell subtypes during IDD progression, aiding in a more comprehensive exploration of the pathogenesis of IDD.
IDD progression associated with imbalance in MΦ M1-M2 polarization
MΦs are reported as a pivotal cell component in regulating NPCs involvement in IDD progression [10]. To further elucidate the role of MΦs in IDD, we initially verified the annotation of MΦs and NPCs. Specifically, marker genes for MΦs demonstrated high expression in Cluster 16 (Figure S3A-B), while NPC marker genes exhibited high expression in Clusters 0, 1, 4, 5, 8, 11, 12, 21, 24, and 28 (Figure S3C-D).
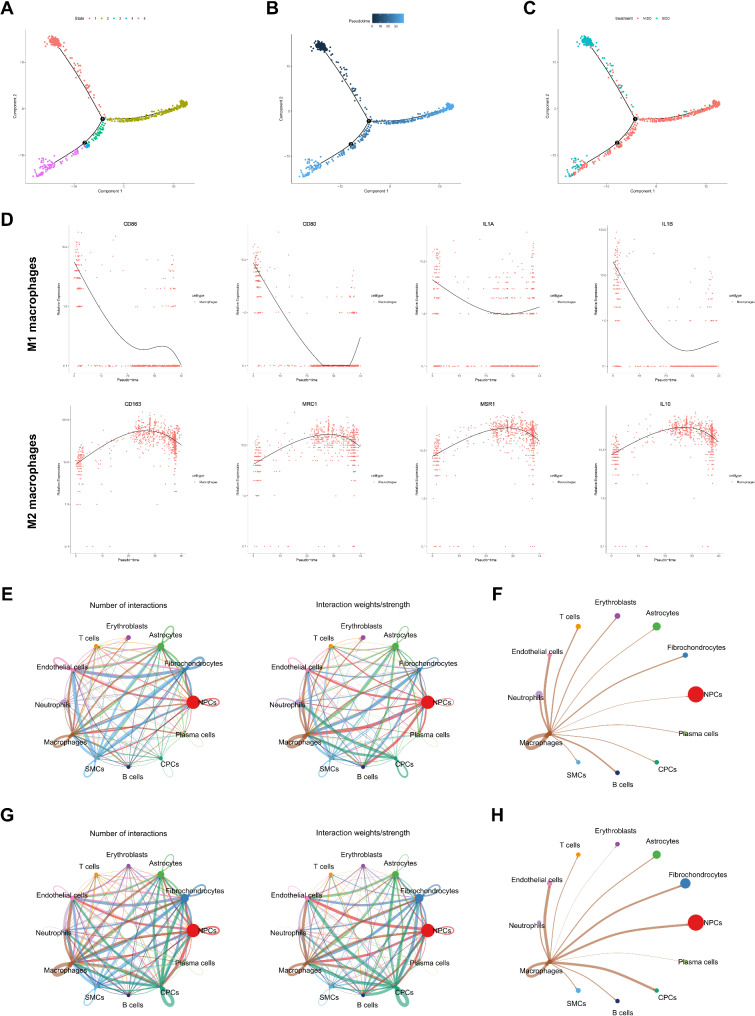
Subsequently, utilizing the “monocle” package, we conducted a pseudo-time analysis to construct a trajectory of cell changes over time within MΦ subtypes, thereby exploring the differential dynamics of MΦs in the MIDD and SIDD groups. The results divided MΦs into five distinct states based on highly variable genes (Fig. 2A). Visualization of pseudotime demonstrated that MΦs transitioned from State 1 to State 5 over time (Fig. 2B). Displaying cells according to different groups revealed that during MΦ recruitment or polarization, States 2, 3, and 4 were unique branches in the MIDD group, with State 1 being specific to the SIDD group; additionally, the SIDD group also exhibited distribution towards the end of the pseudotime axis (Fig. 2A-C). Subsequent plotting of marker gene expression-pseudotime curves for distinct MΦ polarization subtypes indicated significant upregulation of M1 MΦ marker genes CD80, CD86, IL1A, and IL1B, and downregulation of M2 MΦ marker genes CD163, MRC1, MSR1, and IL10 along the pseudotime trajectory. Consistent with this, towards the end of the pseudotime axis, compared to States 2, 3, and 4, M1 marker gene expression was increased, while M2 marker gene expression was decreased (Fig. 2D), suggesting a close association between MΦ skewing towards M1 polarization and IDD progression.
Fig. 2.
Pseudo-temporal analysis of MΦ subtypes and intercellular communication among cell subtypes. Note: (A) Differentiation plot of MΦ cell trajectories, where each color represents a distinct State; (B) Pseudo-temporal visualization of MΦ cell trajectory, with darker colors indicating earlier time points; (C) Visualization of MΦ cell trajectories from MIDD group (n = 4) and SIDD group (n = 3) NP samples, with different colors indicating different groups; (D) Gene expression-pseudo-time curve of M1 and M2 MΦ marker genes, with time on the x-axis and gene expression levels on the y-axis; (E) Network plot of intercellular communication in MΦs from MIDD group NP tissue samples (n = 4), where the thickness of the lines on the left represents the number of pathways and on the right represents interaction strength; (F) Network diagram of intercellular communication among MΦs in MIDD group NP tissue samples (n = 4), with varying line thickness indicating interaction strength; (G) Network plot of intercellular communication in NP tissue samples (n = 3) of the SIDD group, with line thickness on the left representing pathway quantity and on the right representing interaction strength; (H) Network diagram of intercellular communication among MΦs in NP tissue samples (n = 3) of the SIDD group, where line thickness reflects interaction strength
Moreover, to delve deeper into the regulatory mechanisms of IDD progression, we utilized the “CellChat” package to explore cellular communication between MΦs and other cells. The findings revealed a substantial increase in the interactions between MΦs and NPCs in the SIDD group compared to MIDD, suggesting enhanced communication (Fig. 2E-H).
Overall, these results suggest that a critical regulatory mechanism underlying IDD progression may be linked to an increase in M1 polarization and a decrease in M2 polarization in MΦs.
IL4I1: A key gene regulating IDD onset and progression
In-depth exploration of crucial regulatory mechanisms in IDD development and their correlation with MΦ polarization prompted us to acquire two IDD-related transcriptomic datasets (GSE167199 and GSE186542) from the GEO database. To ensure an adequate sample size, we integrated these datasets into a unified one. Subsequently, we utilized the “sva” package to correct and normalize the original data, addressing batch effects between datasets. The results are depicted in Figure S4A-B. Before batch correction, distinct samples from different datasets were segregated, indicating batch effects among them (Figure S4A). Post-batch correction, samples from different datasets were randomly dispersed on the coordinate axis, signifying the elimination of batch effects between datasets, with all samples exhibiting good uniformity (Figure S4B). The merged dataset comprised 6 normal NP samples and 6 IDD NP samples.
Differential analysis on the merged dataset using a threshold of |log2FC| > 1 and p-value < 0.05 identified 553 differentially expressed genes in the merged dataset (M-DEGs), with 160 upregulated and 393 downregulated in the IDD group (Figure S4C-D). Furthermore, GO enrichment analysis revealed that M-DEGs primarily enriched in biological processes (BP) like regulation of hormone levels, cellular components (CC) including synaptic membrane, and molecular functions (MF) such as ion channel activity (Figure S4E). KEGG enrichment analysis pointed out that M-DEGs were mainly enriched in pathways like Cholesterol metabolism (Figure S4F).
Next, to delineate IDD’s disease-specific genes, we employed machine learning algorithms for feature gene selection. The LASSO algorithm identified 7 feature genes (Fig. 3A), SVM-RFE selected 8 feature genes (Fig. 3B), and random forest recognized 3 feature genes (Fig. 3C). The intersection of these results derived through Venn diagrams pinpointed IL4I1 as a core gene (Fig. 3D). Further analysis compared the expression of these feature genes in IDD with normal tissues, highlighting reduced expression of IL4I1 in the IDD group (Fig. 3E). Additionally, the distinct State 1 branch in the SIDD group MΦ subtypes and reduced IL4I1 expression at the end of the pseudotime axis compared to the MIDD group were revealed (Figs. 2A-C and 3F).
Fig. 3.
Selection and Expression of Core M-DEGs in the GEO Dataset. Note: (A) LASSO selection plot for M-DEGs; (B) SVM-RFE analysis results for M-DEGs; (C) Results from the Random Forest algorithm for M-DEGs; (D) Venn diagram showing the intersection of results from LASSO, SVM-RFE, and Random Forest algorithms; (E) Expression levels of IL4I1 in NP samples from each group (n = 6) in the merged dataset; (F) Pseudo-time gene expression curve of IL4I1 in MΦ subtypes from scRNA-seq data, with time on the x-axis and gene expression level on the y-axis; ** P < 0.01 compared between the two groups
These findings suggest that IL4I1 may influence IDD onset and progression through the regulation of MΦ M1-M2 polarization.
IL4I1 regulates MΦ polarization
To validate the findings from our previous bioinformatics analysis, we initially established mild (Mild Model) and severe IDD mouse models (Figure S5A). Micro-CT analysis of the mouse tail IVDs revealed that compared to the Sham group, the DHI percentage was significantly reduced in the Mild Model group, with a further decrease observed in the Severe Model group (Figure S5B). Histological analysis with H&E staining after 4 weeks post-puncture showed that in the Sham group, the IVD structure was intact, the AF was without fissures, NPCs were abundant, and the boundary between AF and NP tissues was clear. Conversely, the IVD structure in the Mild Model group was compromised, with disorganized and ruptured AF, atrophied and fragmented NP tissue, a noticeable decrease in NPCs, and unclear boundaries between AF and NP tissues. The pathological degeneration in the Severe Model group was further exacerbated compared to the Mild Model group (Figure S5C). Safranin-O/Fast green staining revealed that the AF and NP tissue structures in the Sham group were clear, with abundant red staining indicating a rich content of proteoglycans within the IVD. In the Mild Model group, a significant reduction in the central NP of mouse IVDs was observed, accompanied by the replacement of original cartilage-like structures with bone-like tissue, leading to IVD deterioration and collapse. Additionally, the content of proteoglycans was markedly decreased. These degenerative changes in the IVD were more pronounced in the Severe Model group (Figure S5C). Furthermore, the histological scores were significantly higher in the Mild Model group, which further increased in the Severe Model group (Figure S5D). These results collectively indicate the successful establishment of mouse IDD models with varying degrees of degeneration.
The imbalance in the synthesis/degradation metabolism of the extracellular matrix (ECM) in IVD is a crucial event in the progression of IDD [10]. Therefore, we further evaluated the changes in ECM metabolism in the IDD models. Immunohistochemistry results showed that in the IVDs of IDD model mice, the positive staining area of ECM synthesis markers Collagen II and Aggrecan decreased as the degeneration severity increased, while the positive staining area of matrix degradation markers ADAMTS5 and MMP13 increased with degeneration progression (Figure S5E). These findings indicate that worsening IDD is associated with increased ECM degradation metabolism and suppressed synthesis metabolism, disrupting the balance of ECM metabolism.
Moreover, WB and RT-qPCR results demonstrated that the mRNA expression levels of IL4I1 protein decreased with the severity of IVD degeneration (Figure S6A-B). To further investigate the polarization of MΦ in IDD mice, we isolated MΦ from the tail IVDs using flow cytometry (Figure S6C). Consistent with expectations, as degeneration worsened, the pro-inflammatory M1 MΦ significantly increased, while the anti-inflammatory M2 MΦ notably decreased (Figure S6D-F). Correspondingly, levels of pro-inflammatory cytokines TNF-α and IL-1β in the tail IVD significantly increased, while anti-inflammatory cytokines TGF-β and IL-10 levels decreased significantly (Figure S6G). These results suggest that with worsening IDD in mice, IL4I1 expression is downregulated, and there is an increase in M1 polarization along with a decrease in M2 polarization. This aligns with our previous bioinformatics results.
To further validate the expression changes of IL4I1 during MΦ polarization, we induced MΦ derived from human monocytic cell line THP-1 towards M1 or M2 polarization by treating them with LPS or IL-4, respectively (Figure S7A). The results showed that LPS induced MΦ polarization toward M1, while IL-4 induced polarization toward M2 (Figure S7B-D). Significantly, M1 polarization increased the suppression of IL4I1 expression, whereas M2 polarization promoted IL4I1 expression (Figure S7E-F).
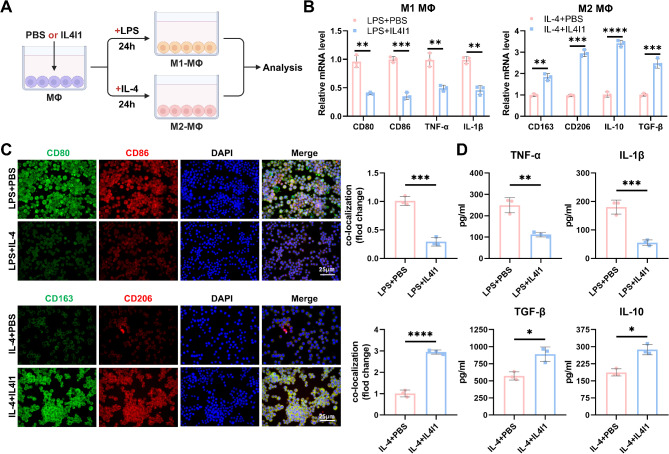
Finally, to explore the correlation between IL4I1 and MΦ polarization, we co-treated MΦ undergoing polarization induction with recombinant human IL4I1 or PBS (Fig. 4A). Treatment with IL4I1 downregulated the expression of M1 MΦ marker genes and upregulated M2 MΦ marker genes (Fig. 4B). Immunofluorescence results indicated that IL4I1 treatment reduced the fluorescence intensity of CD80 and CD86 co-localization while increasing CD163 and CD206 co-localization fluorescence intensity (Fig. 4C). Moreover, ELISA detection revealed that IL4I1 inhibited the secretion of pro-inflammatory cytokines IL-1β and TNF-α while promoting the secretion of anti-inflammatory cytokines TGF-β and IL-10 (Fig. 4D).
Fig. 4.
Influence of IL4I1 on M1-M2 Polarization of MΦs. Note: (A) Schematic representation of IL4I1 treatment on MΦ induced M1 or M2 polarization; (B) RT-qPCR analysis of expression of M1 and M2 MΦ marker genes in different groups of MΦ; (C) Representative immunofluorescence images of MΦ in different groups (scale bar = 25 μm), quantification of fluorescence intensities of CD80 and CD86, CD163 and CD206 co-localization; (D) Levels of IL-1β, TNF-α, TGF-β, and IL-10 in the supernatant of cultured MΦ in each group; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, all experiments were repeated thrice
In summary, IL4I1 can inhibit MΦ polarization towards M1 and promote their polarization towards M2.
MΦ membrane biomimetic NPs targeting MΦ with IL4I1-MNPs
Consequently, we fabricated PLGA NPs loaded with IL4I1 (IL4I1-NPs) and isolated MΦCM to encapsulate IL4I1-NPs, resulting in the IL4I1-MNPs (Fig. 5A).
Fig. 5.
Preparation, characterization, and In Vitro Uptake of IL4I1-NPs and IL4I1-MNPs. Note: (A) Schematic depiction of the preparation process for IL4I1-NPs and IL4I1-MNPs; (B) Representative images of IL4I1-NPs and IL4I1-MNPs using TEM (scale bar = 200 nm); (C-D) DLS analysis of the size and zeta potential of IL4I1-NPs and IL4I1-MNPs; (E) SDS-PAGE protein analysis of cell lysates, MΦCM, IL4I1-NPs, and IL4I1-MNPs; (F) Loading capacity of IL4I1 in IL4I1-NPs and IL4I1-MNPs; (G) The loading capacity and encapsulation efficiency of IL4I1 within IL4I1-NPs and IL4I1-MNPs, respectively, with monitoring of encapsulation rates within 72 h at 4 °C; (H) In vitro cumulative release curve of IL4I1 from IL4I1-NPs and IL4I1-MNPs in PBS at 37 °C; (I) Particle size detection of IL4I1-MNPs in deionized water, 1×PBS, and 50% FBS over three days; (J) Schematic diagram of the preparation process for DiR-NPs and DiR-MNPs; (K) Flow cytometry analysis of fluorescence intensity of DiR in MΦ after incubation with DiR-NPs and DiR-MNPs for 1, 2, and 4 h; (L) Representative images (scale bar = 15 μm) and quantitative analysis of fluorescence intensity of MΦ uptake of DiR-NPs and DiR-MNPs; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, all experiments were conducted in triplicate
Transmission electron microscopy (TEM) results revealed a characteristic core-shell structure of IL4I1-MNPs, where a membrane surrounded the PLGA core, distinctly different from IL4I1-NPs, which lacked membrane coverage (Fig. 5B). DLS analysis indicated that the size of IL4I1-NPs was approximately 120.8 ± 1.1 nm, increasing to 132.9 ± 0.8 nm after encapsulation with MΦCM, with a shift in surface zeta potential from − 26.1 ± 0.6 to -16.8 ± 1.2 mV (Fig. 5C-D). Subsequent SDS-PAGE analysis of IL4I1-NPs, cellular lysates, MΦCM, and IL4I1-MNPs confirmed the presence of MΦCM on IL4I1-MNPs. Notably, IL4I1-NPs displayed no protein expression due to the lack of membrane coverage (Fig. 5E), whereas MΦCM and IL4I1-MNPs exhibited similar protein bands, likely attributed to the membrane proteins and intracellular content present in cellular lysates, confirming the successful encapsulation of MΦCM on IL4I1-MNPs.
The loading capacity of IL4I1 in IL4I1-NPs and IL4I1-MNPs was determined to be 0.376 ± 0.015 µg/mg and 0.384 ± 0.012 µg/mg, respectively (Fig. 5F). Correspondingly, the encapsulation efficiencies were calculated as 69.2 ± 1.3% for IL4I1-NPs and 67.8 ± 1.9% for IL4I1-MNPs. Notably, the encapsulation efficiency of both formulations remained stable over a 72-hour period at 4 °C (Fig. 5G).Moreover, the controlled release capabilities of IL4I1-NPs and IL4I1-MNPs were demonstrated through the analysis of the IL4I1 release profiles at different time points in PBS at 37 °C. IL4I1-NPs exhibited a faster release rate compared to IL4I1-MNPs, with approximately 92% of IL4I1 released after 120 h (Fig. 5H), indicating that the encapsulation of MΦCM extended the release of IL4I1 in IL4I1-MNPs. Notably, when assessing the colloidal stability of IL4I1-MNPs suspended in deionized water, 1×PBS, and 50% FBS at 4 °C, minimal changes in particle size were observed within 72 h, demonstrating the excellent stability of the biomimetic NPs (Fig. 5I).
To evaluate the impact of MΦCM encapsulation on the uptake of NPs by MΦ, fluorescent dye DiR was utilized as a substitute for IL4I1 to prepare fluorescently labeled NPs (DiR-NPs) and MNPs (DiR-MNPs) as described previously (Fig. 5J). By co-culturing these NPs with THP-1-derived MΦ cells and observing the uptake at different time points, it was observed that DiR-NPs were taken up by MΦ in a time-dependent manner. Importantly, at various time points, the fluorescence intensity in MΦ cells treated with DiR-MNPs was significantly higher than that in the DiR-NPs group (Fig. 5K-L), indicating that MΦCM encapsulation enhanced the targeting ability of DiR-MNPs to MΦ, thereby increasing the uptake of DiR-MNPs.
Subsequently, we intravenously injected PBS, DiR-NPs, or DiR-MNPs into IDD mice to assess the biodistribution in the major organs and tail IVD through in vivo and ex vivo fluorescence imaging, as well as flow cytometry to evaluate their targeting of MΦ (Figure S8A). In vivo fluorescence imaging results indicated that a higher fluorescence signal was detected in the tail IVD of mice in the DiR-MNPs group compared to the DiR-NPs group, suggesting that the cell membrane mimetic NPs DiR-MNPs, after encased by MΦCM, could be more effectively delivered to the IDD lesion site (Figure S8B). To further understand the biodistribution of MNPs, we conducted ex vivo fluorescence imaging on the heart, liver, spleen, lungs, kidneys, and tail IVD. As shown in Figure S8C, both treatment groups exhibited notable fluorescence accumulation in the liver and spleen, with DiR-MNPs demonstrating higher fluorescence intensity in the IVD. Flow cytometry analysis revealed that both DiR-NPs and DiR-MNPs were internalized into MΦ in a time-dependent manner, with the fluorescence intensity of DiR in MΦ from the DiR-MNPs group being stronger, indicating a more effective targeting of MΦ (Figure S8D). These findings suggest that cell membrane biomimetic NPs MNPs exhibit specific targeting effects with potential therapeutic benefits for IVD.
Furthermore, 24 h post-tail vein injection, DiR-MNPs exhibited higher fluorescence intensity in the bloodstream (Figure S8E). Subsequently, blood samples were collected at predetermined time points to measure their fluorescence intensity, further evaluating the circulation lifespan of NPs in the blood. Throughout the entire testing period, the concentration of MNPs was higher than that of NPs in the mouse bloodstream (Figure S8F). These results indicate that cell MNPs have a longer circulation time in vivo than NPs.
In summary, our successful preparation of IL4I1-MNPs demonstrates their excellent stability and specific targeting effects.
IL4I1-MNPs improve MΦ polarization imbalance in treating IDD in mice
To investigate the in vivo therapeutic effects of IL4I1, IL4I1-NPs, or IL4I1-MNPs on IDD, we locally injected them into the tail IVD of mice with IDD (Figure S9A). Results from Micro-CT indicated that compared to the Model + PBS group, IL4I1, IL4I1-NPs, and IL4I1-MNPs significantly improved the decreased DHI% in IDD mice, degenerative changes in related tissues (such as AF morphology disruption, reduced NP area, and decreased NPCs), and increased histological scores; notably, the therapeutic effect of IL4I1-MNPs was most significant (Figure S9B-D). Additionally, immunohistochemical results showed that in the Model + PBS group of IDD mice IVD, the expression levels of ECM synthesis markers, Collagen II and Aggrecan, were reduced, while the ECM degradation markers, ADAMTS5 and MMP13, were increased. However, treatment with IL4I1, IL4I1-NPs, and IL4I1-MNPs significantly upregulated the expression of Collagen II and Aggrecan and downregulated the expression of ADAMTS5 and MMP13 (Figure S9E). Among these, the therapeutic effect of IL4I1-MNPs on restoring ECM metabolic balance in IDD mice was most pronounced (Figure S9E).
Subsequently, we further investigated the impact of IL4I1, IL4I1-NPs, or IL4I1-MNPs on MΦ polarization in IDD mice. As expected, in the Model + PBS group, there was a significant increase in the expression and cell proportion of M1 MΦ marker genes and a decrease in the expression and cell proportion of M2 MΦ marker genes. Treatment with IL4I1, IL4I1-NPs, and IL4I1-MNPs significantly reversed this phenomenon, with IL4I1-MNPs showing the most significant inhibition on M1 MΦ polarization in IDD mice (Fig. 6A-C). Consistently, compared to the Sham + PBS group, the levels of pro-inflammatory cytokines TNF-α and IL-1β significantly increased in the tail IVD of the Model + PBS group, while the levels of anti-inflammatory cytokines TGF-β and IL-10 decreased significantly. However, IL4I1, IL4I1-NPs, and IL4I1-MNPs all significantly inhibited the secretion of TNF-α and IL-1β and promoted the secretion of TGF-β and IL-10. Additionally, the effect of IL4I1-MNPs was stronger compared to IL4I1 and IL4I1-NPs (Fig. 6D).
Fig. 6.
Impact of IL4I1-MNPs on MΦ Polarization in IDD Mice In Vivo. Note: (A) RT-qPCR analysis of expression of M1 and M2 MΦ marker genes in IVD MΦs of mice in different groups (n = 6); (B) Percentage of M1 MΦs (CD80+CD86+) in IVD MΦs of mice in different groups (n = 6); (C) Percentage of M2 MΦs (CD163+CD206+) in IVD MΦs of mice in different groups (n = 6); (D) ELISA detection of IL-1β, TNF-α, TGF-β, and IL-10 levels in IVD of mice in different groups (n = 6); * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001
Finally, we evaluated the in vivo safety of IL4I1, IL4I1-NPs, and IL4I1-MNPs. On postoperative day 28 of IDD, we collected blood and major organs from the mice for comprehensive testing. Blood count analysis showed no significant changes in white blood cells, red blood cells, and platelet counts in the IL4I1, IL4I1-NPs, and IL4I1-MNPs treatment groups compared to the control group (Figure S10A). Furthermore, in assessing serum biochemical metabolism, we observed no significant changes in markers related to liver (ALB, TBIL, ALT, AST) or kidney (CRE, UREA, UA) function among the groups (Figure S10B). Histopathological analysis of major organs through H&E staining revealed no apparent damage in the heart, liver, spleen, lungs, or kidneys in the IL4I1, IL4I1-NPs, and IL4I1-MNPs treatment groups compared to the control group (Figure S10C). These results confirm the in vivo safety of IL4I1, IL4I1-NPs, and IL4I1-MNPs.
Overall, these results demonstrate that IL4I1, IL4I1-NPs, and IL4I1-MNPs improve MΦ polarization imbalance in treating IDD by inhibiting M1 MΦ polarization and promoting M2 MΦ polarization. Notably, the IL4I1-MNPs exhibit stronger efficacy in IDD therapy due to their prolonged circulation time and enhanced targeting of MΦ.
The gene FGR is identified as a key regulator of MΦ polarization balance by IL4I1-MNPs
To investigate the specific regulatory mechanism of IL4I1-mediated MΦ polarization therapy for IDD using IL4I1-MNPs, we performed transcriptome sequencing of MΦ extracted from the IVD of mice in the Model + PBS group and the Model + IL4I1-MNPs group using flow cytometry (Fig. 7A). The results revealed that in the MΦ of mice in the Model group treated with IL4I1-MNPs, there were a total of 949 significant DEGs, with 439 upregulated and 510 downregulated genes (Fig. 7B-C).
Fig. 7.
Functional clustering, core gene selection, and expression validation of degs from transcriptome sequencing. Note: (A) Schematic representation of transcriptome profiling of IVD MΦs in mice from Model + PBS and Model + IL4I1 groups; (B) Volcano plot depicting differential analysis of MΦ samples in the IVD tissues of mice from Model + PBS group (n = 3) and Model + IL4I1 group (n = 3); (C) Heatmap illustrating DEGs among MΦ samples (n = 3) from the transcriptome sequencing; (D) Bubble chart of KEGG enrichment analysis for the DEGs; (E) Circos plots showing GO enrichment analysis (BP, CC, MF) for the DEGs; (F) LASSO selection plot of the DEGs; (G) Results plot of SVM-RFE analysis for the DEGs; (H) Venn diagram depicting the intersection of results from LASSO and SVM-RFE algorithms; (I) Expression profile of FGR in MΦ samples (n = 3) from the transcriptome sequencing; (J) RT-qPCR analysis of FGR expression in MΦs of mice from different groups (n = 6); ** P < 0.01, *** P < 0.001
Functional enrichment analysis of the DEGs was conducted using the GO and KEGG databases. KEGG enrichment analysis showed that the DEGs were mainly enriched in signaling pathways such as cytokine-cytokine receptor interaction (Fig. 7D). GO enrichment analysis indicated that the DEGs were primarily enriched in biological processes such as positive regulation of cytokine production, cellular components like receptor complexes, and molecular functions such as cytokine activity (Fig. 7E).
Subsequently, to further identify the characteristic genes regulating MΦ polarization balance in IDD mice by IL4I1-MNPs, we employed machine learning algorithms for feature gene selection. Using the LASSO algorithm, 9 feature genes were identified (Fig. 7F), and the SVM-RFE algorithm selected 27 feature genes (Fig. 7G). The intersection of these genes revealed the core gene FGR (Fig. 7H). It has been reported that the loss of FGR inhibits the pro-inflammatory M1 MΦ polarization while enhancing the anti-inflammatory M2 MΦ polarization [68]. Therefore, we further evaluated the expression of FGR in the MΦ of mice in the Model + PBS group and Model + IL4I1-MNPs group, showing that FGR was downregulated in the MΦ of mice in the Model + IL4I1-MNPs group (Fig. 7I). Consistent results were obtained by RT-qPCR validation of FGR expression (Fig. 7J).
In summary, these results suggest that FGR may serve as a critical gene in regulating the balance of MΦ polarization by IL4I1-MNPs.
IL4I1-MNPs mediate the improvement of NPCs physiological functions through FGR-mediated MΦ polarization balance
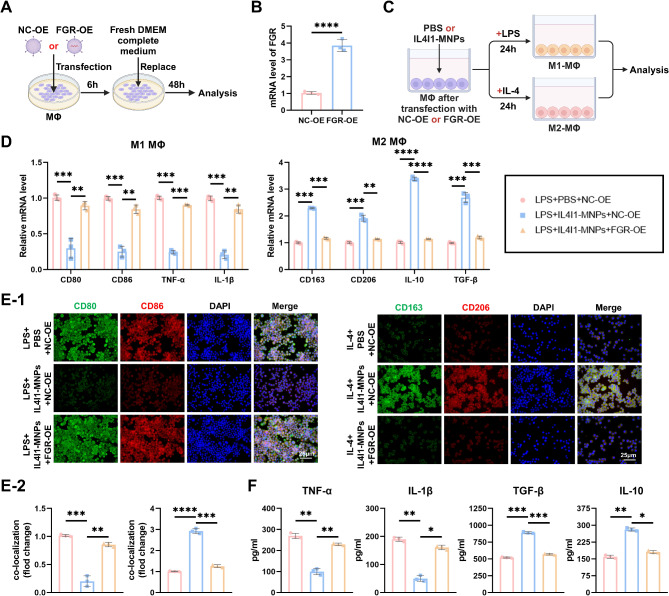
To further investigate the correlation between FGR and IL4I1-MNPs-mediated MΦ polarization balance, we constructed lentiviruses overexpressing human FGR-OE or NC-OE to transfect MΦ (Fig. 8A) and verified successful overexpression through RT-qPCR (Fig. 8B). Subsequently, upon inducing polarization of NC-OE or FGR-OE transfected MΦ, we co-treated them with IL4I1-MNPs or PBS (Fig. 8C). Compared to the control group, under IL4I1-MNPs treatment, the expression of M1 MΦ marker genes was downregulated, while that of M2 MΦ marker genes was upregulated; moreover, FGR overexpression reversed the effects of IL4I1-MNPs (Fig. 8D). Concurrently, IL4I1-MNP treatment reduced the fluorescence intensity of CD80 and CD86 co-localization while increasing that of CD163 and CD206 co-localization, with FGR-OE blocking the effects of IL4I1-MNPs (Fig. 8E). ELISA results indicated that during M1 polarization, IL4I1-MNPs suppressed the secretion of pro-inflammatory cytokines IL-1β and TNF-α, and during M2 polarization, they promoted the secretion of anti-inflammatory cytokines TGF-β and IL-10. FGR-OE treatment counteracted the effects of IL4I1-MNPs (Fig. 8F). These findings suggest that IL4I1-MNPs inhibit the M1 phenotype and promote the M2 phenotype of MΦ by downregulating FGR.
Fig. 8.
Exploration of the Role of FGR in Regulating MΦ Polarization Balance Mediated by IL4I1-MNPs. Note: (A) Illustration of FGR-OE transfection; (B) RT-qPCR analysis of FGR expression in different groups of MΦ; (C) Schematic representation of MΦ polarization induction post-NC-OE or FGR-OE transfection, followed by treatment with PBS or IL4I1-MNPs; (D) RT-qPCR analysis of expression of M1 and M2 MΦ marker genes in different groups of MΦ; (E) Representative immunofluorescence images of MΦ in different groups (scale bar = 25 μm), quantification of CD80 and CD86, CD163 and CD206 co-localization fluorescence intensity; (F) Levels of IL-1β, TNF-α, TGF-β, and IL-10 in the supernatant of cultured MΦ in different groups; *P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, all experiments were repeated three times
Subsequently, we further explored whether the IL4I1-MNPs-mediated FGR-regulated MΦ MΦ polarization process affects the physiological functions of NPCs and thereby influences the development of IDD. We primed NPCs with IL-1β to simulate an inflammatory environment of IDD, then co-cultured differentially treated MΦ with pre-treated NPCs, establishing a Transwell co-culture system (Figure S11A). CCK8 analysis revealed that at 24 and 72 h, compared to the NPCs + PBS + NC-OE group, LPS-induced M1 polarized MΦ significantly reduced the proliferative capacity of NPCs, while IL4I1-MNPs-inhibited M1 polarized MΦ markedly increased NPC proliferation. In the NPCs + LPS + IL4I1-MNPs + NC-OE group, FGR-OE-treated MΦ reversed the promoting effect of IL4I1-MNPs on NPC proliferation (Figure S11B). EdU staining further validated this conclusion (Figure S11C). Results from the Transwell experiment and flow cytometry indicated that LPS significantly reduced the number of migrating NPC cells and promoted their apoptosis, while IL4I1-MNP treatment considerably increased NPC migration and inhibited apoptosis. Relative to the NPCs + LPS + IL4I1-MNPs + NC-OE group, the M1 polarized MΦ induced by FGR overexpression in MΦ obstructed the effects of IL4I1-MNPs (Figure S11D-E).
On the other hand, the results of CCK8 and EdU staining indicated that IL-4-induced M2 MΦ polarization significantly enhanced the proliferation of NPCs compared to the NPCs + PBS + NC-OE group, and this promoting effect was further augmented by IL4I1-MNPs. In contrast to the NPCs + LPS + IL4I1-MNPs + NC-OE group, FGR overexpression in MΦ countered the promoting effect of IL4I1-MNPs on NPC proliferation (Figure S12A-B). Results from Transwell experiments and flow cytometry demonstrated that IL-4 significantly promoted NPC migration and inhibited apoptosis, with IL4I1-MNP treatment further enhancing NPC migration and strengthening the inhibitory effect on apoptosis. Compared to the NPCs + LPS + IL4I1-MNPs + NC-OE group, in the NPCs + LPS + IL4I1-MNPs + FGR-OE group, FGR overexpression-induced M2 MΦ polarization reversed the inhibitory effect of IL4I1-MNPs (Figure S12C-D). These findings suggest that downregulation of FGR1 expression by IL4I1-MNPs mediates an increase in M2 MΦ polarization and a decrease in M1 MΦ polarization, promoting NPC proliferation and migration while inhibiting NPC apoptosis.
Next, we further analyzed the changes in ECM metabolic indicators in NPCs co-cultured with differentially treated MΦ. In comparison to the NPCs + PBS + NC-OE group, LPS-induced M1 MΦ polarization suppressed the expression of ECM synthesis markers, Collagen II and Aggrecan, and promoted the expression of ECM degradation markers, ADAMTS5 and MMP13 (Figure S11F). Conversely, IL-4-induced M2 MΦ polarization had opposite effects on the expression of Collagen II, Aggrecan, ADAMTS5, and MMP13 (Figure S12E). Compared to the NPCs + LPS + PBS + NC-OE group, the NPCs + LPS + IL4I1-MNPs + NC-OE group showed an upregulation of Collagen II and Aggrecan expression in NPCs and a downregulation of ADAMTS5 and MMP13 expression after treating MΦ with IL4I1-MNPs. Subsequently, FGR-OE treatment of MΦ counteracted the effects of IL4I1-MNPs (Figure S11F). Furthermore, compared to the NPCs + IL-4 + PBS + NC-OE group, the NPCs + IL-4 + IL4I1-MNPs + FGR-OE group further upregulated the expression of Collagen II and Aggrecan in NPCs and downregulated ADAMTS5 and MMP13; FGR overexpression in MΦ inhibited the expression of ECM metabolic-related markers regulated by IL4I1-MNPs (Figure S12E). These results indicate that the downregulation of FGR1 expression by IL4I1-MNPs improves the M1-M2 polarization imbalance of MΦ, regulates NPC proliferation, migration, and ECM metabolism balance, and suppresses their apoptosis. Immunofluorescence further confirmed this effect (Figure S11G, Figure S12F).
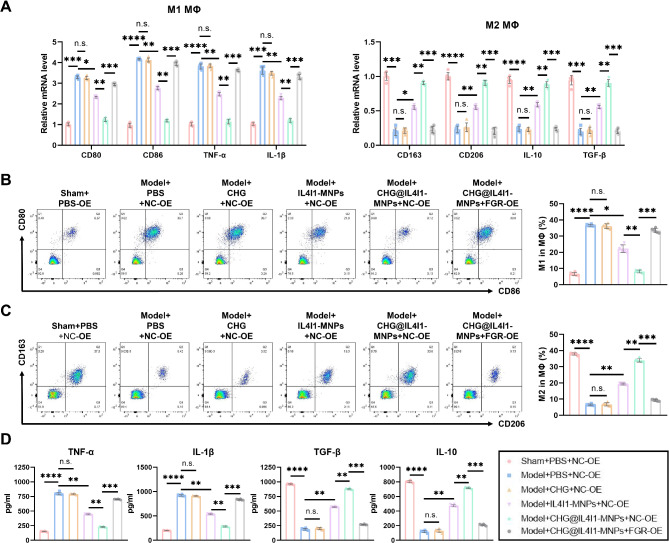
In conclusion, IL4I1-MNPs downregulating FGR1 expression ameliorates the M1-M2 polarization imbalance of MΦ, modulates NPC proliferation, migration, and ECM metabolism balance, and inhibits their apoptosis.
Successful Preparation of CHG@IL4I1-MNPs
Studies suggest that hydrogels, particularly cellulose supramolecular hydrogels, hold significant potential in the field of IDD therapy due to their stability and non-toxic nature, serving as promising delivery platforms for NPs [69, 70]. Therefore, we first prepared CHG by crosslinking cellulose and β-cyclodextrin using epichlorohydrin as a raw material (Figure S13A). The swelling curve of the hydrogel showed a significant increase in the swelling rate in the initial 2 h, followed by a plateau phase (Figure S13B), while the degradation curve exhibited a gradual degradation trend (Figure S13C).
Subsequently, we loaded IL4I1-MNPS into CHG (Figure S13D). SEM images revealed the bioinspired membrane-mimicking NPs, IL4I1-MNPs, binding to the inner surface of hydrogel CHG, maintaining the pore size of the porous hydrogel before and after nanoparticle loading (Figure S13E). Rheological analysis indicated that the storage modulus (G’) of CHG and CHG@IL4I1-MNPs was higher than the loss modulus (G”), suggesting their good stability and viscoelasticity (Figure S13F). Additionally, there was no significant difference in the average storage modulus between CHG and CHG@IL4I1-MNPs, indicating that the inclusion of IL4I1-MNPs had minimal impact on the mechanical properties of hydrogel CHG (Figure S13G). Furthermore, we observed the in vitro release curves of IL4I1 in PBS from IL4I1-MNPs and CHG@IL4I1-MNPs. Compared to IL4I1-MNPs, CHG@IL4I1-MNPs exhibited a slower release of IL4I1, indicating that hydrogel CHG prolonged the drug release, beneficial for long-term in vivo treatment of IDD (Figure S13H). Subsequently, we assessed the cytotoxicity of hydrogels on MΦ using CCK8 assays. The results demonstrated no significant differences in cell viability among the groups (Figure S13I), indicating the non-toxicity of CHG and CHG@IL4I1-MNPs to MΦ cells in vitro.
These results demonstrate the successful preparation of CHG loaded with MΦ membrane-mimicking NPs IL4I1-MNPs, denoted as CHG@IL4I1-MNPs.
CHG@IL4I1-MNPs downregulate FGR to regulate MΦ polarization and treat IDD in mice
To further investigate the potential and mechanism of CHG@IL4I1-MNPs hydrogel in treating IDD in mice, we injected PBS, CHG, IL4I1-MNPs, or CHG@IL4I1-MNPs into the IVD of mice, while simultaneously administering lentiviral vectors carrying the mouse FGR-OE and NC-OE vectors via tail vein injection (Figure S14A). The Micro-CT results indicated that compared to the Model + PBS + NC-OE group, the hydrogel CHG had no significant impact on IDD mice regarding a decreased (DHI%, degenerative tissue pathology changes (e.g., AF morphology disruption, reduction in NP area, and decrease in NPCs), and increased histological scores. However, IL4I1-MNPs and CHG@IL4I1-MNPs markedly reversed these effects, with CHG@IL4I1-MNPs showing the most significant effect. Moreover, FGR overexpression based on the Model + CHG@IL4I1-MNPs + NC-OE group blocked the advantageous effects of CHG@IL4I1-MNPs on IDD (Figure S14B-D). Additionally, immunohistochemistry results showed that in the Model + PBS + NC-OE group of IDD mice, the expression of ECM synthesis markers Collagen II and Aggrecan decreased, while the ECM degradation markers ADAMTS5 and MMP13 increased. In contrast, CHG treatment had no noticeable impact on the expression of ECM synthesis/degradation metabolic markers. Conversely, the Model + IL4I1-MNPs + NC-OE and Model + CHG@IL4I1-MNPs + NC-OE groups revealed that IL4I1-MNPs and CHG@IL4I1-MNPs treatment restored the expression of Collagen II, Aggrecan, ADAMTS5, and MMP13, with CHG@IL4I1-MNPs demonstrating stronger restorative effects. As anticipated, FGR-OE counteracted the effects of CHG@IL4I1-MNPs (Figure S14E). These results indicate that IL4I1-MNPs carried by CHG@IL4I1-MNPs inhibit FGR and effectively treat IDD. CHG@IL4I1-MNPs exhibit a stronger therapeutic effect in treating IDD compared to IL4I1-MNPs due to the prolonged release time of IL4I1.
Furthermore, we investigated the correlation between FGR mediated by CHG@IL4I1-MNPs and MΦ polarization in IDD mice. As expected, the Model + PBS + NC-OE group showed a significant increase in M1 MΦ and a decrease in M2 MΦ. Treatment with IL4I1-MNPs and CHG@IL4I1-MNPs markedly reversed this phenomenon, with CHG@IL4I1-MNPs exhibiting a more pronounced effect. Notably, CHG treatment had no discernible impact on this observation. Additionally, FGR overexpression on the basis of the Model + CHG@IL4I1-MNPs + NC-OE group reversed the effects of CHG@IL4I1-MNPs (Fig. 9A-C).
Fig. 9.
Effect of CHG@IL4I1-MNPs on MΦ Polarization in IDD Mice. Note: (A) RT-qPCR analysis of the expression of M1 and M2 MΦ marker genes in IVD MΦs of mice (n = 6); (B) Percentage of M1 MΦs in IVD MΦs of mice (n = 6); (C) Percentage of M2 MΦs in IVD MΦs of mice (n = 6); (D) ELISA detection of IL-1β, TNF-α, TGF-β, and IL-10 levels in the IVD of mice (n = 6); n.s. P > 0.05 for comparison between groups, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001
Consistent with this, compared to the Sham + PBS + NC-OE group, the Model + PBS + NC-OE group demonstrated a significant increase in pro-inflammatory cytokines TNF-α and IL-1β levels and a notable decrease in anti-inflammatory cytokines TGF-β and IL-10 levels in the tail IVD. The Model + CHG + NC-OE group did not affect the secretion of pro-inflammatory and anti-inflammatory factors in the disc compared to the Model + PBS + NC-OE group. However, IL4I1-MNPs and CHG@IL4I1-MNPs treatments significantly reduced TNF-α and IL-1β levels while increasing TGF-β and IL-10 levels in the disc. CHG@IL4I1-MNPs exerted a stronger regulatory effect on the levels of pro-inflammatory and anti-inflammatory factors. Concurrently, FGR-OE treatment hindered the effects of CHG@IL4I1-MNPs (Fig. 9D). This further confirms that CHG@IL4I1-MNPs effectively ameliorate IDD by inhibiting M1 polarization and promoting M2 polarization.
It is important to note that after 4 weeks post-IDD surgery, there were no significant differences in WBC, RBC, and PLT counts among the mouse groups (Figure S15A). Furthermore, the levels of liver and kidney function-related markers in the various mouse groups did not exhibit statistically significant differences (Figure S15B). Moreover, H&E staining of major organs in differently treated mice revealed that the CHG, IL4I1-MNPs, and CHG@IL4I1-MNPs treatment groups did not manifest apparent tissue damage compared to the control group (Figure S15C). These findings indicate the excellent biocompatibility of CHG, IL4I1-MNPs, and CHG@IL4I1-MNPs.
In summary, CHG@IL4I1-MNPs release IL4I1, thereby downregulating FGR expression, inhibiting M1 MΦ polarization and promoting M2 MΦ polarization to effectively treat IDD.
Discussion
IDD is a prevalent degenerative disease of the IVDs, substantially impacting patients’ quality of life and work efficiency [71, 72]. Traditional treatment methods often fall short in delivering satisfactory results, prompting a growing interest in in-situ biologic repair. MΦs play crucial roles in the pathogenesis of IDD, with their polarization state critical for tissue repair and regeneration [10, 73, 74]. The novel CHG@IL4I1-MNPs, by targeted delivery of IL4I1, can regulate the M1-M2 polarization of MΦs, thus facilitating the restoration of IDD. This study aims to elucidate the mechanisms underlying the therapeutic effects of CHG@IL4I1-MNPs on IDD, with the aspiration of introducing novel therapeutic strategies and theoretical support for the biological repair of IDD. This study utilized CHG@IL4I1-MNPs as a drug delivery system targeting IL4I1, offering an innovative approach for treating IDD. Compared to conventional methods, this nanoparticle drug delivery system enhances the effective delivery of IL4I1, thereby regulating MΦ M2 polarization more effectively and promoting the repair and regeneration of IVD tissue. This paves the way for new avenues in IDD treatment, emphasizing the importance of targeted therapeutic strategies addressing disease-specific mechanisms. To minimize potential off-target effects in clinical applications, strategies such as surface modifications (e.g., PEGylation) and particle size optimization are under consideration to enhance targeting specificity and reduce damage to healthy tissues. Future clinical studies will further validate the safety and efficacy of these NPs, offering robust evidence for broader clinical applications.
The Gene Expression Omnibus (GEO) is a public repository for high-throughput gene expression data, encompassing not only next-generation RNA sequencing data but also microarray and single-cell sequencing data. The sample data are not limited to tumors but are widely used to investigate various biological processes and disease mechanisms [75]. Through single-cell and transcriptome sequencing analysis, the research team revealed the critical role of M1-M2 MΦ polarization imbalance in the development of IDD. This contributes to a deeper understanding of the pathogenesis of IVD tissue diseases, providing vital clues for targeted treatments. In exploring the IL4I1 protein, this study elucidated its role as a key regulator of MΦ polarization, not only clarifying its function in disease progression but also offering valuable insights for designing future therapeutic strategies. This in-depth analysis of molecular mechanisms holds promise for breakthroughs in treating degenerative diseases like IDD.
Through the novel drug delivery system in this study and the in-depth investigation of the mechanism behind MΦ polarization imbalance, not only have new perspectives and hopes been brought to IDD treatment, but a solid foundation has also been laid for fundamental research in related fields. Future research directions could further explore the application effectiveness of this drug system in clinical practice, optimize treatment regimens, and seek broader disease applications to bring greater benefits to human health.
Animal experiments confirmed the therapeutic effects of CHG@IL4I1-MNPs on IDD, further establishing the MΦ polarization regulation mechanism mediated by IL4I1-MNPs. This not only strengthens confidence in treatment strategies but also highlights the significance of targeted drug carriers in therapy. Targeted drug carriers can release therapeutic factors precisely and efficiently in the body, potentially serving as effective tools for developing novel treatment strategies. This study offers valuable insights into exploring more individualized and precise treatment methods, opening new directions for future clinical research and providing more effective treatment choices for patients.
In terms of bioinformatics analysis, the study found that the FGR gene plays a crucial role in MΦ polarization regulation during IL4I1-MNPs treatment, providing new clues for a deeper understanding of the molecular mechanisms. The impact of specific genes on treatment efficacy is emphasized, suggesting that individual differences in treatment response may be related to the expression levels of certain genes. This discovery helps us better comprehend the treatment mechanisms, offering new avenues for gene-regulated therapies and serving as a reference for designing future individualized treatment strategies.
Combining in vitro and in vivo experimental results confirmed the regulatory effect of CHG@IL4I1-MNPs on the balance of MΦ M1-M2 polarization in IDD. This study highlights the crucial role of restoring cell polarization balance in IDD treatment more than previous research has done. It indicates the importance of focusing on restoring the balance of cells during treatment to promote disease repair and maximize treatment outcomes. By regulating the polarization status of MΦs, we may develop more effective treatment strategies, presenting novel perspectives and potential solutions for IDD and similar diseases.
Current treatment approaches for IDD include pharmacotherapy, physical therapy, cellular therapy, and surgical interventions. Traditional pharmacotherapy primarily relies on nonsteroidal anti-inflammatory drugs (NSAIDs), corticosteroids, and analgesics, which can alleviate symptoms but fail to reverse the pathological progression of the disease. Moreover, long-term use of these medications may lead to adverse side effects [76, 77]. Cellular therapies, such as IVD cell transplantation, demonstrate some clinical potential but face challenges such as low cell survival rates and inconsistent therapeutic outcomes [78, 79]. Surgical treatments, such as discectomy, can achieve significant results in severe cases but are associated with high surgical risks, recurrence, and complications [6, 80].
In contrast, the CHG@IL4I1-MNPs nanoparticle system presented in this study represents a novel targeted delivery strategy with distinct advantages. By employing specific targeting mechanisms, this system enables precise delivery of therapeutic agents to IVD tissues, maximizing drug efficacy while minimizing off-target effects. Compared to existing therapies, this approach has the potential to slow disc degeneration, improve the biological microenvironment of the IVD, and achieve disease reversal rather than merely symptom relief. Additionally, the stability and biocompatibility of NPs suggest that this method could provide more durable and controllable therapeutic effects in clinical applications.
The scalability of CHG@IL4I1-MNPs synthesis and the evaluation of its long-term safety are critical for clinical translation. First, regarding synthesis scale-up, CHG@IL4I1-MNPs have been successfully prepared at the laboratory scale, with preliminary validation of their structure and functionality. However, achieving large-scale production will require further optimization of synthesis processes, including control of reaction conditions, selection of raw materials, and cost-effectiveness analysis. To ensure feasibility, stability, and reproducibility assessments of the synthesis process will be essential to guarantee consistent quality, functionality, and safety of the NPs during large-scale production. Second, regarding long-term safety, although CHG@IL4I1-MNPs have demonstrated efficacy and safety in small animal models, potential toxicities and immune responses associated with prolonged use remain a concern. Future research will focus on long-term toxicological and immunological evaluations, assessing nanoparticle accumulation, tissue distribution, and immune responses over extended periods. Additionally, comprehensive studies involving varied doses, administration routes, and treatment cycles will be required to thoroughly evaluate the clinical safety profile of these NPs.
This study has several limitations. First, it was conducted in mouse models, and the efficacy and safety of CHG@IL4I1-MNPs in humans have not yet been validated in clinical trials. This limits the reliability and generalizability of our conclusions. Due to current constraints in time and funding, we did not conduct independent knockdown or overexpression experiments for FGR. Future research will aim to further investigate the role of FGR and its downstream regulatory mechanisms in this process to provide more robust support for our conclusions.
In conclusion, this study suggests that CHG@IL4I1-MNPs, as a novel strategy for targeted delivery of IL4I1 to promote MΦ M2 polarization, holds significant potential value. Despite the promising outlook of the research results, limitations need to be addressed. Future research will focus more on the feasibility of clinical application, long-term treatment outcome evaluations, and risk assessments of side effects, aiming to better serve the health and quality of life of patients and providing further support and insights for the continued development of IDD treatments.
Conclusion
Based on the above results, we can preliminarily conclude the following: we have designed a CHG that loads MΦ cell membrane biomimetic NPs containing IL4I1 (CHG@IL4I1-MNPs), which effectively targets MΦ to release IL4I1, inhibiting M1 polarization and promoting M2 polarization. This process enhances the ECM synthesis/degradation balance of NPCs, ultimately addressing IDD (Figure S16). This study elucidates the therapeutic mechanism of CHG@IL4I1-MNPs in treating IDD, offering new insights into the pathology of IDD.
However, this study has certain limitations. Firstly, it was conducted in a mouse model without confirmation of the efficacy and safety of CHG@IL4I1-MNPs hydrogel in human clinical studies, which affects the reliability and scientific nature of our conclusions. Therefore, further exploration of its mechanism of action and additional animal model studies will provide a more in-depth theoretical basis for optimizing this technology and its clinical application.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Quality Control, Filtering, and PCA of scRNA-seq Data. Note (A) Violin plots showing the number of genes (nFeature_RNA), counts of mRNA molecules (nCount_RNA), and percentage of mitochondrial genes (percent. mt) per cell in scRNA-seq data from NP samples of the MIDD group (n = 4) and SIDD group (n = 3); (B) Scatter plots showing the correlation between post-filtered data, nCount_RNA and percent. Mt, and between nCount_RNA and nFeature_RNA; (C) Variance analysis to filter highly variable genes in samples (red dots represent highly variable genes, black dots represent invariant genes); (D) Cell cycle status of each cell in the scRNA-seq data, where S.Score represents S phase and G2M.The score represents the G2M phase; (E) Distribution of standard deviation of PCs, with important PCs having larger standard deviations; (F) p-values of the top 50 PCs obtained from PCA analysis; (G) Feature genes in the top 2 PCs from PCA analysis; (H) Heatmap of top 15 genes expressed in the first 6 PCs in PCA, where yellow indicates upregulation and purple indicates downregulation; (I) Distribution of cells in PC 1 and PC 2 before batch correction, with each point representing a cell and different colors indicating different samples; (J) Batch correction process diagram of Harmony, with the x-axis representing the number of iterative interactions; (K) Distribution of cells in PC 1 and PC 2 after Harmony batch correction, with each point representing a cell and different colors indicating different samples.
Supplementary Material 3: Expression of MΦ and NPCs Marker Genes. Note (A) FeaturePlot graph of MΦ marker gene expression in various clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3), where darker purple indicates higher average expression levels; (B) Violin plot of MΦ marker gene expression in different clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3); (C) FeaturePlot graph of NPCs marker gene expression in different clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3), with darker purple representing higher average expression levels; (D) Violin plot of NPCs marker gene expression in various clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3).
Supplementary Material 4: Integration, Differential Analysis, and Enrichment Analysis of GEO Transcriptome Dataset. Note(A) PCA distribution of the GEO merged dataset before batch correction, with sample colors representing different datasets; (B) PCA distribution of the GEO merged dataset after batch correction, with sample colors representing different datasets; (C) Volcano plot illustrating differential analysis of NP tissue samples between Normal group (n = 6) and IDD group (n = 6) in the merged dataset; (D) Heatmap of M-DEGs between Normal group (n = 6) and IDD group (n = 6) in the merged dataset; (E) Circos plots of GO enrichment analysis (BP, CC, MF) for M-DEGs; (F) Bubble chart of KEGG enrichment analysis for M-DEGs.
Supplementary Material 5: Establishment and Characterization Assessment of Mouse IDD Models with Varying Degeneration Levels. Note (A) Schematic diagram of IDD mouse model construction; (B) Micro-CT 2D and 3D Datas of the tail IVD in each group of mice (n = 6), along with the DHI%; (C) Representative Datas of H&E staining and Safranin-O/Fast green staining of IVD, AF, and NP tissues of mice (n = 6) in each group; (D) Histological scoring of disc changes in the tail IVD of mice in each group (n = 6); (E) Representative Datas of immunohistochemical staining for Collagen II, Aggrecan, ADAMTS5, and MMP13 in the tail IVD of mice in each group (n = 6), along with quantification of the percentage of positively stained area; * P < 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Material 6: Expression of IL4I1 and MΦ Polarization in Mouse IDD Models with Varying Degeneration Levels. Note (A) WB analysis and quantification of IL4I1 protein expression in the tail IVD of mice in each group (n = 6); (B) RT-qPCR assessment of IL4I1 mRNA expression in the tail IVD of mice in each group (n = 6); (C) Flow cytometry gating strategy for sorting MΦs (single cells were identified using side scatter area (SSA) and height (SSH), and dead cells stained positive for 7-AAD reagent (CST, 72782 S) were excluded. CD45-positive cells were selected, followed by CD11b and F4/80-positive cells. Cells identified as CD45+CD11b+F4/80+ were classified as MΦs); (D) RT-qPCR evaluation of the expression of M1 and M2 MΦ marker genes in MΦs from the tail IVD of mice in each group (n = 6); (E) Percentage of M1 MΦs in MΦs from the tail IVD of mice in each group (n = 6); (F) Percentage of M2 MΦs in MΦs from the tail IVD of mice in each group (n = 6); (G) ELISA measurement of IL-1β, TNF-α, TGF-β, and IL-10 levels in the tail IVD of mice in each group (n = 6); * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, **** indicates P < 0.0001, with all experiments repeated three times.
Supplementary Material 7: Induction of M1 and M2 Polarization in MΦs and Detection of IL4I1 Expression. Note (A) Schematic illustration of the induction of M1 or M2 polarization in THP-1-derived MΦs; (B) Assessment of the expression of M1 and M2 MΦ marker genes in each group of MΦs using RT-qPCR; (C) Representative immunofluorescence Datas of MΦs in each group (scale bar = 25 μm) showing the co-localization fluorescence intensity of CD80 and CD86, CD163 and CD206; (D) Measurement of IL-1β, TNF-α, TGF-β, and IL-10 levels in the cell culture supernatant of MΦs in each group; (E) WB analysis and quantification of IL4I1 protein expression in each group of MΦs; (F) Assessment of IL4I1 mRNA expression in each group of MΦs using RT-qPCR; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, with all experiments repeated three times.
Supplementary Material 8: In Vivo Biodistribution, MΦ Uptake, and Pharmacokinetic Evaluation in IDD Mice. Note (A) Schematic illustration of intravenous injection of DiR-NPs or DiR-MNPs in Model mice; (B) Representative in vivo fluorescence imaging of IDD model mice from different treatment groups (n = 3) at 4 h and 24 h post tail vein injection, with fluorescence intensity color-coded from weak (blue) to strong (red); (C) Ex vivo fluorescence imaging of main organs and tail IVD of mice from each group (n = 3) at 4 and 24 h post tail vein injection; (D) Percentage of DiR-positive cells in MΦs in the tail IVD of mice from each group (n = 3); (E) Ex vivo fluorescence imaging of blood samples from each group of mice (n = 3) at 4 and 24 h post tail vein injection; (F) In vivo circulation kinetics of DiR-NPs and DiR-MNPs (the fluorescence intensity in serum at 0 h was set as 100%, and the ratio of fluorescence values detected at other time points compared to the 0 h value was considered as the fluorescence intensity at that time point); ** P < 0.01, *** P < 0.001.
Supplementary Material 9: Assessment of In Vivo Therapeutic Effects of IL4I1-MNPs in IDD Mice.. Note (A) Schematic representation of in vivo administration of IL4I1, IL4I1-NPs, and IL4I1-MNPs; (B) Micro-CT 2D and 3D Datas of the tail IVD with the DHI percentage for each group of mice (n = 6); (C) Representative Datas of H&E and Safranin-O/Fast green staining of IVD, AF, and NP tissue in each group of mice (n = 6); (D) Histological scores of changes in the tail IVD of mice in each group (n = 6); (E) Representative Datas of immunohistochemistry for Collagen II, Aggrecan, ADAMTS5, and MMP13 in the tail IVD of mice in each group (n = 6) with quantification of the percentage of positively stained areas; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Supplementary Material 10: Safety Evaluation of IL4I1, IL4I1-NPs, and IL4I1-MNPs In Vivo. Note (A) Blood cell count in each group of mice (n = 6); (B) Blood biochemical analysis in each group of mice (n = 6), including albumin (ALB), total bilirubin (TBIL), alanine transaminase (ALT), aspartate transaminase (AST), creatinine (CRE), urea (UREA), and uric acid (UA); (C) H&E staining of major organs in mice from each group (n = 6) under different treatments, scale bar = 100 μm.
Supplementary Material 11: Impact of FGR-Mediated Regulation of M1 MΦ Polarization by IL4I1-MNPs on NPC Phenotype. Note (A) Schematic representation of co-culture of MΦ and NPCs; (B) Cell proliferation assessed by CCK8 in each group of NPCs; (C) Representative Datas of EdU staining and quantitative measure of EdU-positive cells in each group of NPCs (scale bar = 50 μm), where EdU is shown in red and DAPI (nucleus) in blue; (D) Quantification of migrated cells in Transwell experiments of NPC from each group, shown with representative Datas (scale bar = 50 μm); (E) Flow cytometry evaluation of cell apoptosis (Annexin V+/PI−) in NPCs from each group; (F) Protein expression and quantification of Collagen II, Aggrecan, ADAMTS5, and MMP13 in NPCs from each group analyzed by WB; (G) Immunofluorescence staining of Collagen II and MMP13 in NPCs from each group with representative Datas (scale bar = 25 μm) and fluorescence intensity quantification; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, with all experiments conducted thrice.
Supplementary Material 12: The Influence of IL4I1-MNPs-Mediated FGR Regulation on M2 MΦ Polarization Process and its Impact on NPCs Phenotype.. Note (A) Cell Proliferation of NPCs in each group was examined using the CCK-8 assay; (B) Representative Datas of EdU staining in NPCs from each group (scale bar = 50 μm) and quantitative analysis of EdU-positive cell percentage, with EDU (red) and DAPI (cell nuclei, blue); (C) Representative Datas of Transwell experiments in each group of NPCs (scale bar = 50 μm) and quantitative analysis of migrated cell numbers; (D) Cell apoptosis (Annexin V+/PI−) in NPCs from each group detected by flow cytometry; (E) Protein expression and quantification of Collagen II, Aggrecan, ADAMTS5, and MMP13 in NPCs from each group analyzed by WB; (F) Immunofluorescence staining Datas of Collagen II and MMP13 in NPCs from each group (scale bar = 25 μm) and quantification of fluorescence intensity; * P < 0.05 compared between two groups, ** P < 0.01 compared between two groups, *** P < 0.001 compared between two groups, all experiments were repeated 3 times.
Supplementary Material 13: Characteristics of CHG and CHG@IL4I1-MNPs. Note (A) Schematic representation of CHG synthesis; (B) Swelling ratio of hydrogel CHG in PBS at 37 °C; (C) Degradation rate of hydrogel CHG; (D) Schematic representation of IL4I1-MNPs loaded CHG; (E) Representative Datas captured by SEM of CHG or CHG@IL4I1-MNPs (scale bar = 50 μm); (F) Rheological analysis of CHG or CHG@IL4I1-MNPs; (G) Average storage modulus of CHG and CHG@IL4I1-MNPs; (H) Cumulative release curve of IL4I1 in IL4I1-MNPs and CHG@IL4I1-MNPs; (I) Cell viability of MΦ 24 h after different treatments examined by CCK-8 assay; n.s. Indicates P > 0.05 compared between two groups; all experiments were repeated 3 times.
Supplementary Material 14: The Impact of CHG@IL4I1-MNPs-Mediated FGR on Characterization of IDD Mice. Note (A) Schematic representation of CHG, IL4I1-MNPs, CHG@IL4I1-MNPs, and FGR-OE treatments; (B) Micro-CT two-dimensional and three-dimensional Datas of IVD in each group of mice (n = 6) and intervertebral DHI percentage; (C) Representative Datas of H&E staining and Safranin-O/Fast green staining of tail IVD, AF, and NP tissues in each group of mice (n = 6); (D) Histological scores of tail IVD changes in each group of mice (n = 6); (E) Immunohistochemical representative Datas of Collagen II, Aggrecan, ADAMTS5, and MMP13 in tail IVD of each group of mice (n = 6) (scale bar = 200 μm) and quantitative analysis of positive staining area percentage; n.s. indicates P > 0.05 compared between two groups, * P < 0.05 compared between two groups, ** P < 0.01 compared between two groups, *** P < 0.001 compared between two groups, **** P < 0.0001 compared between two groups.
Supplementary Material 15: In Vivo Safety Assessment of CHG and CHG@IL4I1-MNPs. Note (A) Blood cell counts in each group of mice (n = 6); (B) Blood biochemistry analysis in each group of mice (n = 6), including ALB, TBIL, ALT, AST, CRE, UREA, UA; (C) H&E staining of major organs in each group of mice (n = 6) under different treatments, scale bar = 100 μm.
Supplementary Material 16: Schematic Representation of the Molecular Mechanism of CHG@IL4I1-MNPs in Regulating MΦ Polarization for the Treatment of IDD
Acknowledgements
Not applicable.
Author contributions
Q.L. and J.L. conceived and designed the study. J.L., G.J., S.C., and H.W. performed the experiments. G.J., S.C., and H.W. analyzed the data. J.L. and G.J. wrote the manuscript. Q.L. supervised the study. All authors reviewed and approved the final version of the manuscript.
Funding
This study was supported by Science and Technology Plan Joint Plan (Natural Science Foundation Project) of Liaoning Province (2024-MSLH-445) and Special Project of Career Development Support Program for Young and Middle-aged Teachers of Shenyang Pharmaceutical University (ZQN2018024).
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All animal care procedures and surgical operations followed the guidelines outlined in the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health. Ethical approval for all animal experiments in this study was obtained from the ethics review committee of Shengjing Hospital of China Medical University (Approval no. 2024PS1228K), and all procedures were strictly conducted following the regulations for animal welfare and experimental practices.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sakai D, Schol J, Watanabe M. Clinical Development of Regenerative Medicine Targeted for Intervertebral Disc Disease [Internet]. Medicina. MDPI AG; 2022 [cited 2025 Jan 27]. p. 267. Available from: 10.3390/medicina58020267 [DOI] [PMC free article] [PubMed]
- 2.Foreman M, Patel A, Nguyen A, Foster D, Orriols A, Lucke-Wold B. Management Considerations for Total Intervertebral Disc Replacement [Internet]. World Neurosurgery., Elsevier BV. 2024 [cited 2025 Jan 27]. pp. 125–36. Available from: 10.1016/j.wneu.2023.09.100 [DOI] [PubMed]
- 3.Cao G, Yang S, Cao J, Tan Z, Wu L, Dong F et al. The Role of Oxidative Stress in Intervertebral Disc Degeneration [Internet]. Oxidative Medicine and Cellular Longevity. Hindawi Limited; 2022 [cited 2025 Jan 27]. pp. 1–16. Available from: 10.1155/2022/2166817 [DOI] [PMC free article] [PubMed]
- 4.Liu F, Chao S, Yang L, Chen C, Huang W, Chen F et al. Molecular mechanism of mechanical pressure induced changes in the microenvironment of intervertebral disc degeneration [Internet]. Inflamm. Res. Springer Science and Business Media LLC; 2024 [cited 2025 Jan 27]. pp. 2153–64. Available from: 10.1007/s00011-024-01954-w [DOI] [PubMed]
- 5.Donnally CJIII, Patel PD, Canseco JA, Divi SN, Goz V, Sherman MB, Current incidence of adjacent segment pathology following lumbar fusion versus motion-preserving procedures: a systematic review and meta-analysis of recent projections [Internet]. The Spine Journal., Elsevier BV et al. 2020 [cited 2025 Jan 27]. pp. 1554–65. Available from: 10.1016/j.spinee.2020.05.100 [DOI] [PubMed]
- 6.Chen S, Li Y, Liu H, Wu T, Huang K, Yao M et al. Overloaded Vertebral Body Following Consecutive Three-Level Hybrid Surgery Comparing with Anterior Cervical Discectomy and Fusion [Internet]. Orthopaedic Surgery. Wiley; 2024 [cited 2025 Jan 27]. pp. 3036–46. Available from: 10.1111/os.14242 [DOI] [PMC free article] [PubMed]
- 7.Palmisciano P, Balasubramanian K, Scalia G, Sagoo NS, Haider AS, Bin Alamer O et al. Posterior epidural intervertebral disc migration and sequestration: A systematic review [Internet]. Journal of Clinical Neuroscience. Elsevier BV; 2022 [cited 2025 Jan 27]. pp. 115–26. Available from: 10.1016/j.jocn.2022.01.039 [DOI] [PubMed]
- 8.Hu C, Lin B, Li Z, Chen X, Gao K. Spontaneous regression of a large sequestered lumbar disc herniation: a case report and literature review [Internet]. J Int Med Res. SAGE Publications; 2021 [cited 2025 Jan 27]. Available from: 10.1177/03000605211058987 [DOI] [PMC free article] [PubMed]
- 9.Koroth J, Buko EO, Abbott R, Johnson CP, Ogle BM, Stone LS et al. Macrophages and Intervertebral Disc Degeneration [Internet]. IJMS. MDPI AG; 2023 [cited 2025 Jan 27]. p. 1367. Available from: 10.3390/ijms24021367 [DOI] [PMC free article] [PubMed]
- 10.Li X-C, Luo S-J, Fan W, Zhou T-L, Tan D-Q, Tan R-X et al. Macrophage polarization regulates intervertebral disc degeneration by modulating cell proliferation, inflammation mediator secretion, and extracellular matrix metabolism [Internet]. Front. Immunol. Frontiers Media SA; 2022 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2022.922173 [DOI] [PMC free article] [PubMed]
- 11.Teng Y, Huang Y, Yu H, Wu C, Yan Q, Wang Y, Nimbolide targeting SIRT1 mitigates intervertebral disc degeneration by reprogramming cholesterol metabolism and inhibiting inflammatory signaling [Internet]. Acta Pharmaceutica Sinica B., Elsevier BV et al. 2023 [cited 2025 Jan 27]. pp. 2269–80. Available from: 10.1016/j.apsb.2023.02.018 [DOI] [PMC free article] [PubMed]
- 12.Guo S, Yan M, Li X, Zhang S, Liu Z, Li K et al. Single-cell RNA-seq analysis reveals that immune cells induce human nucleus pulposus ossification and degeneration [Internet]. Front. Immunol. Frontiers Media SA; 2023 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2023.1224627 [DOI] [PMC free article] [PubMed]
- 13.Djuric N, Lafeber GCM, Vleggeert-Lankamp CLA. The contradictory effect of macrophage-related cytokine expression in lumbar disc herniations: a systematic review [Internet]. Eur Spine J. Springer Science and Business Media LLC; 2019 [cited 2025 Jan 27]. pp. 1649–59. Available from: 10.1007/s00586-019-06220-w [DOI] [PubMed]
- 14.Liang H, Yang X, Liu C, Sun Z, Wang X. Effect of NF-kB signaling pathway on the expression of MIF, TNF-α, IL-6 in the regulation of intervertebral disc degeneration. J Musculoskelet Neuronal Interact Greece. 2018;18:551–6. [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng P, Li S, Chen H. Macrophages in Lung Injury, Repair, and Fibrosis [Internet]. Cells. MDPI AG; 2021 [cited 2025 Jan 27]. p. 436. Available from: 10.3390/cells10020436 [DOI] [PMC free article] [PubMed]
- 16.Wu H, Zheng J, Xu S, Fang Y, Wu Y, Zeng J et al. Mer regulates microglial/macrophage M1/M2 polarization and alleviates neuroinflammation following traumatic brain injury [Internet]. J Neuroinflammation. Springer Science and Business Media LLC; 2021 [cited 2025 Jan 27]. Available from: 10.1186/s12974-020-02041-7
- 17.Chen M, Menon MC, Wang W, Fu J, Yi Z, Sun Z et al. HCK induces macrophage activation to promote renal inflammation and fibrosis via suppression of autophagy [Internet]. Nat Commun. Springer Science and Business Media LLC; 2023 [cited 2025 Jan 27]. Available from: 10.1038/s41467-023-40086-3 [DOI] [PMC free article] [PubMed]
- 18.Ni X, Wu W, Sun X, Ma J, Yu Z, He X et al. Interrogating glioma-M2 macrophage interactions identifies Gal-9/Tim-3 as a viable target against PTEN -null glioblastoma [Internet]. Sci. Adv. American Association for the Advancement of Science (AAAS); 2022 [cited 2025 Jan 27]. Available from: 10.1126/sciadv.abl5165 [DOI] [PMC free article] [PubMed]
- 19.Wang Y, Groeger S, Yong J, Ruf S. Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation [Internet]. IJMS. MDPI AG; 2023 [cited 2025 Jan 27]. p. 3117. Available from: 10.3390/ijms24043117 [DOI] [PMC free article] [PubMed]
- 20.Cutolo M, Campitiello R, Gotelli E, Soldano S. The Role of M1/M2 Macrophage Polarization in Rheumatoid Arthritis Synovitis [Internet]. Front. Immunol. Frontiers Media SA; 2022 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2022.867260 [DOI] [PMC free article] [PubMed]
- 21.Zhuang H, Lv Q, Zhong C, Cui Y, He L, Zhang C et al. Tiliroside Ameliorates Ulcerative Colitis by Restoring the M1/M2 Macrophage Balance via the HIF-1α/glycolysis Pathway [Internet]. Front. Immunol. Frontiers Media SA; 2021 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2021.649463 [DOI] [PMC free article] [PubMed]
- 22.Wang Y-N, Wang Y-Y, Wang J, Bai W-J, Miao N-J, Wang J. Vinblastine resets tumor-associated macrophages toward M1 phenotype and promotes antitumor immune response [Internet]. J Immunother Cancer. BMJ; 2023 [cited 2025 Jan 27]. p. e007253. Available from: 10.1136/jitc-2023-007253 [DOI] [PMC free article] [PubMed]
- 23.Stone TW, Williams RO. Modulation of T cells by tryptophan metabolites in the kynurenine pathway [Internet]. Trends in Pharmacological Sciences. Elsevier BV; 2023 [cited 2025 Jan 27]. pp. 442–56. Available from: 10.1016/j.tips.2023.04.006 [DOI] [PubMed]
- 24.Zhang G, Xie Z, Jiang J, Zhao Y, Lei K, Lin Z et al. Mechanical confinement promotes heat resistance of hepatocellular carcinoma via SP1/IL4I1/AHR axis [Internet]. Cell Reports Medicine. Elsevier BV; 2023 [cited 2025 Jan 27]. p. 101128. Available from: 10.1016/j.xcrm.2023.101128 [DOI] [PMC free article] [PubMed]
- 25.Lin H, Fu L, Li P, Zhu J, Xu Q, Wang Y, Fatty acids metabolism affects the therapeutic effect of anti-PD-1/PD-L1 in tumor immune microenvironment in clear cell renal cell carcinoma [Internet]. J Transl Med. Springer Science and Business, Media LLC et al. 2023 [cited 2025 Jan 27]. Available from: 10.1186/s12967-023-04161-z [DOI] [PMC free article] [PubMed]
- 26.Ye F, Wang L, Li Y, Dong C, Zhou L, Xu J. IL4I1 in M2-like macrophage promotes glioma progression and is a promising target for immunotherapy [Internet]. Front. Immunol. Frontiers Media SA; 2024 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2023.1338244 [DOI] [PMC free article] [PubMed]
- 27.Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease [Internet]. Immunity. Elsevier BV; 2021 [cited 2025 Jan 27]. pp. 1883–1900.e5. Available from: 10.1016/j.immuni.2021.07.007 [DOI] [PubMed]
- 28.Zeitler L, Murray PJ. IL4i1 and IDO1: Oxidases that control a tryptophan metabolic nexus in cancer [Internet]. Journal of Biological Chemistry. Elsevier BV; 2023 [cited 2025 Jan 27]. p. 104827. Available from: 10.1016/j.jbc.2023.104827 [DOI] [PMC free article] [PubMed]
- 29.Qu F, Geng R, Liu Y, Zhu J. Advanced nanocarrier- and microneedle-based transdermal drug delivery strategies for skin diseases treatment [Internet]. Theranostics. Ivyspring International Publisher; 2022 [cited 2025 Jan 27]. pp. 3372–406. Available from: 10.7150/thno.69999 [DOI] [PMC free article] [PubMed]
- 30.Zhao X, Zhao R, Nie G. Nanocarriers based on bacterial membrane materials for cancer vaccine delivery [Internet]. Nat Protoc. Springer Science and Business, Media LLC. 2022 [cited 2025 Jan 27]. pp. 2240–74. Available from: 10.1038/s41596-022-00713-7 [DOI] [PubMed]
- 31.Luo W, Bai L, Zhang J, Li Z, Liu Y, Tang X et al. Polysaccharides-based nanocarriers enhance the anti-inflammatory effect of curcumin [Internet]. Carbohydrate Polymers. Elsevier BV; 2023 [cited 2025 Jan 27]. p. 120718. Available from: 10.1016/j.carbpol.2023.120718 [DOI] [PubMed]
- 32.Li Z, Ye D, Dai L, Xu Y, Wu H, Luo W et al. Single-Cell RNA Sequencing Reveals the Difference in Human Normal and Degenerative Nucleus Pulposus Tissue Profiles and Cellular Interactions [Internet]. Front. Cell Dev. Biol. Frontiers Media SA; 2022 [cited 2025 Jan 27]. Available from: 10.3389/fcell.2022.910626 [DOI] [PMC free article] [PubMed]
- 33.Tang X, Lin S, Luo H, Wang L, Zhong J, Xiong J et al. ATG9A as a potential diagnostic marker of intervertebral disc degeneration: Inferences from experiments and bioinformatics analysis incorporating sc-RNA-seq data [Internet]. Gene. Elsevier BV; 2024 [cited 2025 Jan 27]. p. 148084. Available from: 10.1016/j.gene.2023.148084 [DOI] [PubMed]
- 34.Li Z, Sun Y, He M, Liu J. Differentially-expressed mRNAs, microRNAs and long noncoding RNAs in intervertebral disc degeneration identified by RNA-sequencing [Internet]. Bioengineered. Informa UK Limited; 2021 [cited 2025 Jan 27]. pp. 1026–39. Available from: 10.1080/21655979.2021.1899533 [DOI] [PMC free article] [PubMed]
- 35.Zhao F, Guo Z, Hou F, Fan W, Wu B, Qian Z, Magnoflorine Alleviates. M1 Polarized Macrophage-Induced Intervertebral Disc Degeneration Through Repressing the HMGB1/Myd88/NF-κB Pathway and NLRP3 Inflammasome [Internet]. Front. Pharmacol. Frontiers Media SA; 2021 [cited 2025 Jan 27]. Available from: 10.3389/fphar.2021.701087 [DOI] [PMC free article] [PubMed]
- 36.Gao X-W, Hu H-L, Xie M-H, Tang C-X, Ou J, Lu Z-H. CX3CL1/CX3CR1 axis alleviates inflammation and apoptosis in human nucleus pulpous cells via M2 macrophage polarization [Internet]. Exp Ther Med. Spandidos Publications; 2023 [cited 2025 Jan 27]. Available from: 10.3892/etm.2023.12058 [DOI] [PMC free article] [PubMed]
- 37.Tan Q, He L, Meng X, Wang W, Pan H, Yin W et al. Macrophage biomimetic nanocarriers for anti-inflammation and targeted antiviral treatment in COVID-19 [Internet]. J Nanobiotechnol. Springer Science and Business Media LLC; 2021 [cited 2025 Jan 27]. Available from: 10.1186/s12951-021-00926-0 [DOI] [PMC free article] [PubMed]
- 38.Bhattacharyya S, Ghosh SS, Transmembrane. TNFα-Expressed Macrophage Membrane-Coated Chitosan Nanoparticles as Cancer Therapeutics [Internet]. ACS Omega. American Chemical Society (ACS); 2020 [cited 2025 Jan 27]. pp. 1572–80. Available from: 10.1021/acsomega.9b03531 [DOI] [PMC free article] [PubMed]
- 39.Li J-D, Yin J. Interleukin-10-alveolar macrophage cell membrane-coated nanoparticles alleviate airway inflammation and regulate Th17/regulatory T cell balance in a mouse model [Internet]. Front. Immunol. Frontiers Media SA; 2023 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2023.1186393 [DOI] [PMC free article] [PubMed]
- 40.Duan Y, Zhou J, Zhou Z, Zhang E, Yu Y, Krishnan N et al. Extending the In Vivo Residence Time of Macrophage Membrane-Coated Nanoparticles through Genetic Modification [Internet]. Small. Wiley; 2023 [cited 2025 Jan 27]. Available from: 10.1002/smll.202305551 [DOI] [PubMed]
- 41.Wang H, Yao Q, Zhu W, Yang Y, Gao C, Han C, Biomimetic Antidote Nanoparticles: a Novel Strategy for Chronic Heavy Metal Poisoning [Internet]. AAPS PharmSciTech. Springer Science and Business, Media LLC et al. 2022 [cited 2025 Jan 27]. Available from: 10.1208/s12249-022-02466-8
- 42.Cao X, Tan T, Zhu D, Yu H, Liu Y, Zhou H, et al. Paclitaxel-Loaded Macrophage Membrane Camouflaged Albumin Nanoparticles for Targeted Cancer Therapy [Internet].IJN. Informa UK Limited; 2020 [cited 2025 Jan 27]. pp. 1915–28. Available from: 10.2147/ijn.s244849. [DOI] [PMC free article] [PubMed]
- 43.Liu X, Liu F. Bimetallic (AuAg, AuPd and AgPd) nanoparticles supported on cellulose-based hydrogel for reusable catalysis [Internet]. Carbohydrate Polymers. Elsevier BV; 2023 [cited 2025 Jan 27]. p. 120726. Available from: 10.1016/j.carbpol.2023.120726 [DOI] [PubMed]
- 44.Zeng R, Lv B, Lin Z, Chu X, Xiong Y, Knoedler S et al. Neddylation suppression by a macrophage membrane-coated nanoparticle promotes dual immunomodulatory repair of diabetic wounds [Internet]. Bioactive Materials. Elsevier BV; 2024 [cited 2025 Jan 27]. pp. 366–80. Available from: 10.1016/j.bioactmat.2023.12.025 [DOI] [PMC free article] [PubMed]
- 45.Cheng H, Guo Q, Zhao H, Liu K, Kang H, Gao F et al. An Injectable Hydrogel Scaffold Loaded with Dual-Drug/Sustained-Release PLGA Microspheres for the Regulation of Macrophage Polarization in the Treatment of Intervertebral Disc Degeneration [Internet]. IJMS. MDPI AG; 2022 [cited 2025 Jan 27]. p. 390. Available from: 10.3390/ijms24010390 [DOI] [PMC free article] [PubMed]
- 46.Gong Y, Wang Y, Qu Q, Hou Z, Guo T, Xu Y et al. Nanoparticle encapsulated core-shell hydrogel for on-site BMSCs delivery protects from iron overload and enhances functional recovery [Internet]. Journal of Controlled Release. Elsevier BV; 2020 [cited 2025 Jan 27]. pp. 381–91. Available from: 10.1016/j.jconrel.2020.01.029 [DOI] [PubMed]
- 47.Jiang W, Ma C, Bai J, Du X, RETRACTED BV. 2022 [cited 2025 Jan 27]. p. 102432. Available from: 10.1016/j.redox.2022.102432
- 48.Psachoulia K, Chamberlain KA, Heo D, Davis SE, Paskus JD, Nanescu SE et al. IL4I1 augments CNS remyelination and axonal protection by modulating T cell driven inflammation [Internet]. Brain. Oxford University Press (OUP); 2016 [cited 2025 Jan 27]. pp. 3121–36. Available from: 10.1093/brain/aww254 [DOI] [PMC free article] [PubMed]
- 49.Namangkalakul W, Nagai S, Jin C, Nakahama K, Yoshimoto Y, Ueha S et al. Augmented effect of fibroblast growth factor 18 in bone morphogenetic protein 2-induced calvarial bone healing by activation of CCL2/CCR2 axis on M2 macrophage polarization [Internet]. J Tissue Eng. SAGE Publications; 2023 [cited 2025 Jan 27]. Available from: 10.1177/20417314231187960 [DOI] [PMC free article] [PubMed]
- 50.Li W, Zhou P, Yan B, Qi M, Chen Y, Shang L et al. Disc regeneration by injectable fucoidan-methacrylated dextran hydrogels through mechanical transduction and macrophage immunomodulation [Internet]. J Tissue Eng. SAGE Publications; 2023 [cited 2025 Jan 27]. Available from: 10.1177/20417314231180050 [DOI] [PMC free article] [PubMed]
- 51.Li L, Wei K, Ding Y, Ahati P, Xu H, Fang H et al. M2a Macrophage-Secreted CHI3L1 Promotes Extracellular Matrix Metabolic Imbalances via Activation of IL-13Rα2/MAPK Pathway in Rat Intervertebral Disc Degeneration [Internet]. Front. Immunol. Frontiers Media SA; 2021 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2021.666361 [DOI] [PMC free article] [PubMed]
- 52.Luo J, Jin G, Cui S, Wang H, Liu Q. Regulatory mechanism of FCGR2A in macrophage polarization and its effects on intervertebral disc degeneration [Internet]. The Journal of Physiology. Wiley; 2024 [cited 2025 Jan 27]. pp. 1341–69. Available from: 10.1113/jp285871 [DOI] [PubMed]
- 53.Ji M, Jiang H, Zhang X, Shi P, Li C, Wu H, Preclinical development of a microRNA-based therapy for intervertebral disc degeneration [Internet]. Nat Commun. Springer Science and Business, Media LLC et al. 2018 [cited 2025 Jan 27]. Available from: 10.1038/s41467-018-07360-1 [DOI] [PMC free article] [PubMed]
- 54.Xia X, Guo J, Lu F, Jiang J. SIRT1 Plays a Protective Role in Intervertebral Disc Degeneration in a Puncture-induced Rodent Model [Internet]. Spine. Ovid Technologies (Wolters Kluwer Health); 2015 [cited 2025 Jan 27]. pp. E515–24. Available from: 10.1097/brs.0000000000000817 [DOI] [PubMed]
- 55.Zhang S, Wang P, Hu B, Liu W, Lv X, Chen S et al. HSP90 Inhibitor 17-AAG Attenuates Nucleus Pulposus Inflammation and Catabolism Induced by M1-Polarized Macrophages [Internet]. Front. Cell Dev. Biol. Frontiers Media SA; 2022 [cited 2025 Jan 27]. Available from: 10.3389/fcell.2021.796974 [DOI] [PMC free article] [PubMed]
- 56.Chen C, Song M, Du Y, Yu Y, Li C, Han Y et al. Tumor-Associated-Macrophage-Membrane-Coated Nanoparticles for Improved Photodynamic Immunotherapy [Internet]. Nano Lett. American Chemical Society (ACS); 2021 [cited 2025 Jan 27]. pp. 5522–31. Available from: 10.1021/acs.nanolett.1c00818 [DOI] [PubMed]
- 57.Wang J, Wu R, Liu Z, Qi L, Xu H, Yang H et al. Core–Shell Structured Nanozyme with PDA-Mediated Enhanced Antioxidant Efficiency to Treat Early Intervertebral Disc Degeneration [Internet]. ACS Appl. Mater. Interfaces. American Chemical Society (ACS); 2024 [cited 2025 Jan 27]. pp. 5103–19. Available from: 10.1021/acsami.3c15938 [DOI] [PubMed]
- 58.Chen Z, Liao Z, Liu M, Lin F, Chen S, Wang G et al. Nucleus Pulposus-Targeting Nanocarriers Facilitate Mirna‐Based Therapeutics for Intervertebral Disc Degeneration [Internet]. Adv Healthcare Materials. Wiley; 2023 [cited 2025 Jan 27]. Available from: 10.1002/adhm.202301337 [DOI] [PubMed]
- 59.Hou Y, Shi J, Guo Y, Shi G. DNMT1 regulates polarization of macrophage-induced intervertebral disc degeneration by modulating SIRT6 expression and promoting pyroptosis in vivo [Internet]. Aging. Impact Journals, LLC; 2023 [cited 2025 Jan 27]. pp. 4288–303. Available from: 10.18632/aging.204729 [DOI] [PMC free article] [PubMed]
- 60.Kang H, Dong Y, Peng R, Liu H, Guo Q, Song K et al. Inhibition of IRE1 suppresses the catabolic effect of IL-1β on nucleus pulposus cell and prevents intervertebral disc degeneration in vivo [Internet]. Biochemical Pharmacology. Elsevier BV; 2022 [cited 2025 Jan 27]. p. 114932. Available from: 10.1016/j.bcp.2022.114932 [DOI] [PubMed]
- 61.Chen F, Lei L, Chen S, Zhao Z, Huang Y, Jiang G et al. Serglycin secreted by late-stage nucleus pulposus cells is a biomarker of intervertebral disc degeneration [Internet]. Nat Commun. Springer Science and Business Media LLC; 2024 [cited 2025 Jan 27]. Available from: 10.1038/s41467-023-44313-9 [DOI] [PMC free article] [PubMed]
- 62.Francisco V, Pino J, González-Gay MÁ, Lago F, Karppinen J, Tervonen O, A new immunometabolic perspective of intervertebral disc degeneration [Internet]. Nat Rev Rheumatol. Springer Science and Business, Media LLC et al. 2021 [cited 2025 Jan 27]. pp. 47–60. Available from: 10.1038/s41584-021-00713-z [DOI] [PubMed]
- 63.Hu C, Liu Y, Cao W, Li N, Gao S, Wang Z et al. Efficacy and Mechanism of a Biomimetic Nanosystem Carrying Doxorubicin and an IDO Inhibitor for Treatment of Advanced Triple-Negative Breast Cancer [Internet]. IJN. Informa UK Limited; 2024 [cited 2025 Jan 27]. pp. 507–26. Available from: 10.2147/ijn.s440332 [DOI] [PMC free article] [PubMed]
- 64.Amoozgar Z, Kloepper J, Ren J, Tay RE, Kazer SW, Kiner E et al. Targeting Treg cells with GITR activation alleviates resistance to immunotherapy in murine glioblastomas [Internet]. Nat Commun. Springer Science and Business Media LLC; 2021 [cited 2025 Jan 27]. Available from: 10.1038/s41467-021-22885-8 [DOI] [PMC free article] [PubMed]
- 65.Hao Y, Zhu G, Yu L, Ren Z, Zhou W, Zhang P et al. FOXO3-Activated HOTTIP Sequesters miR-615-3p away from COL2A1 to Mitigate Intervertebral Disc Degeneration [Internet]. The American Journal of Pathology. Elsevier BV; 2024 [cited 2025 Jan 27]. pp. 280–95. Available from: 10.1016/j.ajpath.2023.10.011 [DOI] [PubMed]
- 66.Li X, An Y, Wang Q, Han X. The new ceRNA crosstalk between mRNAs and miRNAs in intervertebral disc degeneration [Internet]. Front. Cell Dev. Biol. Frontiers Media SA; 2022 [cited 2025 Jan 27]. Available from: 10.3389/fcell.2022.1083983 [DOI] [PMC free article] [PubMed]
- 67.Zhang Y, Li S, Hong J, Yan J, Huang Z, Wu J, et al. Chromobox homolog 4 overexpression inhibits TNF-α-induced matrix catabolism and senescence by suppressing activation of NF-κB signaling pathway in nucleus pulposus cells [Internet]. ABBS. China Science Publishing & Media Ltd.; 2022 [cited 2025 Jan 27]. pp. 1021–9. Available from:10.3724/abbs.2022063. [DOI] [PMC free article] [PubMed]
- 68.Acín-Pérez R, Iborra S, Martí-Mateos Y, Cook ECL, Conde-Garrosa R, Petcherski A et al. Fgr kinase is required for proinflammatory macrophage activation during diet-induced obesity [Internet]. Nat Metab. Springer Science and Business Media LLC; 2020 [cited 2025 Jan 27]. pp. 974–88. Available from: 10.1038/s42255-020-00273-8 [DOI] [PMC free article] [PubMed]
- 69.Jiang Y, Wang J, Sun D, Liu Z, Qi L, Du M et al. A hydrogel reservoir as a self-contained nucleus pulposus cell delivery vehicle for immunoregulation and repair of degenerated intervertebral disc [Internet]. Acta Biomaterialia. Elsevier BV; 2023 [cited 2025 Jan 27]. pp. 303–17. Available from: 10.1016/j.actbio.2023.08.023 [DOI] [PubMed]
- 70.Meis CM, Salzman EE, Maikawa CL, Smith AAA, Mann JL, Grosskopf AK et al. Self-Assembled, Dilution-Responsive Hydrogels for Enhanced Thermal Stability of Insulin Biopharmaceuticals [Internet]. ACS Biomater. Sci. Eng. American Chemical Society (ACS); 2020 [cited 2025 Jan 27]. pp. 4221–9. Available from: 10.1021/acsbiomaterials.0c01306 [DOI] [PMC free article] [PubMed]
- 71.Song C, Xu Y, Peng Q, Chen R, Zhou D, Cheng K et al. Mitochondrial dysfunction: a new molecular mechanism of intervertebral disc degeneration [Internet]. Inflamm. Res. Springer Science and Business Media LLC; 2023 [cited 2025 Jan 27]. pp. 2249–60. Available from: 10.1007/s00011-023-01813-0 [DOI] [PubMed]
- 72.Zhao X, Ma H, Han H, Zhang L, Tian J, Lei B et al. Precision medicine strategies for spinal degenerative diseases: Injectable biomaterials with in situ repair and regeneration [Internet]. Materials Today Bio. Elsevier BV; 2022 [cited 2025 Jan 27]. p. 100336. Available from: 10.1016/j.mtbio.2022.100336 [DOI] [PMC free article] [PubMed]
- 73.Yu P, Mao F, Chen J, Ma X, Dai Y, Liu G et al. Characteristics and mechanisms of resorption in lumbar disc herniation [Internet]. Arthritis Res Ther. Springer Science and Business Media LLC; 2022 [cited 2025 Jan 27]. Available from: 10.1186/s13075-022-02894-8 [DOI] [PMC free article] [PubMed]
- 74.Wang L, He T, Liu J, Tai J, Wang B, Zhang L et al. Revealing the Immune Infiltration Landscape and Identifying Diagnostic Biomarkers for Lumbar Disc Herniation [Internet]. Front. Immunol. Frontiers Media SA; 2021 [cited 2025 Jan 27]. Available from: 10.3389/fimmu.2021.666355 [DOI] [PMC free article] [PubMed]
- 75.Clough E, Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF et al. NCBI GEO: archive for gene expression and epigenomics data sets: 23-year update [Internet]. Nucleic Acids Research. Oxford University Press (OUP); 2023 [cited 2025 Jan 27]. pp. D138–44. Available from: 10.1093/nar/gkad965 [DOI] [PMC free article] [PubMed]
- 76.Zhang Y, Li H, Chen Y, Li C, Ye H, Qiu J et al. Nordihydroguaiaretic acid suppresses ferroptosis and mitigates intervertebral disc degeneration through the NRF2/GPX4 axis [Internet]. International Immunopharmacology. Elsevier BV; 2024 [cited 2025 Jan 27]. p. 113590. Available from: 10.1016/j.intimp.2024.113590 [DOI] [PubMed]
- 77.Yang S, Zhu Y, Shi Y, Su S, Liang H, Li S et al. Screening of NSAIDs library identifies Tinoridine as a novel ferroptosis inhibitor for potential intervertebral disc degeneration therapy [Internet]. Free Radical Biology and Medicine. Elsevier BV; 2024 [cited 2025 Jan 27]. pp. 245–56. Available from: 10.1016/j.freeradbiomed.2024.05.040 [DOI] [PubMed]
- 78.Ukeba D, Ishikawa Y, Yamada K, Ohnishi T, Tachi H, Tha KK et al. Bone Marrow Aspirate Concentrate Combined with Ultra-Purified Alginate Bioresorbable Gel Enhances Intervertebral Disc Repair in a Canine Model: A Preclinical Proof-of-Concept Study [Internet]. Cells. MDPI AG; 2024 [cited 2025 Jan 27]. p. 987. Available from: 10.3390/cells13110987 [DOI] [PMC free article] [PubMed]
- 79.Sang P, Li X, Wang Z. Bone Mesenchymal Stem Cells Inhibit Oxidative Stress–Induced Pyroptosis in Annulus Fibrosus Cells to Alleviate Intervertebral Disc Degeneration Based on Matric Hydrogels [Internet]. Appl Biochem Biotechnol. Springer Science and Business Media LLC; 2024 [cited 2025 Jan 27]. pp. 8043–57. Available from: 10.1007/s12010-024-04953-z [DOI] [PubMed]
- 80.Wu R, Dong Y, Yu Y. Percutaneous Endoscopic Lumbar Discectomy Combined with Annular Suture: Technical Video [Internet]. World Neurosurgery. Elsevier BV; 2025 [cited 2025 Jan 27]. pp. 158–9. Available from: 10.1016/j.wneu.2024.10.132 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Quality Control, Filtering, and PCA of scRNA-seq Data. Note (A) Violin plots showing the number of genes (nFeature_RNA), counts of mRNA molecules (nCount_RNA), and percentage of mitochondrial genes (percent. mt) per cell in scRNA-seq data from NP samples of the MIDD group (n = 4) and SIDD group (n = 3); (B) Scatter plots showing the correlation between post-filtered data, nCount_RNA and percent. Mt, and between nCount_RNA and nFeature_RNA; (C) Variance analysis to filter highly variable genes in samples (red dots represent highly variable genes, black dots represent invariant genes); (D) Cell cycle status of each cell in the scRNA-seq data, where S.Score represents S phase and G2M.The score represents the G2M phase; (E) Distribution of standard deviation of PCs, with important PCs having larger standard deviations; (F) p-values of the top 50 PCs obtained from PCA analysis; (G) Feature genes in the top 2 PCs from PCA analysis; (H) Heatmap of top 15 genes expressed in the first 6 PCs in PCA, where yellow indicates upregulation and purple indicates downregulation; (I) Distribution of cells in PC 1 and PC 2 before batch correction, with each point representing a cell and different colors indicating different samples; (J) Batch correction process diagram of Harmony, with the x-axis representing the number of iterative interactions; (K) Distribution of cells in PC 1 and PC 2 after Harmony batch correction, with each point representing a cell and different colors indicating different samples.
Supplementary Material 3: Expression of MΦ and NPCs Marker Genes. Note (A) FeaturePlot graph of MΦ marker gene expression in various clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3), where darker purple indicates higher average expression levels; (B) Violin plot of MΦ marker gene expression in different clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3); (C) FeaturePlot graph of NPCs marker gene expression in different clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3), with darker purple representing higher average expression levels; (D) Violin plot of NPCs marker gene expression in various clusters of NP samples from the MIDD group (n = 4) and SIDD group (n = 3).
Supplementary Material 4: Integration, Differential Analysis, and Enrichment Analysis of GEO Transcriptome Dataset. Note(A) PCA distribution of the GEO merged dataset before batch correction, with sample colors representing different datasets; (B) PCA distribution of the GEO merged dataset after batch correction, with sample colors representing different datasets; (C) Volcano plot illustrating differential analysis of NP tissue samples between Normal group (n = 6) and IDD group (n = 6) in the merged dataset; (D) Heatmap of M-DEGs between Normal group (n = 6) and IDD group (n = 6) in the merged dataset; (E) Circos plots of GO enrichment analysis (BP, CC, MF) for M-DEGs; (F) Bubble chart of KEGG enrichment analysis for M-DEGs.
Supplementary Material 5: Establishment and Characterization Assessment of Mouse IDD Models with Varying Degeneration Levels. Note (A) Schematic diagram of IDD mouse model construction; (B) Micro-CT 2D and 3D Datas of the tail IVD in each group of mice (n = 6), along with the DHI%; (C) Representative Datas of H&E staining and Safranin-O/Fast green staining of IVD, AF, and NP tissues of mice (n = 6) in each group; (D) Histological scoring of disc changes in the tail IVD of mice in each group (n = 6); (E) Representative Datas of immunohistochemical staining for Collagen II, Aggrecan, ADAMTS5, and MMP13 in the tail IVD of mice in each group (n = 6), along with quantification of the percentage of positively stained area; * P < 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Material 6: Expression of IL4I1 and MΦ Polarization in Mouse IDD Models with Varying Degeneration Levels. Note (A) WB analysis and quantification of IL4I1 protein expression in the tail IVD of mice in each group (n = 6); (B) RT-qPCR assessment of IL4I1 mRNA expression in the tail IVD of mice in each group (n = 6); (C) Flow cytometry gating strategy for sorting MΦs (single cells were identified using side scatter area (SSA) and height (SSH), and dead cells stained positive for 7-AAD reagent (CST, 72782 S) were excluded. CD45-positive cells were selected, followed by CD11b and F4/80-positive cells. Cells identified as CD45+CD11b+F4/80+ were classified as MΦs); (D) RT-qPCR evaluation of the expression of M1 and M2 MΦ marker genes in MΦs from the tail IVD of mice in each group (n = 6); (E) Percentage of M1 MΦs in MΦs from the tail IVD of mice in each group (n = 6); (F) Percentage of M2 MΦs in MΦs from the tail IVD of mice in each group (n = 6); (G) ELISA measurement of IL-1β, TNF-α, TGF-β, and IL-10 levels in the tail IVD of mice in each group (n = 6); * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, **** indicates P < 0.0001, with all experiments repeated three times.
Supplementary Material 7: Induction of M1 and M2 Polarization in MΦs and Detection of IL4I1 Expression. Note (A) Schematic illustration of the induction of M1 or M2 polarization in THP-1-derived MΦs; (B) Assessment of the expression of M1 and M2 MΦ marker genes in each group of MΦs using RT-qPCR; (C) Representative immunofluorescence Datas of MΦs in each group (scale bar = 25 μm) showing the co-localization fluorescence intensity of CD80 and CD86, CD163 and CD206; (D) Measurement of IL-1β, TNF-α, TGF-β, and IL-10 levels in the cell culture supernatant of MΦs in each group; (E) WB analysis and quantification of IL4I1 protein expression in each group of MΦs; (F) Assessment of IL4I1 mRNA expression in each group of MΦs using RT-qPCR; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, with all experiments repeated three times.
Supplementary Material 8: In Vivo Biodistribution, MΦ Uptake, and Pharmacokinetic Evaluation in IDD Mice. Note (A) Schematic illustration of intravenous injection of DiR-NPs or DiR-MNPs in Model mice; (B) Representative in vivo fluorescence imaging of IDD model mice from different treatment groups (n = 3) at 4 h and 24 h post tail vein injection, with fluorescence intensity color-coded from weak (blue) to strong (red); (C) Ex vivo fluorescence imaging of main organs and tail IVD of mice from each group (n = 3) at 4 and 24 h post tail vein injection; (D) Percentage of DiR-positive cells in MΦs in the tail IVD of mice from each group (n = 3); (E) Ex vivo fluorescence imaging of blood samples from each group of mice (n = 3) at 4 and 24 h post tail vein injection; (F) In vivo circulation kinetics of DiR-NPs and DiR-MNPs (the fluorescence intensity in serum at 0 h was set as 100%, and the ratio of fluorescence values detected at other time points compared to the 0 h value was considered as the fluorescence intensity at that time point); ** P < 0.01, *** P < 0.001.
Supplementary Material 9: Assessment of In Vivo Therapeutic Effects of IL4I1-MNPs in IDD Mice.. Note (A) Schematic representation of in vivo administration of IL4I1, IL4I1-NPs, and IL4I1-MNPs; (B) Micro-CT 2D and 3D Datas of the tail IVD with the DHI percentage for each group of mice (n = 6); (C) Representative Datas of H&E and Safranin-O/Fast green staining of IVD, AF, and NP tissue in each group of mice (n = 6); (D) Histological scores of changes in the tail IVD of mice in each group (n = 6); (E) Representative Datas of immunohistochemistry for Collagen II, Aggrecan, ADAMTS5, and MMP13 in the tail IVD of mice in each group (n = 6) with quantification of the percentage of positively stained areas; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Supplementary Material 10: Safety Evaluation of IL4I1, IL4I1-NPs, and IL4I1-MNPs In Vivo. Note (A) Blood cell count in each group of mice (n = 6); (B) Blood biochemical analysis in each group of mice (n = 6), including albumin (ALB), total bilirubin (TBIL), alanine transaminase (ALT), aspartate transaminase (AST), creatinine (CRE), urea (UREA), and uric acid (UA); (C) H&E staining of major organs in mice from each group (n = 6) under different treatments, scale bar = 100 μm.
Supplementary Material 11: Impact of FGR-Mediated Regulation of M1 MΦ Polarization by IL4I1-MNPs on NPC Phenotype. Note (A) Schematic representation of co-culture of MΦ and NPCs; (B) Cell proliferation assessed by CCK8 in each group of NPCs; (C) Representative Datas of EdU staining and quantitative measure of EdU-positive cells in each group of NPCs (scale bar = 50 μm), where EdU is shown in red and DAPI (nucleus) in blue; (D) Quantification of migrated cells in Transwell experiments of NPC from each group, shown with representative Datas (scale bar = 50 μm); (E) Flow cytometry evaluation of cell apoptosis (Annexin V+/PI−) in NPCs from each group; (F) Protein expression and quantification of Collagen II, Aggrecan, ADAMTS5, and MMP13 in NPCs from each group analyzed by WB; (G) Immunofluorescence staining of Collagen II and MMP13 in NPCs from each group with representative Datas (scale bar = 25 μm) and fluorescence intensity quantification; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, with all experiments conducted thrice.
Supplementary Material 12: The Influence of IL4I1-MNPs-Mediated FGR Regulation on M2 MΦ Polarization Process and its Impact on NPCs Phenotype.. Note (A) Cell Proliferation of NPCs in each group was examined using the CCK-8 assay; (B) Representative Datas of EdU staining in NPCs from each group (scale bar = 50 μm) and quantitative analysis of EdU-positive cell percentage, with EDU (red) and DAPI (cell nuclei, blue); (C) Representative Datas of Transwell experiments in each group of NPCs (scale bar = 50 μm) and quantitative analysis of migrated cell numbers; (D) Cell apoptosis (Annexin V+/PI−) in NPCs from each group detected by flow cytometry; (E) Protein expression and quantification of Collagen II, Aggrecan, ADAMTS5, and MMP13 in NPCs from each group analyzed by WB; (F) Immunofluorescence staining Datas of Collagen II and MMP13 in NPCs from each group (scale bar = 25 μm) and quantification of fluorescence intensity; * P < 0.05 compared between two groups, ** P < 0.01 compared between two groups, *** P < 0.001 compared between two groups, all experiments were repeated 3 times.
Supplementary Material 13: Characteristics of CHG and CHG@IL4I1-MNPs. Note (A) Schematic representation of CHG synthesis; (B) Swelling ratio of hydrogel CHG in PBS at 37 °C; (C) Degradation rate of hydrogel CHG; (D) Schematic representation of IL4I1-MNPs loaded CHG; (E) Representative Datas captured by SEM of CHG or CHG@IL4I1-MNPs (scale bar = 50 μm); (F) Rheological analysis of CHG or CHG@IL4I1-MNPs; (G) Average storage modulus of CHG and CHG@IL4I1-MNPs; (H) Cumulative release curve of IL4I1 in IL4I1-MNPs and CHG@IL4I1-MNPs; (I) Cell viability of MΦ 24 h after different treatments examined by CCK-8 assay; n.s. Indicates P > 0.05 compared between two groups; all experiments were repeated 3 times.
Supplementary Material 14: The Impact of CHG@IL4I1-MNPs-Mediated FGR on Characterization of IDD Mice. Note (A) Schematic representation of CHG, IL4I1-MNPs, CHG@IL4I1-MNPs, and FGR-OE treatments; (B) Micro-CT two-dimensional and three-dimensional Datas of IVD in each group of mice (n = 6) and intervertebral DHI percentage; (C) Representative Datas of H&E staining and Safranin-O/Fast green staining of tail IVD, AF, and NP tissues in each group of mice (n = 6); (D) Histological scores of tail IVD changes in each group of mice (n = 6); (E) Immunohistochemical representative Datas of Collagen II, Aggrecan, ADAMTS5, and MMP13 in tail IVD of each group of mice (n = 6) (scale bar = 200 μm) and quantitative analysis of positive staining area percentage; n.s. indicates P > 0.05 compared between two groups, * P < 0.05 compared between two groups, ** P < 0.01 compared between two groups, *** P < 0.001 compared between two groups, **** P < 0.0001 compared between two groups.
Supplementary Material 15: In Vivo Safety Assessment of CHG and CHG@IL4I1-MNPs. Note (A) Blood cell counts in each group of mice (n = 6); (B) Blood biochemistry analysis in each group of mice (n = 6), including ALB, TBIL, ALT, AST, CRE, UREA, UA; (C) H&E staining of major organs in each group of mice (n = 6) under different treatments, scale bar = 100 μm.
Supplementary Material 16: Schematic Representation of the Molecular Mechanism of CHG@IL4I1-MNPs in Regulating MΦ Polarization for the Treatment of IDD
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.