Abstract
Background
The case-only design is a powerful approach to identify gene gene and gene environment interactions for complex traits. It has been demonstrated that for the case-only design to be valid the genetic and environmental factors must be independent in the population. Additionally, there is a rare disease assumption for the case-only design, but the impact of disease prevalence and other factors, e.g., size of main effects, on type I and II error rates has not been investigated.
Methods
Through theoretical and extensive simulation studies, we investigated type I error, power, and bias of interaction term for a wide variety of disease prevalences, main and interaction effect sizes, sample sizes, and variant and environmental exposure frequencies.
Results
For diseases with prevalence 4%, the case-only design usually has well controlled type I error rates and is substantially more powerful to detect interactions than the case–control design, but for higher disease prevalences both type I and II error rates can be inflated and the estimate of interaction term biased. However, when one or both main effects are large there can be inflated type I error rate even for low disease prevalences, e.g., 1%, but if there is no or only one main effect, type I error rate is controlled regardless of the disease prevalence. Additionally, type I error rate can increase with sample size.
Conclusions
We determined the upper bound of the disease prevalence in order not to violate the rare disease assumption for the case-only design. To verify that a case-only design study does not have increased type I error rate, the bias of the interaction term should be estimated. Although the case-only design is a powerful method to detect interactions, prevalences for some complex traits are too high to implement this method without increasing type I error rates.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-025-11318-1.
Keywords: Case-only design, Complex traits, Gene gene and gene environment interactions, Rare disease
Background
Genome-wide association studies (GWAS) have detected associations for thousands of complex traits with millions of single-nucleotide variants (SNVs). Most GWAS are designed to detect main effects, while interactions either between genetic variation and environmental factors (G E) or genetic variations (G G) may explain “missing heritability”. In the past due to insufficient sample sizes most studies were underpowered to detect interactions, however large biobanks that include genetic and environmental data, e.g., UK Biobank, are making it possible to potentially detect interactions [1].
One method to detect G E interactions, the case-only design, has received considerable attention since Piegorsch et al. proposed it [2]. It can also be applied to study G G interactions, aka epistasis [3]. Piegorsch et al. stated that two assumptions must be met when applying the case-only design to estimate the interaction effect: (1) the disease should be rare (2) G and E or G and G are independent. The second assumption of independence is well studied [4–6] and has been evaluated using control data [2, 7]. For the case-only design the effect of deviation from Hardy–Weinberg equilibrium (HWE) in controls was also examined [8, 9]. Additionally, it was shown that population stratification can also introduce a bias [4]. However, the rare disease assumption has not been investigated. In practice the case-only design has been applied to complex traits with a wide range of disease prevalences, including breast, prostate and ovarian cancers, Crohn's disease, and rheumatoid arthritis [10–14]. For these studies it is not clear whether the rare disease assumption was met and if it was appropriate to use the case-only design.
Using theoretical analysis and simulation studies, we evaluated the role of disease prevalence for the case-only design, assuming G and E are independent in the overall population. We determined that there is no set disease prevalence where type I error rates are always well controlled, since not only does disease prevalence impact type I error rate, but also main effects and sample sizes and to a lesser extent variant and environmental exposure frequencies. Generally, for diseases with a prevalence of 4% the case-only design has well controlled type I error rate. When main effects are large, e.g., odds ratio () 5.0, the disease prevalence must be 1% to control type I error rates. Contrarily, when there is no or only one main effect, the disease prevalence can be high, e.g., 20% and type I error rates are controlled. The estimate of the interaction bias can be used to evaluate if type I error rate is controlled. To facilitate this evaluation, we provide the CaseOnly R code, which simulates and analyses data to assess type I error rates and statistical power in case-only designs, thereby offering researchers a robust tool for detecting interactions. The case-only design is a powerful method to detect interactions, and it is advantageous to apply it when type I error rate is controlled.
Methods
Simulation study
Data were generated for a genetic variant under a dominant and additive model and for a binary environmental exposure. To evaluate type I error, the genetic variant and environmental exposure have no interaction effects. The main effects for the genetic variant ranged from to for the dominant model, and for the additive model for the carrier of one risk allele. For the environmental exposure the main effects ranged from to . We also tested when both main effects were protective to , to ], as well as when one main effect was protective and the other increased risk. We also evaluated type I error when neither the genetic nor environmental factor had a main effect and when only one factor had a main effect. Data were also generated under the alternative where there was an interaction, . The minor allele frequency (MAF) of the genetic variant and the frequency of the environmental exposure was varied between 0.05 and 0.50. We also varied the disease prevalence between 1 and 20%. Using a uniform distribution and a random number generator, genetic variant and exposure data were generated for the th sample and a logistic regression model,
| 1 |
was used to assign the th sample as a case or control. For case–control design, we generated samples of 10,000 cases and 10,000 to 30,000 controls to obtain a ratio () between controls and cases (, and ). We also generated samples of different number of cases (2,500, 5,000, 10,000, 20,000, and 50,000) to evaluate how sample size impacts type I error rate for the case-only design, with disease prevalence levels varying from 1% to 20%.
For the case-only design, the following formula was applied to estimate the interaction effect where is odds ratio:
| 2 |
For case–control design, a logistic regression model was used to estimate the interaction effect . The p-value ( for both the case-only and case–control designs was computed using the Wald test for the coefficient of in the logistic regression model.
When estimating type I error under the null of no interaction (), 1,000,000 replicates were generated and analysed. Type I error was calculated for the different significance levels () varying from 0.001 to 0.05. Quantile–quantile (QQ) plots were also generated. For evaluating power under the alternative ], 100,000 replicates were generated and analysed. Power was estimated as the proportion of replicates with . For data generated under the null and alternative the distribution of was obtained using all replicates.
Theoretical and analytical studies
We developed a theoretical framework to demonstrate the relationship between the interaction effect and the between G and E in the cases and controls. As stated in Piegorsch et al. [2], under the logistic model (1), the exponential value of interaction effect approximately equals to the between G and E among the cases when the disease prevalence is sufficiently low and G and E are independent in the population. Here we explicitly write the relationship as follows:
| 3 |
When the disease prevalence is sufficiently low, is approximately the between G and E in the population (Supplementary Methods Sections 1, 2.1, and 2.2).
Besides, by comparing in formula (2) and the true in formula (3), we show the analytical bias given the true , and and the baseline prevalence of the disease (Supplementary Methods Sect. 2.3):
| 4 |
Results
Simulation studies
Type I error—case-only and case–control designs
Simulation studies were used to evaluate type I error when testing for interactions () for the case-only and case–control designs. Tables 1a and Supplementary Table S1 show the type I error for the case-only and case–control designs for and when there are main effects, i.e., and under the dominant model. Similar results were observed for the additive model (Supplementary Table S2).
Table 1.
Type I error rates for case-only and case–control designs
| Disease prevalence | 1.0% | 1.5% | 2.0% | 2.5% | 3.0% | 3.5% | 4.0% | 4.5% | 5.0% | 10.0% | 15.0% | 20.0% | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (a) Two main effects | |||||||||||||
| case–control | 0.001 | 0.000962 | 0.000970 | 0.001041 | 0.001009 | 0.000983 | 0.000958 | 0.000994 | 0.000983 | 0.000970 | 0.000976 | 0.000978 | 0.000997 |
| case-only | 0.001 | 0.001015 | 0.000942 | 0.000983 | 0.001038 | 0.000990 | 0.000995 | 0.001098 | 0.001067 | 0.001083 | 0.001390 | 0.001667 | 0.002091 |
| case–control | 0.05 | 0.050330 | 0.049824 | 0.049887 | 0.049905 | 0.050113 | 0.050161 | 0.049653 | 0.050160 | 0.049735 | 0.049845 | 0.049912 | 0.050371 |
| case-only | 0.05 | 0.050132 | 0.050603 | 0.049854 | 0.050842 | 0.050614 | 0.051195 | 0.051387 | 0.052100 | 0.052221 | 0.057378 | 0.063536 | 0.070049 |
| (b) Only a genetic main effect | |||||||||||||
| case–control | 0.001 | 0.001005 | 0.000929 | 0.001021 | 0.001002 | 0.000976 | 0.000999 | 0.001016 | 0.001030 | 0.000981 | 0.000958 | 0.000973 | 0.001068 |
| case-only | 0.001 | 0.001009 | 0.000969 | 0.000979 | 0.000990 | 0.000995 | 0.000940 | 0.001023 | 0.001030 | 0.001014 | 0.000966 | 0.000986 | 0.000984 |
| case–control | 0.05 | 0.049911 | 0.049822 | 0.050093 | 0.050012 | 0.049857 | 0.049930 | 0.049642 | 0.050276 | 0.050018 | 0.049894 | 0.049908 | 0.050146 |
| case-only | 0.05 | 0.050315 | 0.049506 | 0.050062 | 0.049971 | 0.049997 | 0.049994 | 0.049793 | 0.050205 | 0.049727 | 0.049807 | 0.049933 | 0.049700 |
| (c) Only an environment main effect | |||||||||||||
| case–control | 0.001 | 0.000998 | 0.000981 | 0.000997 | 0.000971 | 0.001026 | 0.000980 | 0.000973 | 0.000951 | 0.001022 | 0.001015 | 0.000960 | 0.000984 |
| case-only | 0.001 | 0.000952 | 0.000998 | 0.000936 | 0.000968 | 0.000975 | 0.001012 | 0.000962 | 0.001046 | 0.000995 | 0.000998 | 0.000961 | 0.000940 |
| case–control | 0.05 | 0.050296 | 0.050185 | 0.050248 | 0.049903 | 0.050320 | 0.049931 | 0.049868 | 0.049853 | 0.049658 | 0.049894 | 0.049753 | 0.049896 |
| case-only | 0.05 | 0.049769 | 0.049906 | 0.049880 | 0.050149 | 0.049980 | 0.050189 | 0.049935 | 0.049731 | 0.049396 | 0.049929 | 0.049621 | 0.050153 |
| (d) No main effects | |||||||||||||
| case–control | 0.001 | 0.001010 | 0.000981 | 0.000929 | 0.001040 | 0.000940 | 0.000984 | 0.000959 | 0.000999 | 0.001019 | 0.001012 | 0.000977 | 0.001056 |
| case-only | 0.001 | 0.000997 | 0.000904 | 0.000899 | 0.000930 | 0.000952 | 0.000959 | 0.000953 | 0.000959 | 0.001036 | 0.000958 | 0.000941 | 0.001027 |
| case–control | 0.05 | 0.049769 | 0.050132 | 0.049922 | 0.050361 | 0.050121 | 0.050052 | 0.049907 | 0.050104 | 0.050338 | 0.049919 | 0.049874 | 0.050470 |
| case-only | 0.05 | 0.049667 | 0.049608 | 0.049764 | 0.049975 | 0.050096 | 0.050202 | 0.049683 | 0.050152 | 0.050234 | 0.050031 | 0.049877 | 0.050120 |
Data were generated under the null with no interaction for four scenarios: (a) two main effects [ and ]; (b) only a genetic main effect [ and ]; (c) only an environmental main effect [ and ]; and (d) no main effects . The MAF for the genetic variant is 0.2 and the frequency of the environmental exposure is 0.1. The sample sizes are N = 10,000 cases for both the case-only and case–control designs and additionally for the case–control design there are N = 10,000 controls. Type I error is calculated as the proportion of instances that the null hypothesis is rejected among all 1,000,000 replicates. Type I errors exceeding 5% of the alpha level are highlighted in bold in the table
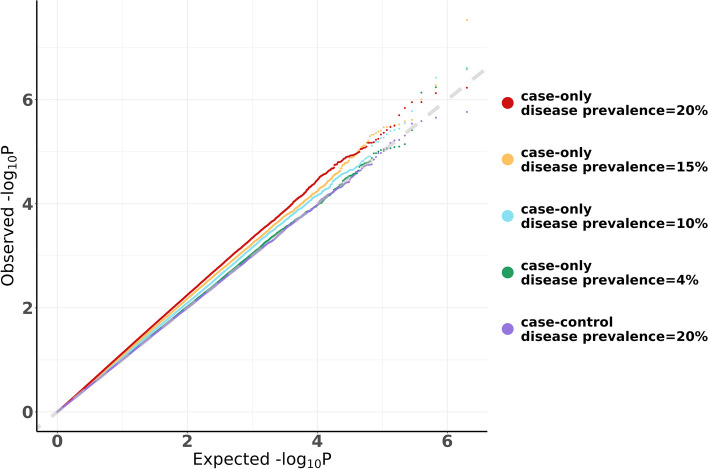
As the disease prevalence increases, type I error rate generally becomes higher in this setting and the type I error rate is inflated when disease prevalence is 4%. When disease prevalence is 4%, type I error for case-only design is 0.001098 and 0.051387 when and , respectively. When disease prevalence is 20%, type I error rate is greatly inflated to 0.002091 and 0.070049, for and , respectively. As observed in the QQ plot it is evident that type I error rate for the case-only design increases with increasing disease prevalence, but for the case–control design the type I error rate is well controlled even when the disease prevalence is 20% (Fig. 1 and Supplementary Fig. 1a). When one or both main effects are absent, the case-only design has well controlled type I error rate even when the disease prevalence is high, e.g., 20% (Table 1b-d and Supplementary Figure S1b-d).
Fig. 1.
Quantile–Quantile plot for the case-only and case–control designs. Data were generated under the null with no interaction when there are two main effects [ and ]. The MAF for the genetic variant is 0.2 and the frequency of the environmental exposure is 0.1. A total of 1,000,000 replicates were generated. The sample sizes are N = 10,000 cases for both the case-only and case–control designs and additionally for the case–control design there are N = 10,000 controls
Main effects and type I error for the case-only design
We also evaluated type I error for a variety of main effects (Table 2a), the results suggest that if one or both main effects are strong, the disease prevalence should be 4% for well controlled type I error rate. If is increased from to and remains , and the disease prevalence is 1%, type I error rate increases from 0.050132 to 0.056403. When both main effects are strong and ], even when the disease prevalence is as low as 1%, type I error is very inflated, i.e., 0.141321 for . Interestingly, if the main effects are protective, the case-only design can be applied to diseases with higher prevalences without inflated type I error issues, e.g., when both main effects are strongly protective, e.g., and , type I error is 0.048999 when the disease prevalence is 5%. If the main effects are in the opposite directions but both strong, even for a disease prevalence of 1%, type I error is still inflated (Supplementary Table S3a). If both main effects are weak, then the disease prevalence can be 4% for well controlled type I error rate, e.g., if and , type I error is well controlled (0.049666) even when the disease prevalence is 20%.
Table 2.
Impact of main effects on type I error and analytical bias for the case-only design
| Disease prevalence | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.0% | 1.5% | 2.0% | 2.5% | 3.0% | 3.5% | 4.0% | 4.5% | 5.0% | 10.0% | 15.0% | 20.0% | ||
| (a) Type I error | |||||||||||||
| 0.141321 | 0.252332 | 0.396712 | 0.551859 | 0.695653 | 0.810552 | 0.891836 | 0.942837 | 0.972389 | 0.999998 | 1.000000 | 1.000000 | ||
| 0.056403 | 0.063479 | 0.072751 | 0.085891 | 0.101210 | 0.119065 | 0.138098 | 0.160377 | 0.183840 | 0.480285 | 0.735506 | 0.873881 | ||
| 0.050132 | 0.050603 | 0.049854 | 0.050842 | 0.050614 | 0.051195 | 0.051387 | 0.052100 | 0.052221 | 0.057378 | 0.063536 | 0.070049 | ||
| 0.049890 | 0.049729 | 0.049665 | 0.049657 | 0.049644 | 0.049992 | 0.049896 | 0.049923 | 0.050019 | 0.050016 | 0.049992 | 0.049666 | ||
| 0.049816 | 0.049991 | 0.049856 | 0.049738 | 0.049618 | 0.050020 | 0.050054 | 0.049950 | 0.049860 | 0.049651 | 0.049931 | 0.050423 | ||
| 0.050030 | 0.049658 | 0.049459 | 0.049875 | 0.049607 | 0.049515 | 0.049339 | 0.049723 | 0.049573 | 0.049940 | 0.051270 | 0.052579 | ||
| 0.048962 | 0.048521 | 0.048793 | 0.048931 | 0.049388 | 0.049655 | 0.050008 | 0.050319 | 0.051327 | 0.063964 | 0.088547 | 0.125843 | ||
| 0.047237 | 0.047101 | 0.046880 | 0.047278 | 0.047025 | 0.047459 | 0.047611 | 0.048081 | 0.048999 | 0.063687 | 0.096078 | 0.152817 | ||
| (b) Analytical bias | |||||||||||||
| −0.039119 | −0.057894 | −0.076150 | −0.093890 | −0.111119 | −0.127843 | −0.144066 | −0.159797 | −0.175042 | −0.302652 | −0.391307 | −0.449137 | ||
| −0.012563 | −0.018684 | −0.024697 | −0.030605 | −0.036407 | −0.042105 | −0.047698 | −0.053188 | −0.058576 | −0.106964 | −0.145864 | −0.175993 | ||
| −0.001667 | −0.002479 | −0.003277 | −0.004061 | −0.004831 | −0.005588 | −0.006330 | −0.007060 | −0.007775 | −0.014227 | −0.019476 | −0.023631 | ||
| .−0.000048 | −0.000072 | −0.000095 | −0.000118 | −0.000141 | −0.000164 | −0.000186 | −0.000208 | −0.000230 | −0.000434 | −0.000612 | −0.000764 | ||
| −0.000044 | −0.000066 | −0.000087 | −0.000109 | −0.000130 | −0.000151 | −0.000171 | −0.000192 | −0.000212 | −0.000404 | −0.000574 | −0.000724 | ||
| −0.000928 | −0.001389 | −0.001847 | −0.002303 | −0.002756 | −0.003207 | −0.003655 | −0.004101 | −0.004544 | −0.008831 | −0.012836 | −0.016536 | ||
| −0.005304 | −0.007952 | −0.010597 | −0.013240 | −0.015878 | −0.018514 | −0.021146 | −0.023774 | −0.026399 | −0.052388 | −0.077767 | −0.102296 | ||
| −0.008789 | −0.013196 | −0.017611 | −0.022035 | −0.026467 | −0.030907 | −0.035354 | −0.039810 | −0.044273 | −0.089276 | −0.134820 | −0.180639 | ||
Data (N = 10,000 cases) were generated under the null with no interaction (). The MAF for the genetic variant is 0.2 and the frequency of the environmental exposure is 0.1. Type I error is calculated as the proportion of instances that the null hypothesis is rejected among all 1,000,000 replicates (. Analytical bias was calculated using formula (4). Type I errors exceeding 5% of the alpha level are highlighted in bold in the table
Exposure frequencies and type I error for the case-only design
Genetic variant and environmental exposure frequencies also affect type I error for the case-only design. Type I error rate first increases as variant and environmental exposure frequencies become greater, then decreases as the minor allele becomes the major allele or > 50% of the population are exposed. For example, when the disease prevalence is 5% and frequency of environmental exposure is 10%, type I error is 0.049830 (MAF = 0.05), 0.052221 (MAF = 20%), and 0.051684 (MAF = 50%), respectively. However, the impact of MAF and the environmental exposure frequency on type I error rate is limited compared to the influence of main effects and disease prevalence (Table 3a).
Table 3.
Impact of variant and environmental exposure frequencies on type I error and analytical bias for the case-only design
| Disease prevalence | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | Frequency of environmental exposure | 1.0% | 1.5% | 2.0% | 2.5% | 3.0% | 3.5% | 4.0% | 4.5% | 5.0% | 10.0% | 15.0% | 20.0% |
| (a) Type I error | |||||||||||||
| 0.05 | 0.10 | 0.049544 | 0.049293 | 0.049588 | 0.049806 | 0.049691 | 0.049811 | 0.050086 | 0.049976 | 0.049830 | 0.051979 | 0.054398 | 0.056482 |
| 0.20 | 0.05 | 0.049945 | 0.050021 | 0.049959 | 0.049814 | 0.050021 | 0.050543 | 0.050531 | 0.050543 | 0.051266 | 0.054281 | 0.057564 | 0.061556 |
| 0.20 | 0.20 | 0.049905 | 0.050096 | 0.050687 | 0.051004 | 0.051012 | 0.051657 | 0.051967 | 0.052483 | 0.052905 | 0.060343 | 0.069782 | 0.079699 |
| 0.50 | 0.10 | 0.049891 | 0.050474 | 0.050255 | 0.050288 | 0.050658 | 0.050898 | 0.050997 | 0.051125 | 0.051684 | 0.055462 | 0.060344 | 0.065009 |
| 0.50 | 0.50 | 0.049928 | 0.050109 | 0.050259 | 0.050434 | 0.049975 | 0.050435 | 0.050898 | 0.050810 | 0.050750 | 0.055206 | 0.060188 | 0.066779 |
| (b) Analytical bias | |||||||||||||
| 0.05 | 0.10 | −0.00175 | −0.002601 | −0.003436 | −0.004255 | −0.005058 | −0.005845 | −0.006618 | −0.007375 | −0.008117 | −0.014751 | −0.020063 | −0.024195 |
| 0.20 | 0.05 | −0.001744 | −0.002591 | −0.003422 | −0.004238 | −0.005037 | −0.005821 | −0.006590 | −0.007344 | −0.008082 | −0.014685 | −0.019975 | −0.024098 |
| 0.20 | 0.20 | −0.001532 | −0.002282 | −0.003020 | −0.003748 | −0.004464 | −0.005169 | −0.005864 | −0.006548 | −0.007221 | −0.013374 | −0.018518 | −0.022706 |
| 0.50 | 0.10 | −0.001557 | −0.002318 | −0.003066 | −0.003803 | −0.004529 | −0.005242 | −0.005945 | −0.006636 | −0.007315 | −0.013503 | −0.018644 | −0.022809 |
| 0.50 | 0.50 | −0.00115 | −0.001718 | −0.002282 | −0.002840 | −0.003394 | −0.003944 | −0.004488 | −0.005028 | −0.005563 | −0.010640 | −0.015208 | −0.019246 |
Data (N = 10,000 cases) were generated under the null with no interaction [, , and ]. Type I error is calculated as the proportion of instances that the null hypothesis is rejected among all 1,000,000 replicates (). Analytical bias was calculated using formula (4). Type I errors exceeding 5% of the alpha level are highlighted in bold in the table
Sample size and type I error for the case-only design
We also evaluated the role of sample size plays on type I error for the case-only design. When the disease prevalence is 4% and there are two main effects [ and ], type I error is 0.050325, 0.051155, and 0.058031 for 2 500, 10,000, and 50,000 cases for . If the sample size is 20,000 the disease prevalence should be 2% for well controlled type I error rate. For a disease prevalence of 2% and a sample size of 50,000 cases, type I error is inflated (0.051977). On the other hand, if the sample size is 2 500, type I error is still well controlled (0.050102) when the disease prevalence is 5% (Table 4).
Table 4.
Impact of sample size on type I error rates for the case-only design
| Disease prevalence | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cases | 1.0% | 1.5% | 2.0% | 2.5% | 3.0% | 3.5% | 4.0% | 4.5% | 5.0% | 10.0% | 15.0% | 20.0% | |
| 2,500 | 0.001 | 0.000983 | 0.000958 | 0.000942 | 0.001019 | 0.000959 | 0.000911 | 0.000962 | 0.000956 | 0.001024 | 0.001057 | 0.001097 | 0.001216 |
| 0.05 | 0.049557 | 0.049826 | 0.049762 | 0.050058 | 0.050021 | 0.049818 | 0.050325 | 0.050030 | 0.050102 | 0.051262 | 0.053111 | 0.054364 | |
| 5,000 | 0.001 | 0.000996 | 0.001050 | 0.000976 | 0.000976 | 0.000972 | 0.001000 | 0.000949 | 0.001010 | 0.001060 | 0.001153 | 0.001275 | 0.001413 |
| 0.05 | 0.049919 | 0.050110 | 0.049596 | 0.050317 | 0.050346 | 0.050453 | 0.050565 | 0.050731 | 0.050874 | 0.053727 | 0.056344 | 0.059515 | |
| 10,000 | 0.001 | 0.000994 | 0.001004 | 0.001058 | 0.001037 | 0.001057 | 0.001106 | 0.001115 | 0.001068 | 0.001097 | 0.001357 | 0.001731 | 0.001966 |
| 0.05 | 0.050155 | 0.050387 | 0.050449 | 0.050325 | 0.051138 | 0.050999 | 0.051155 | 0.051520 | 0.052183 | 0.057479 | 0.063428 | 0.070083 | |
| 20,000 | 0.001 | 0.000980 | 0.000998 | 0.001012 | 0.001052 | 0.000992 | 0.001114 | 0.001162 | 0.001186 | 0.001267 | 0.001838 | 0.002487 | 0.003475 |
| 0.05 | 0.049976 | 0.050759 | 0.050546 | 0.051070 | 0.051843 | 0.052863 | 0.053234 | 0.053472 | 0.054632 | 0.065503 | 0.078536 | 0.090791 | |
| 50,000 | 0.001 | 0.001034 | 0.000988 | 0.001076 | 0.001160 | 0.001208 | 0.001294 | 0.001373 | 0.001544 | 0.001647 | 0.003449 | 0.005998 | 0.008934 |
| 0.05 | 0.050299 | 0.051092 | 0.051977 | 0.053040 | 0.054681 | 0.055883 | 0.058031 | 0.059773 | 0.061797 | 0.089890 | 0.122479 | 0.154596 | |
Data were generated under the null with no interaction but two main effects [ and ]. The MAF for the genetic variant is 0.2 and the frequency of the environmental exposure is 0.1. Type I error is calculated as the proportion of instances that the null hypothesis is rejected among all 1,000,000 replicates. Type I errors exceeding 5% of the alpha level are highlighted in bold in the table
Statistical power—case-only and case–control designs
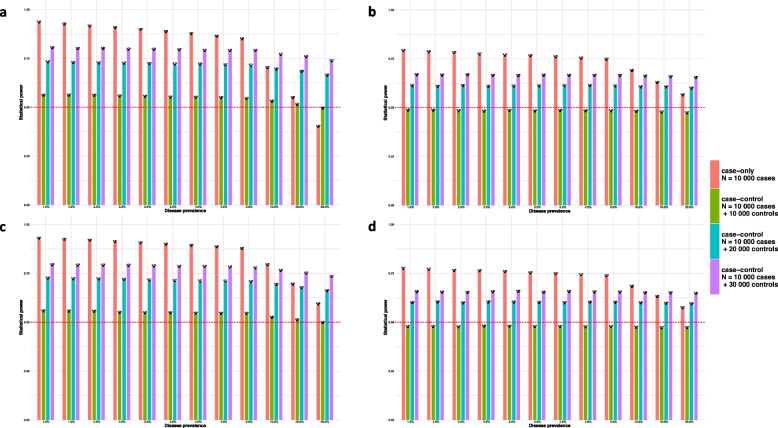
When there are two main effects [ and ] under the dominant model with disease prevalence 5%, the case-only design has higher power than case–control designs even when . Similar results were observed for the additive model (Supplementary Figure S2). When disease prevalence is 1%, the power for case-only and case–control () designs are 0.93 and 0.56, respectively, and even when disease prevalence increases to 4%, the power of case-only design (0.87) is 1.58 × greater than case–control design (0.55). The power of case-only design drops substantially when disease prevalence increases to 10%, but it is not until the disease prevalence reaches 20% that case–control design () has greater power (Fig. 2a).
Fig. 2.
Statistical power of the case-only and case–control designs Data were generated under the alternative with for (a) two main effects [ and ]; (b) only a genetic main effect [ and ]; (c) only an environmental main effect [ and ]; and (d) no main effects (). The MAF for the genetic variant is 0.2 and the frequency of the environmental exposure is 0.1. Power is calculated as the proportion of instances that the null hypothesis is rejected among all 100,000 replicates ()
When one or both main effects are absent, the power for the case-only design is also significantly higher than for the case–control design, even though the former has a much smaller sample size, e.g., N = 10,000 for case-only and N = 20,000 for case–control () designs. For a disease prevalence of 4%, when there is only a main genetic effect [ and ], the power is 0.76 and 0.48 for the case-only and case–control designs, respectively. As the disease prevalence increases to 20% the power for the case-only design is still greater than for the case–control design but the gain in power is not as great, i.e., 0.56 vs. 0.47. The result is similar when there is only one main environmental effect [ and ] or no main effects [ and ] (Fig. 2b-d).
Higher MAF and frequency of environmental exposure may also have an impact on power. For example, when MAF remains 0.2 and disease prevalence is 4%, the power for the case-only design is 0.65, 0.87, 0.97 for frequency of environmental exposure being 0.05, 0.10, 0.20, respectively. However, when MAF increases from 0.05 to 0.20 to 0.50 with frequency of environmental exposure fixed as 10%, under the dominant model, the power will first increase from 0.53 to 0.87, then decrease to 0.73 (Supplementary Figure S3).
Bias—analytical results
In formula (4), we derived the analytical bias of given the baseline risk (), main effects ( and ), and true interaction effect (). Under the null, when one or both main effects are absent, the bias of equals 0. However, when the main effects either both increase or decrease the risk of developing disease, underestimates the true interaction effect , leading to a negative bias of , and the bias is greater with higher disease prevalence. For example, when , for the following disease prevalences 1%, 4%, and 20% the biases are −0.001667, −0.006330, and −0.023631, respectively. Stronger main effects also lead to greater bias of , e.g., when both main effects are strong [ and ], the bias of reaches −0.144066 even when the disease prevalence is only 4%. When main effects ( and ) are in opposite directions, for the case-only design overestimates (a positive bias), with the bias increasing as the disease prevalence increases. As long as there are two non-zero main effects, is biased (Table 3b, Supplementary Table S3, Supplementary Figure S4 and Supplementary Sect. 2.3).
Under the alternative [], even when there are no main effects, the bias of is negative, indicating that the case-only design underestimates the interaction effect. When , and , the bias is −0.005613, −0.021628, and −0.089011 for disease prevalences of 1%, 4%, and 20%, respectively. The bias of is smaller when and are in opposite directions compared to when and are either both positive or negative, e.g., when , , and the disease prevalence is 20%, the bias is −0.089011 when , but only −0.008341 when . However, the bias still increases as the disease becomes more prevalent regardless of the main effects (Supplementary Table S4 and Supplementary Figure S5).
The bias of is closely related to type I and II error rates for the case-only design. Under the null, when there are two main effects the bias of always leads to an increase in type I error rate, and for the case-only design either underestimates or overestimates . For the case-only design, as the disease prevalence increases, the bias of is greater, leading to higher type I error rates. The case-only design may suffer from inflated type I error rate if the absolute value of the analytical bias of 0.006 (both main effects increase risk), 0.01 (one main effect increases and the other decreases risk), and 0.025 (both main effects are protective) (Tables 3 and 4). Under the alternative, if the bias of is in the same direction as , this will boost the power of the case-only design. However, it should be noted even when and the bias of are in opposite directions, e.g., is positive and the bias of is negative, the power for the case-only design is greater than when a case–control sample is analysed for disease prevalences 15% (Fig. 2 and Supplementary Table S4).
Discussion
We evaluated the case-only design through theoretical and simulation analysis, and showed that several factors including disease prevalence, main effects, variant and environmental exposure frequencies, and sample size may impact bias of the interaction term and type I and II error rates. While previous studies stated that for the case-only design a rare disease assumption is necessary, they often lack clarity on what constitutes a "rare" disease. Our simulations investigated various disease prevalence thresholds, from 1% to 20%, to assess their effects on type I error rates and bias in estimating interaction terms. Compared with the conventional case–control design, the power of the case-only design can be a magnitude greater, but it should only be applied when type I error rate is controlled. Generally, under the assumption of independence between G and E, for disease prevalences 4% type I error rate of the case-only design is not inflated, but for higher disease prevalences type I error rate can be high and the estimation of interaction effect biased. When disease prevalence is 20% the power for the case-only design can be lower than analysing a case–control sample. When one or both main effects are absent, the disease prevalence does not impact type I error rate. However, caution is required since there may be a failure to detect main effects even when they exist. The analytical bias can be calculated using formula (4), to aid in evaluating if the case-only design is appropriate to use. Besides the analytical bias, when the sample size is large ( 10,000 cases), the case-only design requires lower disease prevalence to avoid inflation in type I error rate.
Stronger main effects may lead to a greater bias of the interaction estimate and higher type I error rate. Though it is uncommon for complex traits to have a genetic risk factor with >1.5 there are several environmental exposures that have large main effects [15–17]. We recommend applying the case-only design to test for interactions for complex traits with low prevalences (e.g., ovarian cancer and celiac disease), and limit testing of interactions to variants and environmental exposures that do not have strong main effects.
Because of the bias of may be in the same or opposite direction of itself, type I and II error rates can both increase with increasing disease prevalence. Therefore, for the case-only design unlike most statistical tests as type I error rate increases power decreases.
Yang et al. claimed that no assumption about disease prevalence is required stating that the cross-product term of the table (presence or absence of the risk allele for genes 1 and 2 among cases) measures the departure from the multiplicative joint effects of relative risk aka the risk ratio () (not ) [3]. The is used for observational cohort studies that have incidence cases and only approximate the for diseases with low prevalence. Therefore, using to define interaction is inaccurate and both estimated by the case-only design and the “interaction” defined by have high type I error rates when the disease is prevalent. In fact, the case-only design measures the “interaction” effect defined by (Supplementary Methods Sect. 2.4) and they are both biased estimates of (Supplementary Figures S4 and S5).
In addition to multiplicative interaction, there are other types of interactions such as additive [18] and sufficient-cause interactions [9]. Interaction on an additive scale, often uses the index of relative excess risk due to interaction, which should be used with caution due to use of the relative risk as discussed in the paragraph above. Sufficient-cause interaction comprises a set of conditions or events that lead to a specific outcome, that is equivalent to the scenario when the main effects for G and E are both absent.
Conclusions
Although the case-only design is a powerful method to detect interaction due to increased type I error rates for a variety of scenarios, e.g., main effects, allele and environmental exposure frequencies are not sufficiently low, type I error rate should be evaluated. Our research contributes to the existing literatures by establishing clear guidelines on the acceptable thresholds for disease prevalence that maintain type I error rates under control. This can be done by analytically examining the bias [formula (4)] and performing simulation studies by implementing the R code, CaseOnly, that simulates and analyses data to evaluate type I error rate. CaseOnly can also be used to evaluate power. Although the case-only design is a powerful method to detect interactions it should be used with caution.
Supplementary Information
Acknowledgements
Not applicable.
Authors' contributions
RD, GTW and SML designed the theoretical and simulation analysis, performed data interpretation, and RD and SML drafted the manuscript. SML directed the study’s implementation. ATD and SML obtained the funding. All authors provided critical review for important intellectual content of the manuscript and approved the final manuscript for submission.
Funding
This work was supported by grant R01DC017712 to SML and ATD from National Institute and other Communications Disorders (NIDCD).
Data availability
The source code used to simulate and analyze data in this study is available in the CaseOnly repository (https://github.com/RuiDongDR/CaseOnly), and the simulations can be replicated through running the Rscript CaseOnly.R.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang H, Zhang F, Zeng J, Wu Y, Kemper KE, Xue A, et al. Genotype-by-environment interactions inferred from genetic effects on phenotypic variability in the UK Biobank. Sci Adv. 2019; [DOI] [PMC free article] [PubMed]
- 2.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–62. [DOI] [PubMed] [Google Scholar]
- 3.Yang Q, Khoury MJ, Sun F, Flanders WD. Case-Only Design to Measure Gene-Gene Interaction: Epidemiology. 1999;10:167–70. [PubMed] [Google Scholar]
- 4.Wang L-Y, Lee W-C. Population Stratification Bias in the Case-Only Study for Gene-Environment Interactions. Am J Epidemiol. 2008;168:197–201. [DOI] [PubMed] [Google Scholar]
- 5.Gatto NM. Further development of the case-only design for assessing gene-environment interaction: evaluation of and adjustment for bias. Int J Epidemiol. 2004;33:1014–24. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee S, Wang Z, Ciampa J, Kraft P, Chanock S, Yu K, et al. Using Principal Components of Genetic Variation for Robust and Powerful Detection of Gene-Gene Interactions in Case-Control and Case-Only Studies. Am J Hum Genet. 2010;86:331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoury MJ, Flanders WD. Nontraditional Epidemiologic Approaches in the Analysis of Gene Environment Interaction: Case-Control Studies with No Controls! Am J Epidemiol. 1996;144:207–13. [DOI] [PubMed] [Google Scholar]
- 8.Lee W-C, Wang L-Y, Cheng KF. An easy-to-implement approach for analyzing case-control and case-only studies assuming gene-environment independence and hardy-weinberg equilibrium. Stat Med. 2010;29:2557–67. [DOI] [PubMed] [Google Scholar]
- 9.Lee W-C. Testing for sufficient-cause gene-environment interactions under the assumptions of independence and hardy-weinberg equilibrium. Am J Epidemiol. 2015;182:9–16. [DOI] [PubMed] [Google Scholar]
- 10.Lash TL, Bradbury BD, Wilk JB, Aschengrau A. A case-only analysis of the interaction between N-acetyltransferase 2 haplotypes and tobacco smoke in breast cancer etiology. Breast Cancer Res. 2005;7:R385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neslund-Dudas C, Levin AM, Rundle A, Beebe-Dimmer J, Bock CH, Nock NL, et al. Case-only gene–environment interaction between ALAD tagSNPs and occupational lead exposure in prostate cancer. Prostate. 2014;74:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Mukherjee B, Lee S, Lee AW, Wu AH, Bandera EV, et al. Robust Tests for Additive Gene-Environment Interaction in Case-Control Studies Using Gene-Environment Independence. Am J Epidemiol. 2018;187:366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helbig KL, Nothnagel M, Hampe J, Balschun T, Nikolaus S, Schreiber S, et al. A case-only study of gene-environment interaction between genetic susceptibility variants in NOD2 and cigarette smoking in Crohn’s disease aetiology. BMC Med Genet. 2012;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke GM, Pettersson FH, Morris AP. A comparison of case-only designs for detecting gene × gene interaction in rheumatoid arthritis using genome-wide case-control data in Genetic Analysis Workshop 16. BMC Proc. 2009;3:S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuomi T, Huuskonen MS, Virtamo M, Tossavainen A, Tammilehto L, Mattson K, et al. Relative risk of mesothelioma associated with different levels of exposure to asbestos. Scand J Work Environ Health. 1991;17:404–8. [DOI] [PubMed] [Google Scholar]
- 16.Van Hylckama VA, Helmerhorst FM, Vandenbroucke JP, Doggen CJM, Rosendaal FR. The venous thrombotic risk of oral contraceptives, effects of oestrogen dose and progestogen type: results of the MEGA case-control study. BMJ. 2009;339:b2921–b2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloemenkamp KWM, Rosendaal FR, Büller HR, Helmerhorst FM, Colly LP, Vandenbroucke JP. Risk of Venous Thrombosis With Use of Current Low-Dose Oral Contraceptives Is Not Explained by Diagnostic Suspicion and Referral Bias. Arch Intern Med. 1999;159:65. [DOI] [PubMed] [Google Scholar]
- 18.Lee W-C. Sample size calculations for additive interactions. Epidemiology. 2013;24:774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code used to simulate and analyze data in this study is available in the CaseOnly repository (https://github.com/RuiDongDR/CaseOnly), and the simulations can be replicated through running the Rscript CaseOnly.R.