Abstract
Background:
Leptospirosis is an important zoonotic infection worldwide. Diagnosis of leptospirosis is challenging given its nonspecific clinical symptoms that overlap with other acute febrile illnesses and limitations with conventional diagnostic testing. Alternative advanced diagnostics, such as microbial cell-free DNA (mcfDNA), are increasingly being used to aid in the diagnosis of infections and can be applied to pathogens with public health importance such as Leptospira, a nationally notifiable disease.
Methods:
The Karius Test uses plasma mcfDNA sequencing to detect and quantify DNA-based pathogens. This test offered through the Karius lab detected 4 cases of Leptospira santarosai during a 5-month period across the United States in 2021 and were clinically reviewed.
Results:
In our case series, 4 adolescents with recent travel to Central America (Costa Rica, n = 3 and Belize, n = 1) from April to August 2021 were diagnosed with leptospirosis. While a large workup was performed in all cases, mcfDNA testing was the first test to detect L. santarosai as the microbiological diagnosis in all cases.
Conclusions:
Results of the Karius Test enabled rapid, noninvasive diagnosis of leptospirosis allowing for targeted therapy. Use of mcfDNA can be utilized for diagnosis of pathogens where conventional testing is challenging or limited. This in turn can enable quick diagnosis for targeted treatment and potentially aid in supporting case definitions of reportable diseases of public health concern.
Keywords: leptospirosis, Leptospira santarosai, microbial cell-free DNA sequencing
Leptospirosis is a zoonotic infection with potential to cause significant morbidity and mortality. It is caused by Leptospira spp., an obligate aerobic spirochete, which has 39 pathogenic species.1–5 Outbreaks of Leptospira spp. have been reported worldwide with important epidemiological and public health implications.6–9
Animals such as rodents, dogs and livestock serve as reservoirs. Human infection occurs via direct or indirect contact of mucosal surfaces or breaks in skin with bodily fluids, such as urine, from infected animals.1–4 Leptospirosis is endemic in tropical and subtropical climates and becomes more common particularly after rainfall. Based on modeling methods, it is estimated that approximately one million cases and 58,000 deaths occur worldwide annually; however, the true incidence of leptospirosis is likely underreported given its varied manifestations and challenges in diagnosis.2,8
First reported in the 19th century, leptospirosis causes nonspecific, protean clinical manifestations.4 The severity of infections can range from subclinical to life-threatening illness. Anicteric leptospirosis commonly presents as a biphasic illness where individuals present with nonspecific symptoms such as fever, myalgias, gastrointestinal symptoms or rash during a “septicemia” phase. A more pathognomonic symptom is conjunctival suffusion but does not occur in all patients. Patients then recover and several days later may experience return of fever with additional symptoms including aseptic meningitis during an “immune” phase. Icteric leptospirosis is the more severe form of infection and often monophasic in nature. Classically known as Weil disease, patients exhibit jaundice, renal dysfunction and hemorrhage and may rapidly decompensate. Severe illness carries a mortality rate of 5%–15%, but up to 75% in the setting of pulmonary hemorrhage.1,3,4,10
Leptospirosis can be self-limited. However, antibiotic treatment can shorten illness course in mild infection. Targeted and early treatment is also important in the management of severe infections to shorten duration of symptoms and decrease morbidity and mortality. Penicillin is the antibiotic of choice for severe infections, but more mild-to-moderate leptospirosis can be treated with other beta lactams, doxycycline, macrolides or fluoroquinolones.1–4 As such, prompt recognition and treatment are important and it is recommended to start empiric treatment if leptospirosis is suspected while awaiting testing. However, as discussed, leptospirosis can be hard to recognize given its nonspecific symptoms that may overlap with other acute febrile illnesses.11 Furthermore, typical laboratory manifestations are also nonspecific, including leukocytosis, thrombocytopenia, hyponatremia and elevated transaminases, bilirubin and inflammatory markers.4,12
Diagnosis of leptospirosis can also be challenging even with the availability of various methods for microbiological diagnosis including microscopy, culture, molecular testing and serological testing.2,4,12,13 Direct field microcopy and immunostaining has poor sensitivity and specificity and this method is operator dependent requiring skilled technical staff to perform.12,13 Culture is also less often used, it is time consuming and labor intensive and requires use of specialized media. Further, organisms may take up to 3 months to grow in culture, making this an impractical option given the need for a prompt diagnosis.1,4,13,14 Molecular testing utilizing polymerase chain reaction (PCR) has been used to detect Leptospira in clinical specimens, including blood, serum, urine and cerebrospinal fluid (CSF). However, large quantities of leptospires are needed for detection, can be only intermittently detected in urine or can be cleared from blood and not detected following antibody production if specimens are not obtained early.13
More commonly, serologic testing is utilized for diagnosis of leptospirosis and performed via different techniques. Microscopic agglutination test (MAT) is the gold standard for serological diagnosis.2,14 A significant challenge with this method is the labor-intensive nature of the test and need for a laboratory capable of maintaining the live strains used in testing.12,14 While the specificity of MAT is high (>95%), sensitivity of MAT is low if collected during the first week of illness (~40%) but increases to >95% after the fourth week of illness.12,15 Serological testing can also be performed through enzyme-linked immunoassay, latex agglutination, complement fixation or immunofluorescent assay. However, like MAT serological testing, these techniques have limitations. Given the transient nature, delay in development during the first week of illness as antibodies start to develop during the second week, and absence in certain patients, Leptospira IgM may have false negatives. Further, IgM can persist for months to years and can have high false positive rates.1,13,14
Given the challenges in the diagnosis of leptospirosis and importance of early recognition and timely treatment to mitigate severe infection and potential complications, alternative methods should be considered. The use of rapid diagnostics, particularly plasma cell-free DNA metagenomic sequencing, is an emerging area that has potential to expand in the diagnosis of infectious diseases and supporting case definitions. Utilization of metagenomic and targeted sequencing has been used in some reported cases of leptospirosis with detection in primarily bronchoalveolar lavage fluid. In many of these cases, conventional testing with serology and cultures were negative, which led to utilization of metagenomic sequencing results to influence management.16–21
Specific to this case series, the Karius Test (KT, Redwood City, CA) utilizes plasma microbial cell-free DNA (mcfDNA) sequencing for detection and quantification of DNA-based pathogens. It is offered through the Karius lab, a Clinical Labatory Improvement Amendments certified/College of American Pathologists accredited lab (Redwood City, CA). It detects and quantifies mcfDNA in molecules/μL (MPM) from over 16,000 organisms, reporting on over 1000 clinically relevant pathogens.22,23 This test has been used in patients whose illness is diagnostically challenging and can be applied to pathogens such as Leptospira spp. Here we report a case series of 4 travel-related cases of leptospirosis due to Leptospira santarosai infection where the KT was useful in making a timely diagnosis of leptospirosis.
METHODS
We reviewed 4 cases of pediatric patients presenting with febrile illnesses, in whom the KT detected L. santarosai from April to August 2021. These were the only 4 cases of L. santarosai detected by the Karius lab during this time, in which over 300 institutions utilize the KT. In 3 of the 4 institutions where cases were identified, infectious disease approval and/or consultation is required for ordering the KT. Infectious diseases were consulted in all cases in which the KT was sent as part of provider-directed testing. Samples from cases were run on the KT V3.8 (1 sample) and V3.9 (3 samples). The preferred method of collecting the KT sample is the collection of 5 mL of whole blood in a plasma preparation tube. Specimens are then centrifuged and sent to the Karius lab. Specimens that are not sent within 96 hours of collection are frozen after centrifugation and can subsequently be sent to the Karius lab. Specimens are then processed at the Karius lab and undergo quality control and analysis. Pathogens with plasma DNA higher than background thresholds are typically reported 24 hours after receipt of the specimen. Additionally, consultations are available through Karius infectious diseases physicians and clinical microbiologists for support in results interpretation.22
Review of cases were performed by pediatric infectious diseases consultants. Institutional review board exemption or approval status was obtained in all participating institutions as required by each institution and consent/assent was obtained from all patients.
RESULTS
Four adolescents (3 males, 1 female) with recent foreign travel presented with nonspecific febrile illnesses from April to August 2021 throughout the United States (Table 1). All subjects were ultimately diagnosed with leptospirosis secondary to L. santarosai via the KT (Table 2).
TABLE 1.
Clinical History and Differential Diagnoses of Leptospirosis Cases
| Case 1 | Case 2 | Case 3 | Case 4 | |
|---|---|---|---|---|
| Demographics | ||||
| Sex | M | M | M | F |
| Age range (yr) | 15–18 | 15–18 | 15–18 | 15–18 |
| Clinical features | ||||
| Signs and symptoms | Fevers, chills, malaise, polyarthralgia, myalgias, diarrhea, nonpurulent conjunctivitis | Fevers, chest pain, myalgias, headache, photophobia, eye burning sensation, subsequent rash | Fever, nausea, vomiting, diarrhea, chills, headache, photophobia | Fever, chills, myalgias, emesis, diarrhea, headache, conjunctivitis, malaise, night sweats, presyncope, photophobia, eye pain, oliguria, dark urine |
| Maximum temperature (°C) | 38.3 | 40.2 | 40.0 | 38.9 |
| Travel history and exposures | ||||
| Epidemiological risk or potential exposure | Water rafting in Costa Rica | Waterfall and parturient mouse exposure in Costa Rica | Jungle, caves and river water exposure in Belize | River water exposure in rural Costa Rica |
| Symptom onset relative to return from travel (d) | 4 | 12 | 6 | 4 |
| Differential diagnosis | ||||
| Presumptive diagnoses | Sepsis, leptospirosis, malaria, dengue, chikungunya, Zika, West Nile virus | Rickettsial disease, arboviral infection (domestic and travel associated), Colorado tick fever, dengue, chikungunya virus, typhoid fever | Dengue, leptospirosis, Zika, chikungunya, Rocky Mountain spotted fever, multisystem inflammatory syndrome in children, other viral infection (eg, Epstein-Barr virus, adenovirus) | Leptospirosis, typhoid/enteric fever, dengue |
| Initial suspicion of Leptospira spp. as a clinical or microbiologic cause of presentation | Expected | Unexpected | Expected | Expected |
F indicates female; M, male.
TABLE 2.
Workup and Management of Leptospirosis Cases
| Case 1 | Case 2 | Case 3 | Case 4 | |
|---|---|---|---|---|
| Laboratory manifestations* | ||||
| WBC (×103/μL) with % neutrophils (N) | 7.5 w/77.6%N | 4.42 w/83.6%N | 12.8 w/79%N | 3.88 w/84%N |
| Hgb (g/dL)/Hct (vol%) | 14.7/43 | 15.3/44.1 | 14.8/41.9 | 11.4/35.1 |
| Platelets (×103/μL) | 170 | 165 | 96 | 143→97 |
| Na (mEq/L) | 137 | 135 | 134 | 141 |
| BUN (mg/dL)/Cr (mg/dL) | 16/0.86 | 16/1.01 | 21/1.28 | 7/0.75 |
| ALT (U/L)/AST (U/L) | 74/78→353/204 | 11/30→46/115 | 167/143 | 50/40 |
| ESR (mm/h)/CRP (mg/dL) | NA/37.2 | 17/8→NA/18.1 | 42/16.5 | 58/26.55 |
| Procalcitonin (ng/mL) | 4.030 | 1.46 | NA | 0.25 |
| Infectious studies | ||||
| Blood cultures | Negative | Negative | Pseudomonas oryzihabitans (follow-up × 2: negative) | Negative |
| Diagnostic tests | Monospot; RPR; blood smears; EBV, Zika, chikungunya serologies/PCRs; WNV and dengue serologies | Thick/thin smears; chikungunya, dengue, and EBV serologies; SARS-CoV-2 antibody; HIV Ag/Ab; respiratory pathogen and gastrointestinal panels | Dengue, Zika, chikungunya virus serologies/PCRs; arbovirus panel, Rickettsia rickettsii, WNV, EBV, Bartonella, Lyme serologies; RPR, SARS-CoV-2 antibody; HIV, blood smear | Blood Plasmodium PCR; blood smear; stool culture; urine culture; dengue, Anaplasma phagocytophilum, Ehrlichia chaffeensis and Rickettsia (genus and Rickettsia typhi) serologies |
| Initial management | ||||
| Empiric antibiotic therapy | Ceftriaxone | Penicillin | Ceftriaxone | Doxycycline, ceftriaxone, ciprofloxacin |
| Antibiotic pretreatment duration before the KT (d) | 1 | 0 | 2 | 5 |
| The Karius Test | ||||
| Karius Test (mcfDNA sequencing) result† | Prevotella melaninogenica 171 MPM (RI: 2.4 MPM); | |||
| Leptospira santarosai 284 MPM (RI: 0 MPM) | L. santarosai 378 MPM (RI: 0 MPM) | L. santarosai 28‡ MPM (RI: 0 MPM) | L. santarosai, 4‡ MPM (RI: 0 MPM) | |
| Version of KT | V3.8 | V3.9 | V3.9 | V3.9 |
| Day of illness collected (d) | 6 | 5 | 10 | 7 |
| Turnaround time from collection/sample receipt (d) | 3/2 | 2/1 | 3/1 | 2/1 |
| Leptospirosis serological testing | ||||
| Leptospira serology (IgM) | Positive | Negative | Equivocal | Positive |
| Day of illness collected (d) | 10 | 5 | 12 | 14 |
| Turnaround time from collection (d) | 3 | 4 Convalescent serology (31 d after symptom onset): Positive |
2 | 6 |
| Subsequent management and outcome | ||||
| Targeted antibiotic therapy after diagnosis | Azithromycin | Doxycycline | Doxycycline | Ciprofloxacin |
| Hospitalization duration (d) | 5 | 6 | 5 | 7 |
| Outcome | Full recovery | Full recovery | Full recovery | Full recovery |
General reference ranges (slight variations may be seen with some parameters performed in different laboratories): WBC, 5.2–9.7k × 103/μL; Hgb, 11.8–15.8 g/dL; Hct, 34–46 vol%; Plt, 150–500 × 103/μL; Na, 134–143 mEq/L; BUN, 8–21 mg/dL; Cr, 0.42–0.9 mg/dL; ALT, 11–26 U/L; AST, 15–40 U/L; ESR, 0–15 mm/h; CRP, 0.0–0.9 mg/dL; PCT, <0.10 ng/mL.
RI, reference interval that denotes the 97.5%ile MPM of a specific pathogen’s mcfDNA in a healthy cohort of 768 subjects.
Detection below the Karius commercial threshold, disclosed in clinical consultation.
Ab indicates antibody; Ag, antigen; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; Cr, creatinine; CRP, C-reactive protein; EBV, Epstein-Barr virus; ESR, erythrocyte sedimentation rate; Hct, hematocrit; Hgb, hemoglobin; HIV, human immunodeficiency virus; %N, neutrophil percentage; NA, not applicable; Na, sodium; PCT, procalcitonin; Plt, platelet; RI, reference interval; RPR, rapid plasma reagin; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WBC, white blood cell count; w, with; WNV, West Nile virus.
Case 1
An adolescent male presented to care after developing symptoms 4 days after return from a trip to Costa Rica. He presented with fever, malaise, polyarthralgia, myalgias, conjunctivitis and gastrointestinal symptoms. He reported having gone whitewater rafting while in Costa Rica. A diagnosis of leptospirosis was suspected and he was empirically started on ceftriaxone. In addition to leptospirosis serologic testing with Leptospira IgM, other infectious diseases testing was performed and a KT was sent (collected on day of illness 6). KT returned positive for L. santarosai and ultimately his Leptospira IgM also resulted positive 3 days after it was initially collected. He was eventually discharged with azithromycin and fully recovered.
Case 2
An adolescent male with a recent trip to Costa Rica presented to care with fever, chest pain, myalgias, headache, photophobia and eye burning sensation that began 12 days after return from his trip. He recalled heavy rainfall, visiting a waterfall and parturient mouse exposure during his travel. While admitted, he also developed a rash (Fig. 1). A broad infectious workup was performed including Leptospira IgM. Ultimately, KT detected L. santarosai (collected on day of illness 5). He was then started on penicillin and discharged home on a course of doxycycline with complete recovery. Acute Leptospira IgM resulted negative 4 days after being collected but convalescent IgM obtained 31 days after symptom onset returned positive with a turnaround time of 3 days.
FIGURE 1.
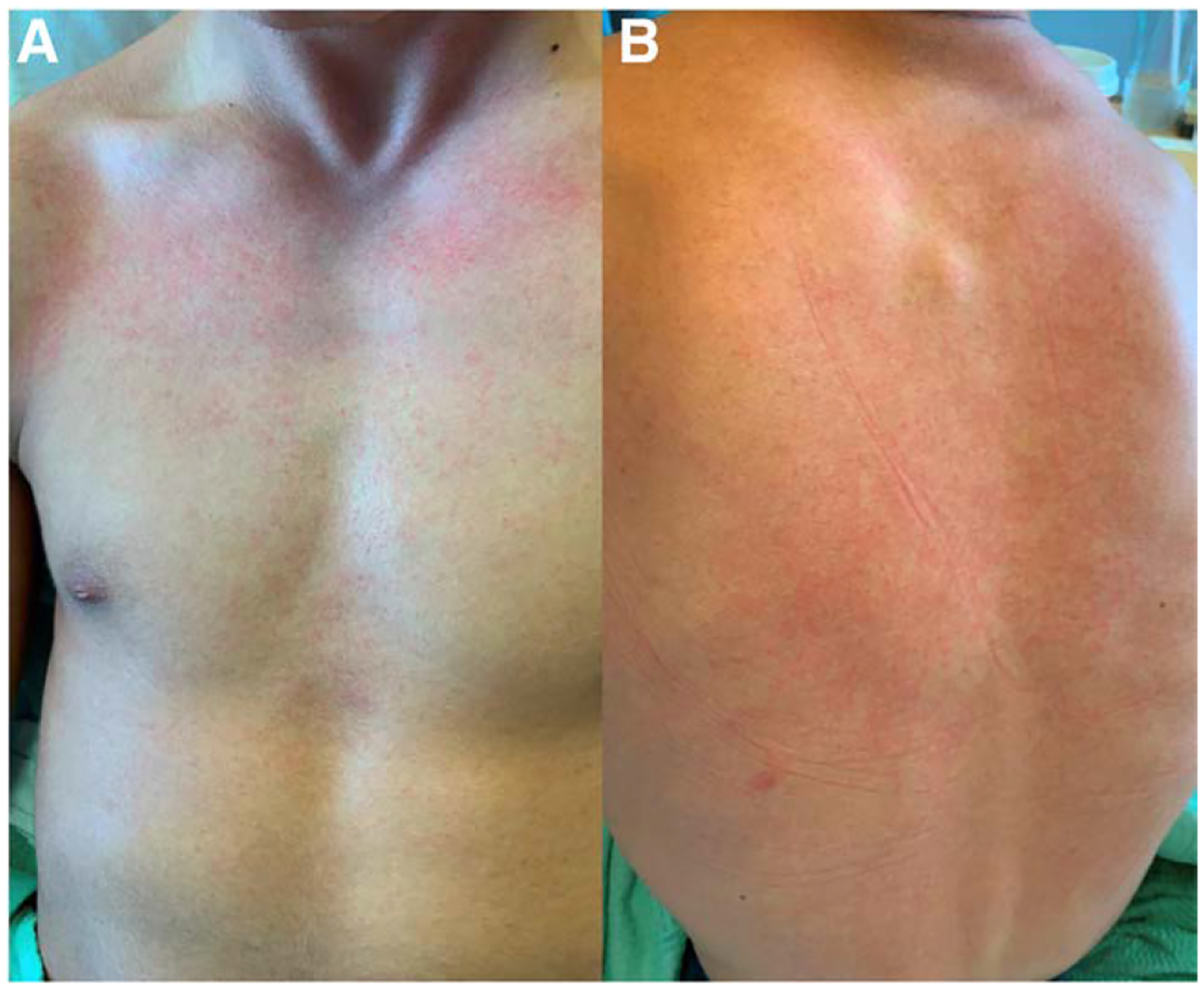
Rash presenting during hospitalization of case 2. A: Present on chest. B: Present on back.
Case 3
An adolescent male presented with fever, gastrointestinal symptoms, headache and photophobia starting 6 days after return from a trip to Belize where he explored the jungle and caves and had river water exposure. Leptospirosis was suspected in which Leptospira IgM was sent and he was started on empiric therapy with ceftriaxone. Additional studies were sent and a KT obtained 10 days after illness onset returned with detection of L. santarosai. Though the level was below the commercial threshold, the result was disclosed following clinical consultation with the Karius lab. Acute phase Leptospira IgM result returned “equivocal” 2 days after being collected; however, given the high suspicion for leptospirosis, he was treated with a course of doxycycline and completely recovered. Of note, initial blood culture was positive for Pseudomonas oryzihabitans but was not thought to be pathogenic given repeat cultures were subsequently negative.
Case 4
An adolescent female presented with fever, chills, myalgias, gastrointestinal symptoms, headache and conjunctivitis that started 4 days after return from Costa Rica where she had exposure to river water. A broad infectious disease workup was performed including Leptospira IgM and ultimately the KT (collected on day of illness 7) returned with L. santarosai and Prevotella melaninogenica from a specimen taken after 5 days of antibiotic treatment. The L. santarosai MPM was below the commercial threshold and was disclosed following clinical consultation. The P. melaninogenica was thought to represent commensal flora rather than a pathogen. She was treated with multiple antibiotics including ceftriaxone, doxycycline and ciprofloxacin during her admission, but was sent home with a course of ciprofloxacin. She fully recovered. Her acute phase Leptospira IgM ultimately returned positive 6 days after being collected.
Each patient in this case series fully recovered. The average time to diagnosis using the KT was 2.5 ± 0.58 days from sample collection, with average L. santarosai mcfDNA concentration of 173.5 ± 186.13 MPM, compared to average turnaround time of Leptospira IgM result from sample collection of 3.75 ± 1.71 days (Table 2).
DISCUSSION
During a 5-month period in 2021, 4 cases of travel-associated leptospirosis secondary to L. santarosai were identified through the KT, which was the first test to identify L. santarosai as the microbiological diagnosis in all cases. While leptospirosis was suspected in some of these cases, a broad workup was still performed in each case, given the nonspecific symptoms and laboratory manifestations, highlighting the challenges with the diagnosis of leptospirosis.
In all cases, Leptospira IgM was also sent; however, the turnaround time from sample collection was longer compared with the KT. Results of the KT allowed for prompt and targeted treatment with full recovery in all patients and therefore the KT may be more advantageous in certain cases compared to more traditional testing with longer turnaround times, particularly when early treatment is crucial. While the sensitivity and specificity of the KT for the detection Leptospira is unknown, in our case series, the KT was able to detect Leptospira both within the first week of illness and later in the second week of illness. As Leptospira antibodies typically develop later and thus the sensitivity is lower during the first week of illness and Leptospira PCR is better detected early with lower sensitivity later in illness, the KT also provides potential benefit in being able to detect Leptospira in both early and later courses of ilness.4
In all 4 cases, the species L. santarosai was identified. This species is commonly found in Latin America but also has been reported in other areas such as Taiwan.17,24–26 Like other species, specific serovars of L. santarosai have been known to cause leptospirosis and severe infections. Utilization of metagenomic sequencing was previously reported in one case where L. santarosai was detected in CSF, rather than in plasma as described in our case series.27,28 Wilson et al reported a case of a 14-year-old male with severe combined immunodeficiency who was status post 2 bone marrow transplants and presented with fever, headaches, seizures and encephalitis. He had travel to Puerto Rico the year prior. Initial studies for leptospirosis, including serological testing through enzyme-linked immunoassay, culture and PCR were negative. Brain biopsy was also unrevealing for infectious pathogens. Ultimately metagenomic sequencing on the CSF was sent and identified L. santarosai, which was subsequently confirmed on repeat PCR testing. Upon initiation of targeted treatment, seizure activity resolved, and he recovered in the following week.28 Given his immunocompromised status, serological testing was unreliable, necessitating use of alternative diagnostic methods such as metagenomic sequencing.
Beyond leptospirosis, plasma mcfDNA sequencing has been increasingly utilized, reported and validated to aid in the diagnosis of infections. In analytical and clinical validation of the KT, there was a 93.7% agreement in blood cultures and KT results in a cohort of 350 patients with a sepsis alert. In a retrospective review of 79 patients in which 100 KTs were sent, the KT had a sensitivity and specificity of 92% and 64%, respectively, for diagnosis of clinically relevant infections compared with 77% and 89% with conventional testing.29,30 In an analysis of patients being evaluated for acute infective endocarditis, the KT identified the same pathogens in the 20 cases with positive cultures.31
Plasma mcfDNA sequencing has also been applied in diagnostic challenges such as evaluation for infection in immunocompromised patients. In this patient population, invasive procedures to obtain source cultures are often not feasible given the risks of these procedures. These patients are also often treated with multiple antimicrobials or immunosuppressive agents that may confound traditional test results. In a cohort of 10 immunocompromised patients, the KT had a positive agreement in pathogen identification compared to more conventional diagnostic methods in 70% of cases. In 2 of the 3 discordant sepsis cases, the KT identified potential microbiological agents that were not detected by culture.32 In addition, plasma mcfDNA sequencing can evaluate for multiple pathogens at once when the differential diagnosis is broad or in diagnostic conundrums where conventional testing is limited or not feasible secondary to time, fastidious nature of pathogens or inability to perform invasive tests.
This platform also has potential to enhance epidemiologic investigation, as results can support case definitions of reportable infections, aiding in surveillance or outbreak investigation. It can also inform regarding the epidemiology of a species. Leptospirosis is a notifiable infection in the United States and in all these cases, patients had travel to Central America. Through use of the KT, we were able to identify this cluster of cases, while geographically distributed in the United States, all linked to a similar area of travel in a similar time frame.
We recognize that this retrospective case series is limited by the small sample of cases. In addition, in 2 of the cases, the MPM was below the commercial threshold. In 1 of these cases, the KT was obtained later in their illness course and in the other case was obtained after several days of antibiotic treatment that could have affected results. However, serologic testing was positive and confirmed diagnosis in 1 of the 2 cases and in the other case, clinical suspicion for leptospirosis was high without other plausible diagnosis. Though the MPM was below commercial reporting threshold, the reference background should not contain Leptospira spp. DNA and disclosure of this result through clinical consultation with Karius were useful in supporting the clinical suspicion and timely diagnosis in both cases. Given the low number of reads detected in case 4, species resolution is made with less confidence; however, each read needs to align at >90% to the species level genomic reference to be reported. Additionally, while the turnaround time from sample collection to report of test result was quicker for the KT as opposed to the Leptospira IgM, this time includes factors other than running the assay in itself (eg, delays with batch testing for Leptospira IgM as tests may only be performed on scheduled days of the week in some reference labs).
Using plasma mcfDNA sequencing, we were able to identify 4 cases of L. santarosai infections in patients with recent travel to Central America over a 5-month period. Plasma mcfDNA sequencing when used in the appropriate clinical context can help shorten time to diagnosis for more targeted treatment, potentially improve prognosis for patients and may have broader implications for epidemiologic or outbreak surveillance.
Footnotes
A.A.A. was an employee of Karius, Inc at the time the manuscript was prepared but no longer works for Karius, Inc. For the remaining authors, there are no conflicts of interest to disclose.
An abstract of this case series was presented as a poster for IDWeek 2022 in October 2022, Washington D.C., USA, and subsequently the abstract was published in Open Forum Infectious Diseases Volume 9, Issue Supplement 2 in December 2022 as a result of the poster presentation. One case described in this case series was presented as an oral case presentation for Pediatrics Fellows’ Day for IDWeek 2022 in October 2022, Washington D.C., USA.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.American Academy of Pediatrics Committee on Infectious Diseases. Leptospirosis Red Book: 2021–2024 Report of the Committee on Infectious Diseases. American Academy of Pediatrics; 2021:475–477. [Google Scholar]
- 2.Centers for Disease Control and Prevention. Leptospirosis. 2019. [cited March 29, 2022]. Available at: https://www.cdc.gov/leptospirosis/index.html. Accessed December 15, 2023.
- 3.Shapiro ED. Leptospira species (leptospirosis). In: Long S, Prober C, Fischer M, eds. Principles and Practice of Pediatric Infectious Diseases. 5th ed. Elsevier; 2018:977–980. [Google Scholar]
- 4.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of Prokaryotic names with Standing in Nomenclature: Genus Leptospira. 2020. [cited November 1, 2022]. Available at: https://www.bacterio.net/genus/leptospira. Accessed December 15, 2023. [DOI] [PMC free article] [PubMed]
- 6.Centers for Disease Control and Prevention (CDC). Outbreak of acute febrile illness among athletes participating in triathlons--Wisconsin and Illinois, 1998. MMWR Morb Mortal Wkly Rep. 1998;47:585–588. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Outbreak of acute febrile illness among participants in EcoChallenge Sabah 2000--Malaysia, 2000. JAMA. 2000;284:1646. [PubMed] [Google Scholar]
- 8.Costa F, Hagan JE, Calcagno J, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS NeglTrop Dis. 2015;9:e0003898-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misao T, Hiroyoshi S, Katsuta K, et al. Canicola fever in Japan. Am J Hyg. 1956;63:294–307. [DOI] [PubMed] [Google Scholar]
- 10.Assimakopoulos SF, Fligou F, Marangos M, et al. Anicteric leptospirosis-associated severe pulmonary hemorrhagic syndrome: a case series study. Am J Med Sci. 2012;344:326–329. [DOI] [PubMed] [Google Scholar]
- 11.Pan American Health Organization. Leptospirosis [cited April 10, 2022]. Available at: https://www.paho.org/en/topics/leptospirosis. Accessed December 15, 2023.
- 12.Musso D, La Scola B. Laboratory diagnosis of leptospirosis: a challenge. J Microbiol Immunol Infect. 2013;46:245–252. [DOI] [PubMed] [Google Scholar]
- 13.Budihal SV, Perwez K. Leptospirosis diagnosis: competancy of various laboratory tests. J Clin Diagn Res. 2014;8:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goris MG, Hartskeerl RA. Leptospirosis serodiagnosis by the microscopic agglutination test. Curr Protoc Microbiol. 2014;32:Unit 12E.5. [DOI] [PubMed] [Google Scholar]
- 15.Bajani MD, Ashford DA, Bragg SL, et al. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. J Clin Microbiol. 2003;41:803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao Q-H, Yu L, Ding J-J, et al. Severe community-acquired pneumonia caused by Leptospira interrogans: a case report and review of literature. World J Clin Cases. 2021;9:1901–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou LF, Chen YT, Lu CW, et al. Sequence of Leptospira santarosai serovar Shermani genome and prediction of virulence-associated genes. Gene. 2012;511:364–370. [DOI] [PubMed] [Google Scholar]
- 18.Dong W-H, Chen Z Leptospirosis with pulmonary haemorrhage and multiple organ failure: a case report and literature review. J Int Med Res. 2021;49:3000605211019665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Hu J, Yu S, et al. Next generation sequencing for diagnosis of leptospirosis combined with multiple organ failure: a case report and literature review. Front Med (Lausanne). 2021;8:756592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng W, Liqiang L, Yuhai B, et al. A severe Leptospira interrogans serovar Copenhageni infection diagnosed by next-generation sequencing and treated with corticosteroids. Arch Clin Microbiol. 2017;08:1–10. [Google Scholar]
- 21.Chen M, Lu W, Wu S, et al. Metagenomic next-generation sequencing in the diagnosis of leptospirosis presenting as severe diffuse alveolar hemorrhage: a case report and literature review. BMC Infect Dis. 2021;21:1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karius. 2022. [cited April 10, 2022]. Available at: https://kariusdx.com/. Accessed December 15, 2023.
- 23.Morales M The next big thing? Next-generation sequencing of microbial cell-free DNA using the Karius Test. Clin Microbiol Newsl. 2021;43:69–79. [Google Scholar]
- 24.Kallel H, Bourhy P, Mayence C, et al. First report of human Leptospira santarosai infection in French Guiana. J Infect Public Health. 2020;13:1181–1183. [DOI] [PubMed] [Google Scholar]
- 25.Peláez Sanchez RG, Lopez J, Pereira MM, et al. Genetic diversity of Leptospira in northwestern Colombia: first report of Leptospira santarosai as a recognised leptospirosis agent. Mem Inst Oswaldo Cruz. 2016;111:737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valverde MLA, Goris MGA, González V, et al. New serovars of Leptospira isolated from patients in Costa Rica: implications for public health. J Med Microbiol. 2013;62:1263–1271. [DOI] [PubMed] [Google Scholar]
- 27.Arnold C. Source code: putting metagenomics to the test in the clinic. Nat Med. 2017;23:645–648. [DOI] [PubMed] [Google Scholar]
- 28.Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuro-leptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4:663–674. [DOI] [PubMed] [Google Scholar]
- 30.Rossoff J, Chaudhury S, Soneji M, et al. Noninvasive diagnosis of infection using plasma next-generation sequencing: a single-center experience. Open Forum Infect Dis. 2019;6:ofz327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah P, Ruffin F, Seng H, et al. 156 direct detection and quantification of bacterial cell-free DNA in patients with infective endocarditis (IE) using the Karius plasma next generation sequencing (NGS) test. Open Forum Infect Dis. 2018;5(Suppl 1):S12–S12. [Google Scholar]
- 32.Camargo JF, Ahmed AA, Lindner MS, et al. Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000Res. 2019;8:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]


