Abstract
RNA secondary structures, e.g. stem–loops that are often found at the 5′ and 3′ ends of mRNAs, are in many cases known to be crucial for transcript stability but their role in prolonging the lifetime of transcripts remains elusive. In this study we show for an essential RNA-stabilizing stem–loop at the 5′ end of rbcL gene transcripts in Chlamydomonas that it neither prevents ribonucleases from binding to the RNA nor impedes their movement along the RNA strand. The stem–loop has a formative function in that it mediates folding of a short sequence around its base into a specific RNA conformation, consisting of a helical and single-stranded region, i.e. the real structure required for longevity of rbcL transcripts in chloroplasts. Disturbing this structure renders transcripts completely unstable, even if the sequence of this element is not altered. The requirement of a specific 5′ sequence and structure for RNA longevity suggests an interaction of this element with a trans-acting factor that protects transcripts from rapid degradation in chloroplasts.
INTRODUCTION
Lifetimes of individual mRNA populations differ considerably in cells, often ranging from a few minutes for short-lived messages to several hours for stable transcripts (1,2). The widespread occurrence of very short-lived mRNAs alongside stable mRNAs shows that the ribonucleases responsible for degradation of mRNAs are capable of degrading transcripts rapidly and that long-lived mRNAs must have features that protect them from ribonucleolytic attack and/or from immediate degradation. Analyses of mRNA stability in prokaryotes and eukaryotes show that key determinants of stability are located predominantly in the 5′- and 3′-untranslated regions (5′- and 3′-UTRs) of transcripts. In eukaryotes, elements important for mRNA longevity have been delineated primarily in the 3′ regions of transcripts (3–5), whereas in prokaryote-type mRNAs, i.e. bacterial and organelle mRNAs, essential determinants of mRNA longevity seem to be located mostly in the 5′-UTR (6,7). Secondary structures at the 5′ ends of prokaryote-type mRNAs have been found to be important for mRNA stability, examples being stem–loops at the 5′ ends of mRNAs in Escherichia coli and Bacillus subtilis (8–10) and in the 5′-UTR of mRNAs in chloroplasts (11–13). Despite the importance of these 5′ secondary structures for mRNA longevity, their exact role in preventing rapid degradation of transcripts has been difficult to assess, even though it is often inferred that these structures function as binding sites for proteins and/or as structural elements that bar ribonucleases from attaching to and/or moving along the mRNA strand.
In this study, we examined in vivo the function of an essential RNA-stabilizing stem–loop that forms at the 5′ end of transcripts of the Chlamydomonas chloroplast rbcL gene. After joining the 5′ region of the rbcL gene to the coding region of the bacterial uidA (GUS) gene, we altered the sequence and secondary structure of the stem–loop in numerous ways and measured accumulation of the resulting transcript variants in Chlamydomonas chloroplast transformants in vivo. The analyses confirm that the 5′-terminal stem–loop is crucial for longevity of rbcL gene transcripts in chloroplasts. However, even though essential for transcript stability, the stem–loop is not the structure that shields transcripts from attack or degradation by ribonucleases. The stem–loop mediates folding of a previously identified sequence element around its base into a specific RNA conformation that our analyses reveal to be the real structure required for protection against degradation.
MATERIALS AND METHODS
Algae and growth conditions
The atpB-deficient mutant strain CC-373 (ac-uc-2-21) of Chlamydomonas reinhardtii—the recipient of all DNA constructs—was obtained from the Chlamydomonas Genetics Center at Duke University, NC, USA. The light-sensitive mutant was grown on a shaker in low light conditions (≈0.05 µmol/s·m2) at room temperature in high salt (HS) medium supplemented with potassium acetate (2.5 g/l) (14). Photosynthetic transformants of CC-373 were grown in high light (≈50 µmol/s·m2) on HS minimal medium in 200 ml tubes in a temperature-controlled water bath at 32°C. Water bath cultures were mixed by continuous bubbling with 2%CO2-enriched air.
Transformation of the Chlamydomonas chloroplast
All DNA constructs were cloned into a chloroplast transformation vector (see below) and precipitated onto 0.6 µm gold particles for bombardment of mutant CC-373 via a particle delivery system (PDS-1000/He; Bio-Rad) as described previously (15,16). Stable transformants were selected on HS agar plates in high light and were screened for the presence of the uidA gene and its transcripts by Southern and northern analyses (16). Only cell lines that were 80–100% homoplasmic were used in the analyses.
Modifications at the 5′ end of the rbcL 5′-UTR
Complementary oligonucleotides (obtained from MWG Biotech, Ebersberg, Germany), containing the sequence or variants of the sequence between the SwaI and BspEI sites of the rbcL 5′-UTR (Figure 1), were annealed, phosphorylated and ligated into the SwaI/BspEI sites of vector +157/SK+ (17) using standard molecular biology techniques (18). The constructs were released from +157/SK+ by digestion with XhoI/XbaI and cloned into the XhoI/XbaI restriction sites of transformation vector pCrc32 (19).
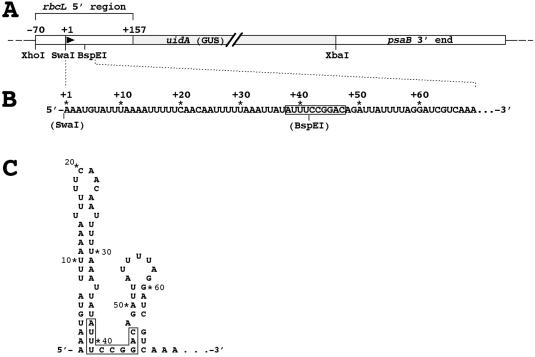
Figure 1.
The chimeric [5′ rbcL:GUS:psaB 3′ end] gene inserted into the Chlamydomonas chloroplast genome and the secondary structure at the 5′ end of its transcripts. (A) The 5′ region of the rbcL gene used in all constructs extends from position −70 to position +157, relative to the start site of transcription at +1. It includes the rbcL gene promoter, the UTR and a portion of the coding region of the rbcL gene containing a transcription-enhancing sequence (data not shown) at around position +126. All restriction sites used in cloning are shown. (B) Original nucleotide sequence at the 5′ end of transcripts of the chimeric reporter gene. The restriction sites shown denote the ends of the portion of the transcripts that were replaced by the modified sequences listed in Figures 2–4. A previously identified transcript-stabilizing sequence is boxed (20). (C) RNA secondary structure at the transcripts' 5′ end as predicted by RNA folding programs and verified in vivo by alkylation with dimethyl sulfate (20).
5′ RNA secondary structures
The native secondary structure of the 5′ end of transcripts of the C.reinhardtii rbcL gene has been determined previously by alkylation with dimethyl sulfate (20). The experimentally determined structure is identical to the structure predicted by version 3.0 of the mfold program (21). Secondary structures shown in Figures 2–4 are the structures predicted by the mfold server at the Burnet Institute (http://mfold.burnet.edu.au/) using a temperature of 32°C and the default settings on the web server. In all cases, except for constructs 4 and 15, folding with the mfold algorithm resulted in just one structure for the rbcL 5′ end. Structures for constructs 4 and 15 given in Figures 2 and 4, respectively, are the most likely structures predicted, i.e. the structures with the lowest free energy value. All structures were re-drawn for uniform and space-saving appearance in the figures. Most of the modified 5′ structures could be predicted without using RNA folding software. For example, the sequence in construct 3 was designed in order to restore the terminal stem–loop that has been destroyed by the changes introduced into the sequence of construct 2. Similarly, the 5′ structures of transcripts from constructs 7, 8, 9, 10, 11, 12, 13 and 14 are easily predictable, when compared with the structure of the original 5′ sequence. Sequences for which our predictions did not match the results obtained with the mfold program were not cloned in order to avoid ambiguous results.
Figure 2.
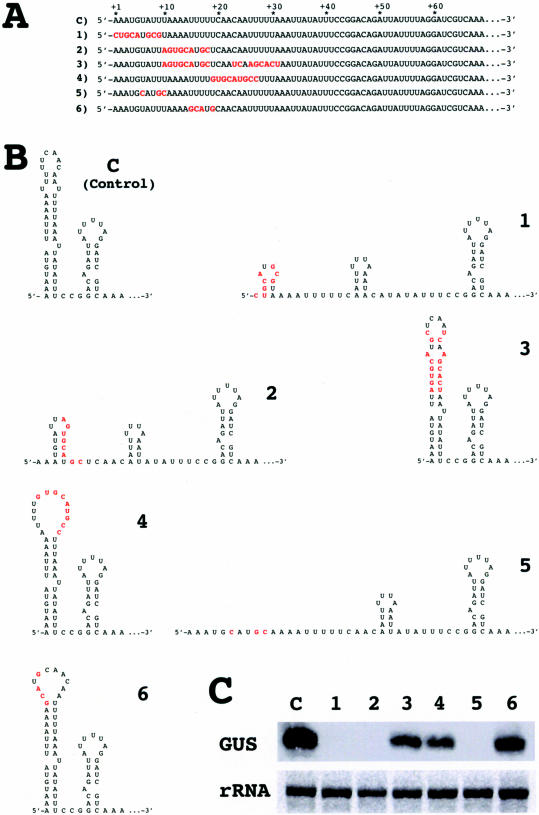
A 5′-terminal stem–loop is required for longevity of transcripts in chloroplasts. (A) Modifications in the sequence at the 5′ end of the rbcL 5′-UTR. C, control. (B) Predicted 5′ RNA secondary structures after changing the nucleotides marked in red in the sequence. Changes preserve, destroy or restore the terminal stem–loop structure without altering the total number of nucleotides in the 5′-UTR. (C) Levels of GUS transcripts in chloroplast transformants harboring constructs 1–6. rRNA, levels of the largest ribosomal RNA band on the RNA gel. Four micrograms of total RNA were loaded in each lane.
Figure 4.
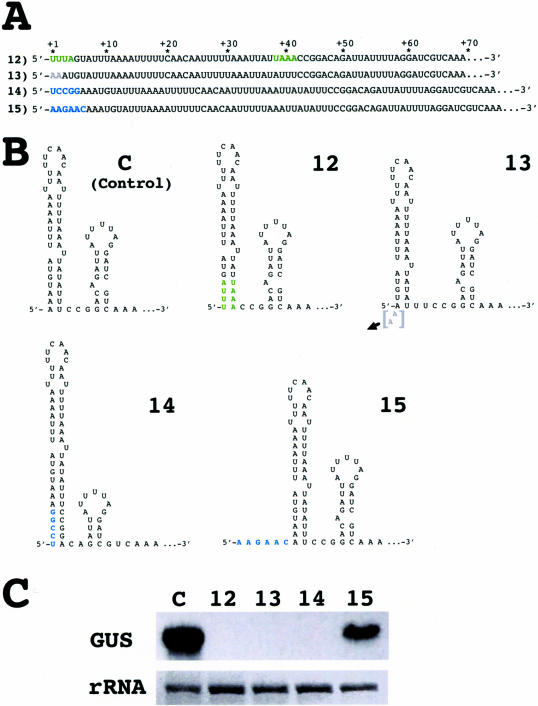
Altering the RNA conformation around the base of the 5′-terminal stem–loop destabilizes transcripts. (A) Modifications in the sequence at the 5′ end of the rbcL 5′-UTR. The modifications are color-coded as described in Figure 3. (B) Predicted RNA secondary structures of the sequences specified in A. (C) Levels of GUS transcripts in chloroplast transformants harboring constructs 12–15.
Southern/northern blots and DNA probes
Genomic DNA and total RNA were isolated from Chlamydomonas cells as described previously (14). DNA was isolated from cells growing in continuous light, whereas RNA was isolated at 11 h in the dark period from cells growing in 12 h light/12 h dark cycles. Sequences of the coding regions of the uidA gene (1.9 kb BamHI–SacI restriction fragment from plasmid pBI221; Clontech, CA, USA) and of the Chlamydomonas chloroplast atpB gene (0.7 kb HpaI–EcoRV restriction fragment) were random primer-labeled with [32P]dCTP and were used as probes in RNA and DNA gel blot analyses as described previously (7).
RESULTS
The chimeric [rbcL 5′ region:uidA (GUS):psaB 3′ end] reporter gene construct, from which all variants analyzed in this study are derived, is shown in Figure 1. The rbcL promoter that directs transcription of the GUS reporter gene is the strongest promoter known of all protein coding genes in Chlamydomonas (16). Transcripts of the reporter gene accumulate to high levels in the chloroplast of Chlamydomonas and are long-lived, having a half-life of ∼5 h (7). It has been shown previously (17) that the rbcL 5′-UTR sequence does not contain a promoter element because significant changes in the sequence of the UTR or altering its distance to the basal promoter sequence located around position −10 relative to the start site of transcription do not abolish transcription. Therefore, levels of GUS transcripts shown in this study reflect essentially their stability in vivo.
In the following section, RNA gel blot (northern) analyses are used to discern 5′ RNA features that result in stable or unstable transcripts. Compared with control levels, levels of stable transcripts of the modified rbcL 5′-UTR:GUS constructs varied in these analyses. We attribute these variations in part to differences in copy numbers of the reporter gene in transformants and to real variations in transcript longevity caused by the sequence modifications. These deviations from control levels were not quantified, nor were half-lives of stable transcript variants determined, as the main objective of the study was to distinguish stable from extremely unstable transcripts, i.e. transcripts that do accumulate from transcripts that do not accumulate in chloroplast transformants, and not to distinguish stable from less stable transcripts.
Requirement of a 5′ stem–loop for RNA stability
Transcripts of the Chlamydomonas rbcL gene are predicted to fold at their 5′ end into two stem–loop structures that are separated by a short single-stranded region (Figure 1). The single-stranded region is part of a previously identified RNA-stabilizing sequence (20) that extends by a few nucleotides on each side into the bottom of both stem structures. The presence of the predicted 5′ structure has been verified in vivo by alkylation with dimethyl sulfate (20). Recent studies suggested that the large 5′-terminal stem–loop is crucial for stability of rbcL transcripts in the Chlamydomonas chloroplast (17).
To investigate the RNA-stabilizing function of the5′-terminal stem–loop structure, a number of sequence variants of the rbcL 5′-UTR were constructed (Figure 2A) that either retained (constructs 4 and 6), destroyed (constructs 1, 2 and 5), or restored (construct 3) the stem–loop (Figure 2B) without altering the total number of nucleotides in the 5′-UTR. Transcripts from these constructs accumulated in chloroplast transformants only when the 5′-terminal stem–loop could possibly form (Figure 2C; constructs 3, 4 and 6). In all cases in which the nucleotide sequence did not allow folding into a 5′-terminal stem–loop, transcripts of the GUS reporter gene were below levels detectable by conventional northern blot analysis. Unambiguous evidence for the requirement of a 5′-terminal stem–loop for transcript stability comes from comparing the accumulation of GUS transcripts in transformants carrying constructs 2 or 3. The changes introduced in construct 2 hinder formation of the native stem–loop structure, and transformants carrying this construct do not accumulate GUS transcripts (Figure 2B and C). Additional complementary changes introduced in construct 3 restore the structure and result in accumulation of transcripts.
Role of sequence and shape of the stem–loop
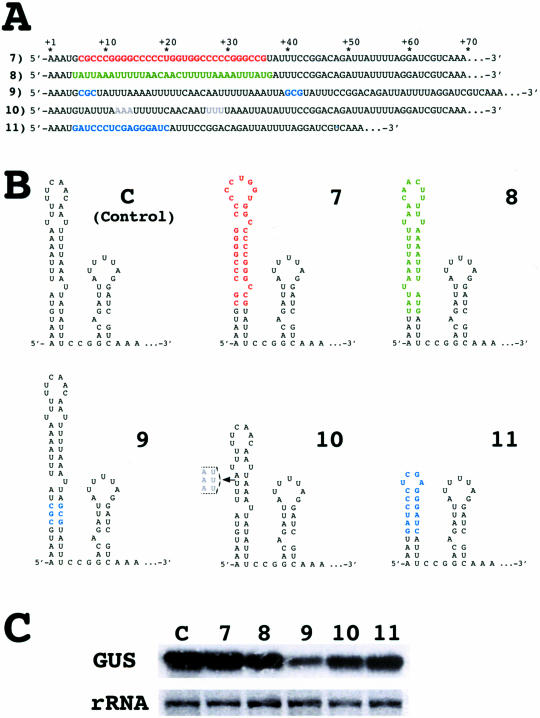
To find out whether the native terminal stem–loop of the rbcL 5′-UTR can be replaced by a different stem–loop structure regardless of sequence and shape, the size, the shape and most of the nucleotide sequence of the stem–loop were altered (Figure 3A and B). GUS transcripts accumulated in all transformants carrying constructs with altered stem–loops (Figure 3C) despite significant differences in their sequences, size and shape. In construct 7 all the purine (A) and pyrimidine (U, C) nucleotides between positions +6 and +36 were replaced by alternative purine (G) and pyrimidine (C, U) nucleotides. In construct 8 the sequence between positions +5 and +37 was inverted. In both cases, the different stem–loop sequences—that also result in slightly different shapes of the terminal stem–loop—had no drastic effect on transcript accumulation (Figure 3C). Extending the stem of the5′-terminal stem–loop by 3 bp (construct 9) or shortening it by 3 bp (construct 10) did not prevent accumulation of GUS transcripts indicating that the length of the stem is not crucial for transcript stability. Finally, GUS transcripts accumulated even when a large portion of the native 5′-terminal stem–loop was replaced with a short but perfect hairpin structure (Figure 3B; construct 11) that has been shown to stabilize transcripts in E.coli (22). These results essentially exclude any specific role of the main portion of the stem–loop in stabilizing transcripts in the Chlamydomonas chloroplast.
Figure 3.
Sequence, size and shape of the 5′-terminal stem–loop are not crucial for RNA stability. (A) Modifications in the sequence at the 5′ end of the rbcL 5′-UTR. The modifications are color-coded as follows: red, change of nucleotide; green, inversion of sequence; blue, insertion or addition of foreign sequence; dim grey, deletion of sequence (in construct 11 the entire sequence from positions +5 to +37 is replaced by a foreign sequence). Changes preserve a stem–loop structure but alter its sequence, shape and/or size. (B) Predicted 5′ RNA secondary structures after changing, adding or deleting the nucleotides marked by color. (C) Levels of GUS transcripts in chloroplast transformants harboring constructs 7–11.
Formative function of the 5′-terminal stem–loop
In all variants of the 5′-terminal stem–loop described above, the 4 bp at the bottom of the stem were deliberately left untouched because previous studies have shown that single nucleotide changes in this region render transcripts unstable (20). The results presented above led us to conclude that any 5′-terminal stem–loop structure suffices to stabilize transcripts of the reporter gene in the Chlamydomonas chloroplast. However, analyses of another set of reporter gene constructs revealed that, even in the presence of a 5′-terminal stem–loop, transcripts fail to accumulate if the RNA conformation around the base of the stem is disturbed (Figure 4). Swapping the nucleotides at positions +1 to +4 with their complementary nucleotides at positions +38 to +41 (Figure 4A; construct 12), which changes the sequence but not the structure at the bottom of the terminal stem–loop (Figure 4B), abolishes transcript accumulation completely (Figure 4C). Deletion of the first 2 nt of the transcript, resulting in a shorter stem and altering the conformation of the sequence at its base (construct 13), also renders transcripts unstable. In this case, shortening the stem can be excluded as a cause of degradation of the transcripts because transcripts with a shorter stem at their 5′ end were even found to be stable (Figure 3; construct 10). Extending the 5′-terminus of the transcripts by 5 nt that pair with nt 42–46 further downstream (construct 14) leads also to destabilization of the transcripts, despite the presence of a stem–loop at the 5′-terminus that is more stable than its native counterpart. It is important to note that the sequence and conformation of the original stem–loop is contained in transcripts of construct 14, these transcripts differing from control transcripts only in the incorporation of nt 42–46 into a helical conformation at the base of the stem–loop 14 (Figure 4B). As shown by accumulation of transcripts from construct 15, which adds six unpaired nucleotides to the RNA's 5′-terminus, it is not the addition of nucleotides to the transcripts' 5′ end that destabilizes transcripts of construct 14 but the conformational change at the base of the stem–loop (Figure 4B and C).
In conclusion, transcripts of chimeric [rbcL 5′-UTR:GUS: psaB 3′ end] genes are stable in the chloroplast of C.reinhardtii only when folding of the nucleotides around position +40 of the 5′-UTR into a specific conformation, consisting of a helical and a single-stranded region, is possible. The 5′-terminal stem–loop contributes to folding of the RNA sequence into this conformation by providing the helical portion of the specific secondary structure but, by itself, does not prevent RNA degradation.
DISCUSSION
Identification of cis-acting elements in RNA sequences
Proper and accurate identification of cis-acting elements is a prerequisite for further analyses of their role in gene expression. For RNA sequences, correct identification of cis-acting elements should ideally be complemented with information on their in vivo conformation because of the potential importance of secondary structures for the function of the elements. Native RNA secondary structures can be critical in in vivo and in vitro assays, e.g. RNA mobility shift assays that are widely used to determine interactions of RNA sequences with trans-acting factors. Unless accompanied by additional analyses, deletions and conventional cloning techniques, which often introduce extra nucleotides into nucleic acid sequences, do not seem to be adequate methods for locating precisely cis-acting elements in RNA sequences. For example, without further analysis, the nucleotides in positions +10 to +18 of construct 2 or positions +6 to +9 of construct 5 (Figure 2) can easily be regarded to belong to a cis-acting RNA-stabilizing element because altering these nucleotides destabilizes transcripts. Additional analyses reveal (Figure 3) that these nucleotides have no specific role in stabilization of transcripts. In light of these observations, we consider it possible that several of the cis-acting elements found in RNA sequences to date do not have the primary function ascribed to them. Revealing their true function requires analyses beyond deletions and a limited number of mutations, which, however, in most cases may be very difficult or impossible to perform.
Role of the 5′-terminal stem–loop
The finding that chloroplast transcripts ending in a stable 5′ stem–loop can be degraded rapidly (Figure 4; constructs 12, 13 and 14) implies that a 5′-terminal stem–loop is generally not a structural impediment for mRNA-degrading ribonucleases in chloroplasts. This seems to be in contrast to the situation in bacteria where 5′-terminal stem–loop structures appear to have a direct effect on ribonuclease binding and mRNA decay (22,23). This study shows for the rbcL 5′-terminal stem–loop that its role is to provide a stable RNA secondary structure that places the bottom nucleotides of the stem and the following nucleotides—those sequences corresponding to a previously identified RNA-stabilizing sequence (20)—into a specific RNA conformation, consisting of a helical and a single-stranded portion, required for longevity of rbcL transcripts in the Chlamydomonas chloroplast. This specific sequence and structure requirement for stability of rbcL transcripts strongly suggests an mRNA-stabilizing mechanism that involves binding of a trans-acting factor to this element.
Trans-acting factors and RNA longevity in chloroplasts
The existence of a number of nuclear mutants of Chlamydomonas that fail to accumulate individual mRNA populations in the chloroplast seems to support the idea of protective transcript-specific RNA-binding proteins (24–27). A couple of proteins that complement RNA stability mutants have been isolated to date (28,29) but direct binding of these proteins to a known stabilizing sequence in 5′-UTRs could not be demonstrated. In addition to proteins that complement RNA stability mutants, several other proteins that bind to 5′-UTRs of chloroplast transcripts, either individually or as part of multi-protein complexes, have been identified (30). Most of these proteins appear to function primarily in translational control of chloroplast gene expression although ancillary roles in RNA stabilization cannot be excluded. Difficulties in linking already known RNA-binding proteins directly to cis-acting RNA-stabilizing sequences may be due to the requirement of specifically folded 5′ RNA secondary structures that may be hard to establish in in vitro RNA/protein binding assays.
Considering the apparent lack of a sequence and a structural consensus among 5′ sequences of chloroplast mRNAs in general, and in particular, among the cis-acting transcript-stabilizing elements that have been delineated to date (20), it appears possible that different chloroplast mRNA species contain different sequences and structures at their 5′ ends that are involved in stabilizing transcripts. This would imply that putative trans-acting factors that might bind to these elements are also transcript-specific. These trans-acting factors may be part of multi-protein complexes that are proposed to control initiation of translation of transcripts (30,31) or be separate entities that function independently of translation. Although most models to date favor a coupled control of RNA stability and translation (30), independent control of mRNA longevity might occur in some mRNAs, e.g. transcripts of the atpB gene (20), in which the distance between an RNA-stabilizing element at the 5′ end and the ribosome-binding site, around which the proteins involved in translational regulation are supposed to assemble, seems to exclude a direct interaction of trans-acting factors associated with the two sites. Conceivably, independent or relaxed control of translation and RNA stability would allow more variability and flexibility in regulation of gene expression than strict coupling of the two processes. Regarding the differences in longevity of different mRNA populations in chloroplasts, it appears feasible that inherent differences in folding and stability of cis-acting RNA secondary structures and/or in binding of putative trans-acting factors to these elements can be responsible for the differences in lifetimes of transcripts in chloroplasts.
Acknowledgments
This study was supported by grant BMC2003-03209 from the Ministerio de Ciencia y Tecnologia, Spain. Funding to pay the Open Access publication charges for this article was provided by Department of Molecular Biosciences, University of Oslo.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang Y., Liu C.L., Storey J.D., Tibshirani R.J., Herschlag D., Brown P.O. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hambraeus G., von Wachenfeldt C., Hederstedt L. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics. 2003;269:706–714. doi: 10.1007/s00438-003-0883-6. [DOI] [PubMed] [Google Scholar]
- 3.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 4.Gutiérrez R.A., MacIntosh G.C., Green P.J. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 1999;4:429–438. doi: 10.1016/s1360-1385(99)01484-3. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P., Tollervey D. mRNA turnover. Curr. Opin. Cell Biol. 2001;13:320–325. doi: 10.1016/s0955-0674(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 6.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 7.Salvador M.L., Klein U., Bogorad L. 5′ sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast transcripts. Proc. Natl. Acad. Sci. USA. 1993;90:1556–1560. doi: 10.1073/pnas.90.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emory A.E., Bouvet P., Belasco J.G. A 5′-terminal stem–loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 9.Diwa A., Bricker A.L., Jain C., Belasco J.G. An evolutionarily conserved RNA stem–loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- 10.Hambraeus G., Karhumaa K., Rutberg B. A 5′ stem–loop and ribosome binding site but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology. 2002;148:1795–1803. doi: 10.1099/00221287-148-6-1795. [DOI] [PubMed] [Google Scholar]
- 11.Higgs D.C., Shapiro R.S., Kindle K.L., Stern D.B. Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell. Biol. 1999;19:8479–8491. doi: 10.1128/mcb.19.12.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickelsen J., Fleischmann M., Boudreau E., Rahire M., Rochaix J.D. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell. 1999;11:957–970. doi: 10.1105/tpc.11.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Z., Eibl C., Koop H.U. The stem–loop region of the tobacco psbA 5′-UTR is an important determinant of mRNA stability and translation efficiency. Mol. Genet. Genomics. 2003;269:340–349. doi: 10.1007/s00438-003-0842-2. [DOI] [PubMed] [Google Scholar]
- 14.Salvador M.L., Klein U., Bogorad L. Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas. Regulation by transcription and RNA degradation. Plant J. 1993;3:213–219. doi: 10.1046/j.1365-313x.1993.t01-13-00999.x. [DOI] [PubMed] [Google Scholar]
- 15.Boynton J.E., Gillham N.W., Harris E.H., Hosler J.P., Johnson A.M., Jones A.R., Randolph-Anderson B.L., Robertson D., Klein T.M., Shark K.B., et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 16.Blowers A.D., Bogorad L., Shark K.B., Sanford J.C. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell. 1989;1:123–132. doi: 10.1105/tpc.1.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvador M.L., Suay L., Anthonisen I.L., Klein U. Changes in the 5′-untranslated region of the rbcL gene accelerate transcript degradation more than 50-fold in the chloroplast of Chlamydomonas reinhardtii. Curr. Genet. 2004;45:176–182. doi: 10.1007/s00294-003-0470-8. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Blowers A.D., Klein U., Ellmore G.S., Bogorad L. Functional in vivo analyses of the 3′ flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol. Gen. Genet. 1993;238:339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- 20.Anthonisen I.L., Salvador M.L., Klein U. Specific sequence elements in the 5′ untranslated regions of rbcL and atpB gene mRNAs stabilize transcripts in the chloroplast of Chlamydomonas reinhardtii. RNA. 2001;7:1024–1033. doi: 10.1017/s1355838201001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouvet P., Belasco J.G. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E.coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- 23.Mackie G.A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 24.Kuchka M.R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J.-D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell. 1989;58:869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- 25.Rochaix J.-D., Kuchka M., Mayfield S., Schirmer-Rahire M., Girard-Bascou J., Bennoun P. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 1989;8:1013–1021. doi: 10.1002/j.1460-2075.1989.tb03468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieburth L.E., Berry-Lowe S., Schmidt G.W. Chloroplast RNA stability in Chlamydomonas: rapid degradation of psbB and psbC transcripts in two nuclear mutants. Plant Cell. 1991;3:175–189. doi: 10.1105/tpc.3.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monod C., Goldschmidt-Clermont M., Rochaix J.D. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol. Gen. Genet. 1992;231:449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- 28.Boudreau E., Nickelsen J., Lemaire S.D., Ossenbühl F., Rochaix J.D. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 2000;19:3366–3376. doi: 10.1093/emboj/19.13.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaistij F.E., Boudreau E., Lemaire S.D., Goldschmidt-Clermont M., Rochaix J.D. Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2000;97:14813–14818. doi: 10.1073/pnas.97.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickelsen J. Chloroplast RNA-binding proteins. Curr. Genet. 2003;43:392–399. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 31.Zerges W., Auchincloss A.H., Rochaix J.D. Multiple translational control sequences in the 5′ leader of the chloroplast psbC mRNA interact with nuclear gene products in Chlamydomonas reinhardtii. Genetics. 2003;163:895–904. doi: 10.1093/genetics/163.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]