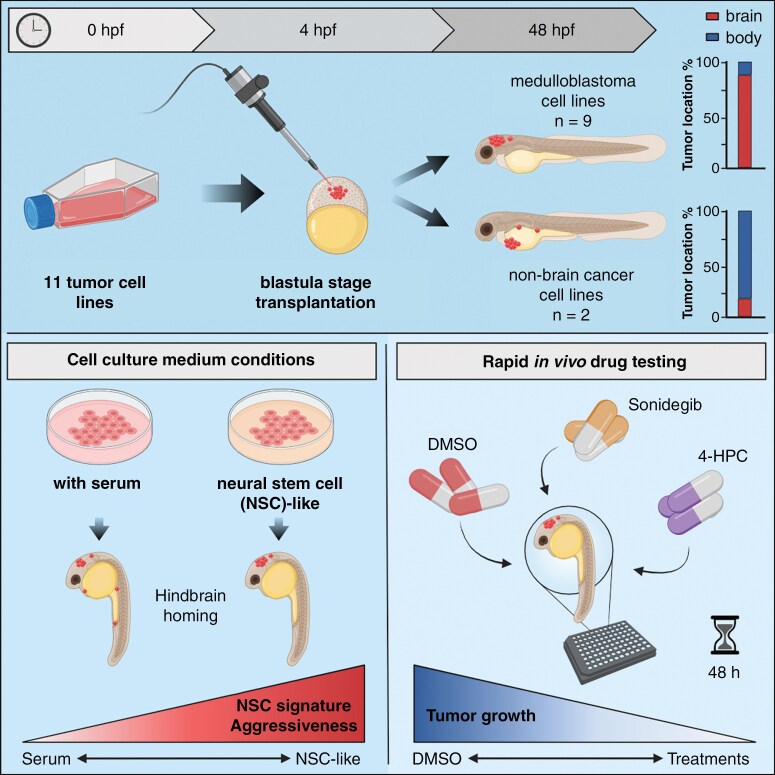
Abstract
Background
Medulloblastoma (MB) is one of the most common malignant brain tumors in children. Current preclinical in vivo model systems for MB have increased our understanding of molecular mechanisms regulating MB development. However, they may not be suitable for large-scale studies. The aim of this study was to investigate if a zebrafish-based xenograft model can recapitulate MB growth and enable rapid drug testing.
Methods
Nine different MB cell lines or patient-derived cells were transplanted into blastula-stage zebrafish embryos. Tumor development and migration were then monitored using live imaging. RNA sequencing was performed to investigate transcriptome changes after conditioning cells in a neural stem cell-like medium. Furthermore, drug treatments were tested in a 96-well format.
Results
We demonstrate here that transplantation of MB cells into the blastula stage of zebrafish embryos leads to orthotopic tumor growth that can be observed within 24 h after transplantation. Importantly, the homing of transplanted cells to the hindbrain region and the aggressiveness of tumor growth are enhanced by pre-culturing cells in a neural stem cell-like medium. The change in culture conditions rewires the transcriptome toward a more migratory and neuronal phenotype, including the expression of guidance molecules SEMA3A and EFNB1, both of which correlate with lower overall survival in MB patients. Furthermore, we highlight that the orthotopic zebrafish MB model has the potential to be used for rapid drug testing.
Conclusions
Blastula-stage zebrafish MB xenografts present an alternative to current MB mouse xenograft models, enabling quick evaluation of tumor cell growth, neurotropism, and drug efficacy.
Keywords: medulloblastoma, neurotropism, patient-derived xenografts, rapid drug testing, zebrafish
Graphical Abstract
Graphical Abstract.
Key Points.
Medulloblastoma (MB) cells home to the hindbrain region in developing zebrafish embryos.
Neural stem cell culture conditions improve the homing capacity of MB tumor cells.
MB-transplanted zebrafish embryos can be used as an in vivo model for rapid drug testing.
Importance of the Study.
One of the challenges of accurately modeling medulloblastoma is the large heterogeneity in tumor characteristics. To accurately model this heterogeneous disease, patient-derived xenograft mouse models are currently the standard. However, such mouse models are labor-intensive, time-consuming, and not suitable for high-throughput studies. Here, we describe a quick and straightforward zebrafish xenograft model that provides a promising alternative to these existing mouse models. We demonstrate that this model can be utilized to study tumor cell growth of several medulloblastoma subgroups. More importantly, our model facilitates rapid drug testing, providing a scalable opportunity for in vivo drug screenings that will support the discovery of novel therapeutic compounds against medulloblastoma.
Medulloblastoma (MB) is one of the most common malignant pediatric brain tumors. Advances in molecular characterization and innovations in the multimodal treatment of MB have drastically increased the survival rate, and today about 70% of patients are cured. However, the prognosis is strongly correlated with the principal molecular MB subgroups (Wingless [WNT], Sonic Hedgehog [SHH], Group 3, and Group 4), with patients with Group 3 MB having the worst prognosis.1 This suggests that more subgroup-specific treatments are needed. Furthermore, the improvement in survival has stalled and remained relatively stable over the past 2 decades.2 In addition, approximately 30% of MB patients (most commonly belonging to the SHH, Group 3, or Group 4 subgroup) still relapse, which is almost always fatal.3 To further improve the survival rate of MB patients and identify novel treatment options, new disease-specific models are needed. Current in vivo models for MB include genetically engineered mouse models, orthotopic transplants, and patient-derived xenograft mouse models.4 While these mouse models accurately represent MB subgroups, they are labor-intensive, time-consuming, and subject to ethical and financial constraints that make them impractical for use in large-scale studies.
Recently, zebrafish (Danio rerio) embryos have emerged as a promising in vivo model for many human cancers, including non-small-cell lung cancer brain metastasis, pilocytic astrocytoma, and glioblastoma.5–7 Zebrafish have proven to be a good model system for high-throughput transplantation studies because of their high fecundity, ex utero embryogenesis, small size, rapid development, and low maintenance and husbandry costs.8 Furthermore, previous studies have shown that cell line-derived (CDXs) and patient-derived xenografts (PDXs) in zebrafish embryos are able to proliferate in vivo and display disease and patient-specific characteristics, suggesting that zebrafish models can be used as an alternative to existing mouse models and could enable rapid identification of patient-specific targeted treatments.7,9–13 These favorable preclinical results have led to the initiation of several trials to study the translational and clinical use of zebrafish PDXs as a model for tumor growth and treatment efficacy (NCT03668418, NCT05289076, NCT05616533). More recently, a SHH-driven MB zebrafish model has been established by CRISPR/Cas9-mediated gene editing.14 However, this model does not facilitate the examination of inter-patient variability in terms of tumor growth, invasion, and drug response. Furthermore, despite all the promising results of zebrafish cancer xenografts, orthotopic zebrafish models have not been explored for MB.
Here, we report the use of zebrafish embryos as an orthotopic model for MB. We demonstrate that MB CDXs and PDXs home primarily to the hindbrain region after transplantation into the blastula stage of developing zebrafish embryos, independently of MB subgroup. This homing mechanism can be further enhanced by conditioning cells to neural stem cell conditions containing growth factors and supplements important for neuronal growth and synaptic plasticity. Furthermore, we show that MB-transplanted zebrafish embryos have the potential to be used as a rapid orthotopic MB zebrafish model for testing drug compounds.
Materials and Methods
Animal Models
Eight- to twelve-week-old female NOD/SCID/IL2Rγ-/- (NSG) mice were used for orthotopic transplantation of 1 × 105 secondary tumor-isolated neuroepithelial stem (tNES) cells and tumor growth was monitored using IVIS SpectrumCT In Vivo Imaging System (PerkinElmer) as previously described.15 Mice were euthanized upon observation of ethical endpoint criteria representing external signs of tumor development. fli1:EGFP zebrafish embryos aged about 4 h old (1k-cell stage) were used for transplantation. Fertilized eggs were obtained from natural spawning and staged according to the standard procedures.16 Transplanted zebrafish embryos were kept for a maximum of 3 days after transplantation. Experiments on embryos younger than 5 days do not require an ethical permit. Zebrafish breeding and housing took place in accordance with the national ethical guidelines and regulations (15591-23). Mouse experiments were approved by Stockholm’s North Ethical Committee of Animal Research (ethical permit 6548/18).
Generation of Tertiary Tumor NES Cell Lines
Tertiary tNES cells were isolated from established tumors from orthotopic transplantation of secondary tNES cells (Supplementary Figure 1). Tumor cell isolation was performed as previously described.15 In brief, tumor tissue was dissociated using digestion buffer (1× PBS with 10 U/mL papain, 200 μg/mL L-cysteine, 250 U/mL DNAse, 1% penicillin-streptomycin, all from Sigma). Cells were triturated and passed through a 100-μm cell strainer to obtain a single-cell suspension. Mouse cell contaminations were removed using the Mouse Cell Depletion Kit (Miltenyi Biotec, 130-104-694). Tertiary tNES cells were cultured in a complete neural stem cell medium (see section Cell Culture) on poly-l-ornithine/laminin-coated plates.
Cell Culture
Tertiary tNES cells were cultured as previously described.15 Briefly, tNES cells were cultured on flasks coated with 20 µg/mL poly-l-ornithine (Sigma, P3655) and 1 µg/mL laminin (Sigma, L2020) in complete neural stem cell medium (DMEM/F12+Glutamax (ThermoFisher, 31331093) supplemented with 1% N2 supplement (ThermoFisher, 17502001), 0.1% B27 supplement (ThermoFisher, 17504044), 10 ng/mL FGF2 (Qkine, Qk053), 10 ng/mL EGF (PeproTech, AF100-15), and 1% penicillin-streptomycin (Sigma, P4333). MB-LU-181, CHLA-01-MED, and CHLA-01R-MED cells were cultured in DMEM/F12+Glutamax with 20 ng/mL (CHLA-01-MED and CHLA-01R-MED) or 40 ng/mL (MB-LU-181) FGF2, 20 ng/mL EGF, 2% B27, and 1% penicillin-streptomycin. DAOY, ONS-76, MDA-MB-231, and HCT116 p53+/+ cells were maintained in DMEM (ThermoFisher, 41966052) supplemented with 10% FBS (ThermoFisher, SV30160.03HI) and 1% penicillin-streptomycin. In addition, medium for DAOY cells contained 1% MEM non-essential amino acids, 1% HEPES, and 1% Glutamax (ThermoFisher, cat. 11140050, SH30237.01, and 35050061, respectively). UW228-3 cells were cultured in RPMI (ThermoFisher, 61870044) and D425 Med cells in DMEM/F12+Glutamax, both supplemented with 10% FBS and 1% penicillin-streptomycin. In this study, we classify the MB tumor cell lines based on the 4 principal molecular MB subgroups (SHH, WNT, Group 3, Group 4).1 Clinical subgroup classification deviates slightly from this subgrouping where MB tumors are classified as either WNT-activated, SHH-activated and TP53-wildtype, SHH-activated and TP53-mutant, or non-WNT/non-SHH (combination of Group 3 and Group 4 MB).17 Clinical information about the cell lines can be found in the Supplementary Data. ONS-76 cells were purchased from Tebu-Bio. DAOY, UW228-3, D425 Med cells, and CHLA-01-MED and CHLA-01R-MED were obtained from ATCC. All cell lines tested negative for mycoplasma and were authenticated by STR analysis at Eurofins (DAOY, D425 Med, CHLA-01-MED, CHLA-01R-MED), ECACC (MDA-MB-231, HCT116 p53+/+), or Multiplexion (ONS-76, UW228-3). BT084 PDX line was previously established by Dr. Till Milde (DKFZ, Germany) and has been maintained as described in ref.18 MB-LU-181 PDX line establishment has previously been described in ref.19
Lentiviral Transduction
Luciferase-expressing cells were generated through lentiviral transduction of pLenti-CMV-Luc-Puro. Lentiviral particles were produced in HEK293T cells using packaging and envelope constructs pCMVΔ8.2 and pMD.G-VSV-G (pCMVΔ8.2 and pMD.G-VSV-G were gifts from Bob Weinberg, Addgene plasmids #8454, #8455), and concentrated using fast-trap virus purification and concentration kit according to the manufacturer’s instructions (Millipore). For transduction, MB cells were seeded into 6-well plates and transduced with a virus at 70% confluency, 24 h after transduction, cells were selected with 2 µg/mL puromycin.
Cell Proliferation Assay
To assess cell proliferation over time, UW228-3 cells were seeded in 12-well tissue culture-treated plates at a density of 5 × 104 cells/well. MDA-MB-231 and D425 Med cells were seeded at a density of 1 × 105 cells/well. Cells were dissociated using TryplE Select (ThermoFisher, A1217701) and counted at defined time points to determine the number of viable cells.
Transplantation of Cells and Treatments
Confluent cell cultures were labeled with Vybrant™ DiI cell-labeling solution (ThermoFisher, V22885) in the corresponding culture medium (1:200) for 15–20 min according to the manufacturer’s instructions. Cells were washed with PBS for 5 min, followed by trypsinization and centrifugation at 300g for 3 min. Cells were resuspended and centrifuged twice to assure the complete removal of the dye. Fluorescent-labeled tumor cells were transplanted in the blastula stage of fli1:EGFP zebrafish embryos as previously described.20 In brief, embryos were lined up on a 1% agarose mold in an E3 medium. Directly before transplantation, cell suspensions were spun down and almost all medium was removed. Concentrated cell suspension was loaded into a microcapillary (World Precision Instruments, TW100-4) connected to a FemtoJet 4x (Eppendorf). About 100–300 cells were injected in the center of the cell mass. After transplantation, the embryos were collected in E3 medium and raised at 33°C. Embryos with intracranial tumors were selected for drug treatment 24 h after transplantation. LDE225 phosphate (sonidegib, Cayman Chemicals, 16263) and 4-hydroxycyclophosphamide (4-HCP, Santa Cruz Biotechnology, sc-206885A) were added directly in E3 medium to a final concentration of 10 µM. DMSO (0.1%, Sigma, D8418) was used as a control.
High-Throughput Imaging of Zebrafish Embryos
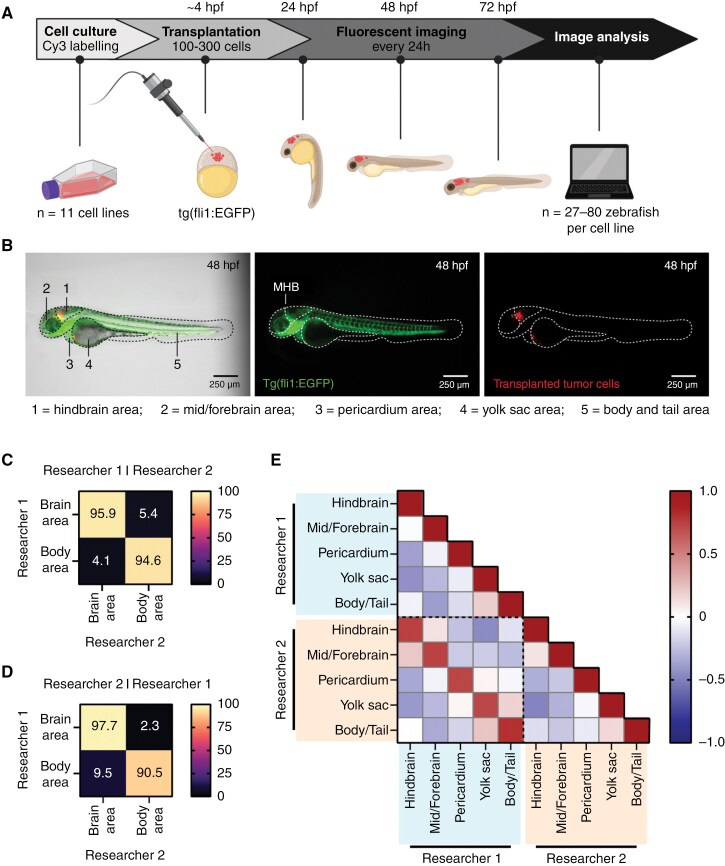
For high-throughput imaging of the transplanted zebrafish embryos, imaging 96-well plates (Ibidi, 89621) with agarose molds were prepared.21 Single embryos were distributed into the wells together with 150 µL of exposure medium (160 µg/mL tricaine, 30 mg/L phenylthiourea in E3 medium). Embryos were manually oriented into position and imaged using ImageXpress Nano (Molecular Devices) every 24 h. After imaging, the plate was moved into a wet chamber inside an incubator at 33°C. Zebrafish embryo images were analyzed for tumor location (hindbrain, mid/forebrain, pericardium, yolk sac, and body/tail area) using ImageJ 1.53. Averages of tumor cell location per region (%) were used to generate schematic heatmaps of tumor cell localization in zebrafish embryos. Unfocused images or dead embryos were excluded from the analysis. All data were analyzed by 2 people individually of which researcher 2 was blinded. To compare the 2 analyzed datasets, each image has been quantitatively classified from area with the highest percentage of tumor cells (= 1) to area with the lowest percentage of tumor cells (= 5).
Luciferase Activity Measurement
To measure luciferase activity, treated embryos were transferred into a white-bottom 96-well plate (Greiner, 655074). Exposure medium was removed before adding 100 µL of freshly prepared lysis buffer containing 10% glycerol, 1% Triton-X 100, 25 mM Tris-phosphate pH 7.8 adjusted with phosphoric acid, 8 mM magnesium chloride, and 1 mM DL-dithiothreitol (Sigma, cat# G012, T8787, T1503, 438081, M8266, 43815, respectively) in dH2O. The plate was placed on a rocking platform and embryos were lysed for 45 min at room temperature. A total of 100 µL of freshly prepared substrate buffer, 25 mM Tris-phosphate pH 7.8, 1 mM DL-dithiothreitol, 0.25 mg/mL d-luciferin (BioThema, BT11-1000K), and 1 mM ATP (ThermoFisher, R0441) in dH2O, was added and incubated for 5 min in the dark at room temperature. Luminescence was measured using the FLUOstar Omega (BMG Labtech).
Transcriptome Sequencing
UW228-3 and D425 Med cells were gradually conditioned toward a complete neural stem cell medium by increasing the concentration of this medium from 25% to 100% over the course of 4 passages. Neural stem cell medium for UW228-3 cells was supplemented with 1% FBS to prevent cell death. Conditioned UW228-3 and D425 Med cells were cultured for at least 2 passages in a complete neural stem cell medium before isolating total RNA for RNAseq. Total RNA was extracted using TRIzol Reagent (ThermoFisher, 15596018) and Direct-zol RNA extraction kit (Zymo Research, R2052) according to the manufacturer’s instructions. RNA sequencing was performed using the Illumina Novaseq 6000 (PE150) platform (Novogene Europe). RNA sequence reads were mapped against the human reference genome GRCh38/hg38 using Hisat2 v2.0.5.22 Differential expression analysis of 2 groups (3 biological replicates per group) was performed using the DESeq2 R package (1.20.0). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value ≤.05 found by DESeq2 were assigned as differentially expressed.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was performed using GSEA v.4.2.3.23 The transcriptome sequencing dataset was analyzed against the MSigDB Hallmark, BioCarta, KEGG, Pathway Interaction Database, Reactome, WikiPathways, and Gene Ontology gene sets according to Reimand et al.24 Pathways enrichment analysis and comparison of multiple transcriptome datasets was performed by using Metascape.25
Statistical Analysis
Data are presented as means ± standard deviation or standard error of the mean (SEM). Statistical significance was analyzed with the Student t-test, 1-way or 2-way ANOVA with Dunnett, Tukey, or Šídák post-hoc test. P-values <.05 were considered statistically significant. All statistical analyses were performed using GraphPad Prism 9.5.0. *P < .05, **P < .01, ***P < .001, ****P < .0001.
Results
Tumor Cell Transplantations Into the Blastula Stage of Zebrafish Embryos as an In vivo Model for Medulloblastoma
Previous studies have observed that human melanoma and glioblastoma cells are able to home to their environment of origin when transplanted into zebrafish embryos.20,26 It was proposed that intrinsic morphogenetic cues in the early developing zebrafish embryo are responsible for this homing mechanism by providing support to these human cells. Based on these results, we speculated that MB cells can migrate to the developing cerebellum after transplantation into the blastula stage of zebrafish embryos (Figure 1A). We transplanted a total of 9 fluorescently labeled MB cell lines (both PDXs and established cell lines) and 2 non-MB cell lines into fli1:EGFP embryos 4 h post-fertilization (hpf). Transgenic fli1:EGFP zebrafish express GFP in vascular endothelial cells. The transplanted zebrafish embryos were imaged every 24 h for 3 days after fertilization to track tumor cell localization. Remarkably, the majority of tumor cells were already present in the region of interest 24 hpf. To quantify tumor localization, images acquired 48 hpf were used in which the developing embryo was divided into 5 areas, namely hindbrain, mid/forebrain, pericardium, yolk sac, and body/tail area (Figure 1B, Supplementary Figure 2A). One of the most distinct veins in the early developing zebrafish embryo is the mid-cerebral vein, which delineates the mid-hindbrain boundary,27 and helps us to separate the hindbrain area from the mid/forebrain area without the need for closeup images of the head.
Figure 1.
Homing of fluorescently labeled tumor cells transplanted in zebrafish embryos can be examined with fluorescent imaging and image analysis. (A) Schematic overview of the experimental workflow for zebrafish embryo transplantations followed by tumor location analysis. (B) Representative image of a tertiary tNES cell transplanted zebrafish embryo 48 h post-fertilization (hpf), displaying the 5 defined locations for location analysis. Left panel: bright field with GFP and Cy3 overlay. Middle panel: expression of enhanced GFP in the vasculature of the fli1:EGFP zebrafish embryo (GFP, green). MHB = mid-hindbrain boundary. Right panel: labeled tumor cells (Cy3, red). (C) Confusion matrices of image classification performed by researcher 1, (D) and researcher 2. (E) Pearson correlation matrix between the analyzed datasets of researcher 1 and researcher 2 revealing high correlation between individually analyzed datasets.
To ensure that the analysis of tumor localization was performed accurately and without bias, all transplanted embryos (n = 756) were analyzed by 2 researchers, the second researcher of whom was blinded to cell line identity. Classification by defining the majority of the tumor cells in either the brain or body area resulted in a high agreement between both researchers (95.5%) (Figure 1C and D), and good accuracy (79.3%) was achieved even with deeper classification into 5 anatomical areas (Supplementary Figure 2B and C). When examining the differently classified images, no major differences in the percentiles of tumor localization were observed (Supplementary Figure 2D), and even embryos with a high spread of tumor localization were analyzed uniformly (Supplementary Figure 2E). Further analysis using Pearson correlation revealed that all 5 anatomical areas showed a strong positive correlation when comparing the 2 analyzed datasets (Figure 1E). As both analyses were very similar, average values were used for further presentation of the data.
Medulloblastoma Tumor Cells Home Toward the Hindbrain Region in Developing Zebrafish Embryos
First, we analyzed tertiary tNES cells transplanted into the blastula stage of zebrafish embryos to test our hypothesis. Tertiary tNES cells represent malignant human SHH-MB and are derived from a stem cell model that we previously established by reprogramming non-cancerous skin cells carrying a germline PTCH1 mutation and differentiating them into patient-derived NES cells.15 These patient-derived NES cells form MB tumors upon orthotopic transplantation into the cerebellum of NSG mice from which tNES cells were isolated. Through serial re-transplantation, more malignant tumor-derived tNES cell lines were generated (Supplementary Figure 1). Interestingly, we observed a high degree of brain localization when tertiary tNES were transplanted, with 87.9 ± 1.6% homing to the brain region (Figure 2A and B). Further analysis revealed that 67.3% of the tertiary tNES cells that were found in the brain were localized in the hindbrain region (Figure 2C, Supplementary Data). To determine whether this ability to localize to the hindbrain is a universal property of SHH-MB cells, we transplanted the SHH-MB PDX BT084, and cell lines ONS-76, DAOY, and UW228-3 cells in blastula-stage embryos. BT084 was derived from a SHH-MB tumor with a TP53 mutation and amplifications in GLI2, TERT, and MYCN, representing one of the most aggressive forms of human SHH MB.28 Like tertiary tNES cells, we observed that the majority of tumor cells were located to the brain for all 4 SHH-MB cell lines at 48 hpf: 60.5 ± 6.2% of ONS-76 cells, 80.6 ± 3.5% of BT084 cells, 72.8 ± 5.7% of DAOY cells, and 59.1 ± 5.4% of UW228-3 cells (Figure 2A and B). Again, a considerable proportion of these brain-localized tumor cells localized to the hindbrain region (58.5%, 58.7%, 57.0%, and 60.6% for ONS-76, BT084, DAOY, and UW228-3 cells, respectively) (Figure 2C, Supplementary Data).
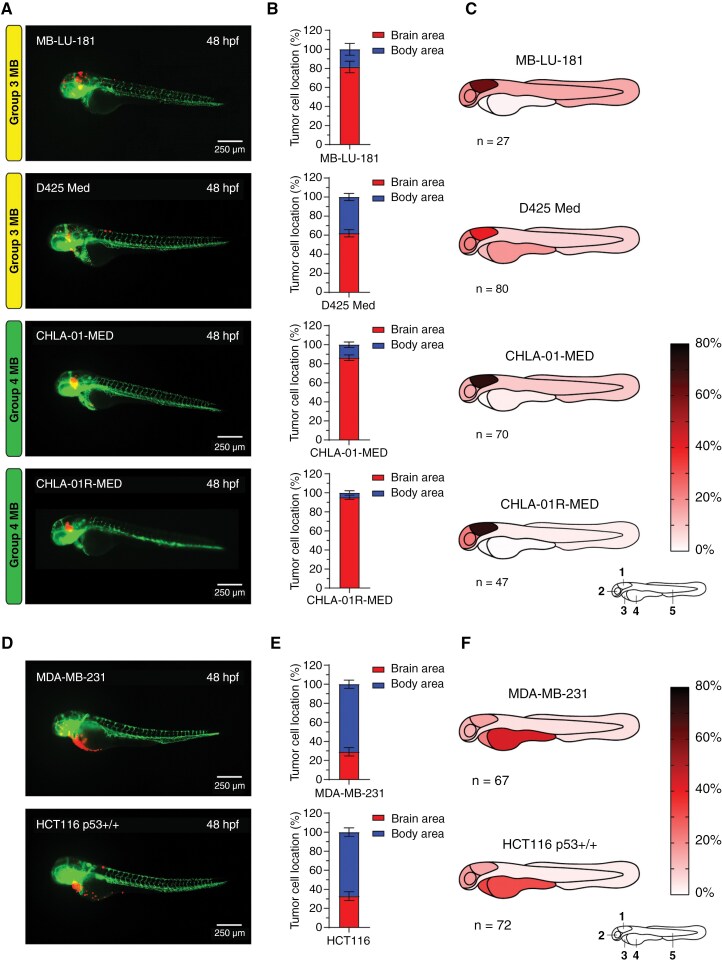
Figure 2.
SHH medulloblastoma cell lines home toward the hindbrain region after transplantation into the blastula stage of zebrafish embryos. (A) Representative images of fli1:EGFP zebrafish embryos 48 hpf transplanted with SHH medulloblastoma (MB) cell lines. (B) Brain/body area distribution of transplanted SHH MB cell lines in 48 h old embryos (tertiary tNES n = 52, ONS-76 n = 45, BT084 n = 42, DAOY n = 48, and UW228-3 n = 65). Mean ± SEM. (C) Schematic heatmaps of the tumor cell location of SHH MB cell lines in 48 h old embryos. 1 = hindbrain area; 2 = mid/forebrain area; 3 = pericardium area; 4 = yolk sac area; 5 = body and tail area. (D) Tumor take rates (%) of all transplanted MB cell lines. (E) Representative images of tumor growth (tertiary tNES and BT084) from 24 hpf to 72 hpf. (F) Tumor growth rate and (G) survival rate analyses of all transplanted MB cell lines. Kaplan Meier curves represent the survival rate of transplanted zebrafish embryos up to 72 hpf.
To further characterize and compare the transplanted cell lines beyond cell homing ability, we examined the tumor take, tumor growth, and survival rates of all transplanted MB cell lines. Tumor take rate was defined as the percentage of transplanted zebrafish embryos with positive tumor growth between 24 and 72 h after transplantation. Previous research utilizing zebrafish embryos for the evaluation of glioblastoma growth has shown that tumor take rates higher than 50% could predict if patient-derived cells are able to initiate tumors in orthotopic mouse xenografts.7 All assessed MB PDX and cell lines demonstrated a tumor take rate above 70% (Figure 2D). This correlates with the extensive amount of research performed using these MB PDX and cell lines in orthotopic mouse xenografts. Tumor growth rate analyses confirmed that all MB tumors displayed exponential growth over a time period of 48 h (Figure 2E and F). However, no significant growth differences were observed between the transplanted MB cell lines. Contrarily, survival differed considerably between PDX and cell lines (Figure 2G, Supplementary Data). PDX, primary tumor-derived, and neurosphere cell lines (MB-LU-181, CHLA-01R-MED, BT084, tNES tertiary, and CHLA-01-MED in order of aggressiveness) demonstrated significantly lower survival rates than established cell lines (ONS-76, UW228-3, DAOY, and D425 Med).
After observing the high degree of hindbrain localization of SHH-MB tumor cells, we speculated whether this is a global MB mechanism. To investigate the localization of other MB subgroups, we transplanted 4 non-SHH MB cell lines, including the Group 3 MB PDX cell line MB-LU-181.19 Importantly, transplantation of these cells also resulted in a high percentage of tumor cells located in the brain area with 81.5 ± 6.1% (Figure 3A and B). The area-specific evaluation showed that 75.0% of these cells were localized in the hindbrain area (Figure 3C, Supplementary Data). Additional transplantation with the Group 3 MB cell line D425 Med yielded comparable results and showed a high proportion of tumor cells localized in the brain area (62.0 ± 3.7% for D425 Med) with the majority of these cells confined within the hindbrain area (64.5% for D425 Med) (Figure 3A–C, Supplementary Data). These findings were even more prominent when we performed transplantations of the Group 4 MB neurosphere cell lines CHLA-01-MED and CHLA-01R-MED.29 86.4 ± 2.9% of CHLA-01-MED and 95.3 ± 2.2% of CHLA-01R-MED cells were localized in the brain area with significantly higher cell proportions localized to the hindbrain; 83.8% for CHLA-01-MED and 75.9% for CHLA-01R-MED (Figure 3A–C, Supplementary Data). To study if the brain-homing mechanism is specific to MB tumor cells or if it is a common process among other cancers, we transplanted the breast cancer cell line MDA-MB-231 and the colon cancer cell line HCT116 p53+/+. Both non-MB cell lines had significantly lower levels of brain localization compared to the MB cell lines, with only 29.1 ± 4.4% of MDA-MB-231 and 32.8 ± 4.6% of HCT116 p53+/+ cells detected in the brain area (Figure 3D and E, Supplementary Figure 3B). Instead, most non-MB cancer cells localized to the non-brain area with preference to the yolk sac, with 43.3 ± 4.4% for MDA-MB-231 and 31.3 ± 4.4% for HCT116 p53+/+ (Figure 3F, Supplementary Data). Intriguingly, different types of embryonal malformations were more commonly observed upon transplantations with non-MB cell lines compared to MB cell lines. Zebrafish embryos transplanted with MB cell lines displayed cranial malformations more often whereas pericardial edema and yolk sac edema were more frequently detected in zebrafish embryos transplanted with non-MB cells (Supplementary Figure 3A). Taken together, these results suggest an intrinsic brain-homing mechanism of MB tumor cells on transplantation in developing zebrafish embryos.
Figure 3.
Group 3 and Group 4 medulloblastoma cell lines display homing toward the hindbrain area upon transplantation into the blastula stage of zebrafish embryos whereas non-medulloblastoma cell lines do not. (A) Representative images of fli1:EGFP zebrafish embryos 48 hpf transplanted with Group 3 (MB-LU-181 and D425 Med) and Group 4 (CHLA-01-MED and CHLA-01R-MED) MB cells, (D) and non-MB cell lines (MDA-MB-231 and HCT116 p53+/+). (B) Brain/body area distribution of transplanted Group 3 and Group 4 MB cell lines MB-LU-181 (n = 27), D425 Med (n = 80), CHLA-01-MED (n = 70), and CHLA-01R-MED (n = 47), and (E) non-MB cell lines MDA-MB-231 (n = 67) and HCT116 p53+/+ (n = 72) in 48 h old embryos. Mean ± SEM. (C) Schematic heatmaps of the tumor cell location of Group 3 MB, Group 4 MB, (F) and non-MB cell lines in 48 h old embryos. 1 = hindbrain area; 2 = mid/forebrain area; 3 = pericardium area; 4 = yolk sac area; 5 = body and tail area.
Neural Stem Cell Culture Conditions Improve Homing Capacity of Tumor Cells
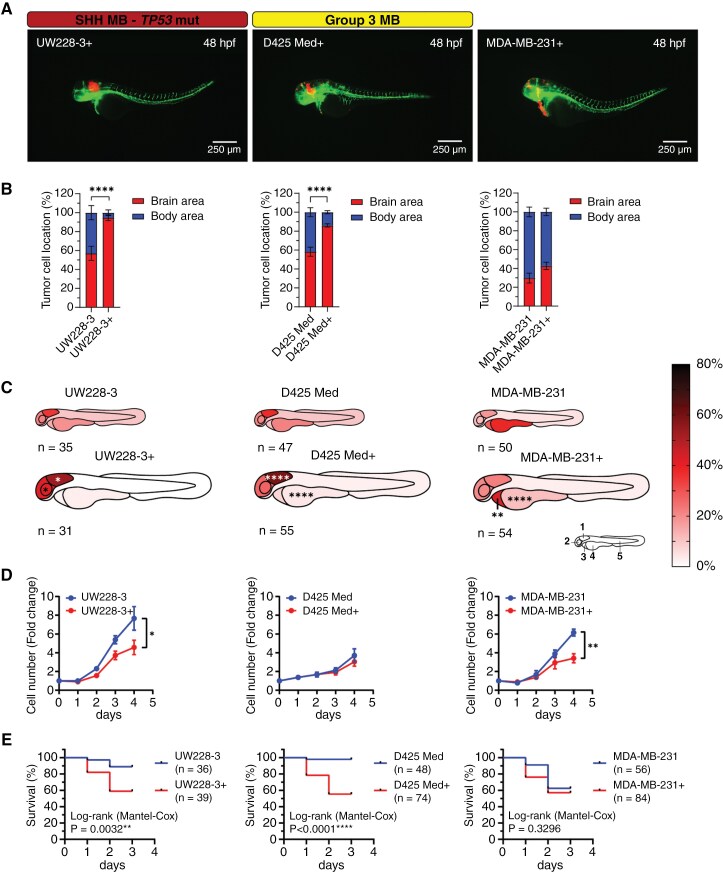
Interestingly, we found that the patient-derived and neurosphere MB cell lines (tNES, BT084, MB-LU-181, CHLA-01-MED, and CHLA-01R-MED) had a significantly higher homing capacity toward the brain compared with the other transplanted MB cell lines (DAOY, UW228-3, ONS-76, and D425 Med) (Supplementary Figure 3B). Moreover, lower tumor take rates (e.g., 70.24% and 72.46% for D425 Med and UW228-3, respectively) and significant differences in survival rates were observed when transplanted established cell lines were compared with PDX and neurosphere cell lines (Figure 2D and G). Because all the PDX and neurosphere cell lines are cultured in neural stem cell conditions, we adapted one SHH-MB cell line (UW228-3) and one Grp3 MB cell line (D425 Med) to similar growth conditions prior to transplantation (Supplementary Figure 4A). Interestingly, MB cells adapted to neural stem cell media (UW228-3+ and D425 Med+) showed significantly higher localization to the brain area and specifically to the hindbrain area compared to cells cultured in standard cell culture conditions (Figure 4A–C). This was specific to MB cells because MDA-MB-231 breast cancer cells adapted to neural stem cell conditions did not show any significant increase in homing to the brain area. Instead, we observed a significant increase of cells localizing to the pericardium area (Figure 4A–C). Switching the cells to neural stem cell conditions resulted in a decrease in cell proliferation in vitro for UW228-3+ and MDA-MB231+ cells (Figure 4D). However, it caused a more aggressive disease progression in vivo in transplanted zebrafish embryos (Figure 4E), resembling the disease progression of the patient-derived MB cell lines (tNES, BT084, and MB-LU-181) (Supplementary Figure 4B–D). In addition, the tumor take rates were considerably higher when cultured under neural stem cell conditions; UW228-3+ (92.31%) and D425 Med (81.08%). These observations suggest that growth conditions play an instrumental role in providing correct homing cues.
Figure 4.
Culture medium composition has a significant effect on tumor cell localization in transplanted zebrafish embryos. (A) Representative images of fli1:EGFP zebrafish embryos 48 hpf transplanted with tumor cell lines cultured in complete neural stem cell medium (UW228-3+, D425 Med+, and MDA-MB-231+). Medium for UW228-3+ was supplemented with 1% FBS for maintenance and removed before transplantation. (B) Brain/body area distribution of tumor cell lines cultured in complete neural stem cell medium (+) compared to normal culture medium in 48 h old embryos. Mean ± SEM, ****P < .0001, Student t-test. (C) Schematic heatmaps of the tumor cell location of tumor cell lines cultured in complete neural stem cell medium (+) compared to normal culture medium in 48 h old embryos. 1 = hindbrain area; 2 = mid/forebrain area; 3 = pericardium area; 4 = yolk sac area; 5 = body and tail area. *P < .05, **P < .01, ****P < .0001, 2-way ANOVA with Šídák post-hoc test. (D) Proliferation rate of UW228-3, D425 Med, and MDA-MB-231 cells cultured in medium with FBS or in neural stem cell medium (+) evaluated by cell counting. Mean ± SD, n = 3 independent experiments. (E) Overall survival analysis of zebrafish transplanted with UW228-3, D425 Med, and MDA-MB-231 cells cultured in medium with FBS or in neural stem cell medium (+). Kaplan–Meier curves depict differences in survival and statistical differences were determined using the Log-rank Mantel-Cox test.
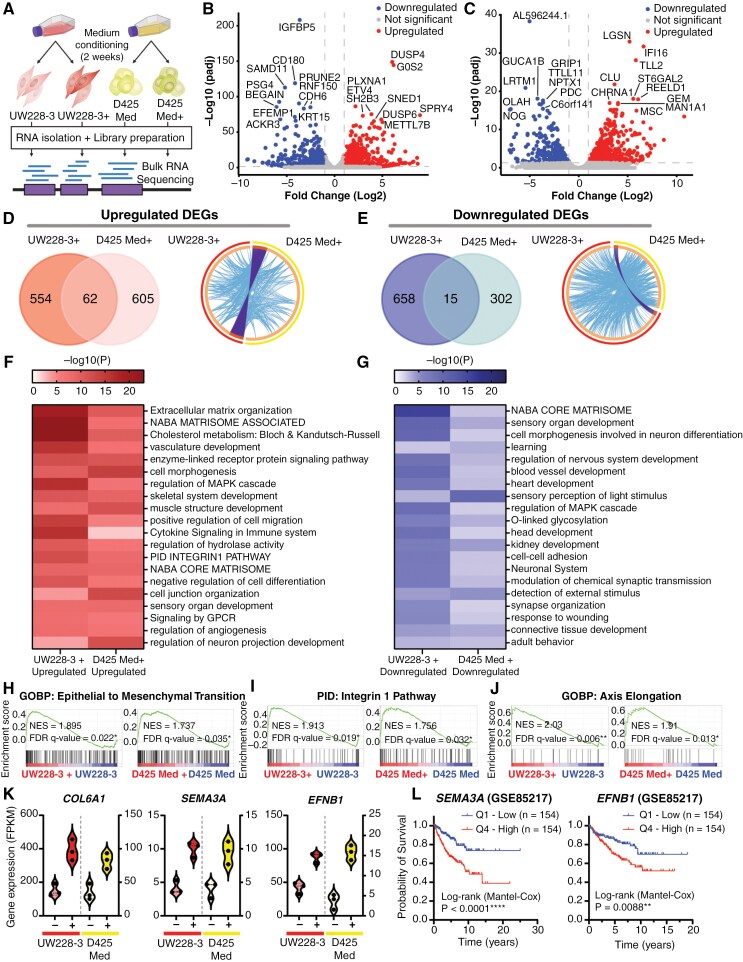
To gain a better understanding of what confers increased homing capacity in cells cultured in neural stem cell conditions, we analyzed transcriptional changes in UW228-3 and D425 Med cultured under standard culture conditions compared with neural stem cell culture conditions (Figure 5A). Differential gene expression identified 1289 genes with significantly altered expression (616 genes upregulated, 673 downregulated, FDR < 0.05, FC > 2) in UW228-3+ cells compared to UW228-3 (Figure 5B, Supplementary Dataset 1), and 984 genes (667 genes upregulated, 317 downregulated, FDR < 0.05, FC > ±2) in D425 Med+ cells compared to D425 Med (Figure 5C, Supplementary Dataset 1).
Figure 5.
Transcriptomic analysis reveals upregulation of genes associated with cancer stemness and migration. (A) Schematic overview of the bulk RNA sequencing protocol. (B) Volcano plot showing all differentially expressed genes (DEGs) upon comparison of the transcriptome of UW228-3 with UW228-3 in complete neural stem cell medium (+), or (C) D425 Med with D425 Med cultured in complete neural stem cell medium (+). Gene names of the top 20 most significant DEGs are shown. Significant DEGs are those with a fold change of ≥ 2 (Log2FC ≥ 1) and an adjusted P-value of ≤ .05. Blue dots (left side) represent downregulated genes and red dots (right side) represent upregulated genes. (D) Left panel: Venn diagram showing overlap between the upregulated and (E) downregulated DEGs in UW228-3+ and D425 Med+. Right panel: Circos plots displaying the gene overlap and shared term level between the gene lists UW228-3 versus UW228-3+ and D425 Med versus D425 Med+. Purple lines link identical genes and blue lines link genes that belong to the same enriched ontology term. The inner circle represents the 2 gene lists, whereas the outside circle represents the group. (F, G) Heatmaps of enriched gene set terms shared across both gene lists, colored by P-values. Each term represents a cluster of multiple associated gene sets. Clusters were named after the most significantly expressed gene set present in both gene lists. (H–J) Epithelial to mesenchymal transition, integrin-1 pathway, and axis elongation are all significantly enriched in both UW228-3 and D425 Med when cultured in complete neural stem cell medium (+) as observed upon GSEA analysis. (K) Violin plots showing the DEGs COL6A1, SEMA3A, and EFNB1 that are significantly enriched in both UW228-3+ and D425 Med+, expressed as fragments per kilobase (FPKM). (L) Kaplan–Meier curves represent the survival of MB patients with low expression (Q1) compared with MB patients with high expression (Q4) of SEMA3A or EFNB1.
Surprisingly, there was little overlap between the differentially regulated genes in UW228-3+ and D425 Med+ cells, with only 62 upregulated genes and 15 downregulated genes shared between them (Figure 5D and E, left panels). However, pathway enrichment analysis identified that UW228-3+ and D425 Med+ share a high degree of genes belonging to the same enriched ontology term (Figure 5D and E, right panels) and converge on a multitude of similar gene sets (Figure 5F and G). Importantly, many of the upregulated pathways were associated with enhanced migratory capacity, such as pathways involved in extracellular matrix organization, matrisome pathways, integrin pathway, and positive regulation of cell migration (Figure 5F, Supplementary Figure 5A, Supplementary Dataset 2). Other upregulated gene set terms were associated with organotypic developmental programs, including pathways related to negative regulation of cell differentiation and regulation of neuron projection development. This suggests that established MB cell lines can be reconditioned to a more progenitor-like state. In addition, the downregulation of pathways involved in the regulation of nervous system development, neuron differentiation, and cell–matrix or cell–cell adhesion may further increase the migratory capacity and promote the progenitor-like state of UW228-3+ and D425 Med+ (Figure 5G, Supplementary Figure 5B, Supplementary Dataset 3). This was further validated using GSEA, which showed that among others epithelial to mesenchymal transition and the integrin-1 pathway are enriched in both UW228-3+ and D425 Med+ cells (Figure 5H and I, Supplementary Dataset 4). Furthermore, the biological process of axis elongation was enriched in neural stem cell culture conditions, suggesting transcriptional changes that assist the MB+ cells to respond to intrinsic signals responsible for developmental growth and organ positioning (Figure 5J, Supplementary Dataset 4). Overall, this suggests that neural stem cell culture conditions affect UW228-3 and D425 Med cells similarly functionally, although not via the exact same genes.
Nevertheless, among the shared upregulated genes, we found COL6A1, COL6A2, SEMA3A, EFNB1, ECE1, IL13RA2, TNC, and TRIB2 (Figure 5K, Supplementary Figure 5C), all of which have been associated with cancer cell stemness, migration, invasion, aggressiveness, and poor prognosis in various cancer types, including glioma,30–37 and are expressed in SHH-MB.38 Notably, SEMA3A functions as an axon guidance factor, playing a pivotal role in navigating the axonal network during neural development, while also regulating synaptogenesis and synaptic plasticity.39 Importantly, autocrine SEMA3A has been demonstrated to modulate substrate adhesion, promoting glioblastoma spread, and acting as a mitogen for glioma stem cells through TGF-signaling activation.40,41 Similarly, EFNB1 promotes MB migration and invasion and plays a crucial role in neural progenitor migration, as well as maintaining the neural progenitor cell state.42–44 Importantly, high SEMA3A and EFNB1 expression correlate with lower overall survival in MB patients (Figure 5L). Altogether, this shows that adapting cells to neural stem cell conditions rewires the transcriptome and confers a more migratory neuronal phenotype, which may explain the increased preference for migrating to the brain area.
Blastula Stage Transplanted Zebrafish Embryos as a Rapid Orthotopic Model for In Vivo Drug Testing in Medulloblastoma
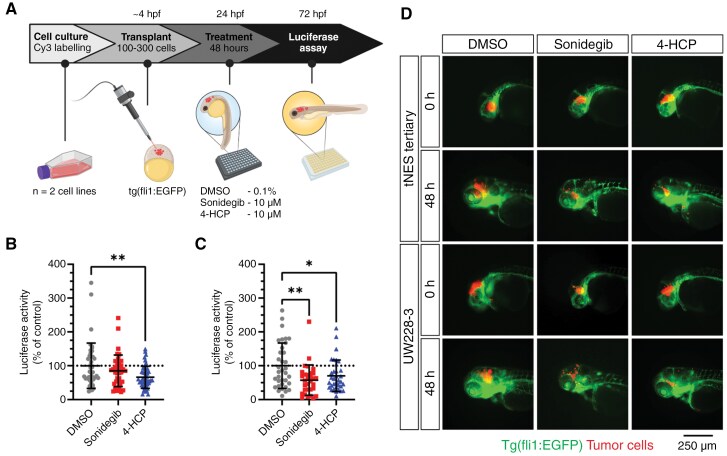
Next, we evaluated the model’s utility for rapid drug efficacy testing. Drug testing for brain tumors faces significant challenges due to the presence of the blood–brain barrier (BBB), limiting access to many drugs. Significantly, zebrafish embryos do not establish a functional BBB until 4 days post-fertilization.45 This provides a unique testing window for assessing the efficacy of drugs that might not cross the BBB, serving as a proof-of-principle platform for a compound’s effectiveness against MB cell growth. To enhance the quantitative analysis of tumor cell growth, we first transduced cells with a luciferase gene, enabling bioluminescence measurement in a plate-format. Zebrafish embryos were transplanted at the blastula stage with either tertiary tNES-Luc or UW228-3-Luc cells. Twenty-four hours post-transplantation, embryos with cells located in the brain area were selected for drug treatment. Forty-eight hours after drug treatment, embryos were imaged and only living embryos were analyzed for bioluminescence activity (Figure 6A). The results demonstrated that the SMO inhibitor Sonidegib, as well as 4-HCP (the active metabolite of cyclophosphamide), significantly reduced the cell viability (Figure 6B and C) and tumor growth of tertiary tNES and UW228-3 cells (Figure 6D). This highlights the zebrafish MB model as a rapid and effective method for assessing drug efficacy in an in vivo setting.
Figure 6.
Transplantation in the blastula stage of zebrafish embryos can be used as an orthotopic medulloblastoma model for rapid in vivo testing of drug compounds. (A) Schematic overview of the treatment protocol. (B) Luciferase activity of transplanted tertiary tNES cells (C) and UW228-3 cells after 48 h of treatment with sonidegib or 4-HCP. Data normalized to the DMSO control and presented as mean ± SD with each point representing one zebrafish embryo of 2 individual experiments. *P < .05, **P < .01, 1-way ANOVA with Dunnett post-hoc test. (D) Representative images of zebrafish upon Sonidegib and 4-HCP treatment. Green indicates the vasculature system of zebrafish (fli1:EGFP) and red (Cy3) indicates the tumor cells transplanted.
Discussion
Here, we present a zebrafish embryo model for MB that can be used to study the kinetics of tumor cell growth, neurotropism, and drug efficacy. The injection of MB cells into blastula-stage zebrafish embryos enables fast and efficient orthotopic transplantation of tumor cells. Importantly, the injected MB cells preferentially home to the brain and especially to the hindbrain region, the part that later develops into the cerebellum. It has previously been shown that the blastula zebrafish embryo contains homing cues that correctly guide transplanted human melanoma and glioma cells.20,26 We demonstrate here the importance of pre-transplantation culture conditions in enhancing the homing capacity of MB cells to the brain region. Many cell lines traditionally grow in culture media containing fetal bovine serum (FBS). However, FBS not only exhibits batch-to-batch variability but also harbors unknown components that may impact experimental outcomes. Thus, opting for a more defined, cell-type-specific media is crucial for enhancing the translatability and reproducibility of experiments. In particular, studies have shown that glioma cells cultured in serum-free media enriched with neuronal supplements B-27 and N2 as well as growth factors EGF and FGF2 show similar proliferation, migration, and invasion properties to the tumors from which they originate.46–48 Similarly, we found that the transcriptome changed significantly when we switched the growth conditions of UW228-3 and D425 Med to a neural stem-cell-like media. Importantly, we observed the upregulation of genes encoding proteins involved in cell–cell and cell–matrix contact, including integrin-1 interacting proteins. Integrins play a central role as a neurogenic factor in regulating the proliferation, stemness, and migration of neural stem/progenitor cells and contribute to the formation of the neurogenic niche.49–51
In addition to primary brain tumors, secondary brain tumors, that is, metastases from other primary malignancies such as lung, breast, melanoma, colon and kidney cancer, occur with an incidence of 10–40% in patients with solid cancers and correlate with poor overall survival.52,53 The preference for metastatic colonization of the brain, neurotropism, depends on the ability of circulating tumor cells disseminated from the primary tumor site to extravasate the vasculature, cross the BBB, and adapt to the brain microenvironment while altering the extracellular matrix (ECM) and resident cells to create their own metastatic niche.54 Interestingly, many of the commonly upregulated genes in UW228-3 and D425 Med cells cultured in neural stem cell media, including IL13RA2, COL6A1, and COL6A2, have been shown to promote or correlate with breast cancer metastasis to the brain,30,31 suggesting that neural stem cell growth condition promote neurotropism. In line with this, we found that neural stem cell culture conditions resulted in the upregulation of guidance molecules SEMA3A and EFNB1, and that high expression of SEMA3A and EFNB1 in MB patients correlated with a poor prognosis. Interestingly, blocking antibodies against SEMA3A have been shown to inhibit glioblastoma progression in a mouse model,55 suggesting that it could be an interesting target for MB to further explore.
Our study represents the first report on orthotopic zebrafish embryo xenografts as a model for MB. We demonstrate that PDX and cell lines derived from SHH, Group 3, and Group 4 MB are able to form orthotopic tumors in the hindbrain of the developing embryo. As a limitation, no experiments were performed in this study utilizing WNT MB models. Available MB WNT PDX models are very difficult to maintain due to their extremely slow growth and subclinical phenotype, resulting in limited availability.
In summary, we demonstrate that early blastula-stage zebrafish embryos are a valuable orthotopic model system for MB. This model is useful not only for assessing short-term drug responses, to quickly determine the potential efficacy of a drug in an in vivo model but also to investigate neurotropism mechanisms associated with both primary brain tumors and brain metastasis. Importantly, we show that patient-derived cells can successfully graft and grow in the zebrafish model, thus allowing for rapid expansion and testing of primary patient samples. This opens opportunities for personalized drug sensitivity screens, allowing rapid testing that can provide tailored treatment strategies for individual patients.
Supplementary material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Acknowledgments
We would like to thank the zebrafish core facility at Karolinska Institutet for their technical expertise and assistance, Dr. Malin Wickström for reagents and helpful discussions, and everyone in the Wilhelm lab for technical assistance and helpful discussions. Figures 1A, 5A, 6A, and Supplementary Figure 1A were created with BioRender.com (https://biorender.com).
Contributor Information
Niek van Bree, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Ann-Sophie Oppelt, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Susanne Lindström, Comparative Medicine, Karolinska Institutet, Stockholm, Sweden.
Leilei Zhou, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Lola Boutin, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Beth Coyle, Children’s Brain Tumour Research Centre, School of Medicine, Biodiscovery Institute, University Park, University of Nottingham, Nottingham, UK.
Fredrik J Swartling, Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden.
John Inge Johnsen, Department of Women’s and Children’s Health, Karolinska Institutet, Stockholm, Sweden.
Lars Bräutigam, Comparative Medicine, Karolinska Institutet, Stockholm, Sweden.
Margareta Wilhelm, Department of Microbiology, Tumor and Cell Biology (MTC), Karolinska Institutet, Stockholm, Sweden.
Funding
This study was supported by Cancerfonden (22 2236 Pj), Barncancerfonden (PR2021-0080), Radiumhemmets Forskningsfonder (#214173), Vetenskapsrådet (2020-1427, 2023-02206), and Karolinska Institutet (2-5586/2017, 2-1060/2018).
Conflict of interest
The authors N.v.B., A.S.O., L.Z., L.Bo., B.C., F.J.S., J.I.J., and M.W. declare no conflict of interest. The authors S.L. and L.Br. received funding from the Karolinska Institutet core facility grant as well as user fees that are paid to the zebrafish core facility.
Authorship statement
Design of the study—N.v.B. and M.W.; Performed research—N.v.B., A.S.O., S.L., L.Z., L.Bo., and L.Br.; Analyzed data—N.v.B., A.S.O.; Contribution of new reagents and materials—M.W., L.Br., J.I.J., F.J.S., B.C.; Writing original draft of manuscript—N.v.B. and M.W.; Revision of manuscript—N.v.B. and M.W.; All authors have reviewed, read, and approved the submitted version of the manuscript.
Data Availability
All data generated or analyzed during this study are included in this manuscript (and its Supplementary Information files). RNA sequencing data have been deposited at Gene Expression Omnibus database with accession number GSE271361.
References
- 1. Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Packer RJ, Gajjar A, Vezina G, et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol. 2006;24(25):4202–4208. [DOI] [PubMed] [Google Scholar]
- 3. Hill RM, Richardson S, Schwalbe EC, et al. Time, pattern, and outcome of medulloblastoma relapse and their association with tumour biology at diagnosis and therapy: a multicentre cohort study. Lancet Child Adolesc Health. 2020;4(12):865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roussel MF, Stripay JL.. Modeling pediatric medulloblastoma. Brain Pathol. 2020;30(3):703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan RY, Wu JQ, Liu YY, et al. Zebrafish xenograft model for studying mechanism and treatment of non-small cell lung cancer brain metastasis. J Exp Clin Cancer Res. 2021;40(1):371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuan M, White D, Resar L, et al. Conditional reprogramming culture conditions facilitate growth of lower-grade glioma models. Neuro Oncol. 2021;23(5):770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Almstedt E, Rosen E, Gloger M, et al. Real-time evaluation of glioblastoma growth in patient-specific zebrafish xenografts. Neuro Oncol. 2022;24(5):726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weintraub A. All eyes on zebrafish. Lab Anim (NY). 2017;46(8):323–326. [DOI] [PubMed] [Google Scholar]
- 9. Blackburn JS, Liu S, Wilder JL, et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell. 2014;25(3):366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin J, Zhang W, Zhao JJ, et al. A clinically relevant in vivo zebrafish model of human multiple myeloma to study preclinical therapeutic efficacy. Blood. 2016;128(2):249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mercatali L, La Manna F, Groenewoud A, et al. Development of a patient-derived xenograft (PDX) of breast cancer bone metastasis in a zebrafish model. Int J Mol Sci. 2016;17(8):1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fior R, Povoa V, Mendes RV, et al. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci USA. 2017;114(39):E8234–E8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Welker AM, Jaros BD, Puduvalli VK, et al. Standardized orthotopic xenografts in zebrafish reveal glioma cell-line-specific characteristics and tumor cell heterogeneity. Dis Model Mech. 2016;9(2):199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casey MJ, Chan PP, Li Q, et al. A simple and scalable zebrafish model of Sonic hedgehog medulloblastoma. Cell Rep. 2024;43(8):114559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Susanto E, Marin Navarro A, Zhou L, et al. Modeling SHH-driven medulloblastoma with patient iPS cell-derived neural stem cells. Proc Natl Acad Sci USA. 2020;117(33):20127–20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF.. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. [DOI] [PubMed] [Google Scholar]
- 17. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer M, Hutter S, Holmberg KO, et al. Humanized stem cell models of pediatric medulloblastoma reveal an Oct4/mTOR axis that promotes malignancy. Cell Stem Cell. 2019;25(6):855–870.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanden E, Dyberg C, Krona C, et al. Establishment and characterization of an orthotopic patient-derived Group 3 medulloblastoma model for preclinical drug evaluation. Sci Rep. 2017;7(1):46366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pudelko L, Edwards S, Balan M, et al. An orthotopic glioblastoma animal model suitable for high-throughput screenings. Neuro Oncol. 2018;20(11):1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wittbrodt JN, Liebel U, Gehrig J.. Generation of orientation tools for automated zebrafish screening assays using desktop 3D printing. BMC Biotechnol. 2014;14(36):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. [DOI] [PubMed] [Google Scholar]
- 23. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14(2):482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee LM, Seftor EA, Bonde G, Cornell RA, Hendrix MJ.. The fate of human malignant melanoma cells transplanted into zebrafish embryos: assessment of migration and cell division in the absence of tumor formation. Dev Dyn. 2005;233(4):1560–1570. [DOI] [PubMed] [Google Scholar]
- 27. Ulrich F, Ma LH, Baker RG, Torres-Vazquez J.. Neurovascular development in the embryonic zebrafish hindbrain. Dev Biol. 2011;357(1):134–151. [DOI] [PubMed] [Google Scholar]
- 28. Purzner T, Purzner J, Buckstaff T, et al. Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and a therapeutic target in medulloblastoma. Sci Signal. 2018;11(547):eaau5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erdreich-Epstein A, Robison N, Ren X, et al. PID1 (NYGGF4), a new growth-inhibitory gene in embryonal brain tumors and gliomas. Clin Cancer Res. 2014;20(4):827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marquez-Ortiz RA, Contreras-Zarate MJ, Tesic V, et al. IL13Ralpha2 promotes proliferation and outgrowth of breast cancer brain metastases. Clin Cancer Res. 2021;27(22):6209–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang L, Wang L, Yang H, Li C, Fang C.. Identification of potential genes related to breast cancer brain metastasis in breast cancer patients. Biosci Rep. 2021;41(10):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turtoi A, Blomme A, Bianchi E, et al. Accessibilome of human glioblastoma: collagen-VI-alpha-1 is a new target and a marker of poor outcome. J Proteome Res. 2014;13(12):5660–5669. [DOI] [PubMed] [Google Scholar]
- 33. Niechi I, Erices JI, Carrillo-Beltran D, et al. Cancer stem cell and aggressiveness traits are promoted by stable endothelin-converting enzyme-1c in glioblastoma cells. Cells. 2023;12(3):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Angel I, Pilo Kerman O, Rousso-Noori L, Friedmann-Morvinski D.. Tenascin C promotes cancer cell plasticity in mesenchymal glioblastoma. Oncogene. 2020;39(46):6990–7004. [DOI] [PubMed] [Google Scholar]
- 35. Xia S, Lal B, Tung B, et al. Tumor microenvironment tenascin-C promotes glioblastoma invasion and negatively regulates tumor proliferation. Neuro Oncol. 2016;18(4):507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang Y, Zekiy AO, Ghaedrahmati F, et al. Tribbles homolog 2 (Trib2), a pseudo serine/threonine kinase in tumorigenesis and stem cell fate decisions. Cell Commun Signal. 2021;19(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeng J, Zhang J, Yang YZ, et al. IL13RA2 is overexpressed in malignant gliomas and related to clinical outcome of patients. Am J Transl Res. 2020;12(8):4702–4714. [PMC free article] [PubMed] [Google Scholar]
- 38. Linke F, Johnson JEC, Kern S, et al. Identifying new biomarkers of aggressive Group 3 and SHH medulloblastoma using 3D hydrogel models, single cell RNA sequencing and 3D OrbiSIMS imaging. Acta Neuropathol Commun. 2023;11(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carulli D, de Winter F, Verhaagen J.. Semaphorins in adult nervous system plasticity and disease. Front Synaptic Neurosci. 2021;13(May):672891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bagci T, Wu JK, Pfannl R, Ilag LL, Jay DG.. Autocrine semaphorin 3A signaling promotes glioblastoma dispersal. Oncogene. 2009;28(40):3537–3550. [DOI] [PubMed] [Google Scholar]
- 41. Jeon HM, Shin YJ, Lee J, et al. The semaphorin 3A/neuropilin-1 pathway promotes clonogenic growth of glioblastoma via activation of TGF-beta signaling. JCI Insight. 2023;8(21):e167049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sikkema AH, den Dunnen WF, Hulleman E, et al. EphB2 activity plays a pivotal role in pediatric medulloblastoma cell adhesion and invasion. Neuro Oncol. 2012;14(9):1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Catchpole T, Henkemeyer M.. EphB2 tyrosine kinase-dependent forward signaling in migration of neuronal progenitors that populate and form a distinct region of the dentate niche. J Neurosci. 2011;31(32):11472–11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qiu R, Wang X, Davy A, et al. Regulation of neural progenitor cell state by ephrin-B. J Cell Biol. 2008;181(6):973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O’Brown NM, Megason SG, Gu C.. Suppression of transcytosis regulates zebrafish blood-brain barrier function. Elife. 2019;8:e47326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 47. Jiang Y, Marinescu VD, Xie Y, et al. Glioblastoma cell malignancy and drug sensitivity are affected by the cell of origin. Cell Rep. 2017;18(4):977–990. [DOI] [PubMed] [Google Scholar]
- 48. Xie Y, Bergstrom T, Jiang Y, et al. The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine. 2015;2(10):1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen YC, Martins TA, Marchica V, Panula P.. Angiopoietin 1 and integrin beta 1b are vital for zebrafish brain development. Front Cell Neurosci. 2023;17:1289794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosa AI, Grade S, Santos SD, et al. Heterocellular contacts with mouse brain endothelial cells via Laminin and alpha6beta1 integrin sustain Subventricular Zone (SVZ) stem/progenitor cells properties. Front Cell Neurosci. 2016;10:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Loulier K, Lathia JD, Marthiens V, et al. beta1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7(8):e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maurya SK, Khan P, Rehman AU, et al. Rethinking the chemokine cascade in brain metastasis: preventive and therapeutic implications. Semin Cancer Biol. 2022;86(Pt 3):914–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aizer AA, Lamba N, Ahluwalia MS, et al. Brain metastases: a Society for Neuro-Oncology (SNO) consensus review on current management and future directions. Neuro Oncol. 2022;24(10):1613–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McDonald B, Barth K, Schmidt MHH.. The origin of brain malignancies at the blood-brain barrier. Cell Mol Life Sci. 2023;80(10):282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee J, Shin YJ, Lee K, et al. Anti-SEMA3A antibody: a novel therapeutic agent to suppress glioblastoma tumor growth. Cancer Res Treat. 2018;50(3):1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript (and its Supplementary Information files). RNA sequencing data have been deposited at Gene Expression Omnibus database with accession number GSE271361.