Abstract
Transferrin receptor (TfR) has been shown to be significantly overexpressed in different types of cancers. We discovered TfR as a target for gambogic acid (GA), used in traditional Chinese medicine and a previously undiscovered link between TfR and the rapid activation of apoptosis. The binding site of GA on TfR is independent of the transferrin binding site, and it appears that GA potentially inhibits TfR internalization. Down-regulation of TfR by RNA interference decreases sensitivity to GA-induced apoptosis, further supporting TfR as the primary GA receptor. In summary, GA binding to TfR induces a unique signal leading to rapid apoptosis of tumor cells. These results suggest that GA may provide an additional approach for targeting the TfR and its use in cancer therapy.
Keywords: rapid apoptosis, caspases, target identification
Transferrin receptor 1 (CD71, TfR), is a type II transmembrane protein with a molecular mass of 85 kDa as a monomer. TfR interacts with two proteins: transferrin (Tf) and hereditary hemochromatosis protein involved in the cellular transport of iron (1, 2).
TfR expression is increased in dividing cells (3), and its overexpression has been reported in different types of cancers such as glioma, pancreatic, and colon cancers (4, 5). Tf and TfR have been targets for therapeutic intervention in certain cancers. These approaches have primarily consisted of using TfR or Tf to direct a toxin or therapeutic molecule to the cancer cells, the use of antisense- or antibody-mediated therapies directed at TfR or Tf, and iron chelator therapies to starve cells of iron (6, 7). Induction of apoptosis or growth inhibition of tumor cells through these approaches has been reported to take several days (8, 9).
Apoptotic cell death is the consequence of a series of precisely regulated events that are frequently altered in tumor cells. The sequence of events that results in the activation of caspases has been broadly categorized into two pathways, the “extrinsic” pathway (10) and the intrinsic mitochondrial pathway (11, 12). In either case, the ultimate cleavage and activation of the executioner caspases ensures destruction of the cell. Using chemical genetics in our drug discovery program, we discovered gambogic acid (GA) as an apoptosis inducer (13).
We have now discovered TfR as the molecular target for GA, a natural product from the resin of the Gamboge Hanburyi tree. The resin has been used in traditional Chinese medicine (14). GA and its active derivatives bind to the purified TfR independent of the Tf binding site and rapidly activate apoptosis in cells. A short exposure to the drug is sufficient to rapidly activate the apoptosis cascade. The discovery and characterization of the GA/TfR interaction in apoptosis may allow us to develop new ways to exploit TfR and other related proteins for therapeutic purposes.
Methods
Preparation of GA and Derivatives. GA was isolated in overall yield of ≈5% from the easily and widely available gamboge resin. It was purified by converting the crude extract from the gamboge resin into pyridine salt, followed by recrystallization. Derivatives of GA were prepared according to the reported procedure in ref. 13. The characterization of GA derivatives is available as Supporting Methods, which is published as supporting information on the PNAS web site.
Cell Lines and Reagents. Jurkat, T47D, Mes, Mes ADR, and 293T cells were purchased from American Type Tissue Culture (American Type Culture Collection, Manassas, VA). Jurkat/Bcl2, WT mouse embryonic fibroblast (MEF), and Apaf1 null MEFs were provided by D.R.G. Primary human umbilical vein endothelial and human mammary epithelial cells were purchased from Cambrex (East Rutherford, NJ). Detailed growth conditions and reagents are available in Supporting Methods. Anti-TfR antibodies and holo-transferrin were used as described in Supporting Methods.
Apoptosis Assays. DAPI staining, propidium iodide viability, and cell cycle analysis were done by using standard methods explained briefly in Supporting Methods. Caspase induction assay was done as described in ref. 15. Cell proliferation assay: The CellTiter 96 AQueous Assay (Promega) was used to determine the 50% growth inhibition (GI50) values for the compounds according to the manufacturer's instructions.
Identification of GA Target. Membrane proteins were solubilized, run on SDS/PAGE, stained, and followed by in-gel tryptic digest according to standard procedures (Supporting Methods). Protein identification was performed by LC/MS/MS (Centre Hospitalier de l'Universite Laval Research Centre, Quebec), and the actual peptides identified are listed in Supporting Methods.
Immunofluorescence, Immunohistochemistry, and Electron Microscopy. For detailed immunofluorescence, immunohistochemistry, and electron microscopy see Supporting Methods. T47D cells were pretreated with 2 μM GA for 15 min at 37°C washed with PBS, and then labeled for 30 min with 1 μg/ml FITC-labeled mouse anti-human TfR (RDI, Flanders, NJ) at 37°C and mounted. For electron microscopy, sections were examined at an accelerating voltage of 60 kV by using a Zeiss EM10C electron microscope core facility (Veterans Hospital, San Diego). More details and methods used for the immunoprecipitations and Western blotting are described in Supporting Methods.
Short Interfering (siRNA) Transfections, cDNA Synthesis, and Real-Time PCR. Chemically synthesized human transferrin receptor and caspase-8 siRNA oligos were used (Ambion, Austin, TX). The target sequence for TfR siRNA was 5′ AAC TTC AAG GTT TCT GCC AGC 3′ and for caspase-8 siRNA was 5′ AAG GAA AGT TGG ACA TCC TGA 3′. The control siRNA oligos, human cyclophilin, and negative control scrambled siRNAs were also from Ambion (16).
Standard procedures were used for cDNA synthesis and quantitative PCR experiments (see Supporting Methods) cDNA was made by using the Retroscript cDNA synthesis kit (Ambion) according to the manufacturer's instructions. Quantitative PCR was done by Sybrgreen incorporation with the Quantitect kit (Qiagen, Valencia, CA) on the LightCycler (Roche Molecular Biochemicals, Mannheim, Germany) by using standard conditions. Data were normalized against the housekeeping gene, cyclophilin. The cells transfected with cyclophilin as a control were normalized against glyceraldehyde phosphate dehydrogenase.
Binding Assays. Jurkat cells were incubated with tritium-GA at 1 μM at 37°C with or without 20 μM unlabeled GA. At indicated time points, bound tritium-GA was quantitated by liquid scintillation counting. TfR-coated wells were incubated with GA-biotin as described above, washed, and incubated with nontagged analogs or binding/washing buffer as a control. Wells were incubated with Europium-Streptavidin, then quantified after incubation with Enhancement Solution (PerkinElmer) by measuring time-delayed fluorescence on a Wallac Victor plate reader (PerkinElmer) according to manufacturer's instructions.
Results
Gambogic Acid Activates Apoptosis and Inhibits Tumor Growth in Vitro. GA triggers morphological changes in cells typical of apoptosis. Membrane blebbing is induced within 15 min in Jurkat cells as observed by electron microscopy (Fig. 1A). To verify that caspases are critical in this pathway, we tested the effect of a pan-caspase inhibitor, MX-1013, on apoptosis (17). We observed that morphological changes (Fig. 7, which is published as supporting information on the PNAS web site), proteolytic cleavage of caspases, and apoptosis (data not shown) depend on activation of caspases.
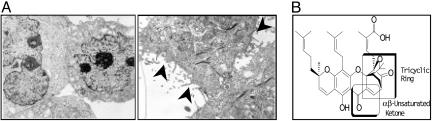
Fig. 1.
GA induces apoptosis. (A) Electron microscopy of T47D cells treated with DMSO (Left) or GA(5 μM, Right). (Scale bar: 1 mm ≈ 0.01 μm.) (B) The structure of GA of which the tricyclic ring and α,β-unsaturated ring (boxed areas) were modified to obtain the derivatives.
GA derivatives were made to elucidate the structure activity relationship (13). The tricyclic ring and the α, β unsaturated ketone, as indicated, are essential for the activity of GA (Fig. 1B). The saturated ketone derivative (inactive GA) does not activate caspases in cells. GA activates caspases in our cell-based assay (15) and inhibits cell growth in various tumor cell lines at submicromolar potencies; this effect is also seen in multidrug resistant human uterine sarcoma cell line (18) as shown in Table 1 where standard chemotherapeutics showed a 100- to 1,000-fold decrease in potency (data not shown).
Table 1. GA inhibits growth in various tumor cell lines.
| Cell | Tissue | Gl50, nM |
|---|---|---|
| T47D | Breast | 630 |
| ZR751 | Breast | 400 |
| HL60 | Lymphocyte | 115 |
| Jurkat | Lymphocyte | 168 |
| Calu1 | Nonsmall cell lung | 550 |
| MES | Uterine | 300 |
| MES ADR* | Uterine | 1,000 |
Multidrug resistance pgp overexpressed (19).
TfR Identified as the Molecular Target. Initial experiments indicated that GA triggered apoptosis through a cell-surface target. We observed from our structure activity relationship studies that certain GA derivatives tolerated different bulky group modifications while maintaining induction of apoptosis. To better characterize the target, we synthesized a biotin-conjugated GA (biotin-GA) for attachment to streptavidin microspheres, fluorescein-conjugated GA (fluorescein-GA), or agarose conjugates. All derivatives retained the activity of GA. Using the equipotency biotinylated derivative (biotin-GA) conjugated to streptavidin FluoSpheres, we tested the ability of GA-conjugated FluoSpheres to activate apoptosis and observed comparable cell death to unconjugated GA at the same concentration (Fig. 2A). We then synthesized a tritium-labeled GA derivative (tritium-GA) to further understand the cellular binding properties of GA. Using tritium-GA, we demonstrated saturable and temperature-dependent binding on Jurkat cells treated at 37°C (Fig. 2B) and 4°C (data not shown) that is competed by cold unlabeled GA (Fig. 2B). To further prove that the GA target was a membrane protein, cell surface biotinylation experiments were performed according to reported procedures (21). Biotinylated cells were treated with fluorescein-GA, immunoprecipitated with anti-FITC antibody, and blotted with anti-biotin antibodies. Identification of a biotinylated protein in the fluorescein-GA affinity experiments that could be competed off by GA and not by an α, β saturated inactive derivative (inactive-GA), further confirmed the presence of target protein on the cell surface (Fig. 2C). These data support the observations that GA mediates apoptosis through a cell surface receptor.
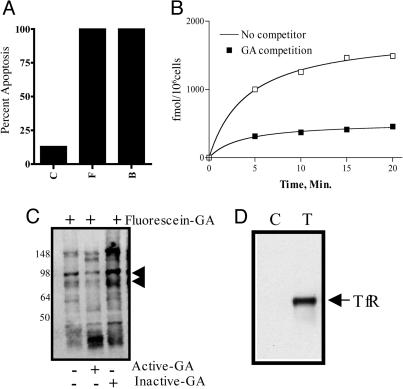
Fig. 2.
GA binds to a cell surface receptor. (A) Biotin-GA conjugated to streptavidin FluoSpheres activates apoptosis. Jurkat cells were treated for 24 h with DMSO (C), 5 μM GA (F), or 5 μM biotin-GA (B) bound to streptavidin FluoSpheres (20). Propidium iodide viability staining was used to quantitate apoptosis. (B) Saturable receptor binding on cells. Jurkat cells were incubated with tritium-GA at 1 μM at 37°C, with or without 20 μM of unlabeled GA. At indicated time points, the amount of bound tritium-GA was determined by liquid scintillation counting. (C) GA binds to a biotinylated cell surface protein. Jurkat cells were surface-biotinylated and treated with either DMSO or 5 μM fluorescein-GA for 30 min at 37°C. Active-GA and inactive-GA were used as controls to determine the specificity of binding. (D) GA binds to TfR. Jurkat cells were treated with either DMSO or 5 μM fluorescein-GA, for 30 min at 37°C. Lysate was immunoprecipitated with anti-FITC antibody and detected by immunoblotting with anti-TfR antibody.
Using an affinity- and specificity-based approach, we identified and sequenced a compound-associated band. Active-GA was used as competitor to further identify the specificity of the bound protein. Membrane fractions from Jurkat cells were used for the affinity-based experiments with the agarose-GA. The bound protein was then isolated from the gel, identified through tryptic digest, and the sequence was determinated by LC/MS/MS methods. The sequence coverage through LC/MS/MS and subsequent ncbinr database search contained 10 peptides from TfR (see Methods) spanning ≈23% of the protein.
In an independent experiment by using treatment of whole cells with fluorescein-GA and LC/MS/MS determination of the bound protein, we also identified TfR peptides. We further confirmed the identification of TfR by Western blotting in immunoprecipitations by using fluorescein-GA (Fig. 2D).
GA Binds to TfR and Does Not Compete with Tf. To evaluate and confirm the binding of GA to TfR, we performed a series of experiments by using purified TfR containing the extracellular domain in a binding assay with biotin-GA. We determined that biotin-GA binds to TfR with a Kd of 2.2 μM (Fig. 3A). Although the Kd is slightly higher than the EC50 values (0.7–1.6 μM) for induction of apoptosis and growth inhibition in cells (0.5–1.5 μM); this result is likely due to the differences across the various assays. GA, but not inactive-GA, competes for specific binding to the receptor with an IC50 of 1 μM (Fig. 3B), also indicating the specificity of binding. To further understand the GA/TfR interaction, we showed that biotin-GA bound to TfR can be displaced by unlabeled GA, suggesting reversibility in the binding of GA to TfR (Fig. 3C).
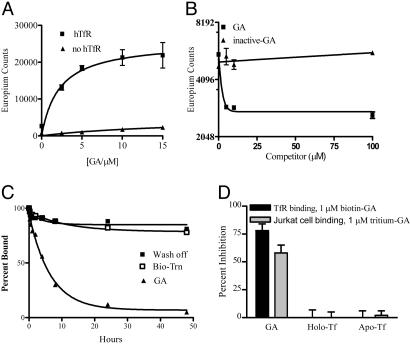
Fig. 3.
GA binds to soluble TfR protein. (A) GA binds TfR in vitro. TfR-coated wells were incubated with increasing concentrations of biotin-GA in binding/washing buffer for 20 min at 30°C, then incubated with Europium labeled-streptavidin (Eu-streptavidin). Amounts of bound Eu-Streptavidin were quantified by measuring time-delayed fluorescence. (B) IC50 of active and inactive GA derivatives. In competition experiments, biotin-GA at 1 μM was premixed with increasing amounts of GA or the inactive-GA as a competitor. Amounts of bound Eu-Streptavidin were quantified by measuring time-delayed fluorescence. (C) GA bound to TfR in vitro can be displaced by active GA derivatives. TfR-coated wells were incubated with biotin-GA as described, washed, and incubated with GA or binding/washing buffer as a wash off control. (D) Binding of biotin-GA and tritium-GA to TfR is not inhibited by either apo-transferrin or holo-transferrin. Binding of biotin-GA and tritium-GA to Jurkat cells (hatched) or in vitro TfR-binding (solid) in the presence of 1 μM GA, 50 μg/ml apotransferrin, or 50 μg/ml holo-transferrin is shown in this graph.
To understand whether GA and Tf bind to the same site on TfR and/or whether Tf binding interferes with GA binding, tritium-GA was used, either with Jurkat cells or on the purified extracellular domain of TfR, in the presence or absence of apotransferrin or holo-transferrin. The two forms of Tf did not affect the binding of tritium-GA indicating that the binding site(s) for GA and Tf on the receptor are independent of each other (Fig. 3D).
To further confirm that TfR is the target for GA, we performed competitive binding assays and compared the inhibitory constants of various GA derivatives in the receptor-binding assay. In an attempt to identify the functional core of the molecule, we initiated a total synthesis of the known essential molecular functional components and synthesized derivatives with just the tricyclic ring with α,β unsaturated ketone backbone or the α,β saturated ketone. These derivatives were then tested in the receptor-binding assay (Table 2). To understand the specificity of the binding to TfR, we selected and evaluated the competition of biotin-GA by a functionally active backbone molecule and its functionally inactive α, β saturated ketone derivative. We observed a good correlation between inhibitory potencies of the various GA derivatives for TfR binding and the EC50 values for caspase activation. The α, β saturated ketone derivatives were inactive in both assays. As expected the compounds that were more potent in activating caspases had also had lower IC50, indicating better binding to the TfR (Table 2).
Table 2. Functional activities of GA derivatives correlate with their binding to TfR.
| Activity in apoptosis induction assay
|
|||
|---|---|---|---|
| Compound | Competition IC50 in TR binding assay, μM | Fold caspase-3 activation | EC50 |
| GA | 4.1 | 18 | 526 |
| Methyl-GA | 3.6 | 24 | 326 |
| Inactive-GA | >30 | 1.2 | ND |
| α, β unsaturated backbone | 12.7 | 10 | 4,016 |
| α, β saturated backbone | >30 | 1 | ND |
ND, not determined.
GA Potentially Interferes with TfR Internalization and Is Independent of Iron Regulation. Because TfR is important for cellular iron regulation and interfering with this pathway in various ways can lead to growth inhibition in cells, we investigated the effects of iron and transferrin-dependent iron regulatory mechanisms during GA-mediated apoptosis. First, we examined whether binding of Tf to the receptor has any effect on GA-induced apoptosis. Even in the presence of saturating levels of holo-Tf, GA was capable of inducing apoptosis in Jurkat cells (Fig. 4A).
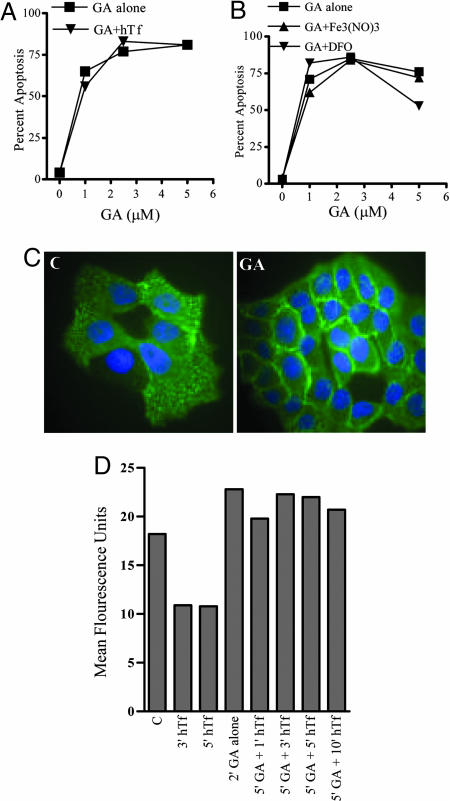
Fig. 4.
Known mechanisms of iron regulation do not overlap with GA-mediated apoptosis. (A) The binding of holo-Tf to TfR has no effect on GA-induced apoptosis. Jurkat cells were pretreated with holo-transferrin for 30 min and subsequently treated with DMSO or 1, 2.5, or 5 μM GA for 4 h, after which cell viability was measured as described. (B) GA-induced apoptosis through TfR is not iron-dependent. Jurkat cells were treated with 10 μM desferrioxamine (DFO) or 50 μM ferric nitrate [Fe3(NO)3] for 1 h and treated as described in A. (C) GA interferes with TfR receptor internalization. T47D cells were treated with DMSO or 2 μM GA for 15 min and further treated with FITC-conjugated anti-TfR for 30 min at 37°C. The cells were then fixed with methanol at –20°C for 5 min, washed with PBS, and mounted with Vectashield mounting medium. Representative of three independently confirmed experiments. (D) GA interferes with TfR internalization as indicated by cell surface TfR expression. Jurkat cells were treated with holo-transferrin alone (50 μg/ml) for 3 or 5 min, GA alone (5 μM) for 2 min, or pretreated for 5 min with GA followed by 1, 3, 5, or 10 min of holo-transferrin treatment. Cells were then stained with FITC-conjugated anti-transferrin receptor antibody for 30 min at 4°C. After washing, cells were analyzed on a Becton Dickinson FACS Calibur. Data are shown as mean fluorescence units. Results were confirmed in three independent experiments.
We also determined whether GA-mediated apoptosis was iron-dependent or sensitive to iron deprivation. We used a clinically tested iron-chelator, desferrioxamine (22) ferric nitrate (Fig. 4B) (23) and ferrous citrate (Fig. 8A, which is published as supporting information on the PNAS web site). Cells were pretreated with these agents for 24–48 h before GA treatment. Cells were also stained for TfR to monitor for cell surface regulation of the receptor. We observed that GA caused apoptosis independent of the iron requirement.
We also examined the effects of GA on receptor internalization by microscopy in T47D cells (Fig. 4C) or by flow cytometry (Fig. 4D) to get a quantitative idea about the cell surface localization of TfR. Although holo-Tf lead to a reduced TfR staining, this effect may be due to interference in their binding sites as indicated by the same experiment repeated at 4°C (Fig. 8C). GA treatment alone for 2 min caused increased cell surface TfR levels. Similar data were also obtained by monitoring for Tf endocytosis by using alexa-fluor-labeled transferrin (Fig. 8B). We observed an increased TfR expression on cell surface on GA treatment. These results suggested that binding of GA to TfR either stabilizes or blocks the receptor from internalization. As a control, another apoptosis-inducing compound, staurosporine, did not have any effects on TfR internalization (data not shown), indicating that activation of apoptosis alone does not block receptor internalization. Taken together, these findings indicate that the binding of GA to TfR and the subsequent induction of apoptosis is unrelated to Tf binding and iron transport.
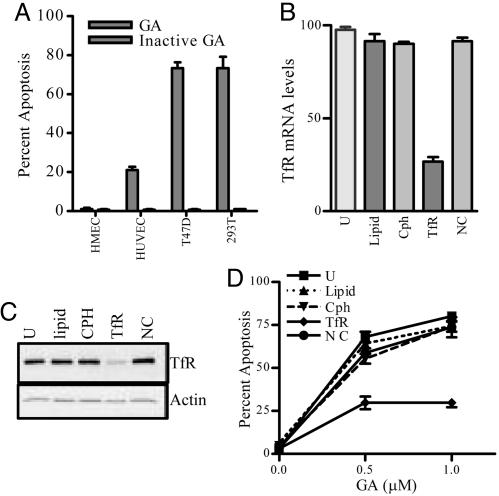
Down-Regulation of TfR in Cells Affects Sensitivity to GA-Induced Apoptosis. To understand the correlation of TfR levels and susceptibility to GA-mediated apoptosis and to determine the therapeutic advantage of GA, we evaluated its effect on tumor cells that overexpress TfR on their cell surface (T47D and 293T) and primary normal human mammary epithelial and human umbilical vein endothelial cells. We observed that tumor cells treated for only 2 h with GA underwent significant apoptosis compared with that observed for primary normal cells (Fig. 5A). There was no correlation between the proliferation rates and sensitivity to GA (data not shown); however, we observed a correlation between the level of TfR expression and rapidity of GA-induced apoptosis (Fig. 5A). Although, 293T cells and T47D cells have a comparable level of apoptosis when treated with 1 μM GA, the latter cells are more sensitive to apoptosis even at lower concentrations of GA (data not shown). To further validate that TfR is the target for GA, we used RNA interference assays with siRNA duplexes to inhibit the expression of TfR protein. Studies were performed to evaluate sensitivity of the transfected cells toward GA. Down-regulation of endogenous TfR after transfection with the siRNA oligonucleotide was confirmed by mRNA analysis (Fig. 5B), Western blot analysis (Fig. 5C), and determination of cell surface receptor expression (data not shown). TfR-specific siRNA transfections showed ≈70% inhibition of TfR expression after 48 h. Down-regulation of TfR by siRNA was specific, as shown by analysis of other cell cycle (24) and apoptosis genes such as p21, cdk2, caspase-8, and clathrin (Fig. 9A, which is published as supporting information on the PNAS web site). After down-regulation of endogenous TfR by using siRNA, 293T cells displayed a significant reduction in apoptosis induced by GA (Fig. 5D). Similar results were obtained by using T47D breast cancer cells (data not shown). The doubling times in the various transfected cells were not significantly altered (Fig. 9B). We also used paclitaxel as a known cytotoxic agent and showed that the down-regulation of TfR did not result in a reduction in apoptosis due to paclitaxel (Fig. 9B), thus indicating that TfR down-regulated cells are not resistant to all apoptotic stimuli.
Fig. 5.
GA displays differential apoptosis potential in normal versus tumor cell lines, and down-regulation of the TfR by using siRNA technology leads to a decrease in apoptosis with GA treatment. (A) Normal and tumor cell lines were treated with DMSO, 1 μM GA, or 1 μM inactive GA for 5 h and assessed for apoptosis. TfR cell surface expression is noted as follows: T47D (++++), 293T (+++), human umbilical vein endothelial cells (HUVEC, ±), and human mammary epithelial cells (HMEC, –). Results are independently confirmed in three experiments. (B) Real-time PCR showing the down-regulation of the TfR at the mRNA level. 293T cells were transfected for 48 h with lipid alone, Cph, TfR, or negative control siRNAs (NC). (C) Western blot representing the down-regulation of TfR in siRNA-transfected cells. Whole-cell lysates of 293T cells after transfection were subjected to SDS/PAGE and Western blotted with anti-TfR antibody. Actin was used as loading control (Lower). (D) Down-regulation of TfR protects cells from GA-induced apoptosis. 293T cells were transfected and treated as described in A. Cells were fixed and analyzed for cell death.
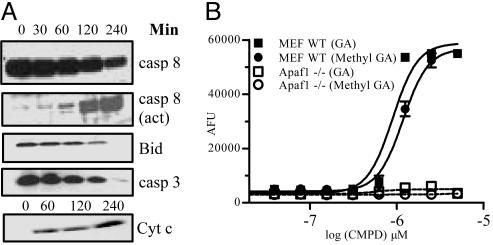
TfR Signaling Mechanism for Apoptosis Induction. GA induces apoptosis through a very robust engagement of the apoptotic pathway. These data include activation of caspase-8, cleavage of Bid, the release of cytochrome c from the mitochondria and activation of caspase-3 (Fig. 6A). Because cleavage of caspase-8 may not empirically imply its enzymatic activation (25), a biotinylated caspase inhibitor was used that binds to the active site of caspases, including caspase-8 and characterized GA-mediated caspase activation. It was observed that GA activates caspase-8 similar to that reported for anti-Fas activation (Fig. 10A, which is published as supporting information on the PNAS web site). However, interfering with the death receptor signaling by using Fas-Fc or TNFR-Fc (data not shown) chimeric proteins did not have any effect on GA-mediated cell death.
Fig. 6.
Signaling pathway of GA-induced apoptosis. (A) A time course of signaling events. Jurkat cells were treated with DMSO or GA (5 μM) for the indicated times. Western blotting was performed with anti-caspase-3, anti-caspase-8, anti-Bid, or anti-cytochrome c antibodies and detected by using ECL. (B) Apaf1 –/– MEFs show a decreased level of caspase activation. Dose–response of GA and methyl-GA induced caspase-3 activities in WT and Apaf –/– MEFs treated for 5 h.
To better define the relative involvement of the extrinsic and intrinsic pathways in GA-mediated apoptosis, a genetic approach was used. Down-regulation of caspase-8 in cells by RNA interference (Fig. 10B) decreased the sensitivity to GA-induced apoptosis. We then used the FADD-negative cell line, Jurkat I2.1 (26), and evaluated whether activation of caspase-8 involves the adaptor protein FADD. These experiments suggest that a lack of FADD has little or no effect in GA-mediated apoptosis (data not shown).
Bcl-2 is an inhibitor of apoptosis (27) and is also overexpressed in many treatment-resistant cancers. To examine the role of Bcl-2 in this pathway, Jurkat cells overexpressing Bcl-2 were used, and in these cells, there was a delay but not inhibition of apoptosis when treated with GA (Fig. 10C). The role of Apaf-1 was evaluated in MEF cells that lack Apaf-1 (19). We observed that there was a significant decrease in the level of caspase activation in the Apaf-1-negative cells than the wild-type MEF cells when treated with GA or one of its active derivative (methyl-GA) for 5 h (Fig. 6B). These data support the conclusions that GA-mediated apoptosis involves caspase-8 and includes the mitochondrial pathway contributing to the amplification of the signaling cascade resulting in the robustness and rapidity of apoptosis. Down-regulation of some of the regulatory molecules in this pathway is able only to delay, but not inhibit, GA-induced apoptosis.
Discussion
We identified TfR as a target of GA and show that it induces apoptosis through a previously unreported mechanism for this receptor. Binding of GA to TfR activates the apoptosis cascade rapidly by using caspase-8 and the mitochondrial pathway. We have observed that high TfR expression levels correlate with sensitivity to GA and contribute to the rate of apoptosis seen. The signaling pathway deciphered indicates activation of caspase-8 as a requirement for apoptosis. Unlike death receptor signaling, we were unable to identify death-inducing signaling complex (DISC) components in TfR immunoprecipitations. A further analysis of DISC involvement or alternate mechanisms for caspase-8 activation (28) in GA-mediated signaling may give further insight into this pathway. Overexpression of Bcl-2 in Jurkat cells caused a delay, but not inhibition, of death. Any role for other members of the Bcl-2 family remains to be studied. Although Jurkat cells are of type II apoptotic phenotype, recent reports (29) on TRAIL-induced apoptosis in these cells warrants further clarification on this topic. Although, caspase-8 activation seems to be required in GA-mediated signaling, the involvement of other initiator caspases, i.e., caspase-10 and caspase-2, cannot be ruled out, especially because of their importance in certain apoptotic pathways (30, 31).
TfR and Tf have been previously identified as targets for cancer therapy. Existing antibody-based approaches may have restricted effectiveness due to inadequate drug delivery and/or immunogenicity issues when using antibody fusion proteins. Iron chelator therapies appear to have clinical activity and are under investigation and in early clinical trials (32). However, iron chelators are reported to act on multiple targets, including those involved in cell cycle progression, which may or may not enhance efficacy or affect toxicity of these compounds. Although TfR has been a target for cancer treatment, the approach reported herein appears to result in a previously unknown mechanism for TfR in the induction of apoptosis.
The specificity of GA/TfR interaction is also demonstrated in activating apoptosis because close analogs of GA that do not compete for receptor binding also do not activate caspases in cells. A GA derivative has also demonstrated significant antitumor efficacy in rodent tumor models with little toxicity (unpublished data). We demonstrate a correlation in TfR expression levels between tumor and normal cells and the sensitivity to GA, which may explain, at least in part, the selectivity seen in vivo. A more detailed evaluation of the pharmacokinetics and biodistribution of the drug and the target expression profile will help to better understand the therapeutic advantage observed by this class of molecules.
These results report the identification and characterization of a molecule (GA) that specifically targets TfR engaging a previously unreported mechanism of action to induce apoptosis. It appears from this body of work that GA interferes with TfR internalization leading to the initial, and rapid, signal for apoptosis. We also demonstrate that GA and Tf bind to independent sites on the receptor, and it appears that GA is not competed by Tf. These studies also suggest a requirement of GA for TfR-mediated rapid apoptosis because the mere down-regulation of TfR does not activate this pathway. Whether GA causes additional conformational changes in the receptor, thereby recruiting the death machinery in an unprecedented manner, remains to be uncovered.
With our continuing efforts, we suggest that the GA/TfR discovery may lead to a new generation of anti-cancer drugs and targeting mechanisms that will be synergistic with existing treatments. Identifying roles for other adaptor proteins or signaling molecules in this GA-induced TfR pathway will possibly identify additional therapeutic targets.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance provided by Candace Crogan-Grundy. This work is supported in part by National Institutes of Health Grant 5R44CA091811-03 (to S.K.).
Author contributions: S.K., K.R.G., S.X.C., D.R.G., and B.T. designed research; S.K., K.A.J., S.M., J.Y.W., N.M.E., L.Q., S.P.A., A.E.P., and N.S. performed research; N.S., S.J., H.-Z.Z., and S.X.C. contributed new reagents/analytic tools; S.K., K.A.J., S.M., J.Y.W., N.M.E., and J.D. analyzed data; and S.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GA, gambogic acid; biotin-GA, biotin-conjugated GA; fluorescein-GA, fluorescein-conjugated GA; LC/MS/MS, liquid chromatograhy tandem mass spectrometry; MEF, mouse embryonic fibroblast; siRNA, short interfering RNA; Tf, transferrin; TfR, transferrin receptor.
References
- 1.Bennett, M. J, Lebron, J. A., Bjorkman, P. J. (2000) Nature 403, 46–53. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence, C., Ray, S., Babyonyshev, M., Galluser, R., Borhani, D. & Harrison, S. (1999) Science 286, 779–782. [DOI] [PubMed] [Google Scholar]
- 3.Chitambar, C. R., Massey, E. J. & Seligman, P. (1983) J. Clin. Invest. 72, 1314–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryschich, E., Huszty, G., Knaebel, H., Hartel, M., Buchler, M. & Schmidt, J. (2004) Eur. J. Cancer 40, 1418–1422. [DOI] [PubMed] [Google Scholar]
- 5.Szekeres, T., Sedlak, J. & Novotny, L. (2002) Curr. Med. Chem. 9, 759–764. [DOI] [PubMed] [Google Scholar]
- 6.Debinski, W. (2002) Cancer Invest. 20, 801–809. [DOI] [PubMed] [Google Scholar]
- 7.Ng, P., Dela Cruz, J., Sorour, D., Stinebaugh, J., Shin, S., Shin, D., Morrison, S. & Penichet, M. (2002) Proc. Natl. Acad. Sci. USA 99, 10706–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White, S., Taetle, R., Seligman, P., Rutherford, M. & Trowbridge, I. (1990) Cancer Res. 50, 6295–6301. [PubMed] [Google Scholar]
- 9.Kawabata, H., Germain, R., Vuong, P., Nakamaki, T., Said, J. & Koeffler, H. (2000) J. Biol. Chem. 275, 16618–16625. [DOI] [PubMed] [Google Scholar]
- 10.Locksley, R. M., Killeen, N. & Lenardo, M. J. (2001) Cell 104, 487–501. [DOI] [PubMed] [Google Scholar]
- 11.Green, D. R. & Reed, J. C. (1998) Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- 12.Desagher, S. & Martinou, J. (2000) Trends Cell Biol. 10, 369–377. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, H. Z., Kasibhatla, S., Wang, Y., Herich, J., Guastella, J., Tseng, B., Drewe, J. & Cai, S. (2004) Bioorg. Med. Chem. 12, 309–317. [DOI] [PubMed] [Google Scholar]
- 14.Asano, J., Chiba, K., Tada, M. & Yoshii, T. (1996) Phytochemistry 41, 815–820. [DOI] [PubMed] [Google Scholar]
- 15.Cai, S., Nguyen, B., Jia, S., Herich, J., Guastella, J., Reddy, S., Tseng, B., Drewe, J. & Kasibhatla, S. (2003) J. Med. Chem. 46, 2474–2481. [DOI] [PubMed] [Google Scholar]
- 16.Brown, D., Jarvis, R., Pallotta, V. & Ford, L. (2002) Ambion TechNotes 9, 3–5. [Google Scholar]
- 17.Yang, W., Guastella, J., Huang, J. C., Wang, Y., Zhang, L., Xue, D., Tran, M., Woodward, R., Kasibhatla, S., Tseng, B., et al. (2003) Br. J. Pharmacol. 40, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harker, W. & Sikic, B. (1985) Cancer Res. 45, 4091–4096. [PubMed] [Google Scholar]
- 19.Milosevic, J., Hoffarth, S., Huber, C. & Schuler, M. (2003) Oncogene 22, 6852–6856. [DOI] [PubMed] [Google Scholar]
- 20.Rabbani, S., Gladu, J., Harakidas, P., Jamison, B. & Goltzman, D. (1999) Int. J. Cancer 80, 257–264. [DOI] [PubMed] [Google Scholar]
- 21.Peirce, M. J., Wait, R., Begum, S., Saklatvala, J. & Cope, A. P. (2004) Mol. Cell Proteomics 3, 56–65. [DOI] [PubMed] [Google Scholar]
- 22.Kemp, J. (1997) Histol. Histopathol. 12, 291–296. [PubMed] [Google Scholar]
- 23.Keyna, U., Nusslein, I., Rohwer, P., Kalden, J. & Manger, B. (1991) Cell Immunol. 132, 411–422. [DOI] [PubMed] [Google Scholar]
- 24.Scacheri, P., Rozenblatt-Rosen, O., Caplen, N., Wolfsberg, T., Umayam, L., Lee, J., Hughes, C., Shanmugam, K., Bhattacharjee, A., Meyerson, M. & Collins, F. S. (2004) Proc. Natl. Acad. Sci. USA 101, 1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boatright, K., Renatus, M., Scott, F. L, Sperandio, S., Shin, H., Pedersen, I. M., Ricci, J. E., Edris, W. A., Sutherlin, D. P., Green, D. R. & Salvesen, G. S. (2003) Mol. Cell. 11, 529–541. [DOI] [PubMed] [Google Scholar]
- 26.Juo, P., Sue-Ann Woo, M., Kuo, C. J., Signorelli, P., Biemann, H. P., Hannun, Y. A. & Blenis, J. (1999) Cell Growth Differ. 10, 797–804. [PubMed] [Google Scholar]
- 27.Kasibhatla, S. & Tseng, B. (2003) Mol. Cancer Ther. 2, 573–580. [PubMed] [Google Scholar]
- 28.Sanchez, I., Xu, C., Juo, P., Kakizaka, A., Blenis, J. & Yuan, J. (1999) Neuron 22, 623–633. [DOI] [PubMed] [Google Scholar]
- 29.Rudner, J., Jendrossek, V., Lauber, K., Daniel, P. T., Wesselborg, S. & Belka, C. (2005) Oncogene 24, 130–140. [DOI] [PubMed] [Google Scholar]
- 30.Chang, D. W., Ditsworth, D., Liu, H., Srinivasula, S. M., Alnemri, E. S. & Yang, X. (2003) J. Biol. Chem. 278, 16466–16469. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer, A., Briand, C. & Grutter, M. G. (2003) J. Biol. Chem. 278, 42441–42447. [DOI] [PubMed] [Google Scholar]
- 32.Chaston, T. B., Lovejoy, D. B., Watts, R. N. & Richardson, D. R. (2003) Clin. Cancer Res. 9, 402–414. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.