Abstract
Relatively little is known about the human genetics of susceptibility to common diseases caused by bacterial pathogens. Tuberculosis, caused by Mycobacterium tuberculosis, is a major cause of morbidity and mortality worldwide. So far, genetic studies of tuberculosis susceptibility have largely been focused on adult patients despite the fact that tuberculosis is highly prevalent among children. To study the host genetic component of pediatric tuberculosis susceptibility, we enrolled 184 ethnically diverse families from the Greater Houston area with at least one child affected by pediatric tuberculosis disease. Using a family-based control design, we found allelic variants of the natural resistance-associated macrophage protein gene 1 (NRAMP1) (alias SLC11A1) significantly associated with tuberculosis disease in this pediatric patient population [P = 0.01; odds ratio = 1.75 (95% confidence interval, 1.10-2.77)]. The association of NRAMP1 with pediatric tuberculosis disease was significantly heterogeneous (P = 0.01) between simplex [P <0.0008; odds ratio = 3.13 (1.54-6.25)] and multiplex families (P = 1), suggesting an interplay between mechanisms of genetic control and exposure intensities. In striking contrast to previous studies in the adult population, we observed that the common alleles of NRAMP1 polymorphisms were risk factors for pediatric tuberculosis disease. To explain the different direction of allelic association between adult and pediatric disease, we hypothesize that NRAMP1 influences the speed of progression from infection to tuberculosis disease.
Keywords: complex traits, host genetics, mycobacterial diseases
The human pathogenic bacterium Mycobacterium tuberculosis, the causative agent of tuberculosis, infects an estimated one-third of the world's population, resulting in >8 million tuberculosis cases and 2 million deaths each year (1). The rate of progression from infection to disease is highly variable, and ≈90% of infected individuals never develop clinical disease. Of the 10% of M. tuberculosis-infected persons who do develop clinically overt disease, approximately half will be diagnosed within <2 years of infection and are considered to be fast progressors of tuberculosis disease. This so-called primary tuberculosis disease is particularly common among children, and the majority of pediatric cases present with primary tuberculosis disease. Tuberculosis patients who progress more slowly from infection to tuberculosis disease and develop clinical disease >2 years after infection are referred to as “reactivation” cases. Little is known about the mechanisms that influence the rate of progression from infection to disease. For example, it is unknown whether different mechanisms of pathogenesis operate in individuals who progress at different rates from infection with M. tuberculosis to clinical tuberculosis disease.
Many lines of evidence support an important role of host genetic variation in tuberculosis susceptibility, including animal models of the disease (2-6), ethnic clustering of tuberculosis cases (7), increased concordance rates of tuberculosis among monozygotic vs. dizygotic twins (8, 9), evidence that certain gene variants are associated or linked with increased risk of tuberculosis (10, 11), and the demonstration that patients with Mendelian disorders of the interleukin 12-IFN-γ axis are hypersusceptible to M. tuberculosis (11, 12). However, the great majority of genetic studies have investigated adult pulmonary tuberculosis cases, whereas few have focused on pediatric tuberculosis disease (13). The exclusion from more intense study of primary tuberculosis disease is surprising, because approximately half of all tuberculosis patients are thought to represent primary disease (14). Moreover, contrasting the results of primary and reactivational disease studies will provide a better understanding of the mechanisms that govern progression from infection to disease in two distinct stages of tuberculosis.
The natural resistance-associated macrophage protein gene 1 (NRAMP1, alias SLC11A1) is the human homologue of the mouse Nramp1 gene that has been shown to be a critical element in the regulation of intracellular membrane vesicle trafficking of macrophages, a principal cell type expressing Nramp1 (15). In the mouse, it has been shown that absence of mature Nramp1 protein is the result of a G169D polymorphism that causes increased susceptibility to several intracellular macrophage pathogens, including Mycobacterium bovis (bacillus Calmette-Guérin), Salmonella typhimurium, and Leishmania donovani (16). In phagocytosing macrophages Nramp1 is rapidly recruited to the membrane of late endosomal-phagosomal vesicles (17, 18). At the phagosome membrane, Nramp1 functions as a divalent cation pump (19, 20), and Nramp1-altered cation fluxes are thought to abrogate pathogen-induced blockage of phagosome maturation (21-23). The mechanism of NRAMP1 action in human macrophages is not known but is thought to follow similar mechanisms.
Polymorphisms in the NRAMP1 gene have been found in a number of genetic studies to be risk factors for the development of tuberculosis among adult populations (24). However, except for the study of a tuberculosis outbreak in a Canadian community (25), no distinction was made between primary and reactivational tuberculosis for the patients enrolled in these previous studies. Such a study design might miss or underestimate genetic control mechanisms that differ in the development of primary and reactivational tuberculosis. Hence, we focused our genetic analysis on pediatric cases where patients present with primary tuberculosis disease. We here report strong association between NRAMP1 alleles and pediatric tuberculosis disease specifically among individuals that are likely to lack previous exposure to M. tuberculosis. We also note an inverse association of NRAMP1 polymorphisms among adult and pediatric tuberculosis disease patients. These results shed light on the role of NRAMP1 in susceptibility to tuberculosis disease and provide a plausible explanation for NRAMP1 genetic heterogeneity in tuberculosis susceptibility.
Materials and Methods
Families. The diagnostic criteria for pediatric cases were culture confirmation of tuberculosis (78 patients) or clear clinical criteria of disease (26, 27). All parental cases were culture-positive. Information regarding bacillus Calmette-Guérin vaccination and previous tuberculosis disease was obtained by interview or by visual inspection of skin scars. Ethnicity was self-reported. Mantoux status of family members was determined as part of routine patient care and contact tracing. Blood (2-10 ml) was obtained by venipuncture and used for extraction of genomic DNA with the Nucleon extraction kit (Amersham Pharmacia). Written informed consent was obtained from all study participants. The study was approved by the Institutional Review Board at Baylor College of Medicine and the Ethics Committee at the Research Institute of the McGill University Health Centre.
Genotyping. The intragenic NRAMP1 polymorphisms 274C/T, 469 + 14G/C, D543N, and 1729 + 55del4 were determined as described (28). The 3′ UTR (N10) insertion/deletion polymorphism was amplified with the 32P-labeled forward primer reported by Buu et al. (29) by using 5′-TCAAGCTCCAGTTTGGAGCCT-3′ as reverse primer and resolved as length variants on 6% polyacrylamide gels. The same conditions were used to genotype the promoter (GT)n (N01) polymorphism except primers 5′-GACATGAAGACTCGCATTAG-3′ and 5′-TACCCCATGACCACACCC-3′ were used as described by Marquet et al. (30).
Markers D543N and 1729 + 55del4 (N09) were also genotyped with Taqman assays. The primers and probes used in the assays were designed by using the software primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The following primers were synthesized: (c.D543N), 5′-CCACCACCACTTCCTGTATG-3′ and 5′-CACGTCATACATGCCACTCC-3′; (c.1729+del4), 5′-GGGAGTGGCATGTATGACG-3′ and 5′-TCTATCCTGCTGCCTGCAC-3′. The following probes were synthesized and labeled with fluorescent dyes: (D543N), 5′-FAM-CCCTTTCTGGTCCTCTTCAAGGA-TAMRA; 5′-TET-CCCTTTCTGGTTCTCTTCAAGGAGC-TAMRA; (c.1729+del4), 5′-FAM-TGGCCTGCTGGATGTGGAGTAMRA, 5′-TET-TGACTGGCCTGCTGGAGAGG-TAMRA. PCR reactions were performed in a volume of 45 μl containing 5 μl of 10 × PCR buffer (Invitrogen); 5 μl of MgCl2 (50 mM) (Invitrogen); 1 μl of dNTPs (10 mM) (Invitrogen); 0.75 μl of forward primer (20 mM), 0.75 μl of reverse primer (20 mM), and 0.60 μl of FAM probe (2 M) (Research Genetics, Huntsville, AL); 0.60 μl of TET probe (2 M) (Research Genetics); 0.1 μl of Platinum Taq polymerase (Invitrogen); and 5 μl of DNA (10 ng/μl). Three nontemplate controls were included on each plate. All PCRs were carried out in transparent 96-well plates with caps (Applied Biosystems). DNAs were amplified in MJ PT-100 machines (MJ Research, Cambridge, MA) under the following conditions: (i) 96°C for 10 min, (ii) 96°C for 25 sec, (iii) 60°C for 1 min, (iv) repeat steps ii-iii 39 times, (v) 72°C for 5 min, and (vi) 10°C ambient time. PCR products were analyzed with an Applied Biosystems 7700 Sequence Detector spectrophotometer equipped with sequence detector, Ver. 1.7 software. Fluorescence readings were exported to a spreadsheet and graphed as a scatter plot.
Markers rs2292555, rs1017698, and rs9076 were genotyped by PCR-RFLP under identical conditions. The PCR reaction mixture included 100 ng of genomic DNA, 1× Buffer (Invitrogen), 2.5 mM MgCl2, 0.09 mM dNTPs, 0.2 μM of each primer (rs2292555: 5′-AGCCAGGGTAGGCAGGATAC-3′; GGCATTCACGATTGCTTTTC-3′; rs1017698: 5′-CCACCATAGCCAAACCATTC-3′, 5′-GGGATGTGATACCCTTCCAG; rs9076: 5′-GTTTTATCCGCAGCCCTTTT-3′, 5′-CCAGTCGGAAGAAACAGCAT-3′), and 1 unit of Taq polymerase. Cycling conditions included an initial denaturation at 95°C for 3 min, followed by 25 cycles of 95°C for 50 sec, 50°C for 50 sec, then 72°C for 50 sec, and a final extension at 72°C for 10 min. For marker rs2292555, a total of 5 μl of PCR product was added to 5 μl of digestion mix, which contained 1 × Buffer 4 (NEB, Beverly, MA) and 0.3 units of DdeI, followed by incubation at 37°C for 4 h. Conditions for DNA restriction were identical for markers rs1017698 and rs9076, except that 0.4 units of BtsI were used for marker rs1017698 and 0.1 units of BsgI and 1× were used for marker rs9076. All banding patterns were resolved on a 2% agarose gel stained with ethidium bromide. Markers rs2104615, rs4324314, and rs4674297 were genotyped on the Orchid Biosciences (Princeton, NJ) UHT genotyping platform (31), as described in detail (32).
Statistical Analysis. The association study was mainly performed by the family-based method implemented in the Family Based Association Test (fbat program (33). The fbat statistic combines the three different methods described in the text [Transmission Disequilibrium Test (tdt), Reconstruction-Combined TDT (rc-tdt), and sib-tdt] and allows the use of an empirical variance-covariance estimator for the statistic that is consistent when sib marker genotypes are correlated (e.g., when the analysis include multiplex families) (34). In addition, empirical P values (Pempirical) can be computed by permutation. Exact P values also were computed by using the rc-tdt software (35). Finally, alleles with evidence for association also were analyzed by conditional logistic regression as described (36), assuming a multiplicative effect of alleles on the disease relative risk. This analysis allowed us to provide odds ratio (OR) estimates and to test for differences in the regression coefficients associated with selected polymorphisms according to five binary criteria described in the text.
To test for heterogeneity of the sample according to a binary criterion (e.g., simplex/multiplex), the analysis was performed on the whole sample (184 families) and separately on the two subsamples (143 simplex and 41 multiplex families). Under the hypothesis of homogeneity, twice the difference between the likelihood of the whole sample and the summed likelihoods of the two subsamples is distributed as a χ2 with one degree of freedom.
The Hardy-Weinberg Equilibrium (HWE) was tested at each SNP for the subset of all parents across ethnicities and for the groups of black and Hispanic parents independently. No significant deviations from HWE were observed. The strength of linkage disequilibrium (LD) between pairs of SNPs was measured as D′ (37) by using haploview (www.broad.mit.edu/mpg/haploview). LD blocks were inferred from the definition proposed by Gabriel et al. (38), as implemented in haploview with D′ confidence bounds of 0.7-0.92.
Results
Description of Patients and Their Families. All families enrolled in the study were from greater Houston. The Houston metropolitan area historically has had a high rate of pediatric tuberculosis cases and is ethnically very diverse (26). To avoid possible confounding of gene-phenotype associations due to inappropriately chosen controls or population substructures, we conducted a family-based association study. This design is particularly robust in an ethnically and racially mixed community like that of the greater Houston area. We enrolled 184 nuclear families with at least one child with pediatric tuberculosis (Table 1). The majority of families (n = 143) were composed of only a single tuberculosis case (simplex families), whereas more than one case was diagnosed in 41 families (multiplex families). In 73 of the 184 families in our sample, one parent was not available for analysis. With regard to ethnicity, we enrolled 136 Hispanic, 69 black, 13 Asian, 7 white, and 9 tuberculosis patients of mixed ethnic origin. Of these 234 tuberculosis cases, 28 were adult cases, and 206 were children. The disease manifestation was classified as pulmonary in 51.3%, extrapulmonary in 37.5%, and mixed pulmonary and extrapulmonary in 11.2% of all pediatric cases. There were no statistically significant differences in the proportion of simplex vs. multiplex families and in pulmonary vs. extrapulmonary involvement across ethnic groups (data not shown).
Table 1. Characteristics of 184 nuclear tuberculosis families comprising 737 individuals enrolled for the present study.
| Tuberculosis-affected patients
|
||||||
|---|---|---|---|---|---|---|
| Subjects | Total | All patients | Children | Mean age (years) at diagnosis (±SD) | Parents | Mean age (years) at diagnosis (±SD) |
| Females | 353 | 118 | 100 | 5.3 ± 5.7 | 18 | 25.4 ± 6.8 |
| Males | 384 | 116 | 106 | 6.2 ± 5.4 | 10 | 29.2 ± 6.3 |
| Combined | 737 | 234 | 206 | 5.7 ± 5.6 | 28 | 28.8 ± 6.6 |
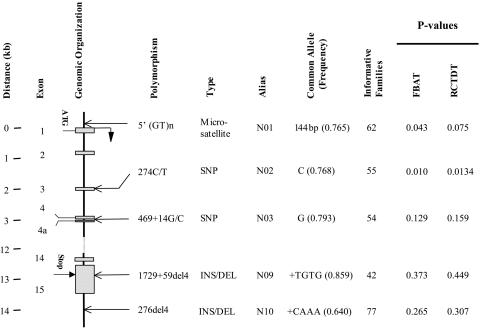
NRAMP1 Is Associated with Pediatric Tuberculosis Disease. Over all families, the common C allele of the NRAMP1 N02 polymorphism was significantly associated with increased risk of pediatric tuberculosis disease (P = 0.01; Fig. 1). Under a multiplicative genetic model, the OR of tuberculosis for C/C homozygotes vs. C/T heterozygotes was equal to the OR of C/T heterozygotes vs. T/T homozygotes and corresponded to 1.75 (95% confidence interval, 1.10-2.77). In addition, there was weaker evidence (0.043 < P < 0.075) in favor of a positive association between the NRAMP1 N01 promoter polymorphism and tuberculosis (Fig. 1). There was no significant association between the NRAMP1 polymorphisms located in the 3′ region of the gene and pediatric tuberculosis disease.
Fig. 1.
Schematic presentation of the NRAMP1 candidate gene and intragenic location of gene polymorphisms. The genomic distance spanned by NRAMP1 in kilobase pairs (kb), the exon numbers, the translational initiation (ATG), and termination sequences (Stop) and the location of distinct gene polymorphisms with respect to the exon-intron organization are given on the left side of the diagram. Designation of gene polymorphisms either adopted names already established in the literature or followed standard nomenclature rules (51). The type of polymorphism-microsatellite repeat, SNP, insertion/deletion polymorphism (INS/DEL), together with a simple polymorphism alias as well as the identity and frequency of the common allele, are also indicated. Finally, the number of families comprising at least one parent heterozygous for the polymorphisms, i.e., a parent for which preferential allele transmission can be monitored, is given. P values indicating evidence for distortion of allele transmissions are given for the Family Based Association Test (FBAT) (33) and the Reconstruction-Combined TDT (RC-TDT) (35) analytical procedures.
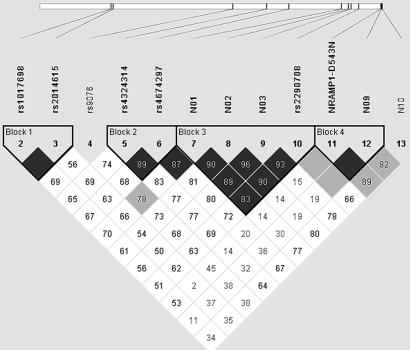
LD Among Markers in the NRAMP1 Genome Region. To better define the observed association of NRAMP1 alleles with pediatric tuberculosis disease, we computed LD, measured as D′, among the five tested markers in the group of Hispanic parents, the largest ethnic group among the enrolled families. We found that the three 5′ markers (N01-N03) were in strong LD among them but observed only weak to moderate LD with markers N09 and N10 in the 3′ NRAMP1 region. To better delimit the LD pattern of the tuberculosis-associated markers N01-N03, we genotyped seven additional markers flanking the 5′ NRAMP1 region. Five of these SNPs were used to tag the MGC5081 ORF (rs4674297 and rs4324314) and the MR-1 gene (rs9076, rs2014615, and rs1017698), the two closest neighbors located 15 and 65 kb upstream of NRAMP1, respectively. The two remaining SNPs were located in intron 6 (rs2290708) and exon 15 (NRAMP1 D543N) of the NRAMP1 gene. Over the entire interval of ≈80 kb, we were able to identify four haplotype blocks (Fig. 2). Although clearly not part of the same haplotype blocks, there was substantial LD between pairs of SNPs among 5′ NRAMP1 markers, the two MGC5081 tag SNPs and, to a lesser degree, the MR-1 located SNPs (Fig. 2). None of the additional markers showed significant evidence for association with pediatric tuberculosis disease and all pediatric tuberculosis-associated SNPs localized to the 5-kb haplotype block 3 (Fig. 2). The variable strength of association with tuberculosis among those markers is likely explained by the fact that LD is not complete between markers of block 3 and by the differences in allele frequencies (especially between N02 and N03). A similar pattern of pairwise D′ values was also observed for the parents of the black families. However, due to the reduced number of informative chromosomes, confidence intervals were too large to allow for the definition of haplotype block structures (data not shown).
Fig. 2.
LD pattern in the Hispanic population between pairs of SNPs spanning the NRAMP1 gene and its upstream genomic region. The NRAMP1 3′ region is located on the right end of the schematic chromosome line indicated on top of the graph. Consequently, the NRAMP1 gene orientation is in the 3′ to 5′ orientation from right to left. The telomere of chromosome 2q is located toward the right. Names of polymorphisms used for the LD matrix of pairs of markers are given, and their chromosomal locations are indicated by solid lines. Haplotype blocks according to Gabriel et al. (38) are indicated, and names of markers that are part of haplotype blocks are indicated in bold. Each square represents the magnitude of pairwise LD. Each pairwise D′ measure is shown as D′ × 102 within the corresponding square. Squares without D′ written on them represent D′of 1.0. Black squares indicate pairwise LD that is strong [lower confidence interval (CI), 0.7, upper CI >0.92], light-gray squares represent intermediate strength LD, and white squares represent weak LD.
NRAMP1 Alleles and Association with Pediatric Tuberculosis Disease. To test whether the association observed between the NRAMP1 N02 polymorphism and tuberculosis was influenced by family or case characteristics, we performed heterogeneity tests in the conditional logistic regression analysis framework (36). Specifically, we tested for differences in the regression coefficient associated with each of the two polymorphisms according to five binary criteria: family structure (simplex/multiplex), ethnicity of family (Hispanic/other), sex of affected child (male/female), anatomic site of tuberculosis (pulmonary/extrapulmonary), and age of onset (≤5years/>5 years). Because of small numbers, Asian and white families could not be tested independently for NRAMP1 N02 association heterogeneity. Only the sex of the pediatric patient and the family structure were found to have significant effects. The association of the NRAMP1 N02 polymorphism and tuberculosis disease was stronger in males [OR for C/C vs. C/T = 2.82 (1.44-5.61)]. The difference in transmission between male and female patients also was significant (P < 0.04).
Next, the association of NRAMP1 with tuberculosis disease was analyzed separately in simplex and multiplex families. Independent of the mode of analysis, there was a highly significant distortion (P < 0.0008) of the NRAMP1 N02 polymorphism transmission in simplex families [OR for C/C vs. C/T and C/T vs. T/T = 3.13 (1.54-6.25)] that was not detected in multiplex families (Table 2). Formal testing of variable strength of association between N02 and pediatric tuberculosis in simplex vs. multiplex families clearly revealed a significant heterogeneity (P < 0.01). This result argues that difference in NRAMP1 N02 transmission to tuberculosis-affected children in simplex and multiplex families represents a true effect and is not simply a reflection of the different numbers of informative simplex and multiplex families leading to loss of significance in the less numerous multiplex families. When focusing only on the 17 informative simplex families with male pediatric patients, the effect of N02 on tuberculosis risk was very highly significant (P < 0.00004) with an estimated OR for C/C vs. C/T of 20.0 (2.69-148). Those 17 families of diverse ethnicity include 20 heterozygous C/T parents who transmitted the C allele to their affected child 19 times.
Table 2. Association between susceptibility to pediatric tuberculosis disease and NRAMP1 alleles stratified by family structure.
| Simplex families
|
Multiplex families
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
P value
|
P value
|
||||||||
| Gene | Polymorphism | Informative families | fbat | rc-tdt | Conditional logistic regression | Informative families | fbat* | rc-tdt† | Conditional‡ logistic regression |
| NRAMP1 | N01 | 39 | 0.020 | 0.048 | ND | 22 | 0.594 | 0.697 | ND |
| N02 | 36 | 0.00045 | 0.00059 | 0.00080 | 19 | 1.000 | 1.000 | ND | |
| N03 | 37 | 0.014 | 0.021 | ND | 17 | 0.7546 | 0.876 | ND | |
Family Exposure to M. tuberculosis and Strength of Association of NRAMP1 with Pediatric Tuberculosis Disease. To follow up on the restriction of NRAMP1 tuberculosis association, we investigated the heterogeneity of family structure in NRAMP1-mediated risk on pediatric tuberculosis disease (Table 2). The majority (26/41) of multiplex families in our family collection included adult infectious cases. Children living in close proximity to adult cases are expected to have increased exposure to M. tuberculosis. Hence, we used purified protein derivative (PPD) skin-test conversion among all unaffected cosibs as a measure of exposure intensity in individual families and found a substantially higher proportion of cosibs tested PPD+ in multiplex families with at least one affected parent (62.5%) as compared with simplex families (36.5%). Because pediatric cases generally have a low infectious potential, there was no significant difference in the proportion of PPD+ cosibs among multiplex families without adult cases (31%) and simplex families.
To test the resulting hypothesis that NRAMP1 effects on tuberculosis disease risk are most readily detectable under conditions of low M. tuberculosis transmission, we selected all simplex families comprising at least one child in addition to the affected sib. Of the available 71 families, 32 included at least one additional PPD+ cosib (”high-exposure families”), whereas among 39 families, no PPD+ cosib was identified (”low-exposure families”). Family size was not a confounding factor for classification into high- and low-exposure families (P > 0.9; Table 3). Among the entire subsample of 71 simplex families with at least one additional cosib, there was strong evidence for an association of NRAMP1 N02 alleles with pediatric tuberculosis disease (Pempirical = 0.006). When separated into high- and low-exposure families, there was less evidence of significant distortion of NRAMP1 N02 allele transmission among high-exposure families (Pempirical = 0.21) as compared with low-exposure families (Pempirical = 0.011). Because <10% of cases had been vaccinated with bacillus Calmette-Guérin, these findings strongly suggest that NRAMP1 alleles have their highest impact on risk of tuberculosis disease under conditions of low transmission/exposure of M. tuberculosis.
Table 3. Family sibship size and proportion of families with at least one unaffected cosib (UCS).
| Number of families | Number of UCS | Number of families with UCS-PPD+ ≥1 | Number of families with only UCS-PPD− |
|---|---|---|---|
| 38 | 1 | 14 | 24 |
| 24 | 2 | 12 | 12 |
| 6 | 3 | 4 | 2 |
| 3 | 4 | 2 | 1 |
UCS-PPD+, unaffected cosibs that are PPD-positive. UCS-PPD−, unaffected cosibs that are PPD-negative.
Discussion
The human NRAMP1 gene has been implicated in increased risk of tuberculosis disease by a number of studies. For example, polymorphisms in the 5′ and 3′ regions of NRAMP1 have been linked or associated with tuberculosis disease susceptibility in Guinea Conakry (39), Japan (40), Korea (41), The Gambia (42, 43), Canada (25), Texas (44), Cambodia (45), Denmark (46), and South Africa (24), but not in Taiwan (47) or Morocco (48). The focus of most studies was on susceptibility to smear-positive tuberculosis disease among adult populations. The design of our study was different from previous investigations, because we analyzed the effect of NRAMP1 alleles on risk of primary tuberculosis disease in a cohort of pediatric tuberculosis disease families. We observed that the NRAMP1 gene, previously implicated in the genetic control of adult tuberculosis, also influences the risk of pediatric tuberculosis disease. Unexpectedly, we discovered the direction of NRAMP1 allele association with pediatric tuberculosis disease to be inverted compared with previous studies in adult pulmonary tuberculosis. Although among adult patients the common 5′ NRAMP1 alleles had been found associated with protection, i.e., depleted among cases, we found that the common alleles are risk factors, i.e., enriched among pediatric tuberculosis disease patients. Consequently, the significant enrichment of the common N02 C allele and, to a lesser degree, the N01 and N03 common alleles in early onset cases and the corresponding depletion of the same allele in late onset patients strongly suggests that the N02 C allele promotes rapid progression from infection to disease.
Importantly, the suggestion that common NRAMP1 alleles are risk factors for early-onset tuberculosis agrees with the results of a previous genetic analysis of tuberculosis in a large Canadian Aboriginal family that experienced a tuberculosis outbreak (25). In this outbreak, individuals had limited prior exposure to mycobacteria, and all tuberculosis cases were diagnosed within a maximum time of 2 years from the index case. The majority of cases occurred within 6-9 months after diagnosis of the index case (49). Consequently, all patients with clinical tuberculosis could be classified as fast progressors or primary tuberculosis disease cases. Segregation analysis revealed that the common NRAMP1 alleles were preferentially transmitted to tuberculosis patients (25). Considering that only ≈10% of individuals infected with M. tuberculosis advance to clinical forms of tuberculosis, the involvement of genes controlling the rate of progression rather than bona fide susceptibility to tuberculosis may offer an effective genetic control of disease risk. Whether the N02 polymorphism is itself the cause of more rapid progression to tuberculosis disease, or whether N02 is in LD with the causative variant is presently not known. However, recent results from our laboratory directly correlated reduced NRAMP1 functional activity with the high-risk C allele of N02 (unpublished work).
Among pediatric tuberculosis disease cases, NRAMP1 alleles have different strength of association among different patient subgroups. The observation that NRAMP1 association with tuberculosis disease is more readily detected among male than female pediatric cases is interesting in light of the known gender-specific differences in frequencies among adult tuberculosis cases. However, because evidence for such a gender-specific effect of NRAMP1 alleles on pediatric disease was significant but weak in our sample, additional studies are required to confirm this observation. By contrast, our data suggest that gene-environment interactions are critical for the appropriate selection of efficient host responses and hence genetic control mechanisms. This is illustrated by our observation that genetic control of tuberculosis disease by NRAMP1 was most easily detected in families that experienced low exposure intensities to M. tuberculosis. Under conditions of increased exposure, the NRAMP1 effect became less strong, suggesting that mechanisms independent of the NRAMP1 gene (e.g., pathogen factors or different sets of genes) are more prominent in this instance.
In the study families, the NRAMP1 gene clearly is a strong risk factor for pediatric tuberculosis disease under conditions of low exposure to M. tuberculosis. Specifically, among simplex families, the population-attributable risk of the NRAMP1 N02 C risk allele is estimated to be 85% (49-96%). Assuming the N02 polymorphism is the only cause of increased tuberculosis in this area, this means that the incidence of pediatric tuberculosis would be 85% lower if the M. tuberculosis-infected Houston population were monomorphic for the protective N02 T allele. Although the actual impact of the N02 polymorphism depends on other risk factors not yet identified, the combination of a relatively strong genetic effect (OR = 3.13) of a risk allele present at high frequency (≈0.76), as described for type II diabetes (50), is expected to make a substantial contribution to the occurrence of pediatric tuberculosis in the Houston area.
The present study is one of an increasing number of positive-association reports between risk of tuberculosis disease and NRAMP1 alleles in populations of vastly different ethnic background. Results obtained in the present investigation provide an important complement for a better understanding of the setting in which NRAMP1 exerts its influence on tuberculosis susceptibility. Specifically, our results suggest that NRAMP1 effects are most pronounced in the absence of prior exposure to mycobacteria, and that NRAMP1 is a modulator of the speed of progression from infection with M. tuberculosis to tuberculosis disease. The detailed molecular events that prevent rapid progression to clinically defined disease are presently unknown. However, a protein that limits multiplication of ingested M. tuberculosis bacilli by antagonizing pathogen-triggered blockage of phagosome maturation offers a reasonable mechanism for containing the spread but not the initial infection by the bacterium.
Acknowledgments
We thank all families that agreed to participate in the study. We also thank our colleagues, J. L. Casanova, K. Morgan, E. Skamene, and P. Gros, for critical comments on earlier versions of the manuscript. This work was supported by National Institutes of Health Grant AI41168-01 (to J.M.M.). L.A. was supported in part by Programme de Recherches Fondamentales en Microbiologie, Maladies Infectieuses et Parasitologie (PRFMMIP) from the Ministère Français de l'Education Nationale de la Recherche et de la Technologie. E.S. is an Investigator of the Canadian Institutes of Health Research.
Author contributions: J.R.S., E.A.G., J.M.M., and E.S. designed research; S.M., H.T., A.P., M.G., G.J.A., and K.C.S. performed research; S.M., L.A., E.A.G., and E.S. analyzed data; L.S., J.R.S., and K.C.S. contributed new reagents/analytic tools; and S.M., L.A., J.M.M., and E.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NRAMP1, natural resistance associated macrophage protein 1; TDT, transmission disequilibrium test; LD, linkage disequilibrium; OR, odds ratio; PPD, purified protein derivative.
References
- 1.World Health Organization Global Tuberculosis Control (2001) WHO Report (World Health Organization, Geneva), WHO/CDS/TB/2001.28.
- 2.Lavebratt, C., Apt, A. S., Nikonenko, B. V., Schalling, M. & Schurr, E. (1999) J. Infect. Dis. 180, 150-155. [DOI] [PubMed] [Google Scholar]
- 3.Kramnik, I., Dietrich, W. F., Demant, P. & Bloom, B. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8560-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitsos, L. M., Cardon, L. R., Fortin, A., Ryan, L., LaCourse, R., North, R. J. & Gros, P. (2000) Genes Immun. 1, 467-477. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez, F., Radaeva, T. V., Nikonenko, B. V., Persson, A. S., Sengul, S., Schalling, M., Schurr, E., Apt, A. S. & Lavebratt, C. (2003) Infect. Immun. 71, 126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan, H., Yan, B. S., Rojas, M., Shebzukhov, Y. V., Zhou, H., Kobzik, L., Higgins, D. E., Daly, M. J., Bloom, B. R. & Kramnik, I. (2005) Nature 434, 767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stead, W. W., Senner, J. W., Reddick, W. T. & Lofgren, J. P. (1990) N. Engl. J. Med. 322, 422-427. [DOI] [PubMed] [Google Scholar]
- 8.Kallmann, F. J. & Reisner, D. (1943) Am. Rev. Tuberc. 47, 549-574. [Google Scholar]
- 9.Comstock, G. W. (1978) Am. Rev. Respir. Dis. 117, 621-624. [DOI] [PubMed] [Google Scholar]
- 10.Marquet, S. & Schurr, E. (2001) Drug Metab. Dispos 29, 479-483. [PubMed] [Google Scholar]
- 11.Casanova, J. L. & Abel, L. (2002) Annu. Rev. Immunol. 20, 581-620. [DOI] [PubMed] [Google Scholar]
- 12.Ozbek, N., Fieschi, C., Yilmaz, B. T., de Beaucoudrey, L., Demirhan, B., Feinberg, J., Bikmaz, Y. E. & Casanova, J. L. (2005) Clin. Infect. Dis. 40, 55-58. [DOI] [PubMed] [Google Scholar]
- 13.Hoal-Van Helden, E. G., Epstein, J., Victor, T. C., Hon, D., Lewis, L. A., Beyers, N., Zurakowski, D., Ezekowitz, A. B. & Van Helden, P. D. (1999) Pediatr. Res. 45, 459-464. [DOI] [PubMed] [Google Scholar]
- 14.Comstock, G. W., Livesay, V. T. & Woolpert, S. F. (1974) Am. J. Epidemiol. 99, 131-138. [DOI] [PubMed] [Google Scholar]
- 15.Gros, P. & Schurr, E. (2004) in Susceptibility to Infectious Diseases, ed. Bellamy, R. (Cambridge Univ. Press, Cambridge, U.K.), pp. 221-258.
- 16.Vidal, S., Tremblay, M. L., Govoni, G., Gauthier, S., Sebastiani, G., Malo, D., Skamene, E., Olivier, M., Jothy, S. & Gros, P. (1995) J. Exp. Med. 182, 655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruenheid, S., Pinner, E., Desjardins, M. & Gros, P. (1997) J. Exp. Med. 185, 717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Searle, S., Bright, N. A., Roach, T. I., Atkinson, P. G., Barton, C. H., Meloen, R. H. & Blackwell, J. M. (1998) J. Cell Sci. 111, 2855-2866. [DOI] [PubMed] [Google Scholar]
- 19.Jabado, N., Jankowski, A., Dougaparsad, S., Picard, V., Grinstein, S. & Gros, P. (2000) J. Exp. Med. 192, 1237-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goswami, T., Bhattacharjee, A., Babal, P., Searle, S., Moore, E., Li, M. & Blackwell, J. M. (2001) Biochem. J. 354, 511-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackam, D. J., Rotstein, O. D., Zhang, W., Gruenheid, S., Gros, P. & Grinstein, S. (1998) J. Exp. Med. 188, 351-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuellar-Mata, P., Jabado, N., Liu, J., Furuya, W., Finlay, B. B., Gros, P. & Grinstein, S. (2002) J. Biol. Chem. 277, 2258-2265. [DOI] [PubMed] [Google Scholar]
- 23.Frehel, C., Canonne-Hergaux, F., Gros, P. & De Chastellier, C. (2002) Cell. Microbiol. 4, 541-556. [DOI] [PubMed] [Google Scholar]
- 24.Hoal, E. G., Lewis, L. A., Jamieson, S. E., Tanzer, F., Rossouw, M., Victor, T., Hillerman, R., Beyers, N., Blackwell, J. M., Van Helden, P. D. (2004) Int. J. Tuberc. Lung Dis. 8, 1464-1471. [PubMed] [Google Scholar]
- 25.Greenwood, C. M., Fujiwara, T. M., Boothroyd, L. J., Miller, M. A., Frappier, D., Fanning, E. A., Schurr, E. & Morgan, K. (2000) Am. J. Hum. Genet. 67, 405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starke, J. R. & Taylor-Watts, K. T. (1989) Pediatrics 84, 28-35. [PubMed] [Google Scholar]
- 27.Starke, J. R. (2000) Pediatr. Infect. Dis. J. 19, 1095-1096. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., Fujiwara, T. M., Buu, N. T., Sanchez, F. O., Cellier, M., Paradis, A. J., Frappier, D., Skamene, E., Gros, P., Morgan, K., et al. (1995) Am. J. Hum. Genet. 56, 845-853. [PMC free article] [PubMed] [Google Scholar]
- 29.Buu, N. T., Cellier, M., Gros, P. & Schurr, E. (1995) Immunogenetics 42, 428-429. [DOI] [PubMed] [Google Scholar]
- 30.Marquet, S., Sanchez, F. O., Arias, M., Rodriguez, J., Paris, S. C., Skamene, E., Schurr, E. & Garcia, L. F. (1999) J. Infect. Dis. 180, 1521-1525. [DOI] [PubMed] [Google Scholar]
- 31.Bell, P. A., Chaturvedi, S., Gelfand, C. A., Huang, C. Y., Kochersperger, M., Kopla, R., Modica, F., Pohl, M., Varde, S., Zhao, R., et al. (2002) Biotechniques Suppl., 70-77. [PubMed]
- 32.Mira, M. T., Alcais, A., Van Thuc, N., Moraes, M. O., Di Flumeri, C., Hong Thai, V., Chi Phuong, M., Thu Huong, N., Ngoc Ba, N., Sarno E. N. et al. (2004) Nature 33, 412-415. [Google Scholar]
- 33.Horvath, S., Xu, X. & Laird, N. M. (2001) Eur. J. Hum. Genet. 9, 301-306. [DOI] [PubMed] [Google Scholar]
- 34.Lake, S. L., Blacker, D. & Laird, N. M. (2000) Am. J. Hum. Genet. 67, 1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knapp, M. (1999) Am. J. Hum. Genet. 64, 861-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaid, D. J. & Rowland, C. (1998) Am. J. Hum. Genet. 63, 1492-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewontin, R. C. (1988) Genetics 120, 849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. (2002) Science 296, 2225-2229. [DOI] [PubMed] [Google Scholar]
- 39.Cervino, A. C., Lakiss, S., Sow, O. & Hill, A. V. (2000) Ann. Hum. Genet. 64, 507-512. [DOI] [PubMed] [Google Scholar]
- 40.Gao, P. S., Fujishima, S., Mao, X. Q., Remus, N., Kanda, M., Enomoto, T., Dake, Y., Bottini, N., Tabuchi, M., Hasegawa, N., et al. (2000) Clin. Genet. 58, 74-76. [DOI] [PubMed] [Google Scholar]
- 41.Ryu, S., Park, Y. K., Bai, G. H., Kim, S. J., Park, S. N. & Kang, S. (2000) Int. J. Tuberc. Lung Dis. 4, 577-580. [PubMed] [Google Scholar]
- 42.Bellamy, R., Ruwende, C., Corrah, T., McAdam, K. P. W. J., Whittle, H. C. & Hill, A. V. S. (1998) N. Engl. J. Med. 338, 640-644. [DOI] [PubMed] [Google Scholar]
- 43.Awomoyi, A. A., Marchant, A., Howson, J. M., McAdam, K. P., Blackwell, J. M. & Newport, M. J. (2002) J. Infect. Dis. 186, 1808-1814. [DOI] [PubMed] [Google Scholar]
- 44.Ma, X., Dou, S., Wright, J. A., Reich, R. A., Teeter, L. D. El Sahly, H. M., Awe R. J., Musser, J. M. & Graviss, E. A. (2002) Int. J. Tuberc. Lung Dis. 6, 818-823. [PubMed] [Google Scholar]
- 45.Delgado, J. C., Baena, A., Thim, S. & Goldfeld, A. E. (2002) J. Infect. Dis. 186, 1463-1468. [DOI] [PubMed] [Google Scholar]
- 46.Soborg, C., Andersen, A. B., Madsen, H. O., Kok-Jensen, A., Skinhoj, P. & Garred, P. (2002) J. Infect. Dis. 186, 517-521. [DOI] [PubMed] [Google Scholar]
- 47.Liaw, Y. S., Tsai-Wu, J. J., Wu, C. H., Hung, C. C., Lee, C. N., Yang, P. C., Luh, K. T. & Kuo, S. H. (2002) Int. J. Tuberc. Lung Dis. 6, 454-460. [PubMed] [Google Scholar]
- 48.El Baghdadi, J., Remus, N., Benslimane, A., El Annaz, H., Chentoufi, M., Abel, L. & Schurr, E. (2003) Int. J. Tuberc. Lung Dis. 7, 599-602. [PubMed] [Google Scholar]
- 49.Mah, A. W. & Fanning, E. A. (1991) Can. J. Infect. Dis. 2, 133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altshuler, D., Hirschhorn, J. N., Klannemark, M., Lindgren, C. M., Vohl, M. C., Nemesh, J., Lane, C. R., Schaffner, S. F., Bolk, S., Brewer, C., et al. (2000) Nat. Genet. 26, 76-80. [DOI] [PubMed] [Google Scholar]
- 51.Antonarakis, S. E. (1998) Hum. Mutat. 11, 1-3. [DOI] [PubMed] [Google Scholar]