Abstract
Bacterial evolution toward endosymbiosis with eukaryotic cells is associated with extensive bacterial genome reduction and loss of metabolic and regulatory capabilities. Here we examined the rate and process of genome reduction in the bacterium Salmonella enterica by a serial passage experimental evolution procedure. The initial rate of DNA loss was estimated to be 0.05 bp per chromosome per generation for a WT bacterium and ≈50-fold higher for a mutS mutant defective in methyl-directed DNA mismatch repair. The endpoints were identified for seven chromosomal deletions isolated during serial passage and in two separate genetic selections. Deletions ranged in size from 1 to 202 kb, and most of them were not associated with DNA repeats, indicating that they were formed via RecA-independent recombination events. These results suggest that extensive genome reduction can occur on a short evolutionary time scale and that RecA-dependent homologous recombination only plays a limited role in this process of jettisoning superfluous DNA.
Keywords: bacterial evolution, genome reduction, serial passage
Most bacterial genomes are continuously changing over time with respect to gene content and size, and several processes including deletion, duplication, and lateral gene transfer contribute to this genome evolution (1-4). Bacterial endosymbionts generally have small genomes, and they were probably derived by a reductive process from ancestral free-living bacteria with larger genomes (5, 6). The reduction of genome size is facilitated by the absence of selection for many bacterial functions (e.g., growth factor biosynthesis) due to the availability and utilization of metabolites provided within the host cell. The combination of the presence of population bottlenecks and the absence of lateral gene transfer allows nonselected and weakly selected genes to be inactivated by random mutation and subsequently lost by deletion. This evolutionary scenario has been convincingly inferred from genomic comparisons of free-living bacteria and obligate intracellular bacteria (6-8) and is thought to be responsible for the high numbers of pseudogenes that have been identified in the genome sequences of some of the more specialized bacterial pathogens (9-11). However, the speed and mechanisms responsible for reductive evolution are poorly defined. Furthermore, the genetic factors that constrain the tempo and mode of such reductive evolution have not been determined, and these could include the availability of functional recombination machinery, recombination substrates, and the number and distribution of essential genes.
Materials and Methods
Serial Passage. Bacteria were propagated clonally to avoid lateral gene transfer with a supply of biosynthetic end products such as amino acids, vitamins, bases, and cofactors (hematin agar plates: GC-agar base from Oxoid, Basingstoke, U.K., supplemented with haemoglobin, isovitox, and iron). Slow-growing mutants were allowed to fix in the population by reducing effective population size. This reduction was achieved experimentally by serially passaging the bacteria on agar plates with repeated one-cell bottlenecks, where each day random colonies that had grown from individual bacterial cells were picked from agar plates and streaked on a new plate. This procedure was repeated up to 270 times. For each serial passage, the population expanded from one cell to ≈108 cells in a colony, representing ≈25 generations of growth. To examine the effect of methyl-directed DNA mismatch repair (MMR) on the rate of gene loss, the experiment was performed in both a WT and a mutS mutant background. Several independent lineages of WT (12 lineages) and mutS (60 lineages) bacteria were serially passaged, and their genomes were analyzed by comparative genomic indexing (CGI) with DNA microarrays (12), pulsed-field gel electrophoresis, Southern hybridization, PCR, and DNA sequencing to characterize any deletions in the context of the 4,857,432 bp Salmonella typhimurium LT2 genome (13). At regular intervals, cells were saved and frozen at -80°C. LB broth (14) was used for growth rate measurements.
Identification of Deletions in Serially Passaged Lineages. All lineages were examined by pulsed-field gel electrophoresis for the presence of deletions. Because this procedure mainly detects large deletions, the number of deletions found is thus likely to represent an underestimate of the actual loss rate. DNA was prepared as described (15) and restricted with XbaI. The DNA fragments were then separated on 1% Seakem LE agarose in 0.5 × 90 mM Tris, 64.6 mM boric acid, and 2.5 mM EDTA (pH 8.3). Electrophoresis conditions were 6 V/cm, 6.8-63.8 s with a running time of 22 h at 14°C. CGI:DNA purification and labeling, microarray hybridization, washing, scanning, and data normalization were performed with S. typhimurium LT2 (13) microarrays containing 4,451 protein-coding sequences as described (12, 16). Deletions were identified as >5-fold reduction in hybridization signal. Each putative deletion identified by CGI was confirmed by PCR and Southern hybridization. In addition, we used PCR to identify the deletion endpoints. CGI allowed the last gene remaining on each side of the deletion to be identified. Primers were designed to amplify across the deletion endpoints and the PCR product was sequenced from both directions to identify the endpoints exactly.
Deletion Assays. To determine the frequency of mod-gal deletions, bacteria (S. typhimurium LT2-derived strains DA6192, WT, and DA6194, mutS) were grown in 20 independent 1.5-ml LB cultures and plated on MacConkey agar plates (Difco) supplemented with 0.2% sodium chlorate and 0.2% galactose. The plates were incubated at 37°C anaerobically for 24 h, then aerobically for 6 h to select for chlorate-resistant colonies that carry mod mutations and these were scored for white appearance (gal). For each independent culture the frequency of mod-gal double mutants was calculated as the ratio of white, chlorate-resistant colonies divided by the total number of cells plated (17). The mutation rate was then estimated by the median method (18). The endpoints of the isolated mod-gal deletions were mapped by PCR. Initial mapping was made with primers ≈5 kb apart from each other throughout the mod-gal region. Then more specific primers were used to define the last existing gene for each strain. PCR across the deletion endpoint was done with specific primers on one side of the deletion and with arbitrary primers on the other side. The arbitrary primers used for the first PCR consist of a defined region (20 bp) and an undefined region (15 bp) where bases were inserted randomly. This allowed the primers to bind randomly all over the chromosome. The arbitrary primer used for the second PCR consisted only of the defined region (20 bp) of the arbitrary primers used for the first PCR. For the second PCR, a new internal primer was also used on the specific primer side of the deletion. All primer sequences are available upon request. To isolate deletions in the pocR region, we used strain (DA9437, pocR::Tn10dTet, Kan) to select for tetracycline-sensitive mutants in a Bochner selection (19). The tetracycline-sensitive mutants were subsequently screened for kanamycin sensitivity. The endpoints for the pocR deletion were determined as for the serially passaged lineages.
Compensatory Evolution. For each of the three low-fitness strains examined, five independent lineages were serially passaged to select for mutants with increased growth rate. The bacteria were grown in 2-ml LB cultures, and each day 2 μl of culture (≈106 cells) was transferred to a new tube with 2 ml of fresh LB. At regular intervals after serial passage, the size of the colonies was compared to the size of parental colonies by plating on LB agar. When the majority of the colonies from the serially passaged cultures showed a larger size than the parental strain, the experiment was stopped (usually after six to 10 serial passages, corresponding to 60-100 generations of growth). The culture was stored by freezing at -80°C, and its growth rate was measured by using a Bioscreen C analyzer. Relative fitness was calculated relative to the parental strain (set to one).
Results
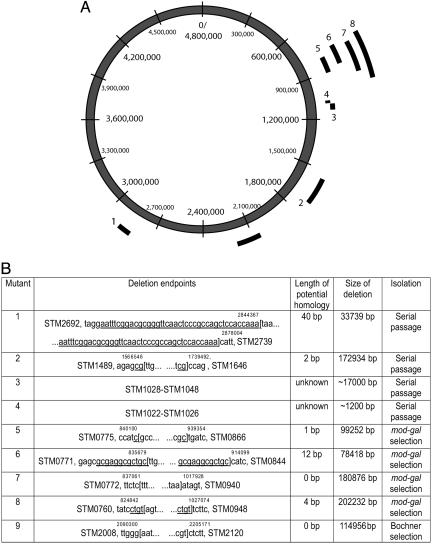
Deletions Identified and Estimation of the DNA Loss Rate in the mutS Strain. Fig. 1 A and B shows the size and location of the deletions identified in the serially passaged lineages (numbers 1-4) by CGI, PCR, and Southern hybridization. Gene loss was only observed in the mutS background and not in the WT strain. Individual chromosomal deletions varied in size from ≈1,200 to 172,934 bp and were located in three different regions of the chromosome (Fig. 1). The initial rate of loss of chromosomal DNA per generation was estimated for the mutS lineages. Sixty lineages were each serially passaged for 1,500 generations, and deletions, totaling 224,873 bp, were found in four of the lineages. From these numbers, we calculated the arithmetic mean DNA loss rate in the mutS lineages to be ≈2.5 bp per chromosome per generation [i.e., 224,873 bp (total amount of DNA deleted)/60 (number of lineages) × 60 (number of serial passages) × 25 (number of generations of growth per serial passage)].
Fig. 1.
Characteristics of identified deletions. (A) Size and location of the nine deletions identified in the chromosome of the mutS+ and mutS- derivatives of S. typhimurium LT2. (B) Endpoints of deletions. S. typimurium (STM) numbers indicate the genes at each end of the deletions. Deleted DNA sequences are flanked by brackets. Numbers indicate nucleotides at deletion endpoints (13), and underlined sequences are repeat sequences that could potentially provide homology during recombination. Deletions 1-4 were obtained in the mutS- strain after 60 cycles of serial passage (≈1,500 generations); deletions 5-8 were obtained in the mutS+ strain after selection for resistance to chlorate and screening for Gal-, and deletion 9 was obtained in the mutS+ strain after selection for loss of a Tn10dTet, Kan transposon inserted into the pocR gene. The endpoints of deletions 3 and 4 could not be determined because of complex chromosomal rearrangements associated with deletion formation.
Estimation of the DNA Loss Rate in the WT. To allow a calculation of the initial gene loss rate in WT bacteria, we compared the deletion rates in WT and mutS bacteria by using a separate assay system (17). A genetic selection was used that allowed determination of the simultaneous loss of two neighboring operons, mod and gal, separated by ≈1 kbp on the chromosome. The deletable region that lacks essential genes surrounding these two operons is at least 200 kbp and thus any deletions that include both mod and gal (>1 kbp) and are smaller than 200 kbp will be detected in this assay. We found the deletion rate to be 50-fold lower for the WT strain than the mutS mutant, 0.5 × 10-8 and 25 × 10-8, respectively. This difference in deletion rate between MMR-proficient and -deficient strains is similar to that observed in other deletion assays in Escherichia coli and S. typhimurium (20, 21) and shows that the MMR system has a strong stabilizing effect on the maintenance of intact chromosome structure (22). This probably reflects the antirecombinational activity of MMR by which the formation of heteroduplex intermediates is impeded or reversed (22). Thus, a defective MMR system is likely to allow increased recombination and deletion-formation between divergent sequences. We used the deletion rate obtained from serial passage of the mutS strain and the 50-fold difference in deletion rate observed between the WT and mutS mutant in the mod-gal assay to estimate the initial loss rate in the WT bacterium to be 0.05 bp per chromosome per generation. These calculated DNA loss rates most likely represent a maximum because each successive deletion will progressively increase the relative fraction of essential genes, and as a result the size of each permitted deletion will be reduced. The deletion rate for the WT strain calculated from the mod-gal selection was used to corroborate the DNA loss rate derived from the serial passage procedure. The arithmetic mean size of the mod-gal deletions was 140 kbp (this number is based on the mean size of four deletions obtained in this selection). Deletion rates at different chromosomal locations may vary considerably, but if we assume that the 4.86 Mbp chromosome is divided into 24 regions of 200 kbp that are deletable with a similar rate as the mod-gal region, we can calculate the chromosomal deletion rate for the WT strain to be equal to 0.017 bp per cell per generation [i.e., 0.5 × 10-8 (deletion rate in mod-gal assay) × 140 kbp (average size of deletions found in mod-gal region) × 24 (size of S. typhimurium chromosome/potentially deletable material in mod-gal region)]. This deletion rate for the WT strain is only 3-fold lower than the rate calculated for the same strain by using the serial passage data (0.05 bp per cell per generation) and provides a partly independent measure of the chromosomal spontaneous deletion rate.
Deletion of Essential Genes. Two of the large deletions isolated (numbers 2 and 8 in Fig. 1 A and B) included several genes that were previously identified as being essential for growth (11 genes for deletion 2 and nine genes for deletion 8) (23). It is possible that these genes can be lost in the context of a large chromosomal deletion, or alternatively, this might reflect differences between S. typhimurium LT2 and the 14028 strain used by Knuth et al. (23) or differences in growth conditions.
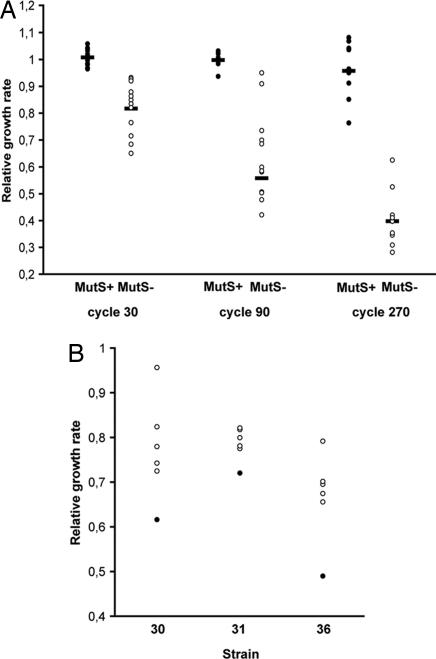
Restoration of Fitness by Compensatory Evolution. Our experimental strategy could be used to generate a smaller genome by a process where gene loss occurs spontaneously rather than by directed recombination events (24, 25). However, a potential limitation could be the continued decrease of fitness observed in these lineages due to the genome-wide accumulation of different types of deleterious mutations (Fig. 2A). To determine whether this reduction of fitness could be prevented by compensatory evolution, we allowed several low-fitness lineages to evolve within large populations in batch cultures. A partial restoration of fitness was observed for all lineages after only 100 generations of evolution (Fig. 2B). This finding suggests that the combination of growth with bottlenecks to allow fixation of deletion mutants with mass propagation to restore fitness by compensatory mutations will allow reiteration of the reductive process.
Fig. 2.
Fitness of serially passaged lineages. (A) Relative growth rate of WT (•) and mutS (○) lineages of S. typhimurium LT2 after 30, 90, and 270 cycles of serial passage on hematin agar plates. The 270 cycles represent ≈6,750 generations of bacterial growth. (B) Relative growth rate of three mutS lineages that were first given 90 cycles of serial passage followed by evolution for 100 generations in large populations to allow selection for faster growth. Slow-growing parental mutS strain (•) and growth-compensated mutants (○) are indicated.
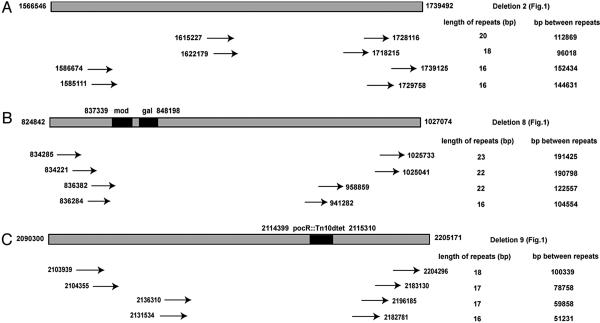
Absence of DNA Repeats at Deletion Endpoints. It has been proposed that endosymbiotic genomes were reduced in size by RecA-dependent homologous recombination between long homologous repeat sequences (6). Using our experimental set-up, we addressed this issue by identifying the endpoints in several of the deletions identified during serial passage (deletions 1-2 in Fig. 1 A and B) and in the mod-gal genetic selection (deletions 5-8 in Fig. 1 A and B) (19). In addition, we used another genetic selection to identify deletions that remove a Tn10 transposon inserted in the pocR gene (deletion 9 in Fig. 1 A and B) (18). The deletable region surrounding pocR is ≈200 kbp. Because the identified deletion endpoints are from four different regions of the Salmonella chromosome, covering ≈600 kbp of deletable nonessential DNA, they are assumed to be representative for the whole chromosome (even though it should be noted that the examined deletions are located in the replication terminus one-half of the chromosome). For five of them, the potential homology at the deletion endpoints used for recombination was shorter than 4 bp. In one mutant, the homology was 12 bp and in one 40 bp. Because efficient RecA-dependent recombination requires at least 25 bp of homology (26), this result suggests that most initial deletion formation and genome reduction occurs via RecA-independent mechanisms. We examined how much homology was present in these deleted regions that could potentially be used for the deletion formation. As shown in Fig. 3, we could find several repeats within each deletable region (only the four longest perfect repeats in each region are shown). It is notable that these identified repeats are all longer than the actual repeats found at the deletion endpoints in the experimentally isolated deletion mutants. This finding underlines the notion that deletion formation does not necessarily require repeats, and that even when repeats are present, deletions are more frequently found at nonrepeat sequences, most likely because the frequency of nonrepeats in the genome is much higher than that of repeats (27).
Fig. 3.
Size and location of repeated sequences present inside three different, experimentally isolated chromosomal deletions. Gray boxes show the largest deletions found by serial passage (A), mod-gal selection (B), and Bochner selection (C). Black boxes indicate the genetic markers used for the selection of deletion mutants. Arrows indicate repeats that could potentially be used for deletion formation. Numbers indicate nucleotides at the outer ends of the repeats.
Discussion
There are several broader implications of our data. The first implication relates to the role of RecA-dependent recombination and DNA repeats in the gene loss process. It has been suggested that the loss of a functional RecA protein and/or the loss of long DNA repeats would gradually slow the rate of gene loss (6, 28). This could ultimately cause evolutionary stasis, e.g., as that observed in the two Buchnera aphidicola genomes that diverged 50 million years ago (28). However, this notion is not easily reconciled with our data because most deletions did not occur via extensive DNA repeats. Because both intra- and interchromosomal RecA-dependent recombination requires at least 25 bp of homology to function efficiently (26, 29), it is likely that six of the seven deletions were formed by RecA-independent recombination processes. Similar RecA independence has been shown in other experimental systems (20, 21, 30-32), suggesting that the absence of RecA and/or DNA repeats does not in fact cause any substantial reduction in deletion rates at the whole genome level.
One potential resolution of this inconsistency is the idea that it is not the absence of RecA and/or DNA repeats that limit endosymbiont recombination rates but rather other defects in the recombination systems, such as those observed for recEFGJNOQR, ruvABC, and sbcDC (33). Alternatively, B. aphidicola genome stasis was not imposed by mechanistic constraints on recombination but rather by selective pressure. Comparisons of the E. coli and S. typhimurium LT2 genomes suggest that selection can conserve gene order and content. These genomes are estimated to have diverged from a common ancestor at least 100 million years ago and, despite the presence of functional RecA-dependent recombination and many DNA repeats, they still show high conservation of synteny (34).
The second implication relates to the gene loss rate calculated here, which suggests that a genome could be extensively reduced in size during a surprisingly short evolutionary time period. For example, assuming that a bacterium grows with a generation time of 1 day and experiences frequent population bottlenecks that allows the fixation of deleterious mutations, a genome with a functional MMR system could be reduced by 1 Mbp in as few as 50,000 years. Furthermore, gene loss occurs ≈50-fold faster when the stabilizing effect of the MMR system is absent. Obviously this estimate is related to our experimental set-up, as the in vivo generation times, the extent of purifying selection, the size of the population bottlenecks and the expected progressive reduction in the loss rate as more DNA is deleted will play an important role. Nevertheless, we have shown that DNA loss rates can be very high and these findings are compatible with the notion that the initial reduction of genome size in endosymbionts occurred by rapid deletion of large blocks of DNA, followed by a gradual decline in the loss rate (28).
Acknowledgments
We thank Matt Rolfe for making the S. typhimurium microarrays, Therese Hall for secretarial assistance, and Diarmaid Hughes for comments on the manuscript. This work was supported by grants from the Swedish Research Council and Uppsala University (to D.I.A.), the Biotechnology and Biological Sciences Research Council (Core Strategic Grant to J.C.D.H.), and European Union Marie Curie Training Site Program QLK2-CT-2001-60081 (to J.C.D.H. and S.E.).
Author contributions: D.I.A. designed research; A.I.N., S.K., and E.K. performed research; J.C.D.H. contributed new reagents/analytical tools; A.I.N., S.K., S.E., and D.I.A. analyzed data; and A.I.N. and D.I.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CGI, comparative genomic indexing; MMR, mismatch repair.
See Commentary on page 11959.
References
- 1.Rocha, E. P. (2003) Genome Res. 13, 1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocha, E. P. (2004) Curr. Opin. Microbiol. 7, 519-527. [DOI] [PubMed] [Google Scholar]
- 3.Ochman, H. (2001) Curr. Opin. Genet. Dev. 11, 616-619. [DOI] [PubMed] [Google Scholar]
- 4.Mira, A., Ochman, H. & Moran, N. A. (2001) Trends Genet. 17, 589-596. [DOI] [PubMed] [Google Scholar]
- 5.Moran, N. A. (2002) Cell 108, 583-586. [DOI] [PubMed] [Google Scholar]
- 6.Klasson, L. & Andersson, S. G. (2004) Trends Microbiol. 12, 37-43. [DOI] [PubMed] [Google Scholar]
- 7.Moran, N. A. & Mira, A. (2001) Genome Biol. 2, RESEARCH0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva, F. J., Latorre, A. & Moya, A. (2001) Trends Genet. 17, 615-618. [DOI] [PubMed] [Google Scholar]
- 9.Andersson, S. G., Zomorodipour, A., Andersson, J. O., Sicheritz-Ponten, T., Alsmark, U. C., Podowski, R. M., Naslund, A. K., Eriksson, A. S., Winkler, H. H. & Kurland, C. G. (1998) Nature 396, 133-140. [DOI] [PubMed] [Google Scholar]
- 10.Cole, S. T., Eiglmeier, K., Parkhill, J., James, K. D., Thomson, N. R., Wheeler, P. R., Honore, N., Garnier, T., Churcher, C., Harris, D., et al. (2001) Nature 409, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 11.McClelland, M., Sanderson, K. E., Clifton, S. W., Latreille, P., Porwollik, S., Sabo, A., Meyer, R., Bieri, T., Ozersky, P., McLellan, M., et al. (2004) Nat. Genet. 36, 1268-1274. [DOI] [PubMed] [Google Scholar]
- 12.Anjum, M. F., Lucchini, S., Thompson, A., Hinton, J. C. & Woodward, M. J. (2003) Infect. Immun. 71, 4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. (2001) Nature 413, 852-856. [DOI] [PubMed] [Google Scholar]
- 14.Maloy, S. R., Stewart, V. J. & Taylor, R. K. (1996) Genetic Analysis of Pathogenic Bacteria (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 15.Liu, S. L. & Sanderson, K. E. (1992) J. Bacteriol. 174, 1662-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. & Hinton, J. C. (2003) Mol. Microbiol. 47, 103-118. [DOI] [PubMed] [Google Scholar]
- 17.Lejeune, P. & Danchin, A. (1990) Proc. Natl. Acad. Sci. USA 87, 360-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lea, D. A. & Coulson, C. A. (1949) J. Genet. 49, 264-285. [DOI] [PubMed] [Google Scholar]
- 19.Bochner, B. R., Huang, H. C., Schieven, G. L. & Ames, B. N. (1980) J. Bacteriol. 143, 926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaaper, R. M. & Dunn, R. L. (1991) Genetics 129, 317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agemizu, Y., Uematsu, N. & Yamamoto, K. (1999) Biochem. Biophys. Res. Commun. 261, 584-589. [DOI] [PubMed] [Google Scholar]
- 22.Schofield, M. J. & Hsieh, P. (2003) Annu. Rev. Microbiol. 57, 579-608. [DOI] [PubMed] [Google Scholar]
- 23.Knuth, K., Niesalla, H., Hueck, C. J. & Fuchs, T. M. (2004) Mol. Microbiol. 51, 1729-1744. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto, M., Ichimura, T., Mizoguchi, H., Tanaka, K., Fujimitsu, K., Keyamura, K., Ote, T., Yamakawa, T., Yamazaki, Y., Mori, H., et al. (2005) Mol. Microbiol. 55, 137-149. [DOI] [PubMed] [Google Scholar]
- 25.Kolisnychenko, V., Plunkett, G., III, Herring, C. D., Feher, T., Posfai, J., Blattner, F. R. & Posfai, G. (2002) Genome Res. 12, 640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, P. & Huang, H. V. (1986) Genetics 112, 441-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura-Masuda, A. & Ikeda, H. (1990) Mol. Gen. Genet. 220, 345-352. [DOI] [PubMed] [Google Scholar]
- 28.Tamas, I., Klasson, L., Canback, B., Naslund, A. K., Eriksson, A. S., Wernegreen, J. J., Sandstrom, J. P., Moran, N. A. & Andersson, S. G. (2002) Science 296, 2376-2379. [DOI] [PubMed] [Google Scholar]
- 29.Chow, S. A. & Radding, C. M. (1985) Proc. Natl. Acad. Sci. USA 82, 5646-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halliday, J. A. & Glickman, B. W. (1991) Mutat. Res. 250, 55-71. [DOI] [PubMed] [Google Scholar]
- 31.Ishiura, M., Hazumi, N., Shinagawa, H., Nakata, A., Uchida, T. & Okada, Y. (1990) J. Gen. Microbiol. 136, 69-79. [DOI] [PubMed] [Google Scholar]
- 32.Albertini, A. M., Hofer, M., Calos, M. P. & Miller, J. H. (1982) Cell 29, 319-328. [DOI] [PubMed] [Google Scholar]
- 33.Silva, F. J., Latorre, A. & Moya, A. (2003) Trends Genet. 19, 176-180. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, J. G. & Ochman, H. (1998) Proc. Natl. Acad. Sci. USA 95, 9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]