Abstract
Enzymes of the blood coagulation pathway enhance the inflammatory response leading to endothelial dysfunction, accounting, in part, for the vascular complications occurring in sepsis and cardiovascular disease. The responses of endothelial cell activation include induction of the expression of tissue factor (TF), a membrane glycoprotein that promotes thrombosis, and of E-selectin, a cell adhesion molecule that promotes inflammation. In this report, we demonstrate synergistic interactions between the coagulation factor Xa (fXa) and the proinflammatory cytokines TNF, IL-1β, and CD40L, leading to enhanced expression of TF and E-selectin in endothelial cells. A detailed analysis of the molecular pathways that could account for this activity of fXa showed that fXa inhibited the cytokine-induced expression of dual specificity phosphatases, MAP kinase phosphatase-L, -4, -5, and -7, blocking a negative regulatory effect on c-Jun N-terminal kinase. The synergistic interaction between fXa and TNF was also involved in the inhibition of A20 and IκBα expression in the IκB kinase-NF-κB pathway. The data indicate that inhibition of negative regulatory signaling accounts for the amplification of cytokine-induced endothelial cell activation by fXa.
Keywords: cytokines, endothelial cells, factor Xa, inflammation
Vascular and inflammatory diseases, such as atherosclerosis and sepsis, manifest thrombosis-driven augmentation of the endothelial response, which, if not attenuated, leads to endothelial dysfunction (1). Early events in the activation of endothelial cells include the induced expression of tissue factor (TF), a membrane glycoprotein that initiates coagulation and promotes thrombosis (2), and of E-selectin, a cell adhesion molecule that promotes inflammation (3). The expression of both TF and E-selectin is highly regulated (4–7) and appears to have coordinated expression after thrombosis and at all stages of the progression of atherosclerosis (8). Inflammatory cytokines, including interleukins (9), TNF (10), and CD40L (11), are well known to induce endothelial cell activation and up-regulate TF and E-selectin expression to various extents (12–17). Expression of E-selectin provides the opportunity for augmenting the endothelial cell response by recruiting monocytes and neutrophils, which release additional cytokines. Although less studied, it is also apparent that two proteases in the coagulation cascade that are generated during thrombosis, factor Xa (fXa) and thrombin, are also capable of inducing endothelial cell activation. The most-studied effectors of these coagulation proteases have been the G-protein-coupled protease-activated receptors (PARs) (18, 19). Endothelial cells express two of the PAR receptors, PAR1 and -2. Thrombin activates PAR1, whereas fXa has been demonstrated to activate both (20, 21). Because TF on activated endothelial cells binds coagulation factor VIIa to initiate the coagulation cascade by converting factor X to fXa, which in turn converts prothrombin to thrombin, it is apparent that the induced expression of TF also provides the opportunity for augmenting the endothelial cell response. The mechanisms by which inflammatory cytokines and coagulation proteases interact to achieve endothelial cell activation are not known.
In this report, we investigate the molecular mechanism by which the coagulation protease, fXa, extends and enhances the inflammatory response of endothelial cells to proinflammatory cytokines. Our results show that in human primary endothelial cells [human coronary artery endothelial cells, human pulmonary microvascular endothelial cells, and human umbilical vein endothelial cells (HUVEC)], fXa increases the induction of endothelial TF and E-selectin by the proinflammatory cytokines, TNF, IL-1β, and CD40L. We further focus on the signaling pathways involved in the interaction between fXa and TNF, in particular the mitogen-activated protein kinase (MAPK) and NF-κB pathways that are important in the regulation of both TF and E selectin promoters (4–7). We show that fXa leads to coordinated reduction in the expression of TNF-induced MAPK phosphatases, PP2A and MKP-L, -4, -5, and -7. This reduced expression most likely delays the termination of TNF-triggered c-Jun N-terminal kinase (JNK) activation. In an analogous manner, we show that fXa inhibits the expression of IκBα and A20, the negative regulators of the IκB kinase-NF-κB pathway, leading to increased activation of transcription factor NF-κB.
Materials and Methods
Reagents and Antibodies. fXa and thrombin were purchased from Haematologic Technologies (Essex Junction, VT), and TNF and IL-1 were from R&D Systems. CD40L was prepared as described (22). PAR peptide agonists were from Bachem, and Argatroban was from SmithKline Beecham. Antibodies used were anti-TF (American Diagnostica, Greenwich, CT), anti-E-selectin, anti-JNK1 (Santa Cruz Biotechnology), anti-p-65 (39331, Active Motif, Carlsbad, CA), anti-pJNK, anti-c-Jun, anti-p-c-Jun, and IκBα (Cell Signaling Technology, Beverly, MA). Antibodies against MKP-5 and -7 were generated in chicken by Washington Biotechnology (Simpsonville, MD) (see Supporting Text, which is published as supporting information on the PNAS web site).
Cell Cultures and Stimulations. HUVEC, human coronary artery endothelial cells, and human pulmonary microvascular endothelial cells were purchased from Clonetics (San Diego) and grown according to the supplier's directions. Where indicated, fXa (10 nM), TNF-α (0.5 to 5 ng/ml), IL-1β (1 ng/ml), or CD40L (10 μg/ml) was added to the culture. Unless indicated otherwise, stimulations were done for 4 h.
TF-Dependent Coagulation. Cells were washed in PBS and incubated with Tris-buffered saline containing 30 mM Ca2+ for 2 min. Citrate anticoagulated fibrinogen-deficient human plasma (George King Biomedical, Overland Park, KS) was added, followed by incubation for 2 min. To assess TF-initiated thrombin generation, an aliquot was withdrawn, added to specific thrombin substrate (S-2238, Diapharma, West Chester, OH), and changes in absorbance were monitored at 405 nm. All steps were carried out at room temperature. Fold induction was calculated by comparing the thrombin activity of stimulated with untreated cells.
Quantitative Real-Time-PCR (TaqMan) Analysis. Total RNA was isolated from HUVEC by using an RNeasy Kit (Qiagen, Valencia, CA), and cDNA was generated from DNased RNA by using the Reverse Transcriptase Kit (Applied Biosystems).
Real-time quantitative PCR was performed by using an Applied Biosystems Prism 7700 sequence-detection system with Taqman reagents (Applied Biosystems). The sequences of the specific probes and primers are listed in Supporting Text.
Microscopy. HUVECs were grown on gelatin-coated glass chamber slides and processed as described in Supporting Text.
Results
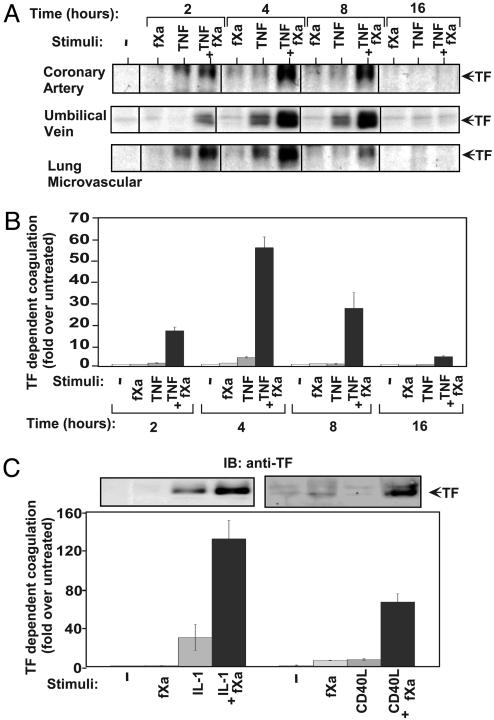
TF Induction by Proinflammatory Cytokines, TNF, IL-1β, and CD40L Is Markedly Increased by fXa. Using the primary endothelial cultures human coronary artery endothelial cells, HUVEC, and human pulmonary microvascular endothelial cells, we assessed the relationship between coagulation protease fXa and endothelial activation by observing the effect of fXa on TF induction. We stimulated cells with fXa alone and with a combination of fXa and TNF (Fig. 1A). TNF has been shown to play a key role in the transition of endothelial cells from basal to activated state and therefore served as a positive control (4, 10, 12). fXa had no substantial effect on TF expression, and TNF alone had a modest effect. However, the combination of TNF and fXa produced very high levels of TF expression in all endothelial cell types (Fig. 1A). The time course reveals the transient nature of TF induction, which was detected as early as 2 h poststimulation, peaked between 4 and 8 h, and down-regulated after 16 h (Fig. 1A).
Fig. 1.
Coagulation protease, fXa, up-regulates endothelial TF expression induced by proinflammatory cytokines. (A) Time course of TF expression as detected by TF immunoblotting in endothelial cells from different origins stimulated by fXa (10 nM), TNF (1 ng/ml), or both. (B) Time course of TF-dependent coagulation in HUVEC stimulated by fXa (10 nM), TNF (1 ng/ml), or both. (C) TF induction by fXa (10 nM) combined with IL-1β (1 ng/ml) or CD40L (10 μg/ml). Expression was measured by anti-TF immunoblotting (Upper) and by TF-dependent coagulation assay (Lower). Representative data from three independent experiments are reported as average ± standard deviation.
It should be noted that the synergistic activity between fXa and TNF was very prominent at TNF concentrations between 0.5 and 5ng/ml (Fig. 8A, which is published as supporting information on the PNAS web site). TNF concentrations of this level (≈5 ng/ml) have been reported in animal models of inflammation and atherosclerosis and also in clinical studies (23, 24). The synergistic effect was observed over a range of TNF concentrations, up to 20 ng/ml, that have been reported to be effective in the activation of endothelial cells (Fig. 8A).
To evaluate the biological activity of TF on the surface of these cells, we performed a TF-dependent functional assay. To verify that this assay was indeed TF-dependent, we included specific TF-neutralizing antibody in the reaction. The antibody completely abolished the detection of thrombin generated in this assay (Fig. 9, which is published as supporting information on the PNAS web site). We then studied the kinetics of TF-dependent coagulation that was exhibited in cells stimulated with fXa and TNF, alone or in combination. The results indicated a comparable synergistic effect between fXa and TNF (HUVEC shown in Fig. 1B; for others, data not shown) that correlate well with the effect we observed on TF expression (Fig. 1A). In addition, real-time PCR analysis indicated a corresponding synergistic enhancement of TF RNA expression with matching kinetics (data not shown).
To learn whether the synergy extends to other functionally related proinflammatory cytokines, we stimulated HUVECs with either IL-1β or CD40L combined with fXa. The combination resulted in a strong and synergistic TF expression (Fig. 1C Upper) and a corresponding enhancement of TF-dependent coagulation activity (Fig. 1C Lower).
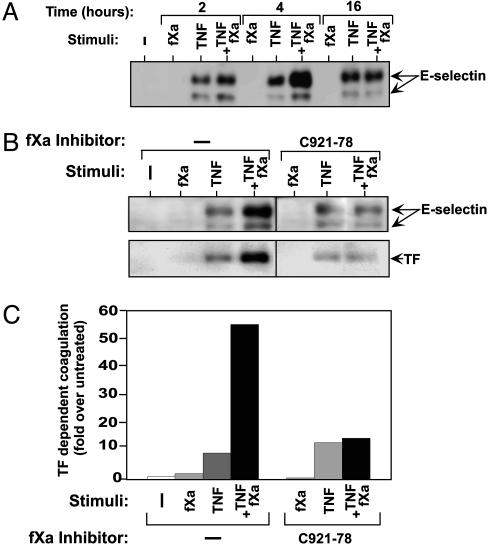
The Expression of E-Selectin Is also Up-Regulated by fXa and Requires fXa Proteolytic Activity. E-selectin is an important inflammatory protein that facilitates the recruitment of leukocytes during endothelial activation. We studied the effect triggered by fXa, alone or combined with TNF, on the expression of E-selectin in endothelial cells (Fig. 2A). TNF induced the expression of E-selectin as early as 2 h after stimulation and sustained this effect up to 16 h. fXa had no effect on its own but, when combined with TNF, it transiently up-regulated TNF-induced E-selectin, with expression peaking 4 h after stimulation (Fig. 2A). This result further substantiates the conclusion that the coagulation protease, fXa enhances both an inflammatory response and an endothelial activation. To investigate whether the proteolytic activity of fXa was required to produce this enhancement, we used the specific fXa inhibitor, C921-78 (25). The presence of C921-78 during stimulation of endothelial cells with fXa and TNF abolished the increase in expression of E-selectin (Fig. 2B Upper) and TF (Fig. 2B Lower). fXa enhancement of TF-dependent coagulation was also abolished by C921-78 (Fig. 2C). To verify that the enhancement was due to stimulation by fXa and not to thrombin generated in the experimental system, we preincubated fXa with a specific thrombin inhibitor (argatroban) and used the enzyme/inhibitor combination for cell stimulation studies with TNF (26). The expression of TF was unchanged as compared with control experiments in the absence of argatroban (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 2.
The expression of E-selectin is up-regulated by fXa. (A) Time course of E-selectin expression as detected by immunoblotting in HUVECs stimulated by fXa (10 nM), TNF (5 ng/ml), or both. (B) fXa-induced up-regulation of E-selectin or TF requires its proteolytic activity. C921-78 (100 nM), a fXa-specific inhibitor, was added to HUVECs during fXa (10 nM) and TNF (1 ng/ml) stimulation. The expression of E-selectin (Upper) or TF(Lower) was detected by immunoblotting. (C) TF-dependent coagulation assay in the presence of C921-78 (100 nM) during the stimulation of HUVECs by fXa (10 nM) and TNF (1 ng/ml). Representative data from three independent experiments are reported as average ± standard deviation.
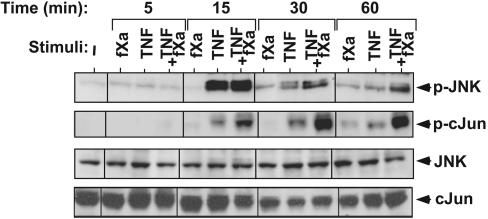
fXa Enhances TNF-Triggered Activation of JNK. To delineate the signaling mechanisms underlying the ability of coagulation proteases to support endothelial response, we assessed fXa- and TNF-triggered signaling pathways in endothelial cells. JNK is the MAPK family member that is preferentially activated by proinflammatory cytokines such as TNF, IL-1, and CD40L (27). Initially, we studied its activation in HUVECs that had been stimulated by fXa, TNF, or a combination of the two (Fig. 3 Top). On its own, fXa had very little effect, but in combination, it substantially prolonged TNF-induced activation of JNK. JNK phosphorylation peaked at 15-min poststimulation by TNF and was reduced back to basal level after 60 min. However, JNK phosphorylation was prolonged and remained substantially enhanced at 60 min, when TNF and fXa were combined (Fig. 3). JNK phosphorylates the transcriptional factor c-Jun on Ser-63 and -73, thereby increasing its transcriptional potential (28). Subsequently, we analyzed the phosphorylation of endogenous c-Jun (Fig. 3, second blot). fXa alone had no effect on c-Jun phosphorylation but, when combined with TNF, phosphorylation was significantly prolonged. c-Jun activation was slightly delayed when compared with JNK, an observation that is compatible with the presumed role of JNK in c-Jun activation. These results strongly suggest that the inflammatory effect of fXa is not independently sustained but instead is coupled to the signaling that is prompted by TNF and possibly by other competent cytokines. The inflammatory function of fXa is apparent in the dramatic prolongation of the JNK signaling cascade.
Fig. 3.
Effect of fXa on TNF-induced JNK signaling pathway. Time course of JNK and c-Jun phosphorylation as detected by immunoblotting with anti-p-JNK and anti-p-c-Jun, respectively (Top) and the control antibodies anti-JNK and anti-c-Jun, respectively (Bottom). Cells were incubated with fXa (10 nM), TNF (5 ng/ml), or both for the indicated times. Representative data from three independent experiments are reported.
fXa Decreases TNF-Triggered Expression of MAPK Phosphatases. The observation that fXa can substantially prolong the activity of JNK led us to hypothesize that it might act by inhibiting the negative regulation associated with TNF. MAPKs, like JNK, are activated by dual phosphorylation of conserved threonine and tyrosine residues by the upstream MAPK kinases (29). Dual-specific phosphatases can reverse the phosphorylation of serine, threonine, and tyrosine residues, thereby terminating the activation of MAPKs (30, 31). A number of phosphatases have been reported to possess the ability to dephosphorylate JNK, and many of them are transcriptionally regulated (31). To test our hypothesis, we first studied the endothelial expression of functionally relevant phosphatases. We investigated the phosphatases whose expression is significantly up-regulated by TNF stimulation, postulating they are the ones most likely to play a role in the negative regulation of TNF-induced JNK signaling (Table 1, which is published as supporting information on the PNAS web site). The expression of several dual-specificity phosphatases was substantially stimulated by TNF and included MKP-1, -4, -L, -5, and -7, as well as the serine/threonine phosphatase, PP2A. We then analyzed the effect of fXa on the TNF induction of these phosphatases: the induction of MKP-1 by TNF was not affected by the addition of fXa (data not shown). In contrast, the expression of all other phosphatases induced by TNF, namely PP2A and MKP-L, -4, -5, and -7, were inhibited by the addition of fXa to TNF (Fig. 4 A–E). PP2A induction by TNF was most transient, appearing after 15 min of stimulation and decreasing back to basal at 30 min (Fig. 4A). At the 15-min time point, fXa alone slightly reduced PP2A expression, and the combination of fXa with TNF, at this time point, abolished PP2A induction by TNF (Fig. 4A). PP2A was previously shown to mediate the inhibition of TNF induced JNK (32, 33). The kinetics of PP2A induction and its inhibition by fXa (Fig. 4A) distinctly correlates with fXa enhancement of both JNK and c-Jun phosphorylation at 15 and 30 min of TNF stimulation (Fig. 3 Top).
Fig. 4.
fXa decreases TNF-induced expression of specific MAPK phosphatases. Time course of relative expression of the phosphatases in HUVECs stimulated by fXa (10 nM), TNF (5 ng/ml), or both, as detected by real-time PCR by using specific primers for (A) PP2A, (B) MKP-L, (C) MKP-4, (D) MKP-5, or (E) MKP-7. (F) Effect of fXa (10 nM) on TNF-induced (5 ng/ml) protein expression of MKP-5 as detected by immunofluorescent staining in HUVECs stimulated for 60 min. a–d represent immunofluorescent staining using anti-MKP-5, whereas e–h represent DAPI staining of the corresponding field. (G) Effect of fXa (10 nM) on TNF-induced (5 ng/ml) protein expression of MKP-7 as detected by immunoblotting with anti-MKP-7 in HUVECs stimulated for 60 min.
The induction of the dual phosphatases MKP-L, -4, -5, and -7 is apparent at 60 min of TNF induction and is reduced by the costimulation with fXa (Fig. 4 B–E). The kinetics of this effect correlates well with the most prominent enhancement of JNK and c-Jun activation by fXa at 60 min of TNF stimulation (Fig. 3 Top). MKP-4 and -L are relatively nonselective phosphatases and can inactivate the MAPKs JNK, p38, and ERK (34). MKP-7 and -5 selectively inactivate JNK (35–37). The inhibition of MKP-5 by fXa is particularly noteworthy, because MKP-5 was recently shown to play a principal role in innate and adaptive immune responses. JNK activity is selectively increased in MKP-5-deficient cells that produce greatly enhanced levels of proinflammatory cytokines (38).
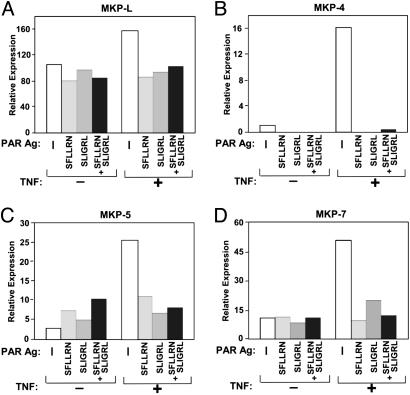
To confirm the inhibitory effect of fXa on TNF-induced expression of these MKPs, we raised antibodies against MKP-5 and -7 and analyzed the expression of MKP-5 and -7 in cells stimulated with fXa and TNF (Fig. 4 F and G). In agreement with the real-time PCR data (Fig. 4 D and E), up-regulation of TNF-induced MKP-5 and -7 was inhibited when fXa was added to cells (Fig. 4 F and G). Taken together, these results strongly suggest that the coordinated inhibition of specific TNF-induced MAPK phosphatases is the mechanism by which the coagulation protease, fXa, extends and enhances JNK activation, leading to increased expression of responsive genes such as those for TF and E-selectin. fXa has been demonstrated (20, 21) to elicit its cellular effects through activation of PAR1 and -2 receptors. Thus, we were interested in determining whether PAR1 and -2 agonists could mimic the inhibitory effects of fXa (Fig. 5). To activate PARs, we used the synthetic peptides, SFLLRN, which selectively activates PAR1 and SLIGRL, a potent agonist of PAR2 and a weaker agonist of PAR1 (39). Indeed, the treatment of endothelial cells with the agonists abolished the expression of these phosphatases (Fig. 5) in a manner similar to the inhibition triggered by fXa (Fig. 4).
Fig. 5.
PAR1 and -2 agonists mimic the inhibitory effects of fXa, decreasing TNF-induced expression of the dual specificity phosphatases. Relative expression of the phosphatases (A) MKP-L, (B) -4, (C) -5, or (D) -7 in HUVECs stimulated for 60 min by TNF (5 ng/ml) in the presence of 100 μM of the indicated PAR peptide agonists.
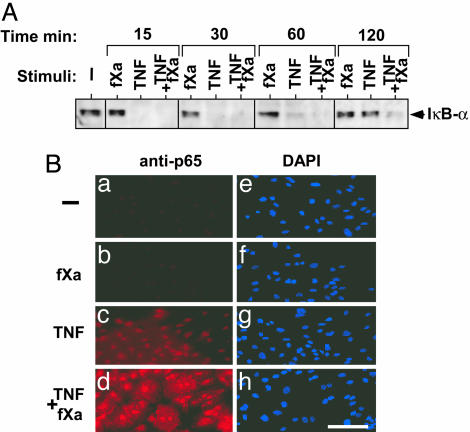
Enhancement of IκB kinase-NF-κB Signaling by fXa. In addition to JNK, a major effect of TNF-induced signaling is the activation of transcription factor NF-κB (40). In unstimulated cells, NF-κB is bound to regulatory proteins called IκBs and is sequestered in the cytoplasm. TNF stimulation leads to the phosphorylation of IκBs by IκB kinases, which targets them for proteosomal degradation (41). After IκB degradation, liberated NF-κB translocates to the nucleus and enhances the transcription of several genes, including those encoding cytokines, chemokines, and adhesion molecules (42). Because fXa had shown a synergistic effect on TNF-induced expression levels, we investigated whether it would have an effect on TNF-induced NF-κB activation in endothelial cells. We studied the pattern of IκBα degradation and synthesis, which is indicative of NF-κB activation and inhibition. fXa, as a sole stimulus, did not affect the pattern of IκBα degradation but fXa, added to TNF, prolonged IκBα degradation, delaying its reappearance (Fig. 6A). The degradation of IκBα is evident after stimulation with both TNF alone and combined with fXa. After 60 min, the accumulation of newly synthesized IκBα is apparent in TNF-treated cells, and it returns to basal level at 120 min. In contrast, at 60 and 120 min of fXa plus TNF treatment, the accumulation of newly synthesized IκBα was substantially reduced. In agreement, using immunofluorescent staining (Fig. 6B), we observed enhanced nuclear staining for p65 in cells stimulated by TNF and fXa for 120 min. Similar results were obtained by using the NF-κB EMSA assay, where TNF-induced DNA binding was enhanced and prolonged in cells that were costimulated with fXa (data not shown).
Fig. 6.
fXa enhances TNF-induced NFκB activation. (A) Time course of IκBα degradation in HUVECs incubated with fXa (10 nM), TNF (5 ng/ml), or both, as detected by anti-IκBα immunoblotting. (B) p65 nuclear localization as detected by p65 immunostaining: HUVECs were incubated for 120 min with fXa (10 nM), TNF (5 ng/ml), or both. a–d represent immunofluorescent staining using anti-p65, whereas e–h are DAPI staining of the corresponding field. Representative data from three independent experiments are reported.
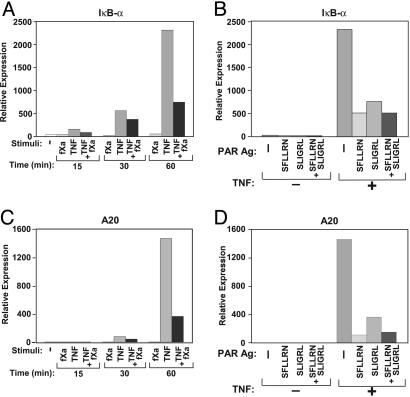
fXa Inhibits TNF-Triggered Expression of IκBα and A20. The total level of IκBα in the cell is a result of the balance between degradation of the phosphorylated IκBα and its de novo synthesis. The transcription of new IκBα, similar to its degradation, is stimulated by TNF and is a part of the negative regulatory loop of NF-κB activation (43, 44). Because the addition of fXa to TNF resulted in delaying the reappearance of the newly synthesized IκBα (Fig. 6), we studied the effect of fXa on TNF-triggered transcription of IκBα (Fig. 7A). In HUVECs, the induced expression of IκBα was observed 60 min after TNF stimulation and was dramatically reduced when fXa was added, indicating that fXa inhibited the TNF-induced expression of IκBα (Fig. 7A). Addition of PAR1 and -2 agonists to TNF reduced the expression of IκBα (Fig. 7B), indicating that this inhibition was also mediated by PAR1 and -2.
Fig. 7.
Inhibition of TNF-induced expression of IκBα and A20 by fXa or PAR agonists, as detected by real-time-PCR. (A) Time course of IκBα expression in HUVECs stimulated for the indicated length of time with fXa (10 nM), TNF (5 ng/ml), or both. (B) IκBα expression in HUVECs incubated for 60 min with 100 μM peptide agonists with or without the addition of TNF (5 ng/ml). (C) Time course of A20 expression in HUVECs stimulated for the indicated length of time with fXa (10 nM), TNF (5 ng/ml), or both. (D) A20 expression in HUVECs incubated with 100 μM peptide agonists with or without the addition of TNF (5 ng/ml) for 60 min. Representative data from at least three independent experiments are reported.
More recently, another cellular inhibitor of TNF signaling was characterized: the zinc finger protein A20 (45). A20 acts upstream in the TNF-induced signaling cascade, blocking TNF-induced activation of NF-κB (46) and JNK (47). A20 is recruited to the TNF receptor-1 signalosome and interferes with its function (48). A20 expression is induced by proinflammatory cytokines including TNF, IL-1, and CD40L, suggesting that it is involved in the negative regulatory feedback of inflammatory signaling (49, 50). Indeed, A20-deficient mice die prematurely as a result of severe inflammation (51). Consequently, we were interested in studying whether fXa had an effect on TNF-induced expression of A20. Using real-time-PCR, we observed that expression of A20 in HUVECs, which was strongly up-regulated after 60 min of TNF treatment, was substantially inhibited by the addition of fXa (Fig. 7C). The addition of PAR1 and -2 agonists also reduced the TNF-induced expression of A20 (Fig. 7D).
Overall, our results demonstrate the ability of the coagulation protease, fXa to orchestrate a synchronized decrease in TNF-induced down-regulation of the inflammatory signaling. This down-regulation is most likely mediated by PAR1 and -2 and occurs at levels affecting both upstream TNF receptor-1 signaling, which is mediated by the A20 protein, and the more downstream pathway, which is mediated by inhibitors such as IκBα and MKPs.
Discussion
In this paper, we present data suggesting that the mechanism used by the coagulation protease, fXa, to enhance expression of TF and E-selectin in endothelial cells relies on fXa's ability to down-regulate the inhibition of the inflammatory process. We demonstrate that crosstalk between fXa and TNF leads to enhanced activation of the MAPK, JNK, and the IκB kinase-NF-κB pathway and propose that the molecular targets include JNK phosphatases, PP2A and MKP-L, -4, -5, and -7, as well as A20 and IκBα. We show that fXa-triggered inhibition of these phosphatases could account for the prolonged activation of JNK by TNF. In parallel, the fXa-triggered inhibition of IκBα and A20 by fXa would account for the enhanced duration of TNF-induced NF-κB activation. These signaling events might subsequently lead to a coordinated synergistic up-regulation of inflammatory genes, which include a compatible combination of AP1 and NF-κB regulatory sites in their promoters.
Sites of thrombosis within the vasculature provide the ideal milieu for the synergistic activity of inflammatory cytokines with fXa. Proinflammatory cytokines, such as TNF, IL-1, and CD40L, are provided by lymphocytes and macrophages, present in atherosclerotic lesions, prompting the initial expression of TF and E-selectin. TF initiates coagulation, resulting in an increased local concentration of fXa and thrombin. The presence of fXa creates a positive feedback mechanism where inflammatory cytokines and active coagulation proteases induce a vast increase in expression of TF and E-selectin, with further induction of coagulation and recruitment of leukocytes. The inflammatory response is most likely further augmented by the recruitment of platelets, which generates additional CD40L (52) and provides additional sites for the generation of procoagulant complexes.
Because PAR1 and -2 agonists mimic fXa response (Figs. 5 and 7 and ref. 20), we propose that the cellular effectors of endothelial activation by fXa are most likely PAR1 and -2. It is not known whether EPR, a reported fXa receptor in endothelial cells (53), is also involved in this activity. Our results also indicate that thrombin (Fig. 8B) is capable of pairing with TNF to lead to functional expression of TF. Because fXa activates both PAR2 and-1 (20, 21), and thrombin activates PAR1, it would be interesting to speculate on the relative contributions of the two receptors (PAR1 and -2) to the observed synergistic activity of fXa. Based on previous work by Coughlin (18) and Ossovskaya and Bunnett (19), our initial dose–response studies were carried out in the peptide concentration range of 1–100 μM. It is likely that the agonist peptide concentration (100 μM) used in the present study (Figs. 5 and 7) is at the maximal response range for both receptors. Although future work involving dose titration of PAR1 and -2 agonists will be required to delineate the relative activities of PARs in cytokine synergism, use of selective PAR antagonists and PAR-deficient cells will be required to rank the contributions of the two receptors.
A recent paper (54) has demonstrated that PAR agonist peptides enhance LPS and TNF induction of an additional inflammatory molecule, the cytokine IL-6. The findings described herein outline the underlying mechanisms by which the coagulation factor Xa or direct PAR activation mediate the enhancement of inflammatory target genes, TF and E-selectin. We suggest a coordinated activation by fXa of the JNK and NF-κB signaling pathways that is affected by a negative regulatory response on the pathways. We also identify the molecular targets whose down-regulation results in this effect. It is also important to note that several recent animal studies have demonstrated that fXa and thrombin, as well as PAR1 and -2, can mediate inflammatory responses in vivo (55–57). Although this study focuses on a coagulation protease, it is likely that other proteases that are capable of cleaving PAR1 or -2, such as proteases originating from monocytes or neutrophils, when costimulated with inflammatory cytokines could elicit similar endothelial activation (TF and E-selectin induction). It is particularly intriguing to consider the ability of additional proteases to modulate regulatory inhibition in the context of pathologies resulting from excessive inflammation, such as sepsis or rheumatoid arthritis, where infiltrating immune cells concomitantly release both proteases and cytokines.
Supplementary Material
Acknowledgments
We acknowledge the contributions made by Nicole Avitahl, Alan Carpino, Christine Matson, Kristine Burke, and Susan Edwards.
Author contributions: A.H.-Y. and U.S. designed research; A.H.-Y., P.W.W., N.B.-L., L.G.K., K.S.S.P., and D.R.P. performed research; A.H.-Y., P.W.W., L.G.K., K.S.S.P., and U.S. analyzed data; and A.H.-Y., D.R.P., and U.S. wrote the paper.
Abbreviations: TF, tissue factor; fXa, factor Xa; PAR, protease-activated receptor; MAPK, mitogen-activated protein kinase; JNK, c-Jun N-terminal kinase; HUVEC, human umbilical vein endothelial cells.
References
- 1.Esmon, C. T. (2001) Thromb. Haemostasis 86, 51–56. [PubMed] [Google Scholar]
- 2.Osterud, B. (1997) Thromb. Haemostasis 78, 755–758. [PubMed] [Google Scholar]
- 3.Ballantyne, C. M. (2001) Clin. Cardiol. 24, III13–III17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schindler, U. & Baichwal, V. R. (1994) Mol. Cell. Biol. 14, 5820–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min, W. & Pober, J. S. (1997) J. Immunol. 159, 3508–3518. [PubMed] [Google Scholar]
- 6.Read, M. A., Whitley, M. Z., Gupta, S., Pierce, J. W., Best, J., Davis, R. J. & Collins, T. (1997) J. Biol. Chem. 272, 2753–2761. [DOI] [PubMed] [Google Scholar]
- 7.Read, M. A., Whitley, M. Z., Williams, A. J. & Collins, T. (1994) J. Exp. Med. 179, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luther, T. & Mackman, N. (2001) Trends Cardiovasc. Med. 11, 307–312. [DOI] [PubMed] [Google Scholar]
- 9.Bevilacqua, M. P., Pober, J. S., Majeau, G. R., Cotran, R. S. & Gimbrone, M. A., Jr. (1984) J. Exp. Med. 160, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway, E. M., Bach, R., Rosenberg, R. D. & Konigsberg, W. H. (1989) Thromb. Res. 53, 231–241. [DOI] [PubMed] [Google Scholar]
- 11.Slupsky, J. R., Kalbas, M., Willuweit, A., Henn, V., Kroczek, R. A. & Muller-Berghaus, G. (1998) Thromb. Haemostasis 80, 1008–1014. [PubMed] [Google Scholar]
- 12.Bevilacqua, M. P., Pober, J. S., Majeau, G. R., Fiers, W., Cotran, R. S. & Gimbrone, M. A., Jr. (1986) Proc. Natl. Acad. Sci. USA 83, 4533–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parry, G. C. & Mackman, N. (1995) Arterioscler. Thromb. Vasc. Biol. 15, 612–621. [DOI] [PubMed] [Google Scholar]
- 14.Miller, D. L., Yaron, R. & Yellin, M. J. (1998) J. Leukocyte Biol. 63, 373–379. [DOI] [PubMed] [Google Scholar]
- 15.Karmann, K., Min, W., Fanslow, W. C. & Pober, J. S. (1996) J. Exp. Med. 184, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gedeit, R. G. (1996) Crit. Care Med. 24, 1543–1546. [DOI] [PubMed] [Google Scholar]
- 17.Raab, M., Daxecker, H., Markovic, S., Karimi, A., Griesmacher, A. & Mueller, M. M. (2002) Clin. Chim. Acta 321, 11–16. [DOI] [PubMed] [Google Scholar]
- 18.Coughlin, S. R. (2001) Thromb. Haemostasis 86, 298–307. [PubMed] [Google Scholar]
- 19.Ossovskaya, V. S. & Bunnett, N. W. (2004) Physiol. Rev. 84, 579–621. [DOI] [PubMed] [Google Scholar]
- 20.Camerer, E., Kataoka, H., Kahn, M., Lease, K. & Coughlin, S. R. (2002) J. Biol. Chem. 277, 16081–16087. [DOI] [PubMed] [Google Scholar]
- 21.Riewald, M., Kravchenko, V. V., Petrovan, R. J., O'Brien, P. J., Brass, L. F., Ulevitch, R. J. & Ruf, W. (2001) Blood 97, 3109–3116. [DOI] [PubMed] [Google Scholar]
- 22.Andre, P., Prasad, K. S., Denis, C. V., He, M., Papalia, J. M., Hynes, R. O., Phillips, D. R. & Wagner, D. D. (2002) Nat. Med. 8, 247–252. [DOI] [PubMed] [Google Scholar]
- 23.Signorelli, S. S., Mazzarino, M. C., Di Pino, L., Malaponte, G., Porto, C., Pennisi, G., Marchese, G., Costa, M. P., Digrandi, D., Celotta, G., et al. (2003) Vasc. Med. 8, 15–19. [DOI] [PubMed] [Google Scholar]
- 24.Waehre, T., Halvorsen, B., Damas, J. K., Yndestad, A., Brosstad, F., Gullestad, L., Kjekshus, J., Froland, S. S. & Aukrust, P. (2002) Eur. J. Clin. Invest. 32, 803–810. [DOI] [PubMed] [Google Scholar]
- 25.Betz, A., Wong, P. W. & Sinha, U. (1999) Biochemistry 38, 14582–14591. [DOI] [PubMed] [Google Scholar]
- 26.White, C. M. (2005) Am. Heart J. 149, S54–S60. [DOI] [PubMed] [Google Scholar]
- 27.Davis, R. J. (2000) Cell 103, 239–252. [DOI] [PubMed] [Google Scholar]
- 28.Pulverer, B. J., Kyriakis, J. M., Avruch, J., Nikolakaki, E. & Woodgett, J. R. (1991) Nature 353, 670–674. [DOI] [PubMed] [Google Scholar]
- 29.Ichijo, H. (1999) Oncogene 18, 6087–6093. [DOI] [PubMed] [Google Scholar]
- 30.Farooq, A. & Zhou, M. M. (2004) Cell Signalling 16, 769–779. [DOI] [PubMed] [Google Scholar]
- 31.Camps, M., Nichols, A. & Arkinstall, S. (2000) FASEB J. 14, 6–16. [PubMed] [Google Scholar]
- 32.Shanley, T. P., Vasi, N., Denenberg, A. & Wong, H. R. (2001) J. Immunol. 166, 966–972. [DOI] [PubMed] [Google Scholar]
- 33.Avdi, N. J., Malcolm, K. C., Nick, J. A. & Worthen, G. S. (2002) J. Biol. Chem. 277, 40687–40696. [DOI] [PubMed] [Google Scholar]
- 34.Muda, M., Boschert, U., Smith, A., Antonsson, B., Gillieron, C., Chabert, C., Camps, M., Martinou, I., Ashworth, A. & Arkinstall, S. (1997) J. Biol. Chem. 272, 5141–5151. [DOI] [PubMed] [Google Scholar]
- 35.Masuda, K., Shima, H., Watanabe, M. & Kikuchi, K. (2001) J. Biol. Chem. 276, 39002–39011. [DOI] [PubMed] [Google Scholar]
- 36.Tanoue, T., Yamamoto, T., Maeda, R. & Nishida, E. (2001) J. Biol. Chem. 276, 26629–26639. [DOI] [PubMed] [Google Scholar]
- 37.Theodosiou, A., Smith, A., Gillieron, C., Arkinstall, S. & Ashworth, A. (1999) Oncogene 18, 6981–6988. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., Blattman, J. N., Kennedy, N. J., Duong, J., Nguyen, T., Wang, Y., Davis, R. J., Greenberg, P. D., Flavell, R. A. & Dong, C. (2004) Nature 430, 793–797. [DOI] [PubMed] [Google Scholar]
- 39.Scarborough, R. M. (2003) Curr. Med. Chem. 1, 73–82. [DOI] [PubMed] [Google Scholar]
- 40.Mercurio, F. & Manning, A. M. (1999) Curr. Opin. Cell Biol. 11, 226–232. [DOI] [PubMed] [Google Scholar]
- 41.Regnier, C. H., Song, H. Y., Gao, X., Goeddel, D. V., Cao, Z. & Rothe, M. (1997) Cell 90, 373–383. [DOI] [PubMed] [Google Scholar]
- 42.Li, Q. & Verma, I. M. (2002) Nat. Rev. Immunol. 2, 725–734. [DOI] [PubMed] [Google Scholar]
- 43.Tzen, C. Y., Cox, R. L. & Scott, R. E. (1994) Exp. Cell Res. 211, 12–16. [DOI] [PubMed] [Google Scholar]
- 44.Hanada, T. & Yoshimura, A. (2002) Cytokine Growth Factor Rev. 13, 413–421. [DOI] [PubMed] [Google Scholar]
- 45.Beyaert, R., Heyninck, K. & Van Huffel, S. (2000) Biochem. Pharmacol. 60, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 46.Cooper, J. T., Stroka, D. M., Brostjan, C., Palmetshofer, A., Bach, F. H. & Ferran, C. (1996) J. Biol. Chem. 271, 18068–18073. [DOI] [PubMed] [Google Scholar]
- 47.Ferran, C., Stroka, D. M., Badrichani, A. Z., Cooper, J. T., Wrighton, C. J., Soares, M., Grey, S. T. & Bach, F. H. (1998) Blood 91, 2249–2258. [PubMed] [Google Scholar]
- 48.He, K. L. & Ting, A. T. (2002) Mol. Cell. Biol. 22, 6034–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krikos, A., Laherty, C. D. & Dixit, V. M. (1992) J. Biol. Chem. 267, 17971–17976. [PubMed] [Google Scholar]
- 50.Sarma, V., Lin, Z., Clark, L., Rust, B. M., Tewari, M., Noelle, R. J. & Dixit, V. M. (1995) J. Biol. Chem. 270, 12343–12346. [DOI] [PubMed] [Google Scholar]
- 51.Lee, E. G., Boone, D. L., Chai, S., Libby, S. L., Chien, M., Lodolce, J. P. & Ma, A. (2000) Science 289, 2350–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henn, V., Slupsky, J. R., Grafe, M., Anagnostopoulos, I., Forster, R., Muller-Berghaus, G. & Kroczek, R. A. (1998) Nature 391, 591–594. [DOI] [PubMed] [Google Scholar]
- 53.Cirino, G., Cicala, C., Bucci, M., Sorrentino, L., Ambrosini, G., DeDominicis, G. & Altieri, D. C. (1997) J. Clin. Invest. 99, 2446–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi, L., Li, Y., Stehno-Bittel, L., Gao, J., Morrison, D. C., Stechschulte, D. J. & Dileepan, K. N. (2001) J. Interferon Cytokine Res. 21, 231–240. [DOI] [PubMed] [Google Scholar]
- 55.Cunningham, M. A., Rondeau, E., Chen, X., Coughlin, S. R., Holdsworth, S. R. & Tipping, P. G. (2000) J. Exp. Med. 191, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindner, J. R., Kahn, M. L., Coughlin, S. R., Sambrano, G. R., Schauble, E., Bernstein, D., Foy, D., Hafezi-Moghadam, A. & Ley, K. (2000) J. Immunol. 165, 6504–6510. [DOI] [PubMed] [Google Scholar]
- 57.Vergnolle, N. (1999) J. Immunol. 163, 5064–5069. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.