Abstract
Metazoan arrestins bind to seven-transmembrane (7TM) receptors to regulate function. Aspergillus nidulans PalF, a protein involved in the fungal ambient pH signaling pathway, contains arrestin N-terminal and C-terminal domains and binds strongly to two different regions within the C-terminal cytoplasmic tail of the 7TM, putative pH sensor PalH. Upon exposure to alkaline ambient pH, PalF is phosphorylated and, like mammalian β-arrestins, ubiquitinated in a signal-dependent and 7TM protein-dependent manner. Substitution in PalF of a highly conserved arrestin N-terminal domain Ser residue prevents PalF-PalH interaction and pH signaling in vivo. Thus, PalF is the first experimentally documented fungal arrestin-related protein, dispelling the notion that arrestins are restricted to animal proteomes. Epistasis analyses demonstrate that PalF posttranslational modification is partially dependent on the 4TM protein PalI but independent of the remaining pH signal transduction pathway proteins PalA, PalB, and PalC, yielding experimental evidence bearing on the order of participation of the six components of the pH signal transduction pathway. Our data strongly implicate PalH as an ambient pH sensor, possibly with the cooperation of PalI.
Keywords: seven-transmembrane receptor, signal transduction, ubiquitination, Aspergillus, endocytosis
Arrestins play key roles in signal transduction through interaction with cytoplasmic domains of seven-transmembrane (7TM) receptors, regulating activity after receptor activation and phosphorylation by specific kinases (1, 2). Of the four known mammalian arrestins, visual and cone arrestins are expressed in photoreceptors, regulating rhodopsin and cone opsins, respectively, whereas ubiquitously expressed β-arrestins 1 and 2 regulate many 7TM receptors. First characterized as blocking signal transduction by preventing interaction between receptor cytoplasmic domains and heterotrimeric G proteins, arrestins are now known to participate in endocytosis and signaling of 7TM receptors, e.g., by linking them to the endocytic internalization machinery (1, 2). Importantly, both ubiquitination and dephosphorylation of β-arrestins appear essential to agonist-dependent receptor endocytosis (3, 4). Although fungal 7TM receptors have been characterized in fungi (e.g., ref. 5), experimentally documented arrestins have only been identified in metazoa.
In the fungus Aspergillus nidulans, alkaline ambient pH results in two-step proteolytic processing activation of the transcription factor PacC, mediating gene regulation by ambient pH (6, 7). pH signal transduction, largely conserved in the fungal kingdom, involves six components: PalA, PalB, PalC, PalF, PalH, and PalI. PalH (Fig. 1 A) is a 7TM protein candidate for ambient pH sensor (8), possibly in cooperation with the 4TM PalI (7). PalA is a BRO1 domain-containing protein, which binds two YPXL/I (X = any amino acid) motifs in PacC and is required for the ambient pH-dependent first PacC proteolytic cleavage, probably catalyzed by the calpain-like cysteine protease PalB (9-11). The roles of PalC and PalF remain elusive (7).
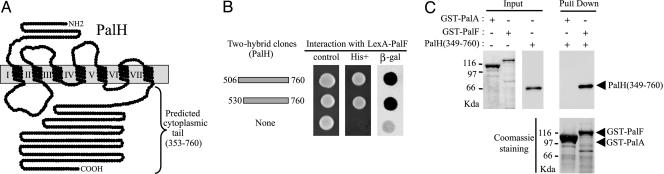
Fig. 1.
Interaction of PalF with the PalH C-terminal cytoplasmic tail. (A) Predicted PalH seven-pass topology and C-terminal cytoplasmic tail. (B) Overlapping fragments of the PalH cytoplasmic tail obtained in two-hybrid screening by using PalF bait. Positive interactions were revealed by growth of S. cerevisiae TAT7 on histidine-free medium containing 5 mM 3-aminotriazole (His+) and β-galactosidase lift filter assays (β-gal). (C) In vitro binding of PalF to the PalH cytoplasmic tail. GST-PalF or GST-PalA was immobilized on glutathione-Sepharose beads and incubated with in-vitro-synthesized [35S]PalH (349-760). Pulled down proteins were electrophoresed on 10% SDS/polyacrylamide and detected by autoradiography (Upper) and Coomassie staining (Lower). Input: purified GST fusion proteins (Coomassie) or [35S]PalH (349-760) (autoradiography) used for binding experiments (20% of the total reaction mixture).
Endocytic trafficking, plasma membrane to endosome, is almost certainly involved in fungal ambient pH signal transduction. PalA and its yeast Rim20 ortholog interact with Vps32 (10, 12), a component of endosomal sorting complex required for transport (ESCRT)-III, one of three multiprotein complexes associating with the membrane of the late endosome to mediate sorting of ubiquitinated cargoes into the multivesicular body pathway (13). In Saccharomyces cerevisiae, many multivesicular body pathway components, including Vps32, are required for pH signaling (12). The links between ambient pH signal reception and pH signaling multivesicular body components are unknown.
Here we show that PalF is a non-metazoan member of the arrestin family and that it interacts with the C-terminal cytoplasmic tail of PalH to mediate pH signaling. Like mammalian β-arrestins, PalF is phosphorylated and ubiquitinated, both posttranslational modifications being regulated in a signal-dependent manner. These data implicate PalF as the possible link between ambient pH sensor(s) and late endosome components.
Materials and Methods
A. nidulans Techniques. A. nidulans strains carried markers in standard use (14). Phenotypic testing of palH mutations followed (15). In pH shift experiments, mycelia were grown for 17 h at 37°C in Aspergillus minimal medium (16) containing 1% (wt/vol) d-glucose and 5 mM ammonium(+)-tartrate, buffered at pH 4 with 50 mM sodium citrate, and shifted for 30 min to the same medium buffered at pH 8.7 with 100 mM Hepes-NaOH. Final pH values before and after the shift were ≈3.8 and 8.4, respectively.
Selection of palF58 and palF60 was described in ref. 15. The palF58 T1084C mutation results in a Ser86Pro substitution. The palF60 T1820A mutation results in Ile331Lys substitution. palF15 (17), used in construction of the palF::HA3 allele by gene replacement, has the T1394G mutation, changing the Leu-189 codon into a TGA stop codon. UV-induced palH420 was selected in a diploid of genotype pabaA1 yA2 aroC660 palH72 camC108; areAr5; pan-toB100/areAr5; inoB2; glrA1; fwA1 as allowing usage of 5 mM γ-aminobutyrate as nitrogen source (see ref. 15). palH420 has the nucleotide sequence change C1186GAG truncating PalH after Leu-368 and out-of-frame encodes RGERSLTVMR before a stop codon (18).
To epitope-tag PalF, a NotI restriction site was engineered after palF codon 185 and used to introduce, as a NotI fragment obtained from pGTEP1 (19), a sequence encoding a triple hemagglutinin (HA) epitope tag. The resulting (HA)3-tagged PalF contains the triple epitope inserted into a region corresponding to the loop connecting β-strands 9 and 10 in bovine visual arrestin (Fig. 2). palF::HA3 was introduced into A. nidulans by replacing the loss-of-function palF15 allele, using direct selection of pal+ transformants at pH 8.5 (8). The homologous recombination event was confirmed by PCR and Southern analysis. PalF-(HA)3 allows normal growth at alkaline pH and WT responses to molybdate and neomycin toxicities (20). pal loss-of-function mutant strains (palH17, palI32, palA1, palB7, and palC4) expressing PalF-(HA)3 were obtained by crossing.
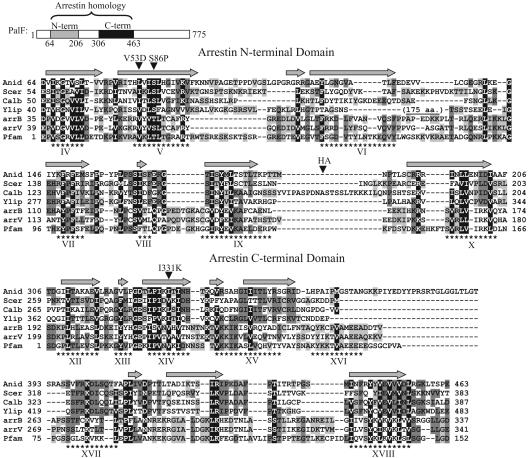
Fig. 2.
Conservation of the PalF family arrestin N-terminal and C-terminal domains. Shown above are positions of the PalF PFAM arrestin N-terminal (shaded bar) and C-terminal (solid bar) domains. The alignment includes amino acid sequences from PalF (Anid, A. nidulans), Rim8 (Scer, S. cerevisiae; Calb, C. albicans; Ylip, Y. lipolytica), human arrestin β1 (arrB), and bovine visual arrestin (arrV) and the consensi for arrestin N-terminal and C-terminal PFAM domains. Conserved (BLOSUM62) residues were shaded according to degree of conservation: black, >90%; dark gray, 50-90%; light gray, 30-50%. Numbers indicate amino acid positions. Dashes indicate gaps. Asterisks indicate positions of β-strands (Roman numerals) in the crystal structure of bovine visual arrestin (27). Gray arrows indicate predicted β-strands in PalF according to the PredictProtein database search engine (www.predictprotein.org). Positions of the Val53Asp (V53D) substitution in arrestin β1, Ser86Pro (S86P), and Ile331Lys (I331K) substitutions in PalF, and the (HA)3 tag in PalF-(HA)3, are indicated.
Sequence Analysis. Amino acid sequence alignments involving arrestin Protein Families Database of Alignments and Hidden Markov Models (PFAM) domains were made with T-Coffee (21). The Uniprot accession nos. for amino acid sequences are P08168 (bovine visual arrestin), P49407 (human β1 arrestin), P78612 and Q9P904 (A. nidulans PalF and PalH, respectively), Q9URQ4 and P48565 (S. cerevisiae Rim8p and Rim21p, respectively), Q9UVF5 and Q9UVF6 (Yarrowia lipolytica Rim8 and Rim21, respectively), and Q9UW13 (Candida albicans Rim8). C. albicans Rim21 was obtained from the Candida Genome Database (available at www.candidagenome.org).
Plasmids. Plasmids encoding GST-PalF or LexA-PalF fusion proteins were constructed by inserting a PCR fragment containing the PalF coding sequence into the EcoRI site of pGEX-2T, pLexA (1-202)+PL (22), or pBTM116 (23). Plasmids encoding GST-PalF mutants carrying Ser86Pro and Ile331Lys PalF substitutions were obtained by mutagenic PCR. A plasmid encoding GST-PalA has been described (10). The construct encoding the PalH cytoplasmic tail tagged with triple Myc [3MYC-PalH (349-760)] was derived from pGBKT7 (Clontech) by inserting a PCR fragment containing the corresponding palH-coding region into its single BamHI site. Plasmids encoding truncated derivatives of LexA-PalF and Gal4 activation domain (GAD)-PalH were constructed by inserting PCR or restriction fragments containing the corresponding palF and palH coding sequences in the polylinker of pLexA (1-202)+PL and pACTII (Clontech), respectively. The palH mutation resulting in the Gly369Ala substitution was obtained by using mutagenic PCR.
Two-Hybrid Analysis. The two-hybrid screen for LexA-PalF-interacting proteins (from a pBTM116 derivative, see above) was carried out in S. cerevisiae TAT7 (MATα ade2-101 his3- Δ200 leu2-Δ1 trp1-Δ901gal4 gal80 LYS2::lexAop-HIS3 URA3::lexAop-lacZ) cotransformed with an A. nidulans cDNA library ligated into pAD-GAL4 (from Stephen Osmani, Ohio State University, Columbus, OH). His+ transformants were selected in the presence of 5 mM 3-aminotriazole and subsequently screened for β-galactosidase activity by using a filter assay (24). Two-hybrid mapping analysis of PalF and PalH interaction domains was carried out in CTY10.5d (Matα ade2-101 his3-Δ200 leu2-Δ1 trp1-Δ901 gal4 gal80 URA3::lexAop-lacZ) with pLexA (1-202)+PL and pACT2 plasmid derivatives. β-galactosidase was quantitatively assayed in permeabilized yeast cells grown to mid-log phase in selective SD (synthetic dextrose) medium (25) and expressed in Miller units (26) as the average of four independent transformants. SEs were <17%. Similar levels of expression for LexA-PalF and LexA-PalF (212-775) were determined by immunoblotting by analysis using anti-LexA rabbit antiserum (Invitrogen) and for GAD-PalH (349-530)G369A and GAD-PalH (349-530) using anti-HA Ab (Roche).
Pull-Down Assays. Recombinant GST-PalF, GST-PaF(S86P), GST-PalF(I331K), and GST-PalA were purified from Escherichia coli DH1 cells as described (10). [35S]-3MYC-tagged PalH (349-760)] was synthesized in vitro by using the Promega TNT system in the presence of [35S]methionine [1,000 Ci/mmol (1 Ci = 37 GBq)]. Sepharose beads loaded with GST fusion proteins were incubated with 5 ml of labeling reaction mixture for 1 h at 4°C in 500 μl of STE buffer (10 mM Tris·HCl, pH 8.0/1 mM EDTA/150 mM NaCl) with 1% (vol/vol) Triton X-100, washed extensively, and boiled in sample buffer. Bound proteins were separated by 7.5% SDS/PAGE and detected by autoradiography (labeled preys) or Coomassie staining (baits).
Immunoblots. Lyophilized A. nidulans mycelia (0.3 g) were ground with a 4-mm ceramic bead for 15 s in a FastPrep apparatus (power setting 4, model BIO101, (Q-BIOgene, Irvine, CA) and resuspended in 1.7 ml of extraction buffer (50 mM Hepes, pH 7.5/150 mM NaCl/0.1% Triton X-100/1 mM DTT/10% glycerol) containing Complete protease inhibitor mixture (Roche) and 5 mM N-ethylmaleimide. Protein extracts were prepared by glass bead homogenization (20 s in a FastPrep apparatus at power setting 6) and clarified after microcentrifugation at 14,000 rpm and 4°C for 30 min in an Eppendorf microcentrifuge. PalF-(HA)3 was immunoprecipitated from crude extracts (4 mg) by incubation with 20 μl of anti-HA affinity matrix (Roche) for 1 h on a rotating wheel and immunoblotted by using anti-HA at 1:1,000 (3F10, Roche) or anti-ubiquitin at 1:1,000 (P4D1, Cell Signaling Technology, Beverly, MA) Ab, peroxidase-coupled secondary Ab, and enhanced chemiluminescence (ECL-plus, Amersham Pharmacia). The λ phosphatase treatment of PalF-(HA)3 immunoprecipitates was carried out for 30 min at 37°C in 100-μl reaction mixtures containing 2 mM MnCl2 and 400 milliunits of λ phosphatase (Biolabs, North-brook, IL). Where indicated, 10 mM Na3VO4 was used as phosphatase inhibitor.
Results
PalF Binds to the C-Terminal Cytoplasmic Tail of the 7TM Protein PalH. Using a LexA-PalF fusion protein bait to screen an A. nidulans two-hybrid cDNA library, we obtained, among 2 × 106 colonies screened, three strong interactors, all containing PalH C-terminal cytoplasmic tail codons 506-760 as well as two slightly weaker interactors containing PalH codons 530-760 (Fig. 1B). Binding of PalF to PalH is direct, as shown by GST pull-down assays by using purified bacterially expressed GST-fusion proteins as baits and in vitro synthesized [35S]-labeled PalH (349-760) as prey (Fig. 1C). In contrast to the efficient GST-PalF bait, a control GST-PalA fusion was ineffective, demonstrating that the PalF-PalH interaction is specific.
PalF Is a Fungal Arrestin-Related Protein. Although previous sequence analyses of PalF family members failed to reveal significant similarity to proteins of known function (7), PFAM database searches revealed the presence of arrestin N-terminal (PFAM 00339) and C-terminal (PFAM 02752) domains in PalF (E = 2.3e-0.6 and 5.7e-18, respectively) (Fig. 2A) and its yeast Rim8 orthologs. Sequence similarity (Fig. 2) is restricted to short stretches of amino acid sequence, mostly corresponding to β-strands in visual arrestin crystal structures (27, 28) and in the predicted secondary structure of PalF. PalF family member regions corresponding to interconnecting loops in arrestins are very divergent in length and sequence, including an ≈175-residue insertion in Y. lipolytica Rim8 (Fig. 2). Together with PalF-PalH interaction data, this sequence similarity strongly supports a structural relationship between PalF family members and mammalian arrestins and thus designation of PalF as a fungal arrestin-related protein.
The PalF-PalH Interaction Involves the Arrestin N-Domain Containing Moiety of PalF and Two Regions of the PalH Cytoplasmic Tail. Analysis of truncated and chimeric mammalian arrestins showed that the N-domain is directly involved in 7TM receptor binding, although additional determinants in the C-domain participate in receptor recognition (29, 30). Two-hybrid mapping showed that the PalF PFAM arrestin N-domain plus 63 additional N-terminal residues are sufficient for PalH interaction whereas a region containing the PFAM arrestin C-domain has no detectable binding activity (Fig. 3A). In contrast, two regions of the PalH cytoplasmic tail (residues 349-385 and 657-760) are able to interact with PalF (Fig. 3B). The more N-terminal 37-residue region, beginning shortly before the end of the seventh predicted transmembrane helix shows significant conservation between PalH, its yeast Rim21 orthologs, and the likely Rim21 paralog Dfg16 (31) within their otherwise highly divergent C-terminal moieties (Fig. 3C and data not shown). Substitution of the completely conserved Gly-369 by Ala, although not affecting immunoblot-determined GAD-PalH (349-530) levels (data not shown), abolished the two-hybrid interaction (Fig. 3B).
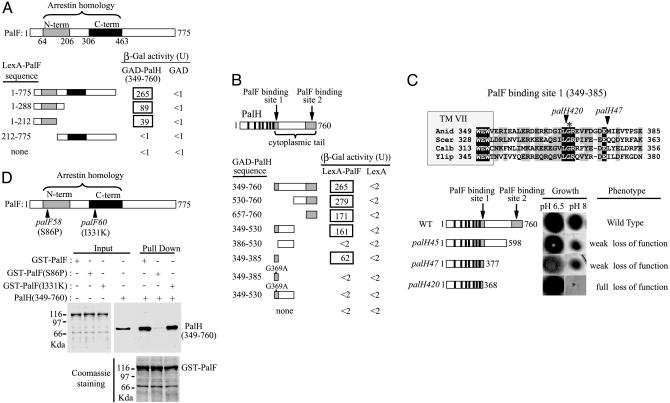
Fig. 3.
Mutational analysis of the interaction between PalF and PalH. (A and B) Two-hybrid mapping of the PalF- and PalH-interacting domains. (A) LexA-PalF fusions containing indicated PalF residues were tested for interaction with GAD-PalH (349-760) or GAD alone. N-term (shaded bar) and C-term (solid bar) indicate PalF PFAM arrestin N-terminal and C-terminal domains, respectively. (B) GAD-PalH fusions containing indicated PalH residues were tested for interaction with LexA-PalF or LexA alone. Solid bars indicate the positions of PalH-predicted transmembrane helix (8), and shaded bars indicate the two PalF-binding sites. (C) Phenotypic analysis of palH mutations affecting the PalF-binding sites. The sequence alignment including A. nidulans PalH and S. cerevisiae, Y. lipolytica, and C. albicans Rim21 shows the PalF-binding site located immediately downstream of the seventh PalH TM helix (TM VII). (Upper) Fully conserved (black shading) or similar (BLOSUM62, gray shading) residues are shown. Arrowheads indicate positions of the truncation sites in palH420 and palH47 mutant proteins. Fully conserved PalH Gly-369, crucial for PalF-PalH interaction, is denoted by an asterisk. Numbers at the ends indicate residue positions. (Lower Right) A phenotypic analysis of palH mutant strains (8, 18) grown on synthetic media at the indicated pH values. (Lower Left) Schematic representation of truncated proteins. Filled boxes and solid bars are as in B. Numbers on the right show the final in-frame encoded residues of the truncated proteins. (D) In vitro binding of palF58 (S86P) and palF60 (I331K) proteins to the C-terminal region of PalH. Pull-down assays were as in Fig. 1.
The Interaction of PalF with PalH Mediates pH Signaling In Vivo. Mutant phenotypes confirm the physiological importance of the PalF-PalH interaction. Mutations abolishing pH signal transduction lead to, among other phenotypic features, inability to grow at alkaline pH (reviewed in ref. 7). As reported (8), PalH truncation after residues 598 (palH45) or 377 (palH47) partially impairs growth at pH 8 (Fig. 3C). These mutations remove the C-terminal PalF-binding site (657-760) but leave fully or mainly intact the N-terminal site (349-385) (Fig. 3C). PalH truncation after residue 368 (palH420), nearly halving the N-terminal site and removing the crucial Gly-369, completely prevents growth at pH 8 (Fig. 3C). Substitution of the highly conserved Ser-86 by Pro (Fig. 2) in the loss-of-function palF58 mutant almost completely prevents PalF-PalH interaction in vitro (Fig. 3D), underlining the physiological importance of the interaction and the PalF arrestin N-domain in enabling it. Substitution of the highly conserved Ile-331 residue in the arrestin C-domain (Fig. 2) by Lys (palF60) did not affect in vitro interaction (Fig. 3D), suggesting that its loss-of-function phenotype is unrelated to the interaction.
PalF Is Phosphorylated and Ubiquitinated in Response to Alkaline pH in a PalH- and PalI-Dependent Manner. Because the function of mammalian β-arrestins in agonist-dependent receptor endocytosis is regulated through phosphorylation and ubiquitination (3, 4), a phenotypically WT (data not shown) epitope (HA)3 tagged, gene replacement palF allele was used to monitor PalF posttranslational modification. Anti-HA Western blot analysis of protein extracts prepared from mycelial cells after mechanical cell wall and membrane disruption detected two immunoreactive bands in acidic grown WT (Fig. 4A, lane 1), which were absent in untagged cell extracts (data not shown). Inspection of the palF 5′-UTR and coding regions indicates that alternative translation initiation sites cannot be responsible. We determined that the faster, less prominent band is almost undetectable when crude protoplast extracts were analyzed (data not shown), indicating that this faster band represents a degradation product. Shifting acidic-grown mycelia to alkaline pH resulted in decreased mobility of both bands (Fig. 4A, lane 2), attributable to phosphorylation as it is reversed by λ phosphatase, as shown by comigration of dephosphorylated species with those preceding alkaline pH activation (Fig. 4B). Alkaline pH-mediated phosphorylation of PalF was abolished by a palH- mutation (Fig. 4A, lanes 3 and 4).
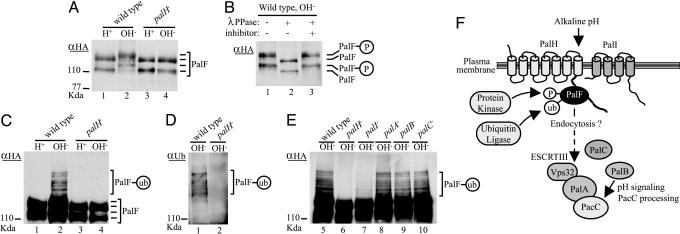
Fig. 4.
PalH- and alkaline ambient pH-dependent phosphorylation and ubiquitination of PalF: A model for pH signaling in A. nidulans. (A-E) WT and indicated pal mutants expressing PalF-(HA)3 were grown under acidic conditions (H+) and shifted for 30 min to alkaline conditions (OH-). Total (A) or anti-(α)-HA-immunoprecipitated (B-E) protein extracts were analyzed by Western blotting. Positions of the different PalF forms (P, phosphorylated; ub, ubiquitinated) are indicated. (A) Immunoblot of extracts with anti-HA Ab. (B) Anti-HA immunoprecipitates from the WT shifted to alkaline conditions were treated with λ phosphatase (λPPase), with or without phosphatase inhibitor. (C and E) Anti-HA immunoprecipitates were immunoblotted with anti-HA Ab. (D) Anti-HA immunoprecipitates were immunoblotted with anti-ubiquitin Ab. pH signaling is impaired in all pal strains, as determined by deficient PacC proteolytic processing at alkaline pH (data not shown). (F) Model for pH signaling in A. nidulans. The predicted 7TM PalH would be a pH sensor activating the signaling pathway possibly with assistance of 4TM protein PalI, in agreement with the accessory role suggested by partial impairment of pH signaling in palI-null mutants (ref. 7 and this work). PalF interacts with the PalH cytoplasmic tail and is phosphorylated and ubiquitinated at alkaline pH by as-yet-unidentified protein kinase and ubiquitin ligase enzymes in a PalH-dependent and partially PalI-dependent manner. Based on the role that β-arrestin dephosphorylation and ubiquitination play in receptor-mediated endocytosis, we propose that PalF phosphorylation/ubiquitination promotes internalization of PalF-PalH complexes to transduce the pH signal to the PalA/PalB PacC-processing machinery acting downstream of PalF (this work). PalA/Rim20 interacts with PacC/Rim101 to mediate its signaling proteolytic cleavage, presumably by the cysteine protease PalB/Rim13 (9-11), and with Vps32, a component of the ESCRT complexes transiently associating to endosomal membranes (10, 12).
Anti-HA immunoblot analysis of PalF-(HA)3 immunoprecipitates from pal+ extracts revealed, after deliberate overexposure, additional PalF forms with lower mobility, migrating as a ladder (Fig. 4C, lane 2), which are still present after λ phosphatase treatment (data not shown). As with phosphorylation, these additional modifications occurred after shifting cells from acidic to alkaline pH and were abolished by a palH- mutation (Fig. 4C). The relative proportions of these modified PalF species and their ladder-like pattern are strongly suggestive of ubiquitination, which was confirmed by immunoblot analysis of PalF-(HA)3 immunoprecipitates with anti-ubiquitin (P4D1) Ab (Fig. 4D). Taking into account the two phosphorylated forms, this pattern would be consistent with addition of one to three ubiquitin moieties.
In contrast to its absolute requirement for PalH function, PalF ubiquitination is partially independent of PalI and completely independent of PalA, PalB, and PalC (Fig. 4E). A similar pattern of dependence was observed for PalF phosphorylation (data not shown).
Discussion
Although the presence of PFAM arrestin domain-containing proteins in fungi was uncovered by genomic sequencing projects, the physiological role of these proteins had not been experimentally documented. Here we show that PalF is a non-metazoan arrestin-related protein, as verified by (i) the presence of arrestin N- and C-terminal PFAM domains; (ii) sequence similarity to mammalian arrestins in the β-strands forming the two seven-stranded β sandwiches, suggesting structural similarity; (iii) PalF, like arrestins, interacts with a 7TM protein, the putative pH sensor PalH, in two-hybrid and pull-down assays, and this interaction is required for pH signaling in vivo; and (iv), like mammalian β-arrestins, the phosphorylation and ubiquitination status of PalF is signal (ambient pH) and 7TM protein (PalH)-dependent, strongly suggesting similarity of regulatory mechanisms. In view of the role that dephosphorylation and ubiquitination of β-arrestins play in mammalian receptor-mediated endocytosis, these findings are consistent with the current model involving endocytic trafficking in fungal pH signal transduction (7, 12).
The loss-of-function Ser86Pro substitution in the PalF arrestin N-domain strongly impairs PalF-PalH binding and occurs within the PalF family and mammalian arrestin highly conserved (L/I/V)X(L/I/V)(S/T)(L/F) motif within arrestin β-strand V (Fig. 2). Similarly the Val53Asp mutant β-arrestin 1, affecting the first residue of this motif (Fig. 2), is impaired in interaction with mammalian 7TM receptors (32), consistent with the essential role of β-strand V in receptor binding (30, 33) and suggesting conservation of the structural basis with the PalF-PalH interaction. However, just as the strongest PalF-PalH interaction was obtained with the full-length PalF protein (Fig. 3A), there is evidence that determinants in arrestins other than the N-domain might be involved in receptor binding (29, 30). The cooperative involvement of the N-terminal and C-terminal PalF-binding sites in the PalH cytoplasmic tail bears comparison with the involvement of two discrete regions at the N-terminal and C-terminal ends of the large third cytoplasmic loop of α2-adrenergic receptors in β-arrestin 2 binding (34). Relatively few studies have addressed the 7TM receptor molecular determinants required for arrestin binding but a β-arrestin binding site also appears to be located at the C-terminal end of the chemokine receptor D6 cytoplasmic tail (35). Thus PalF and β-arrestins might preferentially recognize 7TM protein cytoplasmic domains near transmembrane domains and/or near cytoplasmic tail C termini.
The 7TM receptors usually signal to downstream heterotrimeric G proteins. Although we cannot exclude the possibility that a G protein mutant might have a very pleiotropic phenotype, precluding selection or recognition in pH mutant screens, the failure of extensive genetic screens carried out for the pal pathway in A. nidulans and for the homologous Rim pathways in S. cerevisiae and Y. lipolytica to reveal involvement of G-proteins in pH regulation suggests that these proteins are not required for ambient pH signaling.
The positive role of PalF in pH signal transduction and its mediation by interaction with PalH, together with the apparent absence of G protein involvement in pH signaling, strongly suggests that PalF, contrary to mammalian arrestins, does not play a role in receptor desensitization. However, evidence suggests that the signaling function of PalF is related to endocytic trafficking: (i) fungal pH signaling involves most of the components of the endosomal ESCRT complexes (12), and (ii) PalF is phosphorylated and ubiquitinated in a pH- and PalH-dependent manner, thus resembling many yeast plasma membrane proteins whose endocytosis requires phosphorylation and subsequent ubiquitination (36). The effect of pal mutations on PalF phosphorylation and ubiquitination indicates that PalH and PalI act upstream in the signaling pathway whereas PalA, PalB, and PalC act downstream, which, together with the involvement of endosome ESCRT complexes in pH signaling (12), leads to the model in Fig. 4F.
Despite the mechanistic novelty of the fungal pH signal transduction pathway, this work strengthens the possibility that this pathway and Hedgehog signaling in metazoa share common ancestry (37). Both pathways involve 7TM proteins (PalH and Smoothened), apparently unrelated in amino acid sequence, without compelling evidence for G protein involvement (38). Signaling is mediated by arrestins (PalF and β-arrestin 2) (this work and refs. 39 and 40) and apparently involves endocytic trafficking to the late endosome (12, 41). Finally, signaling in both cases leads to proteolytic processing of the transcription factors (PacC and Cubitus interruptus/Gli), which appear related evolutionarily in their zinc finger DNA-binding domains (37).
Acknowledgments
We thank Joan Tilburn (Imperial College) and Eduardo Espeso (Consejo Superior de Investigaciones Científicas) for helpful discussions and useful materials, E. Reoyo and L. Stanton for technical assistance, and Steve Osmani for his generous gift of the A. nidulans two-hybrid library. This work was supported by Comision Interministerial de Ciencia y Tecnologia Grants BIO2002-00803 (to O.V.) and BIO2003-0077 (to M.A.P.) and Wellcome Trust Grant 067878 (to H.N.A.). O.V. is currently supported by the Ministerio de Ciencia y Tecnología of Spain Ramón y Cajal Program. S.H. held a Plan de Formación de Personal Investigador Studentship.
Author contributions: O.V. designed research; S.H., J.M.R., H.-J.B., H.N.A., and O.V. performed research; J.C.S.-F. contributed new reagents/analytical tools; S.H., J.M.R., H.N.A., M.A.P., and O.V. analyzed data; and H.N.A., M.A.P., and O.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PFAM, Protein Families Database of Alignments and Hidden Markov Models; HA, hemagglutinin; GAD, Gal4 activation domain; 7TM, seven-transmembrane; ESCRT, endosomal sorting complexes required for transport.
References
- 1.Shenoy, S. K. & Lefkowitz, R. J. (2003) Biochem. J. 375, 503-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefkowitz, R. J. & Shenoy, S. K. (2005) Science 308, 512-517. [DOI] [PubMed] [Google Scholar]
- 3.Lin, F. T., Krueger, K. M., Kendall, H. E., Daaka, Y., Fredericks, Z. L., Pitcher, J. A. & Lefkowitz, R. J. (1997) J. Biol. Chem. 272, 31051-31057. [DOI] [PubMed] [Google Scholar]
- 4.Shenoy, S. K., McDonald, P. H., Kohout, T. A. & Lefkowitz, R. J. (2001) Science 294, 1307-1313. [DOI] [PubMed] [Google Scholar]
- 5.Versele, M., Lemaire, K. & Thevelein, J. M. (2001) EMBO Rep. 2, 574-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orejas, M., Espeso, E. A., Tilburn, J., Sarkar, S., Arst, H. N., Jr. & Peñalva, M. A. (1995) Genes Dev. 9, 1622-1632. [DOI] [PubMed] [Google Scholar]
- 7.Peñalva, M. A. & Arst, H. N., Jr. (2004) Ann. Rev. Microbiol. 58, 425-451. [DOI] [PubMed] [Google Scholar]
- 8.Negrete-Urtasun, S., Reiter, W., Díez, E., Denison, S. H., Tilburn, J., Espeso, E. A., Peñalva, M. A. & Arst, H. N., Jr. (1999) Mol. Microbiol. 33, 994-1003. [DOI] [PubMed] [Google Scholar]
- 9.Díez, E., Álvaro, J., Espeso, E. A., Rainbow, L., Suárez, T., Tilburn, J., Arst, H. N., Jr. & Peñalva, M. A. (2002) EMBO J. 21, 1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent, O., Rainbow, L., Tilburn, J., Arst, H. N., Jr. & Peñalva, M. A. (2003) Mol. Cell. Biol. 23, 1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, W. & Mitchell, A. P. (2001) J. Bacteriol. 183, 6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu, W., Smith, F. J., Jr., Subaran, R. & Mitchell, A. P. (2004) Mol. Biol. Cell 15, 5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell. Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- 14.Clutterbuck, A. J. (1993) in Genetic Maps: Locus Maps of Complex Genomes, ed. O'Brien, S. J. (Cold Spring Harbor Lab. Press, Plainview, NY), pp 3.71-3.84.
- 15.Arst, H. N., Jr., Bignell, E. & Tilburn, J. (1994) Mol. Gen. Genet. 245, 787-790. [DOI] [PubMed] [Google Scholar]
- 16.Cove, D. J. (1966) Biochim. Biophys. Acta 113, 51-56. [DOI] [PubMed] [Google Scholar]
- 17.Dorn, G. (1965) Genet. Res. 6, 13-26. [DOI] [PubMed] [Google Scholar]
- 18.Rudnicka, J. D. (2004) Ph.D. thesis (University of London, London).
- 19.Tyers, M., Tokiwa, G., Nash, R. & Futcher, B. (1992) EMBO J. 11, 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caddick, M. X., Brownlee, A. G. & Arst, H. N., Jr. (1986) Mol. Gen. Genet. 203, 346-353. [DOI] [PubMed] [Google Scholar]
- 21.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205-217. [DOI] [PubMed] [Google Scholar]
- 22.Ruden, D. M., Ma, J., Li, Y., Wood, K. & Ptashne, M. (1991) Nature 350, 250-252. [DOI] [PubMed] [Google Scholar]
- 23.Fields, S. & Song, O. (1989) Nature 340, 245-246. [DOI] [PubMed] [Google Scholar]
- 24.Yang, X., Hubbard, E. J. & Carlson, M. (1992) Science 257, 680-682. [DOI] [PubMed] [Google Scholar]
- 25.Rose, M. D., Winston, F. & Hieter, P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 26.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 27.Hirsch, J. A., Schubert, C., Gurevich, V. V. & Sigler, P. B. (1999) Cell 97, 257-269. [DOI] [PubMed] [Google Scholar]
- 28.Granzin, J., Wilden, U., Choe, H. W., Labahn, J., Krafft, B. & Buldt, G. (1998) Nature 391, 918-921. [DOI] [PubMed] [Google Scholar]
- 29.Gurevich, V. V., Dion, S. B., Onorato, J. J., Ptasienski, J., Kim, C. M., Sterne-Marr, R., Hosey, M. M. & Benovic, J. L. (1995) J. Biol. Chem. 270, 720-731. [DOI] [PubMed] [Google Scholar]
- 30.Vishnivetskiy, S. A., Hosey, M. M., Benovic, J. L. & Gurevich, V. V. (2004) J. Biol. Chem. 279, 1262-1268. [DOI] [PubMed] [Google Scholar]
- 31.Barwell, K. J., Boysen, J. H., Xu, W. & Mitchell, A. P. (2005) Eukaryotic Cell 4, 890-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krupnick, J. G., Santini, F., Gagnon, A. W., Keen, J. H. & Benovic, J. L. (1997) J. Biol. Chem. 272, 32507-32512. [DOI] [PubMed] [Google Scholar]
- 33.Han, M., Gurevich, V. V., Vishnivetskiy, S. A., Sigler, P. B. & Schubert, C. (2001) Structure (London) 9, 869-880. [DOI] [PubMed] [Google Scholar]
- 34.DeGraff, J. L., Gurevich, V. V. & Benovic, J. L. (2002) J. Biol. Chem. 277, 43247-43252. [DOI] [PubMed] [Google Scholar]
- 35.Galliera, E., Jala, V. R., Trent, J. O., Bonecchi, R., Signorelli, P., Lefkowitz, R. J., Mantovani, A., Locati, M. & Haribabu, B. (2004) J. Biol. Chem. 279, 25590-25597. [DOI] [PubMed] [Google Scholar]
- 36.Hicke, L. (1999) Trends Cell Biol. 9, 107-112. [DOI] [PubMed] [Google Scholar]
- 37.Arst, H. N., Jr. & Peñalva, M. A. (2003) Trends Genet. 19, 224-231. [DOI] [PubMed] [Google Scholar]
- 38.Kalderon, D. (2005) Curr. Biol. 15, R175-R178. [DOI] [PubMed] [Google Scholar]
- 39.Chen, W., Ren, X. R., Nelson, C. D., Barak, L. S., Chen, J. K., Beachy, P. A., de Sauvage, F. & Lefkowitz, R. J. (2004) Science 306, 2257-2260. [DOI] [PubMed] [Google Scholar]
- 40.Wilbanks, A. M., Fralish, G. B., Kirby, M. L., Barak, L. S., Li, Y. X. & Caron, M. G. (2004) Science 306, 2264-2267. [DOI] [PubMed] [Google Scholar]
- 41.Incardona, J. P., Gruenberg, J. & Roelink, H. (2002) Curr. Biol. 12, 983-995. [DOI] [PubMed] [Google Scholar]