Abstract
Most HIV transmission occurs on the mucosal surfaces of the gastrointestinal and cervicovaginal tracts, both of which are normally coated by a biofilm of nonpathogenic commensal bacteria. We propose to genetically engineer such naturally occurring bacteria to protect against HIV infection by secreting antiviral peptides. Here we describe the development and characterization of Nissle 1917, a highly colonizing probiotic strain of Escherichia coli, secreting HIV-gp41-hemolysin A hybrid peptides that block HIV fusion and entry into target cells. By using an appropriate combination of cis- and transacting secretory and regulatory signals, micromolar secretion levels of the anti-HIV peptides were achieved. The genetically engineered Nissle 1917 were capable of colonizing mice for periods of weeks to months, predominantly in the colon and cecum, with lower concentrations of bacteria present in the rectum, vagina, and small intestine. Histological and immunocytochemical examination of the colon revealed bacterial growth and peptide secretion throughout the luminal mucosa and in association with epithelial surfaces. The use of genetically engineered live microbes as anti-HIV microbicides has important potential advantages in economy, efficacy, and durability.
Keywords: bacterial, microbicide, Nissle 1917, AIDS, prevention
Every day, ≈14,000 people, most of whom live in resource-poor environments, are infected by HIV. Over 80% of these infections occur by the mucosal route during unprotected rectal or vaginal sexual intercourse when HIV enters the body through the surfaces lining the rectum, colon, vagina, or cervix (1). Each of these surfaces is normally populated by a rich and varied commensal microflora that play important roles in host physiology, immune development, and defense against pathogenic bacteria, fungi, and viruses (2, 3).
We propose to genetically engineer such naturally occurring bacteria to provide protection against HIV by secreting compounds that interfere with viral attachment, fusion, entry, or replication. Introduction of such genetically modified organisms under conditions in which they colonized the mucosa and became a stable component of the resident microbiota of uninfected individuals would provide a long-lasting bioshield against HIV.
Previously, a similar strategy was used by Beninati et al. (4) to protect rats against Candida albicans by vaginal colonization with Streptococcus gordonii genetically engineered to secrete a single-chain antiidiotype antibody. Similarly, Kruger et al. (5) showed that Lactobacillus zeae expressing a single-chain antibody against Streptococcus mutans reduced bacterial growth and pathology in a rat model of dental caries. With regard to anti-HIV bacteria, Giomerelli et al. (6) genetically engineered S. gordonii, a member of the normal microbial flora of the human oral cavity that also colonizes the vaginal mucosa of experimental animals to produce cyanovirin-N, a potent HIV-inactivating protein originally isolated from a cyanobacterium. Recently Chang and colleagues (7) described the modification of a natural human vaginal isolate of Lactobacillus jensenii to secrete two-domain CD4 and showed that it inhibited HIV entry into target cells in a dose-dependent manner in vitro.
For the live microbial microbicide strategy to succeed, it is necessary to demonstrate that genetically engineered bacteria can (i) synthesize and secrete sufficient quantities of potent and broadly active HIV inhibitory compounds to block infection by diverse HIV primary isolates; (ii) compete with indigenous microbes for prolonged colonization of mucosal surfaces in the gastrointestinal or the reproductive tract; and (iii) not cause any pathology of the colonized organ. Here we tested the feasibility of this approach by genetically engineering Nissle 1917, a highly colonizing probiotic strain of Eschericia coli, to secrete a potent HIV fusion inhibitor peptide. Our data demonstrate that the resulting bacteria are capable of nonpathogenically colonizing the large intestine of mice and secreting inhibitory concentrations of the anti-HIV peptide onto mucosal surfaces of the gastrointestinal tract in intact animals.
Materials and Methods
To construct the C52-HlyA218 (Hly, hemolysin) expression plasmid, the region of HIVNL4.3-encoding amino acids 114–162 of gp41 was cloned into pEHLYA2-SD, a high-copy-number ampicillin resistance (Apr) plasmid that contains the lac operon promoter, a Shine–Dalgarno sequence, multiple cloning sites, and an epitope tag (Etag) in frame with the C-terminal 218 aa of HlyA (8). The recombinant plasmid was contransformed with the low copy number chloramphenicol resistance (Cmr) plasmid pVDL9.3 (9), which provides the HlyB and HlyD transporters in trans, into Escherichia coli Nissle 1917 (obtained from a commercial preparation of the probiotic Mutaflor from Ardeypharm, Herdecke, Germany). The gene aadA1 was introduced into Nissle 1917 by phage P1 transduction. Other strains and plasmids were obtained by standard methods (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
For peptide expression, Nissle 1917 transformants were grown at 37°C in LB liquid medium containing chloramphenicol and ampicillin to stationar y phase without isopropyl β-d-thiogalactoside (IPTG) or to late log phase with IPTG as indicated. Culture supernatants were analyzed by gel electrophoresis, Western blotting, and complex formation with purified 5-helix protein. The ability of the secreted proteins to block HIV infection in peripheral blood mononuclear cells was tested by using HIV–GFP reporter virus pseudovirions encapsidated with HIVNL4.3, HIVJR-CSF, or murine leukemia virus (MuLV) envelope and flow cytometry (Supporting Materials and Methods).
Animal experiments were conducted in the Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility of the Vaccine Research Center using procedures, protocols, and personnel approved by the Animal Care and Use Committee. For mouse colonization studies, 4- to 8-week-old CD-1 mice (Charles River Laboratories) were fed ampicillin (800 mg/liter) in drinking water for 1 day before and during the study if indicated, then orally or rectally inoculated with 5 × 108 bacteria in 200 μl of PBS. Fresh fecal samples were collected at intervals and assayed for Nissle 1917 strains by plating on Luria agar containing ampicillin and chloramphenicol. The identity of the antibiotic-resistant colonies recovered from feces and tissues was confirmed by analysis of secreted peptides and plasmid DNA. For tissue analysis, samples from duodenum, jejunum, ileum, cecum, colon, rectum, and vagina were isolated from euthanized animals and analyzed by microbiological plating, histopathology, and immunohistochemical staining with the monoclonal antibody 2F5 (obtained from P. Kwong, National Institutes of Health), which recognizes the epitope ELDKWA contained in the C-terminal region of gp41 (10) (Supporting Materials and Methods).
Results
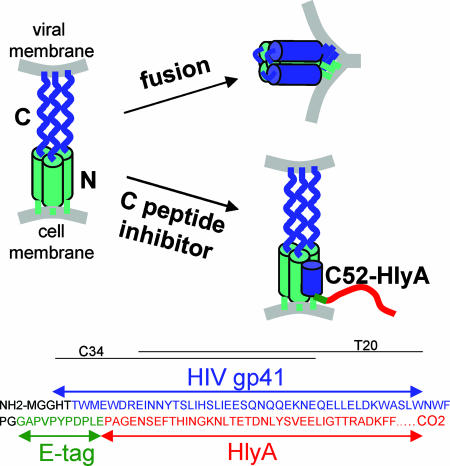
Experimental Design: Choice of Inhibitory Peptide, Secretion System, and Bacterial Strain. Our method depends on the use of a potent anti-HIV peptide, an efficient secretory system, and a highly colonizing yet nonpathogenic bacterial host. As inhibitor, we choose a 52-aa sequence (C52) derived from the C-terminal heptad repeat region of gp41, the transmembrane subunit of the HIV envelope that catalyzes receptor-mediated membrane fusion (11). As diagrammed in Fig. 1, C52 can bind to the N-terminal heptad repeat region of gp41, thereby preventing the formation of the trimer-of-hairpins structure that is essential for membrane fusion and viral entry. HIV C-peptides are potent entry inhibitors that are highly conserved between different isolates, do not require disulfide bonding or other posttranslational modifications, and are active against direct and trans infection of multiple primary cell types (12, 13). One C-peptide, T-20 (enfuvirtide), is currently in clinical use as a salvage antiretroviral therapy (14).
Fig. 1.
Fusion inhibitor design. (Upper) The mechanism of membrane fusion and C-peptide inhibition. In the prehairpin intermediate shown on the left, the C-terminal region of gp41 is anchored in the viral membrane, and the N-terminal region is inserted into the host cell membrane. In the absence of inhibitor, this transient state collapses into a trimer-of-hairpins that brings the N- and C-terminal regions into proximity, promoting fusion of the viral and cellular membranes. In the presence of a C-peptide, fusion is inhibited by binding of the peptide to the exposed N-terminal region of gp41, thereby preventing formation of the trimer-of-hairpins. (Lower) The structure of the C52-HlyA218 peptide and the locations of the C-peptides T20 and C34.
For secretion, we used the E. coli Hly system because it allows direct export from the bacterial cytoplasm into the extracellular medium without a periplasmic intermediate (15, 16). The protein machinery of the Hly type I secretory apparatus consists of two operon-specific inner membrane components, HlyB and HlyD, and the chromosomally encoded outer membrane protein, TolC, which form a protein channel between the inner and outer membranes. The HlyB–HlyD complex recognizes the C-terminal portion of the Hly enzyme (HlyA), thereby allowing the secretion of polypeptides fused to this signal sequence (8, 9).
As host we used Nissle 1917, a commensal strain of E. coli that was isolated in 1917 from the stool of a German soldier who survived an outbreak of enterocolitis. Nissle 1917 is an excellent colonizer in mice and humans and has been detected in infants up to 6 months after initial administration (17–21). Analysis of the genome structure of Nissle 1917 reveals the presence of numerous fitness factors, including multiple iron uptake systems, proteases, bacteriocins, fimbriae, and other adhesions (22). This strain does not produce any known virulence factors and is known to be safe for humans because it has been widely used as a probiotic treatment for intestinal disorders, such as diarrhea, irritable bowel disease, ulcerative colitis, and Crohn's disease (23–26).
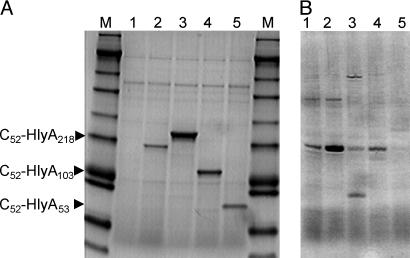
Secretion of Anti-HIV Peptides by Nissle 1917. The HIV C52 peptide coding sequences were cloned into a high-copy-number Apr expression vector containing an Etag linker and the C-terminal 218 aa from HlyA, and the resulting plasmid was introduced into Nissle 1917 expressing the HlyB and HlyD transporters from a low-copy-number Cmr episome. Growth of the resulting Apr Cmr strain in rich medium led to the accumulation of the 32 kDa C52-HlyA218 fusion peptide as the predominant extracellular protein in the culture supernatant (Fig. 2A). The identity of this protein was confirmed by positive Western blot reactions with the anti-C-peptide antibody 2F5 (10) and an anti-Etag antibody and by its ability to form a complex with HIV 5-helix protein, which contains a high-affinity binding site for C-peptides (data not shown) (27).
Fig. 2.
Secretion of anti-HIV peptides by bacteria. (A) Coomassie blue-stained SDS gel of supernatants of log-phase IPTG-induced cultures of E. coli Nissle 1917 transformed with HlyB,HlyD plasmid pVDL9.3 plus no additional plasmid (lane 1), Etag-HlyA218 (lane 2), C52-HlyA218 (lane 3), C52-HlyA103 (lane 4), and C52-HlyA53 (lane 5). The markers (M) have approximate molecular masses of 188, 98, 62, 49, 38, 28, 17, 14, 6, and 3 kDa. (B) Coomassie blue-stained SDS gel of supernatants of overnight cultures of various E. coli strains transformed with pVDL9.3 plus C52-HlyA218. Lanes: 1, Nissle 1917; 2, Nissle 1917 (aadA1); 3, C594.72; 4, C641.72; 5, C105.72.
The amount of C52-HlyA218 peptide secreted by Nissle 1917 was estimated to be 40 mg/liter, representing a peptide concentration of 1.25 μM. Similar expression levels were found in cultures grown to late log phase in the presence or absence of IPTG and in saturated overnight cultures without inducer (Fig. 2B); this lack of dependence on IPTG was expected because of the titration of the available Lac repressor by the high-copy-number Lac operator. Several other commensal strains of E. coli isolated from feces or vagina of normal healthy volunteers (28) also expressed the C52-HlyA218 peptide, but the levels of secreted protein were no greater than or lower than for Nissle 1917 (Fig. 2B).
In anticipation that the length of the HlyA tail might affect the secretion or biological activity of the fusion peptide, plasmids containing progressively shorter C-terminal domains were constructed. C52 fusion peptides containing 103 or 53 aa from the C terminus of HlyA were efficiently secreted into the culture medium, as peptides of 19.5 kDa and 13.7 kDa, respectively, at molar levels similar to those for C52-HlyA218 (Fig. 2 A); however, a construct retaining 43 aa from the C terminus did not show any peptide secretion (data not shown). These experiments define the N-terminal boundary of the HlyA secretion signal sequence at 43–53 aa upstream of the C terminus of the protein.
An important advantage of working with E. coli is the extensive knowledge of the cis- and trans-acting signals that can be manipulated to increase foreign gene expression. We have previously found that gene aadA1 originating from Tn21 transposon, which confers resistance to aminoglycoside antibiotics, increases the expression of a broad spectrum of cellular proteins in bacteria (Kar and Adhya, unpublished results). Introduction of aadA1 into Nissle 1917 up-regulated C52-HlyA218 peptide expression by 3-fold without any adverse effect on the growth of the bacteria (Fig. 2B).
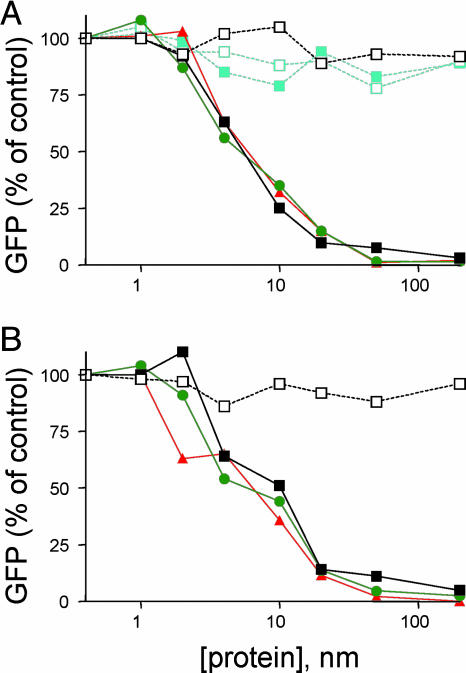
Inhibition of HIV Infection by Secreted Peptides. The ability of the secreted C52-HlyA218 peptides to block HIV infection was demonstrated by using HIV–GFP reporter virus assays in peripheral blood mononuclear cells. Dilutions of the bacterial culture supernatants were directly added to cells, which were then infected with an HIV or heterologous reporter virus and assayed for GFP expression 3 days later. Fig. 3 shows that HIV infection was inhibited in a dose-dependent fashion by each of the C52-HlyA peptides but not by a control Etag-HlyA peptide lacking HIV sequences. This inhibition was specific for the HIV envelope glycoprotein as shown by the lack of any effect on pseudovirions encapsidated with MuLV envelope. Average IC50 values were 5.2 ± 0.5 nM for the laboratory-adapted T-cell (X4-tropic) reporter virus HIVNL4.3–GFP (Fig. 3A) and 5.8 ± 1.2 nM for the macrophage (R5-tropic) reporter virus HIVJR-CSF–GFP (Fig. 3B). Specific inhibition of HIV infection was also observed when the bacteria were cocultured with Jurkat cells in transwell plates before reporter virus infection (data not shown).
Fig. 3.
Inhibition of HIV infection by secreted peptides. Peripheral blood mononuclear cells were incubated with various dilutions of bacterial culture supernatants for 1 h at 37°C, infected with a GFP reporter virus, and assayed 72 h later for GFP reporter expression by FACS. Results are normalized to the percent of GFP+ cells in the absence of inhibitor. (A) Inhibition of the laboratory-adapted CXCR4-dependent HIV isolate NL4.3 and of heterologous MuLV pseudotyped reporter virus. Key: open black squares, Etag-HlyA218 plus HIVNL4.3; filled black squares, C52-HlyA218 plus HIVNL4.3; filled green circles, C52-HlyA103 + HIVNL4.3; filled orange triangles, C52-HlyA53 plus HIVNL4.3; open aqua squares, Etag-HlyA218 plus MuLV; filled aqua squares, C52-HlyA218 plus MuLV. (B) Inhibition of the primary CCR5-dependent HIV isolate JRCSF. Key: open black squares, Etag-HlyA218 plus HIVJR-CSF; filled black squares, C52-HlyA218 plus HIVJR-CSF; filled green circles, C52-HlyA103 plus HIVJR-CSF; filled orange triangles, C52-HlyA53 plus HIVJR-CSF.
There were no consistent differences between the anti-HIV potencies of the C52 peptides containing different lengths of HlyA sequences. Furthermore, the observed IC50 values for the fusion peptides were in the same range reported for the unfused T20 and C34 peptides (11, 27). Thus, it appears that the addition of non-HIV sequences to the C terminus of the C52 peptide does not interfere with its ability to bind to the N-terminal region of gp41 and block viral infection.
Colonization of Mouse Gastrointestinal Tract. The ability of the genetically modified Nissle 1917 to colonize and secrete anti-HIV peptides in intact animals was investigated by using the mouse as a model system. The initial studies focused on the gastrointestinal tract, which is the main reservoir for E. coli in the body and the site for HIV transmission by rectal intercourse. Nissle 1917 expressing C52-HlyA218 was orally or rectally administered to female mice, and the extent of colonization was determined by bacterial plating assays of the feces. Because it is difficult for exogenous human bacteria to compete with the indigenous microflora of the mouse intestine, some of the mice were either pretreated or continuously treated with ampicillin during the experiment.
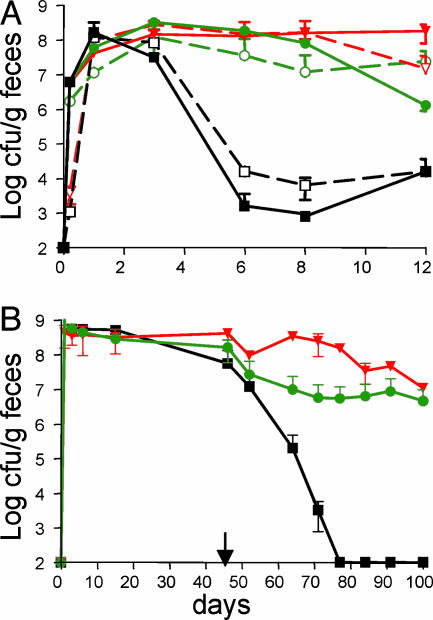
In the presence of continuous ampicillin selection, a single administration of 5 × 108 genetically engineered bacteria led to sustained colonization at levels of 107 to108 colony-forming units (cfu) per g of feces for up to 12 days (Fig. 4A). Substantial colonization was also observed in mice given a single day of pretreatment with ampicillin, although the levels of bacteria dropped to106 to 107 cfu/g by the end of the experiment. In the absence of antibiotic treatment, colonization levels fell rapidly to 103 to 104 cfu/g; however, the genetically engineered bacteria were still present as evidenced by the fact that they returned to levels of 107 cfu/g when the animals were re-administered ampicillin (data not shown). There were no significant differences in colonization efficiency between oral and rectal administration. Analysis of Apr Cmr colonies isolated from the feces of animals colonized by either route demonstrated that they secreted the C52-HlyA218 peptide at similar levels as the starting strain.
Fig. 4.
Mouse colonization. (A) Effects of antibiotic treatment and administration route on short-term colonization. Mice were orally or rectally inoculated with 5 × 108 Nissle 1917 carrying the Cmr HlyB,HlyD plasmid pVDL9.3 and the Apr peptide expression plasmid pC52-HlyA218. The mice were given no antibiotic, 1-day pretreatment with ampicillin, or continuous ampicillin treatment as indicated. At intervals, feces samples were assayed for Apr Cmr cfu. Key: filled black squares, no antibiotic/oral inoculation; open black squares, no antibiotic/rectal inoculation; filled green circles, 1 day of antibiotic/oral inoculation; open green circles, 1 day of antibiotic/rectal inoculation; filled orange inverted triangles, continuous antibiotic/oral inoculation; open orange inverted triangles, continuous antibiotic/rectal inoculation. (B) Long-term colonization. Mice were inoculated orally and rectally with a total of 109 Nissle 1917 carrying pVDL9.3 and the indicated peptide expression plasmid. Mice were treated for 45 days with ampicillin, then taken off antibiotic for the remainder of the experiment (arrow). Feces samples were periodically assayed out to 100 days for Apr Cmr cfu. Key: filled black squares, pC52-HlyA218; filled green circles, pC52-HlyA103; filled orange inverted triangles, pC52-HlyA53.
To investigate the cis- and trans-acting signals involved in the maintenance of gastrointestinal tract colonization, mice were given a single treatment with ampicillin, orally administered various bacterial strains, and then maintained without antibiotic for 25 days. Colonization levels of Nissle 1917 expressing C52-HlyA218 fell >10,000-fold over this period (Table 1). This drop in bacterial levels was not simply due to the amount of peptide secreted, as shown by the somewhat better retention of bacteria that overproduced the peptide because of the introduction of aadA1 gene. The drop was also not exclusively due to the presence of HIV sequences in the secreted peptide, as shown by the poor maintenance of a strain expressing Etag-HlyA218. By contrast, the extent of HlyA C-terminal sequences did appear to play an important role, as demonstrated by the ability of bacteria expressing the C-terminal deletion mutants C52-HlyA103 and C52-HlyA53 to undergo more persistent colonization than C52-HlyA218, with levels at day 25 >100-fold higher than for the full-length construct. This conclusion was confirmed by a competitive colonization experiment in which mice were fed an equal mixture of bacteria expressing C52-HlyA218 and C52-HlyA53 and subsequently analyzed for the ratio of colonies expressing the different length peptides. At day 1, a slight excess of bacteria expressing the HlyA218 construct was excreted, but, by day 8 and thereafter, only bacteria expressing the short HlyA53 construct could be recovered.
Table 1. Effects of cis- and trans-acting signals on mouse colonization.
| Log cfu/g feces
|
||||
|---|---|---|---|---|
| Nissle 1917 | Plasmid | Day 1 | Day 25 | Log diff. |
| WT | pC52-HlyA218 | 8.35 (0.27) | 3.61 (0.16) | 4.74 |
| aadA1 | pC52-HlyA218 | 8.42 (0.52) | 4.62 (0.79) | 3.80 |
| WT | Etag-HlyA218 | 7.84 (0.50) | 3.02 (0.27) | 4.81 |
| WT | C52-HlyA103 | 7.98 (0.36) | 5.99 (0.91) | 1.99 |
| WT | C52-HlyA53 | 8.91 (0.13) | 7.00 (0.91) | 1.91 |
Mice were orally administered 5 × 108 cfu of the indicated strain and maintained without antibiotic. Feces were assayed for Apr Cmr cfu at intervals and the results at day 1 and day 25 are shown as the mean of the log10 cfu per g of feces with standard errors in parentheses. diff., difference.
To examine the potential of the genetically engineered Nissle for long-term, stable colonization in the absence of antibiotics, mice were orally and rectally administered bacteria expressing a C52 peptide, then maintained on ampicillin for 50 days before removal of the antibiotic. The rational for this prolonged initial antibiotic treatment was that it would eliminate much of the competing indigenous microflora while allowing the genetically engineered bacteria to adapt to the nutritional environment of the intestine. In the absence of antibiotics, bacteria expressing C52-HlyA218 were again eliminated from the mice reaching undetectable levels by 77 days (Fig. 4B). However, bacteria secreting C52-HlyA103 and C52-HlyA103 were maintained in the mice at levels of ≈106 cfu per g of feces for up to 50 days after the removal of drug selection. Bacteria recovered from the mouse feces after prolonged colonization were still capable of secreting high levels of the C52 peptides, indicating there was no strong selection against peptide secretion in vivo.
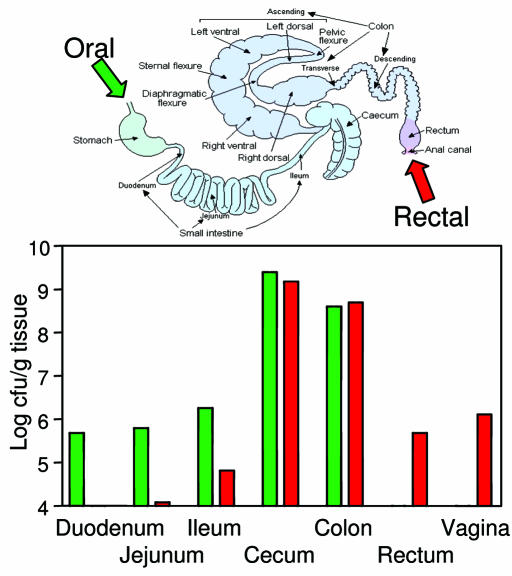
In Vivo Growth Patterns and Peptide Secretion. The distribution of the anti-HIV bacteria in different tissues was examined in mice that has been pretreated with ampicillin to reduce the endogenous microflora then orally or rectally administered Nissle 1917 expressing C52-HlyA53. The highest concentrations of bacteria (108 to 109 cfu/g) were present in the colon and cecum after oral and rectal administration (Fig. 5). Lower levels of bacteria (105 to 106 cfu/g) were also recovered along the upper intestine, including duodenum, jejunum, and ileum, in mice that were orally inoculated, whereas bacteria were recovered from rectum primarily in mice that were rectally inoculated. Nissle 1917 was also recovered from vagina in approximately one third of rectally inoculated animals.
Fig. 5.
Anatomical distribution of bacteria. Mice were given 1 day of ampicillin pretreatment, then orally or rectally inoculated with 5 × 108 Nissle 1917 carrying pVDL9.3 and pC52-HlyA53. After 3 days, the animals were euthanized and dissected, and tissue segments were assayed for Apr Cmr cfu. (Upper) A diagram of the gastrointestinal tract. (Lower) The distribution of genetically engineered bacteria.
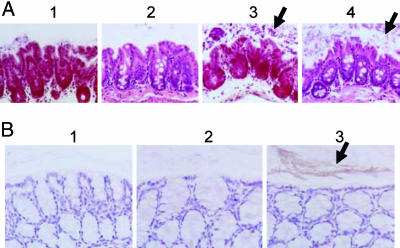
Tissues from colonized animals were also processed for histopathology and immunohistochemistry. None of the target organs demonstrated any inflammation, necrosis, or other pathology. Detailed examination of the colon revealed the presence of numerous monomorphic, hematoxylin–eosin-stained bacterial colonies in animals inoculated with Nissle 1917 expressing C52-HlyA53 (Fig. 6A3) but not in control animals inoculated with PBS (Fig. 6A1). As expected for E. coli, these bacteria were Gram-negative (Fig. 6A4). The bacterial colonies were present throughout the lumen, often in close association with the epithelial surface. In some samples, the colon and cecum of Nissle 1917-inocculated animals demonstrated goblet cell hyperplasia and copious mucus secretion into the lumen that was not evident in control PBS-inoculated animals.
Fig. 6.
Histopathology and immunohistochemistry of colon. Mice were given 1 day of ampicillin pretreatment, then orally inoculated with PBS or with Nissle 1917 carrying pVDL9.3 and pEtag-HlyA218 or pC52-HlyA53. After 3 days, the animals were euthanized, and the colon was dissected, fixed, and sectioned for microscopy. Arrows point to areas of dense bacterial growth and peptide secretion. (A) Histopathology. (1) PBS control mouse, hematoxylin–eosin stain; (2) PBS control mouse, gram stain; (3) Nissle 1917::pC52-HlyA53 mouse, hematoxylin–eosin stain; (4) Nissle 1917::pC52-HlyA53 mouse, Gram stain. (B) Immunohistochemistry using 2F5 antibody. (1) Nissle 1917::pEtag-HlyA218 control mouse; (2) Nissle 1917::pC52-HlyA53 mouse without antibody; (3) Nissle 1917::pC52-HlyA53 mouse with antibody.
To examine peptide secretion, colon samples were subjected to immunohistochemistry using the human monoclonal antibody 2F5, which recognizes an epitope present in the C52 peptide. Clear staining was observed throughout the lumen in samples from animals inoculated with Nissle 1917 expressing pC52-HlyA53 (Fig. 6B3), but not in control samples processed without antibody (Fig. 6B2) or from animals expressing control Etag peptide (Fig. 6B1).
Discussion
There is an urgent need for new approaches to prevent the sexual transmission of HIV, especially in the developing world, where the AIDS epidemic is continuing to spread essentially unabated. Despite considerable effort, attempts to develop an efficacious vaccine have so far been unsuccessful. Condoms and other barriers are effective when available and used consistently, but this is often not the case. Recently there has been increased interest in the use of topical microbicides as a practical and cost-effective alternative method to block HIV transmission during vaginal and anal intercourse (29–32); however, the need for active, repeated application of such agents before sex is likely to limit their use, especially in resource-poor environments.
We propose to use live, genetically modified bacteria as a more durable method to deliver microbicidal agents to the mucosal surfaces on which HIV transmission occurs. Our results using the commensal bacterium Nissle 1917 genetically engineered to secrete an HIV fusion inhibitor peptide demonstrate several potential advantages of using live microbial microbicides for HIV prevention.
First, protection should be long-lasting because of the ability of the bacteria to colonize and replicate in the host. This would greatly improve prevention efforts by eliminating the necessity for administering the microbicide immediately before sex and by giving control to the vulnerable receptive partner in sexual intercourse (32). In the present work, Nissle 1917 expressing HIV-C52-HlyA persisted in mice for periods of weeks to months under antibiotic selection, indicating the ability of this strain to efficiently replicate in the intestine. Microscopy of the colon confirmed the presence of dense bacterial growth and mucosal adherence. Although efficient colonization of mice required antibiotic treatment to reduce competing microflora, colonization in humans may be more effective because Nissle 1917 is a human rather than rodent strain. It should be possible to further improve the retention of the bacteria by selecting for highly colonizing variants or by deliberate manipulation of colonization factor genes.
Second, bacteria are simple and practical to manufacture, store, distribute, and administer, and they are far less expensive than protein-based microbicides. Commercial preparations of Nissle 1917 (Mutaflor) consist of lyophilized bacteria in an enteric coated capsule for oral ingestion, which can be manufactured at low cost and stored at room temperature indefinitely.
Third, our approach could be used to express a variety of peptides that inhibit HIV recognition, fusion, and entry into susceptible host cells either by directly binding to the viral envelope glycoprotein or by blocking cellular receptor and coreceptor sites (30). In the present work, we focused on the C52 peptide because of its potent inhibitory activity against a wide spectrum of HIV isolates because of the strong conservation of the gp41 sequences recognized by C peptides (11). Although Hamburger et al. (33) have reported that adding cargo proteins to the N terminus of a C peptide decreased its potency, this was not the case for addition of HlyA sequences to the C terminus of C52. Because the atomic basis of C–N peptide interaction is known, similar peptide inhibitors can be designed for any related virus of known sequence, including enfuvirtide-resistant variants of HIV or simian immunodeficiency virus.
Fourth, the method could be adapted to deliver peptides to the cervicogenital, oral, and gastrointestinal mucosa by using different microbial hosts; e.g., the vaginal bacteria L. jensenii (7), the oral bacteria S. gordonii (4), or commensal yeasts such as Pichia guelliermondii and Saccharomyces boulardii. Vaginal delivery may also be possible by using E. coli, which is found in the vagina of ≈20% of healthy women (28, 34). In the present work, we found Nissle 1917 in the vagina of approximately one-third of rectally inoculated animals; preliminary data indicates a close correlation between bacterial colonization and the estrous phase of the mice.
Fifth, although our method is primarily intended to prevent new HIV infections, it could also potentially be used as a therapeutic method in combination with standard highly active antiretroviral drug treatment or as a means of preventing viral rebound in individuals who have ceased treatment. This application would require that peptide secreted into the lumen be taken up into the mucosa. Delivery of anti-HIV peptides to the intestine would be especially useful in view of recent evidence that standard antiretroviral treatment does not fully block CD4 T cell depletion in this compartment, which is the major site of HIV replication and pathogenesis at all stages of infection (35, 36).
The particular microbial microbicide described here should be safe for human use because it is based on a commensal strain of bacteria, Nissle 1917, that has been used as an over-the-counter probiotic with an excellent biosafety record (18, 23, 25, 37). We found that administration of these bacteria to mice had no inflammatory effects in the gastrointestinal tract or vagina. Before testing in humans, however, it would be desirable to remove any transferable antibiotic resistance markers from the bacteria, which could be accomplished by integration of the peptide expression cassette into the host cell chromosome.
The most likely use for the genetically engineered Nissle 1917 would be to prevent HIV infection during rectal intercourse, which is the predominant mode of transmission for men who have sex with men; however, rectal intercourse also plays an important albeit often overlooked role in transmission to women; e.g., when used as a form of birth control, to preserve a woman's virginity until marriage, in cases of rape, and by sex workers.
Although the role of the natural microflora in preventing infection by pathogenic microbes is well known, the concept of using genetically engineered organisms for this purpose is a concept that has not yet been tested in human beings. The severity of the worldwide HIV/AIDS epidemic combined with the lack of effective biomedical interventions warrants such an approach.
Supplementary Material
Acknowledgments
We thank Betsy Foxman (University of Michigan, Ann Arbor) for bacterial strains, Michael Root (Thomas Jefferson University, Philadelphia) for 5-helix protein, Shayda Eskandary for protein secretion experiments, George Pavlakis (National Cancer Institute, Frederick, MD) for HIV–GFP indicator viruses, and Peter Kwong for 2F5 antibody.
Author contributions: S.R., S.A., and D.H.H. designed research; S.R., S.H., L.M., K.L., K.H., Q.Z., R.A.F., S.K., and D.H.H. performed research; S.R., S.H., L.M., K.L., K.H., R.A.F., S.K., S.A., and D.H.H. analyzed data; and S.R., S.A., and D.H.H. wrote the paper.
Abbreviations: Hly, hemolysin; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; cfu, colony-forming units; Etag, epitope tag; MuLV, murine leukemia virus; IPTG, isopropyl β-d-thiogalactoside.
References
- 1.Joint United Nations Programme on HIV/AIDS (2004) 2004 Report on the Global AIDS Epidemic (Joint U.N. Prog. HIV/AIDS, Geneva).
- 2.Nester, E. W. (2004) Microbiology: A Human Perspective (McGraw–Hill, Boston).
- 3.Tlaskalova-Hogenova, H., Stepankova, R., Hudcovic, T., Tuckova, L., Cukrowska, B., Lodinova-Zadnikova, R., Kozakova, H., Rossmann, P., Bartova, J., Sokol, D., et al. (2004) Immunol. Lett. 93, 97–108. [DOI] [PubMed] [Google Scholar]
- 4.Beninati, C., Oggioni, M. R., Boccanera, M., Spinosa, M. R., Maggi, T., Conti, S., Magliani, W., De Bernardis, F., Teti, G., Cassone, A., et al. (2000) Nat. Biotechnol. 18, 1060–1064. [DOI] [PubMed] [Google Scholar]
- 5.Kruger, C., Hu, Y., Pan, Q., Marcotte, H., Hultberg, A., Delwar, D., van Dalen, P. J., Pouwels, P. H., Leer, R. J., Kelly, C. G., et al. (2002) Nat. Biotechnol. 20, 702–706. [DOI] [PubMed] [Google Scholar]
- 6.Giomarelli, B., Provvedi, R., Meacci, F., Maggi, T., Medaglini, D., Pozzi, G., Mori, T., McMahon, J. B., Gardella, R. & Boyd, M. R. (2002) AIDS 16, 1351–1356. [DOI] [PubMed] [Google Scholar]
- 7.Chang, T. L., Chang, C. H., Simpson, D. A., Xu, Q., Martin, P. K., Lagenaur, L. A., Schoolnik, G. K., Ho, D. D., Hillier, S. L., Holodniy, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 11672–11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez, L. A., Sola, I., Enjuanes, L. & de Lorenzo, V. (2000) Appl. Environ. Microbiol. 66, 5024–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzschaschel, B. D., Guzman, C. A., Timmis, K. N. & de Lorenzo, V. (1996) Nat. Biotechnol. 14, 765–769. [DOI] [PubMed] [Google Scholar]
- 10.Ofek, G., Tang, M., Sambor, A., Katinger, H., Mascola, J. R., Wyatt, R. & Kwong, P. D. (2004) J. Virol. 78, 10724–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777–810. [DOI] [PubMed] [Google Scholar]
- 12.Ketas, T. J., Frank, I., Klasse, P. J., Sullivan, B. M., Gardner, J. P., Spenlehauer, C., Nesin, M., Olson, W. C., Moore, J. P. & Pope, M. (2003) J. Virol. 77, 2762–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketas, T. J., Klasse, P. J., Spenlehauer, C., Nesin, M., Frank, I., Pope, M., Strizki, J. M., Reyes, G. R., Baroudy, B. M. & Moore, J. P. (2003) AIDS Res. Hum. Retroviruses 19, 177–186. [DOI] [PubMed] [Google Scholar]
- 14.Kilby, J. M., Lalezari, J. P., Eron, J. J., Carlson, M., Cohen, C., Arduino, R. C., Goodgame, J. C., Gallant, J. E., Volberding, P., Murphy, R. L., et al. (2002) AIDS Res. Hum. Retroviruses 18, 685–693. [DOI] [PubMed] [Google Scholar]
- 15.Andersen, C. (2003) Rev. Physiol. Biochem. Pharmacol. 147, 122–165. [DOI] [PubMed] [Google Scholar]
- 16.Gentschev, I., Dietrich, G. & Goebel, W. (2002) Trends Microbiol. 10, 39–45. [DOI] [PubMed] [Google Scholar]
- 17.Cukrowska, B., LodInova-ZadnIkova, R., Enders, C., Sonnenborn, U., Schulze, J. & Tlaskalova-Hogenova, H. (2002) Scand. J. Immunol. 55, 204–209. [DOI] [PubMed] [Google Scholar]
- 18.Lodinova-Zadnikova, R. & Sonnenborn, U. (1997) Biol. Neonate 71, 224–232. [DOI] [PubMed] [Google Scholar]
- 19.Hockertz, S. (1997) Arzneim.-Forsch. 47, 793–796. [PubMed] [Google Scholar]
- 20.Schultz, M., Strauch, U. G., Linde, H. J., Watzl, S., Obermeier, F., Gottl, C., Dunger, N., Grunwald, N., Scholmerich, J. & Rath, H. C. (2004) Clin. Diagn. Lab. Immunol. 11, 372–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruis, W. (2004) Aliment. Pharmacol. Ther. 20, Suppl. 4, 75–78. [DOI] [PubMed] [Google Scholar]
- 22.Grozdanov, L., Raasch, C., Schulze, J., Sonnenborn, U., Gottschalk, G., Hacker, J. & Dobrindt, U. (2004) J. Bacteriol. 186, 5432–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruis, W., Fric, P., Pokrotnieks, J., Lukas, M., Fixa, B., Kascak, M., Kamm, M. A., Weismueller, J., Beglinger, C., Stolte, M., et al. (2004) Gut 53, 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malchow, H. A. (1997) J. Clin. Gastroenterol. 25, 653–658. [DOI] [PubMed] [Google Scholar]
- 25.Rembacken, B. J., Snelling, A. M., Hawkey, P. M., Chalmers, D. M. & Axon, A. T. (1999) Lancet 354, 635–639. [DOI] [PubMed] [Google Scholar]
- 26.Tromm, A., Niewerth, U., Khoury, M., Baestlein, E., Wilhelms, G., Schulze, J. & Stolte, M. (2004) Z. Gastroenterol. 42, 365–369. [DOI] [PubMed] [Google Scholar]
- 27.Root, M. J., Kay, M. S. & Kim, P. S. (2001) Science 291, 884–888. [DOI] [PubMed] [Google Scholar]
- 28.Foxman, B., Manning, S. D., Tallman, P., Bauer, R., Zhang, L., Koopman, J. S., Gillespie, B., Sobel, J. D. & Marrs, C. F. (2002) Am. J. Epidemiol. 156, 1133–1140. [DOI] [PubMed] [Google Scholar]
- 29.D'Cruz, O. J. & Uckun, F. M. (2004) Curr. Pharm. Des. 10, 315–336. [DOI] [PubMed] [Google Scholar]
- 30.Shattock, R. J. & Moore, J. P. (2003) Nat. Rev. Microbiol. 1, 25–34. [DOI] [PubMed] [Google Scholar]
- 31.Stone, A. (2002) Nat. Rev. Drug Discovery 1, 977–985. [DOI] [PubMed] [Google Scholar]
- 32.Harrison, P. F., Rosenberg, Z. & Bowcut, J. (2003) Clin. Infect. Dis. 36, 1290–1294. [DOI] [PubMed] [Google Scholar]
- 33.Hamburger, A. E., Kim, S., Welch, B. D. & Kay, M. S. (2005) J. Biol. Chem. 280, 12567–12572. [DOI] [PubMed] [Google Scholar]
- 34.Larsen, B. & Monif, G. R. (2001) Clin. Infect. Dis. 32, e69–e77. [DOI] [PubMed] [Google Scholar]
- 35.Mehandru, S., Poles, M. A., Tenner-Racz, K., Horowitz, A., Hurley, A., Hogan, C., Boden, D., Racz, P. & Markowitz, M. (2004) J. Exp. Med. 200, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brenchley, J. M., Schacker, T. W., Ruff, L. E., Price, D. A., Taylor, J. H., Beilman, G. J., Nguyen, P. L., Khoruts, A., Larson, M., Haase, A. T. & Douek, D. C. (2004) J. Exp. Med. 200, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruis, W., Schutz, E., Fric, P., Fixa, B., Judmaier, G. & Stolte, M. (1997) Aliment. Pharmacol. Ther. 11, 853–858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.