Abstract
Rhabdoid tumors are aggressive pediatric malignancies for which, currently, there are no effective or standard treatment strategies. Rhabdoid tumors arise because of the loss of the tumor suppressor gene INI1. We have previously demonstrated that INI1 represses Cyclin D1 transcription in rhabdoid cells by directly recruiting histone deacetylase 1 complex to its promoter, leading to G0-G1 arrest. Expression of Cyclin D1 overcomes cell cycle arrest mediated by INI1 and Cyclin D1 overexpression in human rhabdoid tumors is a common phenomenon. However, it is not clear whether Cyclin D1 is a critical downstream target of INI1 in vivo and whether the derepression of this gene is essential for rhabdoid tumorigenesis. To determine the requirement of Cyclin D1 for genesis of rhabdoid tumors in vivo, we developed Ini1 heterozygous mice by targeted disruption. We found that the tumors developed in these Ini1+/- mice are rhabdoid, defective for Ini1 protein, and like the human tumors, express Cyclin D1. We crossed Ini1+/- mice to Cyclin D1-/- mice and found that Ini1+/- mice with Cyclin D1 deficiency did not develop any spontaneous tumors, in contrast to the parental Ini1+/- mice. These results strongly support the hypothesis that Cyclin D1 is a key mediator in the genesis of rhabdoid tumors. Our results provide an in vivo proof of concept that drugs that target Cyclin D1 expression or activity could be potentially effective as novel therapeutic agents for rhabdoid tumors.
Keywords: INI1/hSNF5, cell cycle, atypical teratoid, tumorigenesis
Rhabdoid tumors, including malignant rhabdoid tumors of the kidneys (MRT) and atypical teratoid and rhabdoid tumors (AT/RT) of the brain and soft tissues, are highly aggressive pediatric malignancies (1-3). Prognosis for infants with these tumors is poor and ≈80% of the children die within 1 yr of diagnosis despite intensive treatment. Chemotherapy is effective only transiently in some AT/RT patients and the use of radiotherapy is limited in children under 3 yr of age because of subsequent neurological damage and because surgical resection is challenging (3). In the past several years, there has been significant progress in the cytogenetic and molecular genetic studies, which indicated that the majority of MRT and AT/RT tumors harbor recurrent biallelic alterations in the INI1/hSNF5 gene, located in chromosome 22q11.2 (4-6). Several studies support the model that INI1 is a tumor suppressor. Children who harbor germ-line constitutive mutation in one allele of INI1 develop MRT or AT/RT when the second allele of INI1 is also mutated (5), and families who are carriers of a mutation in this gene are predisposed to rhabdoid syndrome (7). Furthermore, mice heterozygous for Ini1/hSNF5 mutations develop rhabdoid tumors with a high frequency because of loss of heterozygosity at the Ini1 locus (8-10).
INI1/hSNF5 is a component of the chromatin-remodeling mammalian SWI/SNF complex (11). Reintroduction of this gene into rhabdoid cell lines causes G0-G1 arrest and affects the transcription of several cell cycle regulatory genes (12). We have demonstrated that INI1/hSNF5 represses the transcription of the Cyclin D1 gene by recruiting the histone deacetylase complex directly to its promoter and that coexpression of Cyclin D1 from a heterologous promoter overcomes the cell cycle arrest caused by INI1 (12). Immunohistochemical analysis demonstrated that Cyclin D1 is derepressed in human AT/RT tumors that lack functional INI1 (12). Further analysis indicated that INI1 also activates p16INK4a (13) and represses Cyclin E and Cyclin A (12). Furthermore, recent studies suggest that INI1/hSNF5 may be involved in activating the mitotic checkpoint through the p16-Cyclin D1/CDK4-pRb-E2F pathway (14).
Cyclin D1 is overexpressed in many human tumors (15-20). However, in the majority of these cases it is not clear whether Cyclin D1 is actually required for tumorigenesis. Mouse models have demonstrated that Cyclin D1 function is specifically required for the genesis of a subset (e.g., Ras-Neu-induced breast cancer) but not all cancers (21). Cyclin D1 is an important cell cycle protein, required to overcome the restriction point in the G1 stage of the cell cycle (22-24). By binding to cyclin-dependent kinase (cdk) 4/6, Cyclin D1 facilitates the phosphorylation of Rb to mediate G1 to S progression. In addition, a cdk-independent function of Cyclin D1 is involved in modulating the activity of several transcription factors and may be important for tumorigenesis (17, 25-29). It is possible that derepression of Cyclin D1 due to INI1 loss in rhabdoid cells is a key event in the genesis of these tumors. Genetic knockout studies in mice indicate that Cyclin D1 is not essential for survival, probably due to redundancy of D-type cyclins (30). Mice deficient for Cyclin D1 exhibit a few developmental disorders (30), including reduced body size, reduced viability, and symptoms of minor neurological impairment. In addition, these mice exhibit reduction in retinal cells in the adult, a unique pattern of photoreceptor cell death, and lack of breast epithelium that undergoes proliferative changes during pregnancy (30). Interestingly, the D-type cyclins show specificity during development, and the mice lacking specific D-type cyclins show narrow, restricted developmental abnormalities and distinct phenotypes (30, 31). These results indicate that although the D-type cyclins have similar function during cell cycle, their different cell type distribution and specific roles in development attest to their unique function.
In this report we have tested the in vivo requirement of Cyclin D1 for the genesis of rhabdoid tumors. We generated Ini1+/- mice by targeted disruption and found that the tumors developed in these mice are rhabdoid and exhibit derepression of Cyclin D1. Furthermore, we crossed these Ini1+/- mice to the Cyclin D1-/- mice and found that the lack of Cyclin D1 resulted in abrogation of rhabdoid tumor genesis. These striking results establish a direct link between INI1 and Cyclin D1 in vivo. Furthermore, these studies provide a proof of principle to indicate that inhibition of Cyclin D1 or its pathway may be a good therapeutic strategy for treatment of children with rhabdoid tumors.
Methods
Generation of Targeting Construct, Gene Targeting, and Germ-Line Transmission. An Ini1 genomic DNA fragment was isolated by screening the RPCI-22 mouse bacterial artificial chromosome genomic library arrayed on filters, using the exon 7 portion of the mouse Ini1 cDNA as the probe. A 15-kb genomic DNA fragment was cloned into pBluescript vector, and subsequently the targeting vector was generated by replacing the XbaI to StuI fragment that contains exons 6 and 7 regions with a hygromycin cassette. WW6 embryonic stem cells (32) were electroporated with the targeting construct and selected for hygromycin-resistant colonies as per standard techniques. Pools of 400 clones were screened by long-range PCR for the presence of a 4.2-kb fragment, using primers D149 (5′-GAG CCC AGA AAG CGA AGG-3′), which hybridizes to the hygromycin cassette, and mIEK-3′A (5′-GGG CAA GCT GCT GAT ACA AT-3′), which hybridizes to the 3′-flanking region, outside the targeting sequence. Two different homologous recombinant embryonic stem cell (ES cell) clones were injected into C57BL/6 recipient blastocysts. Male transmitting chimeric mice were obtained and crossed with C57BL/6 females to obtain heterozygous Ini1+/Δexon6,7 F1 mice. Ini1+/Δexon6,7 mice were crossed with each other to obtain the F2 generation. For germ-line analysis, DNA from mice was genotyped by PCR using primers specific for the hygromycin cassette: D141, 5′-TGA GCT GGC CCT TAA TTT GG-3′, and J65, 5′-TGT GTA GAA GTA CTC GCC GA-3′.
Generation of Ini1/Cyclin D1 Double Mutant Mice. Cyclin D1-knockout mice (of the C57BL/6J strain) generated by Sicinski et al. (30) were a kind gift of R. G. Pestell (Georgetown University School of Medicine, Washington, DC). Two females and two male heterozygous Cyclin D1+/- mice were mated with Ini1+/Δexon6,7 mice to obtain Ini1+/Δexon6,7 Cyclin D1+/- double heterozygous mice. The double heterozygous mice were mated with littermates to obtain mice with different combinations of genotypes. Ini1 genotyping was done as described above, and Cyclin D1 genotyping was done as described in ref. 30.
Cell Culture and RT-PCR Analysis. INI1-/- cell lines were grown as described (12). MON cells were transfected with pCGN-INI1/hSNF5 expressing HA-INI1 as described in ref. 12. RNA was isolated from transfected cells and subjected to RT-PCR analysis. Primers used for RT-PCR analysis are as follows: for Cyclin D1, 5′-CTT CCT CTC CAA AAT GCC AG-3′ and 5′-AGA GAT GGA AGG GGG AAA GA-3′; for Cyclin D2, 5′-TTC ATT GCA GAC ACC ACC AT-3′ and 5′-GCC AGA TAC CAG AAG CGA AG-3′; for Cyclin D3, 5′-TGA TTT CCT GGC CTT CAT TC-3′ and 5′-AGC TTG ACT AGC CAC CGA AA-3′; for p16, 5′-CGG AAG GTC CCT CAG ACA TC-3′ and 5′-TCA TGA AGT CGA CAG CTT CCG-3′; and for p21, 5′-CTC AGA GGA GGC GCC ATG-3′ and 5′-GGG CGG ATT AGG GCT TCC-3′. The PCR was performed in a 25-μl reaction by using 55°C as the annealing temperature and 1.5 mM MgCl2 and amplification for 25 cycles. PCR using primers specific to the GAPDH gene (5′-GGT GAA GGT CGG AGT CAA CG-3′ and 5′-CAA AGT TGT CAT GGA TGA CC-3′) was also carried out as an internal control, using the same RNA samples and same conditions.
Protein Expression and Western Blotting. Immunoblotting was performed by standard techniques using antibodies specific for the following proteins: Cyclin D1 (rabbit polyclonal from NeoMarkers, Fremont, CA), Cyclin D2 (rabbit polyclonal, catalog no. sc-181, Santa Cruz Biotechnology), Cyclin D3 (mouse monoclonal, catalog no. c-7214, Sigma), affinity-purified Ini1 antibodies (33), α-tubulin (catalog no. T5168, Sigma), and β-actin (mouse monoclonal, catalog no. A 5441, Sigma).
Immunohistochemical Analysis of Tumor Sections. Immunohistochemistry was performed on mouse tumors developed in Ini1+/Δexon6,7 mice and in Ini1+/Δexon6,7 mice with Cyclin D1+/+ or Cyclin D1+/- background and on a sample of normal brain present adjacent to one of the tumors. Affinity-purified Ini1 antibodies were used at 1:50 dilutions. For Cyclin D1 immunostaining, a mouse monoclonal antibody, mAbDCS-6 (Cell Marque, Austin, TX) was used at 1:100 dilution. For vimentin (vimentin V9, Ventana Medical Systems, Tucson, AZ) and smooth muscle actin (Dako), we used 1:100 dilutions. For each case, 5-μm-thick sections were cut from the paraffin blocks, deparaffinized, rehydrated, and stained by standard methods, using the avidin-biotin-horseradish peroxidase complex to localize the antibody bound to antigen, with diaminobenzidine as the final chromogen. Appropriate antigen retrieval was performed in the appropriate buffer. Immunostaining was performed on an automated immunostainer, NexES (Ventana Medical Systems). All immunostained sections were lightly counterstained with hematoxylin. For negative controls, the immune serum was replaced by either PBS or nonimmune serum. The sections were reviewed by the pathologist author (D.Z.).
Results
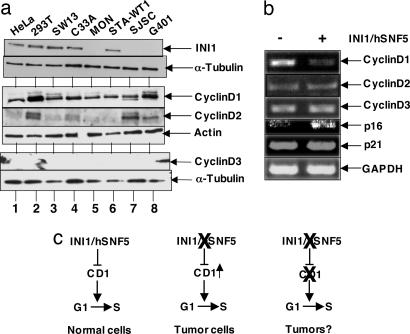
Expression of Cyclin D1 and Repression of D-Type Cyclins by INI1 in Rhabdoid Cell Lines. To determine whether the D-type cyclins are specifically overexpressed in rhabdoid tumors as a consequence of loss of INI1, we first analyzed the levels of Cyclin D1 in cell lines derived from human rhabdoid tumors and then tested the specificity of regulation of transcription of three D-type cyclins by reintroduction of INI1 into these cells. We carried out an immunoblot analysis of four different rhabdoid cell lines defective for INI1, including STA-WT1, a cell line that carries a cytoplasmically localized, inactive form of the protein (33). We found that Cyclin D1 is the only D-type cyclin that was consistently expressed in all these cell lines (Fig. 1a, lanes 5-8). Although D2 expression was not consistent, D3 was undetectable in all of the cell lines examined, indicating that Cyclin D1 expression is a common feature of these rhabdoid cell lines and tumors (Fig. 1a, lanes 5-8). Previously, we demonstrated that INI1 is directly recruited to Cyclin D1 promoter and represses its expression, by using chromatin-immunoprecipitation assays (12). To determine whether the repressive effect of INI1 is restricted to Cyclin D1, we tested the effect of reintroduction of INI1 into the INI1-/- MON cell line on the expression of a panel of genes including the three D-type cyclins. RT-PCR analysis of the mRNA expressed in the presence or absence of INI1 indicated that INI1 specifically repressed Cyclin D1 but not D2 and D3 (Fig. 1b). Furthermore, as reported in ref. 13, INI1 also activated p16INK4a but had no effect on p21Kip1 expression (Fig. 1b). These results indicate the specificity of repression of Cyclin D1 by INI1 in MON cells.
Fig. 1.
INI1 regulates expression of Cyclin D1 but not Cyclin D2 or Cyclin D3. (a) Western analysis of lysates from INI1+/+ and INI1-/- cell lines. Total protein from INI1+/+ cells (HeLa, lane 1; 293T, lane 2; SW13, lane 3; and C33a, lane 4) and INI1-/- cells (MON, lane 5; STA-WT1, lane 6; SJSC, lane 7; and G401, lane 8) were separated by SDS/PAGE and probed with antibodies to INI1, α-tubulin, Cyclin D1, Cyclin D2, β-actin, and Cyclin D3. (b) RT-PCR analysis of the RNA isolated from MON cells transfected with pCGN-INI1/hSNF5 expressing HA-INI1 or just pCGN control. (c) Graphic representation of the hypothesis that abrogation of Cyclin D1 (CD1) may result in loss of rhabdoid tumorigenesis.
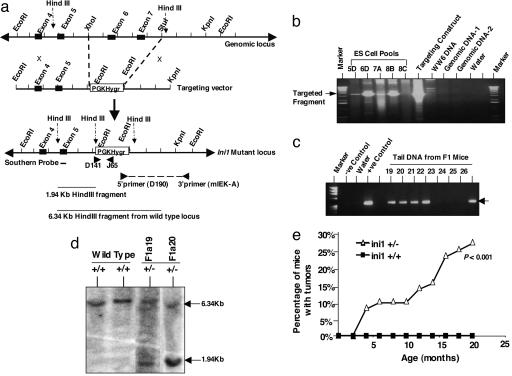
Cyclin D1 Is Derepressed in Mouse Rhabdoid Tumor Models. Based on the observations that (i) Cyclin D1 but not D2 and D3 is specifically repressed by INI1 in rhabdoid cells; (ii) expression of Cyclin D1 from a heterologous promoter is sufficient to overcome the INI1-mediated cell cycle arrest (12); and (iii) Cyclin D1 is expressed in human rhabdoid cell lines and tumors (12), we surmised that Cyclin D1 is a key downstream target of INI1-mediated tumor suppression. We further hypothesized that rhabdoid tumors are critically dependent on Cyclin D1 expression for survival and that ablating Cyclin D1 would result in the lack of or at least reduced frequency of tumors that arise because of the loss of the Ini1 gene (Fig. 1c). To test this hypothesis, we first generated mice carrying heterozygous mutations in Ini1 by targeted disruption. The genomic Ini1 exons 6 and 7, which encode the highly conserved repeats 1 and 2, were replaced with a hygromycin cassette to generate the mutant allele Ini1Δexon6,7 (Fig. 2a). Correct targeting of the Ini1 locus was identified first by screening the pools of ES cell clones and then by screening the individual clones by long-range PCR using a primer pair, one specific for the hygromycin cassette and another spanning the 3′-flanking sequence adjacent to the targeted region (Fig. 2 a and b). These analyses indicated the presence of homologous recombination in ≈25% of ES cell clones. The positive ES cells were microinjected into blastocysts, and germ-line transmission was monitored by PCR analysis using primers specific to the hygromycin cassette (Fig. 2c). Correct targeting was confirmed by Southern analysis of mouse-tail DNA obtained from F1 mice (Fig. 2d).
Fig. 2.
Targeted disruption of Ini1 locus and analysis of tumor formation. (a) The mouse Ini1/Snf5 genomic locus and targeting strategy. (b) Long-range PCR analysis of the pools of ES cell clones to detect the correct targeting. Genomic DNA-1 and -2 are negative controls that harbor the hygromycin cassette in an unrelated locus. The targeting construct was used as a positive control. (c) Germ-line transmission of the correct targeted allele to the F1 generation was determined by PCR of the tail DNA by using primers (D141 and J65) specific to the pPGK-Hygro cassette, which results in the amplification of a 325-bp fragment by PCR. (d) Southern analysis of the DNA isolated from wild-type and heterozygous mice. Correct targeting is indicated by the presence of a 1.9-kb HindIII fragment by using a probe specific to exon 5. (e) Frequency of tumor formation in Ini1+/Δexon6,7 heterozygous mice.
Analysis of the F1 and F2 generations indicated the lack of live-born mice homozygous for the Ini1Δexon6,7 allele, consistent with earlier reports that the Ini1-null genotype is embryonic lethal (8, 10, 34). Immunoblot analysis of total protein isolated from heterozygous mice with an anti-INI1 antibody that recognizes the N-terminal domain demonstrated the presence of only the full-length Ini1 protein (but no truncated protein), indicating that Ini1Δexon6,7 is likely a null allele (data not shown). Ini1+/Δexon6,7 heterozygous mice were obtained in a normal Mendelian ratio and were devoid of any apparent developmental or behavioral disorders. Observation of the cohort of mice for nearly 2 yr revealed the development of spontaneous face and brain tumors, at a very high frequency (≈26%) (Figs. 2e and 3A), and a small percentage of mice developed tumors in the hind limb. The tumors in Ini1+/Δexon6,7 mice developed with an early onset of 3 months and a median onset of ≈11 months of age. They were histopathologically classified as primary brain tumors, undifferentiated sarcomas, ganglioneuromas, and neuroblastomas, but a closer examination revealed the presence of rhabdoid cells in the majority of these tumors (Fig. 3B). These results of development of rhabdoid tumors in Ini1+/Δexon6,7 heterozygous mice confirm the published work of others (8-10).
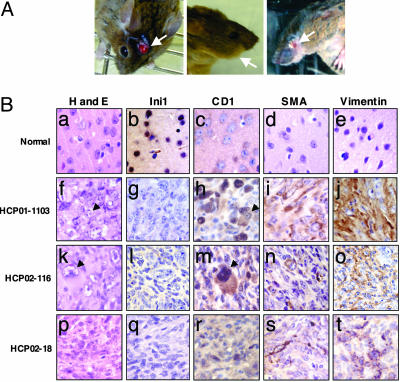
Fig. 3.
Cyclin D1 is overexpressed in tumors developed in Ini1+/Δexon6,7 mice. (A) Ini1+/Δexon6,7 mice develop head and face tumors. (Left) Tumor present in the soft tissue of the brain penetrating the skull. (Center) Enlarged side of the face because of the presence of tumor mass behind the cheek. (Right) Tumor under the cerebellum has dislodged the eye. (B) Immunostain analysis of tumors developed in Ini1+/Δexon6,7 mice. First row: paraffin section of normal brain tissue adjacent to tumor HCP01-1103. Second, third, and fourth rows represent the immunostain analysis of the paraffin sections of three different mouse tumors (HCP01-1103, HCP02-116, and HCP02-18, respectively) developed in Ini1+/Δexon6,7 mice. Column 1, hematoxylin and eosin (H and E) staining (a, f, k, and p). Columns 2-5 represent immunohistochemical staining using anti-INI1 (b, g, l, and q), anti-Cyclin D1 (c, h, m, and r), anti-smooth muscle actin (SMA) (d, i, n, and s); and anti-vimentin (Vim) (e, j, o, and t) antibodies, respectively. The arrows point to rhabdoid cells.
A prerequisite to determining the importance of Cyclin D1 in the genesis of rhabdoid tumors is to demonstrate that this gene indeed is derepressed even in mouse rhabdoid tumors. Therefore, we next characterized the mouse tumors by immunohistochemical analysis using affinity-purified anti-INI1 and anti-Cyclin D1 antibodies, as well as other antibodies against markers characteristic of rhabdoid tumors, such as vimentin and smooth muscle actin (3) (Fig. 3B). The results of these analyses indicated that the mouse tumors are similar to those of human rhabdoid tumors in all respects: They were negative for Ini1 and positive for vimentin and smooth muscle actin. Lack of Ini1 in the tumor tissue is consistent with the fact that these tumors arose because of the loss of heterozygosity at the Ini1 locus. Importantly, Cyclin D1 was well expressed in all of the tumors examined, including the rhabdoid cells present in these tumors (Fig. 3B). These results are consistent with our hypothesis that Ini1 loss leads to the derepression of Cyclin D1. Thus, these knockout mice represent an excellent model system for testing the dependence on Cyclin D1 for genesis of rhabdoid tumors.
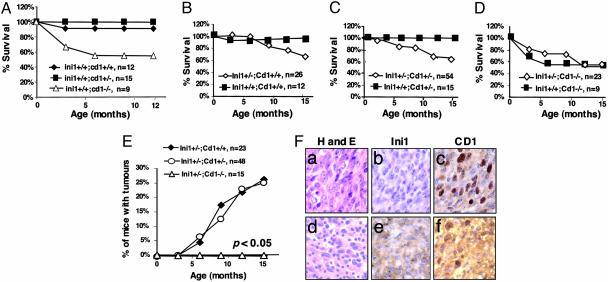
Cyclin D1 Is Necessary for Rhabdoid Tumor Genesis. Because INI1 represses Cyclin D1 and because Cyclin D1 is derepressed in rhabdoid tumors, it is possible that expression of Cyclin D1 is a prerequisite for the genesis of rhabdoid tumors. To test the hypothesis that Cyclin D1 derepression is necessary for the rhabdoid tumors formation in mice, we crossed the heterozygous Ini1+/Δexon6,7 mice with Cyclin D1+/- mice to obtain double heterozygous mice. Mice carrying targeted alleles of Cyclin D1 were generated by Sicinski et al. (30) by replacing one-half of exon 1 and all of exons 2 and 3 with a neomycin cassette. Ini1 and Cyclin D1 heterozygous mice that were obtained from these crosses were mated to each other to generate Ini1+/Δexon6,7 mice with three different Cyclin D1 backgrounds (Cyclin D1+/+, +/-, and -/-). No mice homozygous for Ini1 mutation were found in these crosses, indicating that Cyclin D1 mutation is not epistatic for the embryonic lethality caused by homozygous Ini1 deletions. We obtained two cohorts of mice of six different genetic backgrounds (Ini1+/+ or Ini1+/- mice with three different Cyclin D1 genotypes) and observed them for a period of 15 months. We found that the Ini1+/+ mice with three different Cyclin D1 genotypes exhibited expected phenotypic characteristics as reported earlier (30). Whereas the Cyclin D1+/+ and Cyclin D1+/- mice exhibited normal survival rates, the Cyclin D1-/- mice demonstrated reduced rate of survival because of early neonatal mortality (Fig. 4A). The Cyclin D1-/- mice were smaller than their littermates and exhibited minor neurological and retinal developmental defects in both Ini1+/+ and Ini1+/- backgrounds, indicating that these characteristics are due to the manifestation of the Cyclin D1 lesions and that they were not related to Ini1 heterozygosity (Fig. 4).
Fig. 4.
Analysis of Ini1 mice with different Cyclin D1 genetic backgrounds. (A) Survival profile of mice with three different Cyclin D1 genetic backgrounds. (B-D) Comparison of the survival rate of Ini1+/+ with Ini1+/Δexon6,7 mice with three different Cyclin D1 genotypes. Although survival rate of mice with Cyclin D1+/+ and Cyclin D1+/- background was reduced when Ini1+/Δexon6,7 genotype was present because of tumor formation, the survival rates of mice with Cyclin D1-/- were identical for both Ini1+/+ and Ini1+/Δexon6,7 mice. (E) Tumor formation frequency of Ini1+/Δexon6,7 mice in different Cyclin D1 backgrounds. Ini1+/Δexon6,7; Cyclin D1+/+ mice and Ini1+/Δexon6,7; Cyclin D1+/- mice develop rhabdoid-like tumors (≈25%), whereas Ini1+/Δexon6,7; Cyclin D1-/- do not develop rhabdoid tumors (P < 0.05). (F) Immunohistochemical analysis of tumors developed in Ini1+/Δexon6,7 in Cyclin D1+/- background. Upper HCPO2-713 and Lower HCPO3-710. (Left) Hematoxylin and eosin (H and E) staining. (Center) Anti-INI1 antibody staining. (Right) Anti-Cyclin D1 antibody staining.
Comparative analysis of survival of the cohort of Cyclin D1 mice with two different Ini1 genetic backgrounds is depicted in Fig. 4 B-D. We found that both Cyclin D1+/+ and Cyclin D1+/- mice with the Ini1+/- heterozygous background exhibited a reduced survival rate as compared to the Ini1+/+ mice with similar Cyclin D1 background (Fig. 4 B and C). However, there was no significant difference in the survival rate of Cyclin D1-/- mice with either Ini1+/+ or Ini1+/- background. Both of these mice exhibited early neonatal mortality, but adult mice survived at an equal frequency (Fig. 4D). We next observed Ini1+/- mice with three different Cyclin D1 genetic back-grounds for tumor formation frequency for a period of up to 15 months. The Cyclin D1+/+ or +/- mice developed tumors with a frequency similar to what we observed before (26.1% in Cyclin D1+/+ and 25% in Cyclin D1+/-), with a median onset of ≈9 months of age. The majority of the tumors developed in the face and head. Immunohistopathological characterization of representative tumors from these cohorts of mice indicated that they were rhabdoid in nature. The tumors exhibited the presence of rhabdoid cells with typical morphology, were positive for vimentin and smooth muscle actin, and overexpressed Cyclin D1 (Fig. 4F and data not shown). Interestingly, although one of the tumors we observed was negative for Ini1 staining, another tumor exhibited strong cytoplasmic localization of the Ini1 protein (Fig. 4F). This observation strengthens our previous report that in human tumors, one mechanism of inactivation of INI1 is by mislocalization of the protein in the cytoplasm due to the activation of the masked nuclear export signal (33).
The striking result of these analyses was that none of the Ini1+/- mice that carried the Cyclin D1-/- genotype developed tumors within the observation period (Fig. 4E). These results are significant (P < 0.05) and indicate that Cyclin D1 deletion overcomes the tumorigenic effect of Ini1 loss. This observation is in contrast to the effect of Cyclin D1 deletion on embryonic lethality of Ini1-/- mutation and indicates that tumorigenesis due to Ini1 loss in vivo is exquisitely dependent on the presence of Cyclin D1.
Discussion
Our results provide strong in vivo evidence to support the hypothesis that Cyclin D1 expression is essential for the genesis of rhabdoid tumors. Our previous report indicated that (i) Cyclin D1 is repressed by INI1 in rhabdoid tumor cells; (ii) expression of Cyclin D1 from a heterologous promoter overcomes the cell cycle arrest caused by INI1; and (iii) human rhabdoid tumors express Cyclin D1. In this report we demonstrate that (i) Cyclin D1 is expressed in all of the rhabdoid cell lines, but other D-type Cyclins are not consistently expressed; (ii) INI1 represses transcription of Cyclin D1 but not D2 and D3 genes. Analysis of rhabdoid tumors developed in Ini1 knockout mice generated in our laboratory indicated that, similar to human rhabdoid tumors, mouse rhabdoid tumors also express Cyclin D1. Furthermore, our results demonstrated that Ini1+/- mice lacking Cyclin D1 failed to develop rhabdoid tumors. These results provide in vivo evidence to indicate that Cyclin D1 is at least one of the critical determinants for the genesis of rhabdoid tumors and that deletion of Cyclin D1 is sufficient to eliminate rhabdoid tumor formation.
Cyclin D1 has been implicated to have multiple roles during cell cycling, propagation, and tumorigenesis (15, 17). It is a sensor of mitogenic extracellular stimuli, and it is required to overcome the G1 checkpoint and to facilitate the transition of cells from G1 to S stage of the cell cycle. It has also been implicated to be required for cell growth and proliferation. Furthermore, it is a cellular oncogene thought to cooperate with E1a, Ras, and c-Myc. Because Cyclin D1 is the only D-type cyclin that is consistently expressed in rhabdoid cells, it is possible that the cell cycle progression and/or survival of these cells and tumors are dependent on this critical factor. In mammary carcinogenesis, it has been demonstrated that mammary tumors induced by Neu and Ras but not those induced by c-Myc are dependent on the presence of Cyclin D1. These results in breast carcinogenesis imply that Cyclin D1 is involved in the Neu-Ras pathway. Thus far, there is no information or report in the literature that indicates the involvement of the Ras-Neu pathway in rhabdoid tumorigenesis. Our results provide evidence that Cyclin D1 is required for rhabdoid tumorigenesis. Therefore, it will be interesting in the future to determine whether there is a connection between Cyclin D1 and Ras-Neu during rhabdoid tumorigenesis. One alternative possibility for explaining the observed results is that genetic ablation of Cyclin D1 leads to the elimination of precursors to rhabdoid cells because these cells may require the expression of this gene for propagation. However, at this point, this question cannot be easily addressed because the cell type of origin of rhabdoid tumors is unknown. Furthermore, it is not clear why rhabdoid tumors originate from multiple tissues, making it difficult to test this possibility.
Our studies indicate the exquisite dependence of rhabdoid tumors on Cyclin D1. The fact that Cyclin D1 is repressed by Ini1 in rhabdoid cell lines alone does not provide proof of principle that Cyclin D1 is indeed required for tumorigenesis in vivo, because Ini1 regulates a number of genes in these cell lines. Furthermore, Cyclin D1 is overexpressed in many types of tumors. In these cases (except in a subset of breast cancers), it is not clear whether Cyclin D1 derepression is the cause or just a consequence of an event during tumorigenesis. Therefore, it is important to address this question in vivo, and our results provide a definitive, genetic proof for the essential role of Cyclin D1 in rhabdoid tumorigenesis. Furthermore, our study suggests that Cyclin D1 has the potential to be an excellent drug target. We propose that drugs that specifically target CylinD1 expression and/or its activity could be used to treat rhabdoid tumors. Interestingly, current drug regimens for rhabdoid tumors include neither Cyclin D1 nor cyclin-dependent kinase inhibitors. Thus, a therapeutic strategy targeted to inhibit Cyclin D1 may be effective in treating this malignancy, which typically has a poor prognosis.
Acknowledgments
We thank Drs. V. R. Prasad and J. Lenz for critically reading the manuscript; R. Kucherlapathi for providing the RPCI-22 bacterial artificial chromosome library; Drs. P. Sicinski and R. G. Pestell for providing Cyclin D1-knockout mice; and Dr. R. Russell, H. Hou, K. Challagulla, and D. Carrion for technical assistance. This work was supported by grants from the Children's Brain Tumor Foundation, New York (to G.V.K.), National Cancer Institute Grant CA09060 to the Albert Einstein College of Medicine Cancer Center, and American Cancer Society Grant CCG-104933 (to G.V.K.). G.V.K. and W.E. are Irma T. Hirschl Scholars, and G.V.K. is a Mark Trauner Faculty Scholar in Neuro-oncology.
Author contributions: G.V.K. designed research; M.T., Z.Z., and D.Z. performed research; W.E. contributed new reagents/analytical tools; M.T. and G.V.K. analyzed data; and M.T. and G.V.K. wrote the paper.
Abbreviations: AT/RT, atypical teratoid and rhabdoid tumors; ES cells, embryonic stem cells.
References
- 1.Biegel, J. A., Kalpana, G., Knudsen, E. S., Packer, R. J., Roberts, C. W., Thiele, C. J., Weissman, B. & Smith, M. (2002) Cancer Res. 62, 323-328. [PubMed] [Google Scholar]
- 2.MacDonald, T. J., Rood, B. R., Santi, M. R., Vezina, G., Bingaman, K., Cogen, P. H. & Packer, R. J. (2003) Oncologist 8, 174-186. [DOI] [PubMed] [Google Scholar]
- 3.Packer, R. J., Biegel, J. A., Blaney, S., Finlay, J., Geyer, J. R., Heideman, R., Hilden, J., Janss, A. J., Kun, L., Vezina, G., et al. (2002) J. Pediatr. Hematol. Oncol. 24, 337-342. [DOI] [PubMed] [Google Scholar]
- 4.Biegel, J. A., Allen, C. S., Kawasaki, K., Shimizu, N., Budarf, M. L. & Bell, C. J. (1996) Genes Chromosomes Cancer 16, 94-105. [DOI] [PubMed] [Google Scholar]
- 5.Biegel, J. A., Zhou, J. Y., Rorke, L. B., Stenstrom, C., Wainwright, L. M. & Fogelgren, B. (1999) Cancer Res. 59, 74-79. [PubMed] [Google Scholar]
- 6.Versteege, I., Sevenet, N., Lange, J., Rousseau-Merck, M. F., Ambros, P., Handgretinger, R., Aurias, A. & Delattre, O. (1998) Nature 394, 203-206. [DOI] [PubMed] [Google Scholar]
- 7.Sevenet, N., Sheridan, E., Amram, D., Schneider, P., Handgretinger, R. & Delattre, O. (1999) Am. J. Hum. Genet. 65, 1342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guidi, C. J., Sands, A. T., Zambrowicz, B. P., Turner, T. K., Demers, D. A., Webster, W., Smith, T. W., Imbalzano, A. N. & Jones, S. N. (2001) Mol. Cell. Biol. 21, 3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts, C. W., Leroux, M. M., Fleming, M. D. & Orkin, S. H. (2002) Cancer Cells 2, 415-425. [DOI] [PubMed] [Google Scholar]
- 10.Klochendler-Yeivin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C. & Yaniv, M. (2000) EMBO Rep. 1, 500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, W., Cote, J., Xue, Y., Zhou, S., Khavari, P. A., Biggar, S. R., Muchardt, C., Kalpana, G. V., Goff, S. P., Yaniv, M., et al. (1996) EMBO J. 15, 5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, Z. K., Davies, K. P., Allen, J., Zhu, L., Pestell, R. G., Zagzag, D. & Kalpana, G. V. (2002) Mol. Cell. Biol. 22, 5975-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betz, B. L., Strobeck, M. W., Reisman, D. N., Knudsen, E. S. & Weissman, B. E. (2002) Oncogene 21, 5193-5203. [DOI] [PubMed] [Google Scholar]
- 14.Vries, R. G., Bezrookove, V., Zuijderduijn, L. M., Kia, S. K., Houweling, A., Oruetxebarria, I., Raap, A. K. & Verrijzer, C. P. (2005) Genes Dev. 19, 665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, E. K., Ng, S. C., Arber, N., Begemann, M. & Weinstein, I. B. (1999) Apoptosis 4, 213-219. [DOI] [PubMed] [Google Scholar]
- 16.Katabami, M., Dosaka-Akita, H., Mishina, T., Honma, K., Kimura, K., Uchida, Y., Morikawa, K., Mikami, H., Fukuda, S., Inuyama, Y., et al. (2000) Hum. Pathol. 31, 973-979. [DOI] [PubMed] [Google Scholar]
- 17.Lamb, J. & Ewen, M. E. (2003) Cell Cycle 2, 525-527. [DOI] [PubMed] [Google Scholar]
- 18.Mishina, T., Dosaka-Akita, H., Kinoshita, I., Hommura, F., Morikawa, T., Katoh, H. & Kawakami, Y. (1999) Br. J. Cancer 80, 1289-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Utsunomiya, T., Doki, Y., Takemoto, H., Shiozaki, H., Yano, M., Sekimoto, M., Tamura, S., Yasuda, T., Fujiwara, Y. & Monden, M. (2001) Oncology 61, 226-233. [DOI] [PubMed] [Google Scholar]
- 20.Yamanouchi, H., Furihata, M., Fujita, J., Murakami, H., Yoshinouchi, T., Takahara, J. & Ohtsuki, Y. (2001) Lung Cancer 31, 3-8. [DOI] [PubMed] [Google Scholar]
- 21.Yu, Q., Geng, Y. & Sicinski, P. (2001) Nature 411, 1017-1021. [DOI] [PubMed] [Google Scholar]
- 22.Diehl, J. A. (2002) Cancer Biol. Ther. 1, 226-231. [DOI] [PubMed] [Google Scholar]
- 23.Kato, J. Y., Matsuoka, M., Strom, D. K. & Sherr, C. J. (1994) Mol. Cell. Biol. 14, 2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacey, D. W. (2003) Curr. Opin. Cell Biol. 15, 158-163. [DOI] [PubMed] [Google Scholar]
- 25.Bienvenu, F., Gascan, H. & Coqueret, O. (2001) J. Biol. Chem. 276, 16840-16847. [DOI] [PubMed] [Google Scholar]
- 26.Ewen, M. E. & Lamb, J. (2004) Trends Mol. Med. 10, 158-162. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, K. & Sherr, C. J. (1998) Mol. Cell. Biol. 18, 1590-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb, J., Ramaswamy, S., Ford, H. L., Contreras, B., Martinez, R. V., Kittrell, F. S., Zahnow, C. A., Patterson, N., Golub, T. R. & Ewen, M. E. (2003) Cell 114, 323-334. [DOI] [PubMed] [Google Scholar]
- 29.Zwijsen, R. M., Buckle, R. S., Hijmans, E. M., Loomans, C. J. & Bernards, R. (1998) Genes Dev. 12, 3488-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sicinski, P., Donaher, J. L., Parker, S. B., Li, T., Fazeli, A., Gardner, H., Haslam, S. Z., Bronson, R. T., Elledge, S. J. & Weinberg, R. A. (1995) Cell 82, 621-630. [DOI] [PubMed] [Google Scholar]
- 31.Sicinska, E., Aifantis, I., Le Cam, L., Swat, W., Borowski, C., Yu, Q., Ferrando, A. A., Levin, S. D., Geng, Y., von Boehmer, H., et al. (2003) Cancer Cell 4, 451-461. [DOI] [PubMed] [Google Scholar]
- 32.Ioffe, E., Liu, Y., Bhaumik, M., Poirier, F., Factor, S. M. & Stanley, P. (1995) Proc. Natl. Acad. Sci. USA 92, 7357-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig, E., Zhang, Z. K., Davies, K. P. & Kalpana, G. V. (2002) EMBO J. 21, 31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, C. W., Galusha, S. A., McMenamin, M. E., Fletcher, C. D. & Orkin, S. H. (2000) Proc. Natl. Acad. Sci. USA 97, 13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]