Abstract
Genetic manipulation of hematopoietic stem and progenitor cells is an important tool for experimental and clinical applied hematology. However, techniques that allow for gene targeting, subsequent in vitro selection, and expansion of genetically defined clones are available only for ES cells. Such molecularly defined and, hence, “safe” clones would be highly desirable for somatic gene therapy. Here, we demonstrate that in vitro differentiated ES cells completely recapitulate the growth and differentiation properties of adult bone marrow cells, in vitro and in vivo, when ectopically expressing HOXB4. Myeloid development was enforced and (T) lymphoid development suppressed over a wide range of expression levels, whereas only high expression levels of the transcription factor were detrimental for erythroid development. This indicates a close association between the amounts of ectopic HOXB4 present within a progenitor cell and and the decision to self renew or differentiate. Because HOXB4 mediates similar fates of ES-derived and bone marrow hematopoietic stem cells, the primitive embryonic cells can be considered a promising alternative for investigating hematopoietic reconstitution, in vivo, based on well defined clones. Provided that HOXB4 levels are kept within a certain therapeutic window, ES cells also carry the potential of efficient and safe somatic gene therapy.
Keywords: gene therapy, hematopoiesis, transplantation
The best-defined system for clonally organized organ regeneration is the hematopoietic system. Its central organizer is the hematopoietic stem cell (HSC), which resides in the bone marrow cavity of all adult vertebrates. This rare multipotent and primarily dormant cell type (1, 2) is capable of choosing three different developmental fates when entering the cell cycle: self renewal, differentiation, or apoptosis (3). These different decisions appear to be controlled by a delicate balance of extrinsic signals and the intrinsic genetic program executed within the stem cell (4). Although control of this equilibrium is still poorly understood, it is clear that nonhematopoietic cells of the bone marrow microenvironment, the stem cell niche, seem to be critical for controlling stem cell numbers (5, 6).
Because of the ability of the HSC to restore the entire hematopoietic system, this cell type is of key importance for any clinical application aiming to achieve a life-long cure of the human hematopoietic compartment. Nevertheless, ex vivo culture and manipulation of HSCs without loss of their stem cell characteristics has remained a difficult task. On the other hand, ectopic expression of certain transcription factors normally expressed in HSCs has been shown to support stem cell maintenance and even expansion in vitro and in vivo (7–9). The first gene shown to lead to an expansion of HSCs when ectopically expressed in the mouse model was the human homeodomain transcription factor HOXB4 (10). Meanwhile, it has been repeatedly reported that ectopic expression of HOXB4 can mediate a significant expansion of HSCs of mice and humans in vitro and in vivo (11–17).
Ectopic expression of this protein also enhances hematopoietic development of pluripotent mouse ES cells in vivo (18, 19). This opens avenues for regenerative medicine by allowing efficient correction of genetic defects of the hematopoietic system by homologous recombination with subsequent hematopoietic differentiation and transplantation (19). When using vectors that integrate randomly into the host genome, use of ES cells could give the opportunity to preselect clones in which integration has not altered the activity of known protooncogenes (20, 21). Thus, expansion and transplantation of well defined clones, either ES-cell-derived or from adult HSCs, could increase the safety of gene therapy.
In vivo, HOXB4 is expressed in the stem cell fraction of the bone marrow and subsequently down-regulated during differentiation in humans (22, 23) and mice (24). Dysregulation of HOX genes is a common observation in different types of malignancies (25). However, although no leukemia has been reported after ectopic long-term expression of HOXB4 so far, the influence of HOXB4 expression levels on the balance of HSC self renewal and differentiation has not been investigated extensively. Our group (14) and others (26) have recently reported that increased HOXB4 expression perturbs the differentiation of transduced human CD34+ stem and progenitor cells in vitro and in the NOD/SCID mouse model in vivo.
In this work, we show that in vitro differentiated mouse ES and bone marrow cells of adult mice are almost indistinguishable during extended times of growth in vitro and after transplantation, in vivo, when expressing HOXB4 from a retroviral vector. Furthermore, we show that the amount of ectopically expressed HOXB4 influences differentiation in both systems similarly. HOXB4 enforced myeloid and suppressed (T) lymphoid development over a wide range of expression levels, whereas very high expression levels of HOXB4 attenuated late stages of erythroid development. Thus, HOXB4 enforces similar fates of in vitro differentiated ES cells and adult bone marrow cells.
Methods
Supporting Text. Details of ES cell culture (27), retroviral transduction (13), generation of ES-cell-derived hematopoietic and erythroid progenitor cultures (28), and in vivo experiments are described in Supporting Text, which is published as supporting information on the PNAS web site.
Production of Ecotropic Retroviral Vectors. Phoenix-gp packaging cells (G. Nolan, Stanford University School of Medicine, Palo Alto, CA) were transfected with 10 μg of each murine stem cell virus (MSCV) or spleen focus virus-derived 91 (SF91) derivative together with 10 μg of a murine leukemia virus gag-pol and 5 μg of an ecotropic env expression plasmid. Cell culture supernatants were collected after 24, 48, and 72 h and filtered through 0.45-μm filter units (Millipore). Titers of supernatants were determined on SC-1 fibroblasts by measuring GFP expression after 48 h. The percentage of positive cells was determined for each dilution and the mean value (= titer per ml of supernatant) calculated.
Flow Cytometry. All monoclonal antibodies were purchased (Pharmingen, BD Biosciences); the IL7-R antibody was a kind gift of M. Busslinger (Research Institute of Molecular Pathology, Vienna). Red cells were lysed with hemolytic buffer (150 mM ammonium chloride/12 mM sodium bicarbonate/0.1 mM EDTA). Nucleated cells (5 × 105 to 1 × 106) were washed in staining medium (4% FCS, 1 × PBS) and preincubated in this medium containing 1 μg/106 cells with unconjugated anti-CD16/32 antibodies for 10 min at 4°C. Subsequent staining reactions were incubated for 30 min at 4°C and washed twice. Dead cells were excluded from the analysis by propidium iodide (Sigma). Analysis was performed on a FACScalibur flow cytometer (Becton Dickinson, Immunocytometry Systems, San Jose, CA) by using cellquest software.
Statistical Analysis. For all statistical analyses, prism (Version 3.03, GraphPad, San Diego) software was used. Dispersion is presented as the SEM, unless stated otherwise. Student's t test (two-sided) was applied for paired or nonpaired samples, assuming G distribution. Differences with P < 0.05 were considered statistically significant.
Results
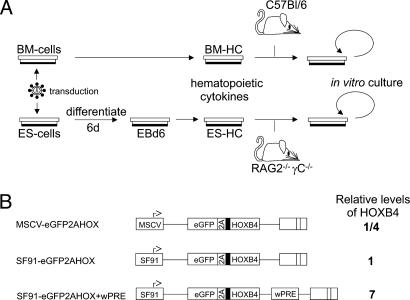
First, we addressed the question whether in vitro differentiated ES-cell-derived hematopoietic cells (ES-HCs) ectopically expressing HOXB4 posses the full functional competence to replace adult bone marrow-derived hematopoietic cells (BM-HCs) as a source for transplantation. We therefore performed the experiments for both the ES-HC and BM-HC system in a similar fashion. The setup is depicted in Fig. 1A.
Fig. 1.
Experimental design for comparison of embryonic and adult stem cell-derived hematopoiesis and retroviral vectors used for HOXB4 expression. (A) Transduction, culture, and transplantation procedure. ES cells (CCE) and prestimulated primary mouse bone marrow (C57BL/6J, CD45.2) were treated with ecotropic retroviral HOXB4 coexpression (and eGFP control) vector particles (as described in Methods). After differentiation to EBs for 6 days, EBs were dissociated and cultured in medium supportive for hematopoiesis. HOXB4 transduced ES-HCs and BM-HCs were transplanted into six irradiated Rag2(–/–)γC(–/–) recipient mice and C57BL/6J mice (CD45.1, 10 mice per vector), respectively. (B) Retroviral expression vector architecture and expression levels of HOXB4. For HOXB4 expression, retroviral backbones of MSCV (30, 31) and SF91 (44) were used, the latter with or without an additional wPRE (45). The coexpression cassette eGFP2AHOXB4 leads to cotranslational separation of eGFP and HOXB4 [with an additional N-terminal hemagglutinin (HA) tag] mediated by the 2A-esterase of foot-and-mouth disease virus, resulting in a constant ratio of both proteins (13). The relative expression levels of HOXB4 as determined by Western blot are indicated (14). The HA tag is symbolized as filled black block.
To follow expression of HOXB4 throughout the in vitro and in vivo experiments in a qualitative and quantitative manner, we used a coexpression system based on cotranslational separation of eGFP and HOXB4 by the 2A esterase of foot-and-mouth disease virus (13, 14, 29). Its activity leads a stable molar ratio of both proteins, thus allowing for an indirect quantitative measurement of HOXB4 at the single cell level via flow cytometrical determination of eGFP expression. Both well characterized retroviral vector backbones MSCV (30, 31) and SF91 (32), with and without the posttranscriptional regulatory element of woodchuck hepatitis virus (wPRE), allowed us to achieve expression levels in HOXB4-transduced cells ranging from one-fourth of up to 7-fold higher levels than in cells transduced by the expression vector used by Humphries and colleagues (10). The expression levels of the vectors used have been tested in vitro and in vivo (13, 14) (Fig. 1B).
ES-HCs and BM-HCs Ectopically Expressing HOXB4 Show Comparable Growth Behavior in Vitro. For differentiation toward HSC and progenitor cells, mouse ES cells were first transduced with retroviral expression vectors. EGFP-positive cells were then sorted by flow cytometry and subsequently differentiated into embryoid bodies (EB) for 6 days. In the EB, retroviral expression was turned off in ≈75% of the transduced cells, possibly as a consequence of DNA methylation (33). Importantly, however, retroviral expression of HOXB4 was stable and did not alter the growth properties of ES cells before differentiation (data not shown). The day 6 EB were dissociated and cultured in serum-free medium containing cytokines supportive for hematopoiesis (see Supporting Text). To keep the culture conditions as defined as possible, no stromal coculture system was involved. These cells, which we hereafter term ES-HCs, were used for comparison with transduced primary bone marrow cells (BM-HCs), in vitro and in vivo.
In both ES-HC and BM-HC bulk cultures, HOXB4 expression mediated a competitive growth advantage to the transduced cells, rapidly leading to a culture consisting solely of cells ectopically expressing HOXB4 (Fig. 5, which is published as supporting information on the PNAS web site). This effect was independent of the expression levels of HOXB4 in the BM-HCs (Fig. 5 Upper). In ES-HCs, growth kinetics were similar, both in bulk cultures and in cultures derived from single ES-HC clones (Fig. 5 Lower Right). In the case of ES-HCs, the eGFP control vector-transduced cells ceased growth under the conditions used, consistent with previously published data (18).
In summary, HOXB4 not only enabled in vitro differentiated ES-cells cells to survive and grow continuously without stromal support under the culture conditions used but also conferred a selective growth advantage to transduced ES-HCs that was comparable to that of transduced adult mouse bone marrow cells.
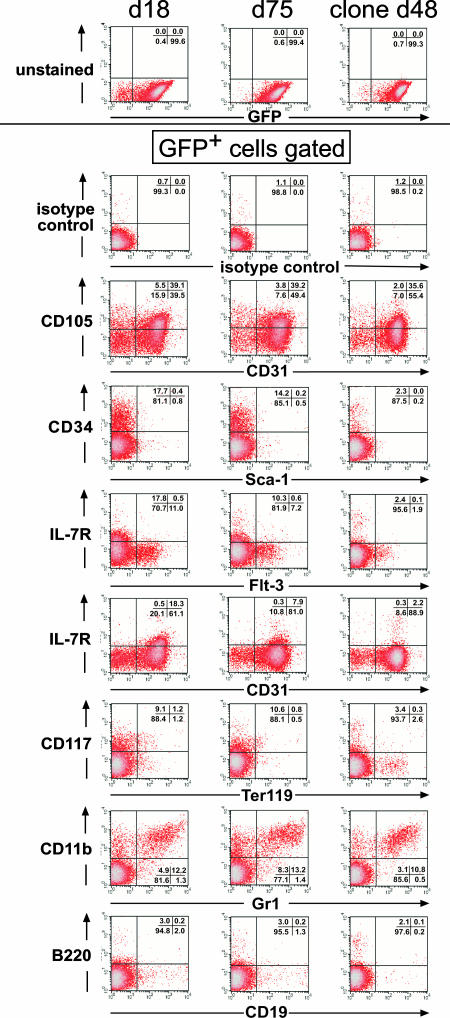
HOXB4-ES-HCs Stably Express Hematopoietic Surface Markers During Culture in Bulk and Clonally in Vitro. To address the important question whether the HOXB4-transduced ES-HCs would maintain their immature presumably multipotent state for longer periods of in vitro culture, we analyzed the expression of certain surface markers associated with immature and more mature hematopoietic cells over time. Fig. 2 clearly shows that the main proportion of transduced cells stably expressed these markers for at least 2 months. The majority of them expressed CD31 (PECAM-1) with a subpopulation also positive for CD105 (endoglin) expression. ES-cell-derived HCs have been shown to express CD105+ and CD31+ (34). Furthermore, endoglin-positive cells have been reported to contain essentially all of the long-term repopulating activity within the “side population” bone marrow cells (35, 36). In addition, the expression of molecules such as CD34, CD117 (c-kit), CD11b, and Gr1 indicates the presence of hematopoietic progenitor cells.
Fig. 2.
FACS analysis of HOXB4 transduced ES-HCs maintained as bulk or clonal cultures, in vitro. SF-HOX vector (SF91-eGFP2AHOXB4+wPRE) transduced bulk ES-HCs grown in vitro for 18 or 75 days or one representative clone, grown for 48 days, were immunostained against the assigned surface antigens. GFP+ and live cells subjected to FACS analysis were gated electronically (Upper) and analyzed for the assigned surface antigen expression. The quantitation of quadrant analysis is shown within each FACS analysis diagram.
The same stable distribution of surface molecule expression was observed not only at different time points during bulk culture but also on single cell clone progeny expanded in vitro (Fig. 2 Right). As expected, erythroid (Ter119+) and B cell progenitors (B220+ and CD19+) were hardly detectable under the culture conditions used. The characterization of the ES-HCs cells growing for 30 days in vitro was extended by semiquantitative RT-PCR analysis for the presence of transcripts associated with erythro-, lympho-, and myelopoiesis and confirmed the presence of progenitor cells in the ES-HC culture (data not shown). These results were additionally verified by colony formation assays and investigation of the morphological appearance of the cells on Giemsa-stained cytospin preparations (Fig. 6, which is published as supporting information on the PNAS web site). Taken together, these results show that ectopically expressed HOXB4 effectively mediates long-term growth of hematopoietic progenitor cells derived from in vitro differentiated ES cells.
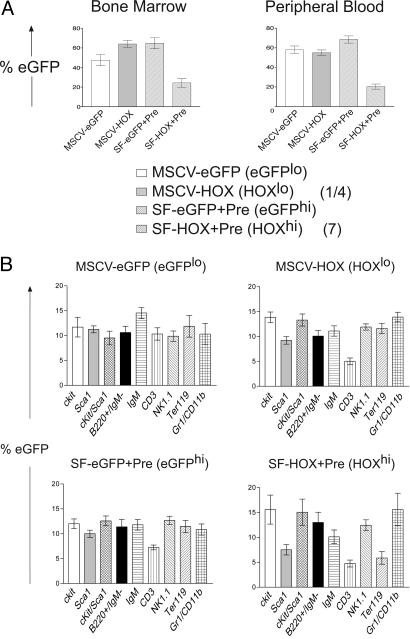
Overall Repopulation Efficiency and Lineage Differentiation Critically Depends on HOXB4 Expression Levels in Vivo. Recently, an inhibition of lymphoid and erythroid differentiation has been reported after transduction of mouse yolk sac and EB cells with HOXB4 (18, 19). Whether these effects were intrinsic properties of the in vitro differentiated ES cells or a result of ectopic HOXB4 expression remained unclear. At least for human HSCs and progenitor cells, we and others have already shown that the expression level of HOXB4 directly influences their differentiation behavior (14, 26). To test whether lineage distribution in mice, in vivo, also depends on the amounts of HOXB4 expressed in certain progenitor populations, we compared the influence of HOXB4 expression levels on hematopoietic differentiation in C57BL/6 mice, in vivo, using two retroviral vectors, MSCV and SF91+wPRE, which mediate a 28-fold difference in HOXB4 protein expression, respectively (14) (Fig. 1B). Thirteen weeks posttransplantation, the influence of HOXB4 expression levels on overall repopulation efficiency and lineage distribution was determined. Although the transduction efficiencies of all vectors, as determined immediately before transplantation, were found to be comparable (≈10%, data not shown), high expression levels of HOXB4 led to a statistically significant reduced engraftment of the transduced cells (SF91-HOX+Pre, Fig. 3A). The reason for this effect was not investigated and is beyond the scope of this work. Fig. 3B shows a comparison of the different HOXB4 expression levels and their influence on the relative contribution of transduced cells to certain lineages. It suggests a high sensitivity of CD3+ T lymphocytes toward a broad range (28-fold) of HOXB4 levels as both MSCV and SF91 retroviral vectors expressing low and high amounts of HOXB4, respectively, led to a significant reduction of transduced cells within this population. In contrast, only high levels of HOXB4 expression led to a significant reduction of the proportion of transduced cells within the Ter119+ erythroid cell compartment. Low levels of HOXB4 expression (MSCV-based) did not interfere with red blood cell differentiation.
Fig. 3.
Influence of HOXB4 expression levels on overall repopulation and lineage distribution in vivo. (A) HOXB4 expression levels affect in vivo repopulation of transduced bone marrow cells. C57BL/6J CD45.2 bone marrow cells were transduced with retroviral expression vectors, which mediate a 28-fold difference in HOXB4 expression levels (14). Thirteen weeks after transplantation, bone marrow and peripheral blood was analyzed for repopulation by GFP+ cells. Engraftment by CD45.2 cells was comparable (≈80%). Repopulation by GFP+ cells was significantly reduced in bone marrow and the periphery of the SF-HOX+Pre (HOXhigh) group (SF91-eGFP vs. SF-HOX+Pre, BM and PB: P < 10–4). HOXB4 expression levels relative to the reference vector described in ref. 10 are shown in parentheses next to the construct designations. (B) Influence of HOXB4 expression levels on the lineage distribution in bone marrow in vivo. Each column represents the relative contribution of cells transduced with different retroviral vectors to a certain lineage in the competitive repopulation assay (see Methods). For statistical calculations, the proportion of GFP+ cells within each lineage was first determined for each mouse group (eGFPlo, eGFPhi, HOXlo, and HOXhi). These values were summarized for each mouse, set to 100%, and the relative contribution of each of the lineages recalculated. The relative contributions of transduced cells were compared pair-wise by the two-sided Student's t test. The significance level was defined as 0.05. Error bars represent standard deviations. Statistics for CD3+ lymphocytes and Ter119+ erythroid cells: CD3+: eGFPlo vs. HOXlo, P = 0.003; eGFPhi vs. HOXhi, P = 0.021; Ter119+: eGFPlo vs. HOXlo, P = 0.920; eGFPhi vs. HOXhi, P = 0.0122; HOXlo vs. HOXhi: P = 0.005.
Directed Erythroid Differentiation of ES Cells in Vitro Is Inhibited by High HOXB4 Expression Levels. The negative influence of high levels of HOXB4 expression on in vivo erythropoiesis was evaluated by using a well defined experimental system for directed differentiation of mouse ES cells into adult definitive erythroid progenitors [ES-cell-derived hematopoietic and erythroid progenitor (ES-EPs); see Methods] (28, 37). Under these conditions, unmanipulated wild-type control cultures efficiently differentiated into Ter119+ proerythroblasts. Nonerythroid CD11b+ cells were hardly detected (Fig. 7A Left, which is published as supporting information on the PNAS web site). In contrast, ES-EPs expressing high amounts of HOXB4 (≈30% of all HOXB4-expressing cells, Fig. 7A Right, left quadrant), as measured via the 2A-based EGFP coexpression strategy, could not completely differentiate toward Ter119+ proerythroblasts and contained a significant amount of cells with myeloid appearance (Fig. 7B Right) that are highly EGFP+ (Fig. 7A Right; reflected by the mean EGFP fluorescence of CD11b+ cells; 7.8% of all live cells).
Thus, hematopoietic lineage differentiation critically depends on the amount of HOXB4 in stem and progenitor cells. Expression of HOXB4 above a certain level most likely explains the reported reduced capability of erythroid differentiation of respective transduced yolk sac and EB cells (18, 19).
HOXB4 Transduced ES-HCs and BM-HCs Show Comparable Properties in Vivo. It has been reported that mouse yolk sac cells and EB cells engineered to ectopically express HOXB4 can repopulate a lethally irradiated recipient animal (18, 19). Nevertheless, it was unclear whether these cells can be used instead of adult bone marrow for complete repopulation because of the reported low level of lymphoid and erythroid engraftment. We therefore tested whether the HOXB4+ ES-HCs can functionally replace adult bone marrow cells after transplantation and compared the outcome of repopulation by BM-HCs and ES-HCs. In the case of BM-HCs, the well established C57BL/6 syngeneic donor/recipient mouse model system was used as a recipient. For ES-HC donor cells, the Rag2(–/–)γC(–/–) immune-deficient model was chosen to avoid graft rejection by natural killer cells (19, 38).
Due to the difficulty of estimating the competitive repopulating unit frequency in the ES-HC population before transplantation, we chose to compare mice from the ES-HC and BM-HC transplanted groups in which a similar overall extent of GFP+ bone marrow cells and, hence, a similar engraftment rate, was measured (39). This was the case 7 months after serial transplantation of 105 bone marrow cells each from primary C57BL/6 recipients, which themselves had been repopulated with a mixture of HOXB4-transduced (or eGFP control) cells and dsRed2-expressing competitor cells for ≈1 year. The primary recipient mice showed only low overall repopulation rates as a result of high expression levels of HOXB4 (Fig. 3A).
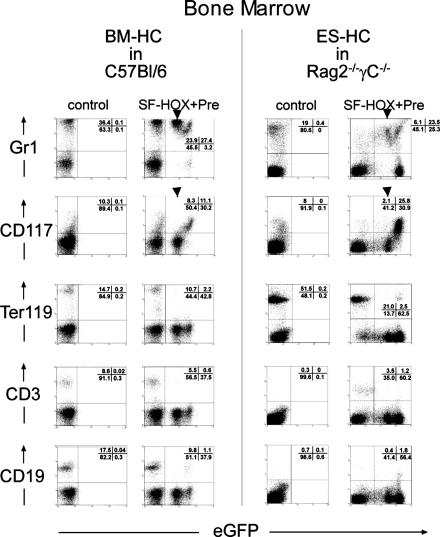
The Rag2(–/–)γC(–/–) mice were analyzed 6 months posttransplantation with HOXB4+ ES-HCs. FACS analysis of the lineage distribution in the bone marrow from a representative mouse of each experimental line is shown in Fig. 4. The results clearly demonstrate that the repopulation pattern of transduced BM-HCs and ES-HCs was almost indistinguishable and, importantly, independent of the different genetic backgrounds of the recipient mice. In summary, there was an obvious dominance of myeloid (Gr1+ cells, Fig. 4, top) and CD117+ (c-kit+) cells in the transduced population. Noteworthy is the comparable surface marker expression height of Gr1 and CD117 in relation to HOXB4 expression (reflected by the EGFP fluorescence intensity). At intermediate HOXB4 expression levels, transduced (GFP+) bone marrow cells were mainly Gr1+ but CD117–, both in C57BL/6 and Rag2–/–γC–/– mice (Fig. 4, top, “intermediate” subpopulations marked with an arrow). However, at higher HOXB4 expression levels, the proportion of CD117+ cells concomitantly increased, with a parallel reduction of Gr1+ myeloid cells. This was most obvious in mice showing an overall higher average HOXB4 expression (Fig. 4 Right, Rag2–/–γC–/–). Lymphoid (CD3, CD19, Fig. 4, bottom) and erythroid repopulation (Ter119) was nearly absent, in accordance to previous reports (18, 19). Thus, the most likely explanation for the low frequency within the transduced population is an incompatibility of high levels of HOXB4 expression with differentiation of certain lineages. In summary, these results demonstrate that HOXB4 confers comparable engraftment qualities both to embryonic-derived hematopoietic and adult bone marrow stem and progenitor cells, in vivo.
Fig. 4.
FACS analysis of bone marrow cells from C57BL/6J and Rag2(–/–)γC(–/–) posttransplantation of HOXB4 expressing BM-HCs or ES-HCs, respectively. In the case of C57BL/6J mice, SF-HOX (SF91-eGFP2AHOXB4+wPRE) or SF-eGFP (SF91-eGFP+wPRE, control) transduced bone marrow cells were transplanted into 10 primary recipient mice each, together with SF-dsRed2 (SF91-dsRed2+wPRE) transduced bone marrow cells. After 54 weeks of repopulation, 103, 104, and 105 cells of each control and HOX mice were serially transplanted each into four secondary recipient animals, which were finally analyzed after 7 months. Rag2(–/–)γC(–/–) mice were transplanted with 2.5 × 106 SF-HOX transduced ES-HCs grown for 20 days in vitro. As a control, the same amount of freshly prepared Rag2(–/–)γC(–/–) bone marrow cells was transplanted. Analysis was performed 6 months posttransplantation. FACS analysis of bone marrow cells from representative animals from both experimental approaches is shown. x axis, eGFP (= HOXB4) expression height; y axis, surface marker. Quadrant analysis, given as percentages, is shown as Insets within the FACS diagrams. Arrows mark cell populations expressing intermediate amounts of HOXB4.
HOXB4 Expression Leads to Myeloid Proliferation (Nonreactive), Subclass Myeloproliferation, in Mice Transplanted Either with Transduced ES-HCs or BM-HCs. Despite the pronounced shift of the HOXB4 transduced and transplanted BM-HCs and ES-HCs toward granulopoiesis in vivo, we never observed any leukemias in any of the transplanted mice, even after long repopulation periods. Nevertheless, we wanted to know whether the apparent bias of the transduced cells toward granulopoiesis correlated with histomorphological alterations in the C57BL/6 or Rag2(–/–)γC(–/–) recipients. Analysis of hematoxylin/eosin-stained organ sections revealed pronounced hematopoiesis in the sternal bone marrow with a dominance of granulopoiesis in almost all animals transplanted with BM-HCs or ES-HCs expressing HOXB4 ectopically from the same retroviral vector (Fig. 8 Lower, which is published as supporting information on the PNAS web site). Even more dramatically, analysis of spleen sections showed metaplasia of spleen with enormously increased granulopoiesis (Fig. 8 Upper). Importantly, all stages of granulocytic differentiation were observed and no leukemic infiltration in other organs detected, arguing against a preleukemic state. None of the control transplanted animals showed any other abnormalities. Furthermore, peripheral white blood cell counts were not increased in any of the transplanted mice (data not shown). According to the defining criteria recently proposed for classification of nonlymphoid neoplasms in mice (40), the phenotype mediated by ectopic HOXB4 expression thus has to be diagnosed as myeloid proliferation (nonreactive), subclass myeloproliferation.
These results demonstrate that HOXB4-transduced bone marrow as well as transduced and in vitro differentiated ES cells behave equivalently after engraftment, in vivo.
Discussion
In this work, we have demonstrated that ES cells differentiated in vitro can mimic adult bone marrow under conditions of HOXB4 overexpression. Furthermore, we show that the manipulated differentiated ES cells fully recapitulate dosage-dependent inhibition of lineage differentiation (lymphoid and erythroid) observed with transplanted adult bone marrow expressing HOXB4. Previously published work has shown that HOXB4 can enhance the hematopoietic potential of transduced ES cells in vitro and in vivo (18, 19). However, functional equivalence to adult bone marrow was uncertain because of the negative effects on differentiation, which has not been reported before for transduced mouse bone marrow cells. It was therefore speculated that the observed differentiation bias of ES cell derivates were due to their primitive nature (41).
By using identical retroviral expression systems, we were able to demonstrate that both HOXB4-transduced adult bone marrow cells (BM-HCs) and ES-HCs behaved almost indistinguishably after transplantation into appropriate recipient mice in vivo. This included the previously unknown side effect of myeloid proliferation (nonreactive) subclass myeloproliferation (40). Comparative use of retroviral vectors that mediate different HOXB4 expression levels in vivo and the use of a recently described in vitro model for erythroid differentiation directly from ES cells (28) enabled us to demonstrate that the inhibitory effects on the differentiation of certain hematopoietic lineages is a direct result of increased amounts of ectopically expressed HOXB4, in vitro as well as in vivo. This was most obvious for red cell development, which was not inhibited by low expression levels, but at high expression levels of this transcription factor. T cell differentiation was inhibited at all levels of HOXB4 tested, suggesting that the minimal dose of inhibition lies below that of erythropoiesis.
The hematopoietic system is an extremely attractive candidate for therapeutic genetic intervention at the stem cell level. However, efficient techniques for their targeted genetic manipulation, which are well established for ES cells, have not yet been established. Instead, retroviral vectors are commonly used for stable and long-term gene expression. These integrate into the host genome in a more or less random fashion and, hence, are insertional mutagens that inherently increase the risk of cellular transformation by activation of protooncogenes (20, 21, 42, 43). To avoid such undesirable events, it is important to develop techniques allowing for selection and expansion of genetically defined HSC clones, which then could subsequently be used for reconstitution of the hematopoietic system.
The results presented here show that retroviral expression of HOXB4 confers a competitive growth advantage to both transduced bone marrow cells (BM-HCs) and in vitro differentiated ES cells (ES-HCs). These cells were capable of being expanded as clones for extended periods without any general change of surface marker expression. Transplanted ES-HC bulk cultures led to long-term repopulation in Rag2(–/–)γC(–/–) mice, whereas in vitro expanded clones, which had been cultured in parallel for the same period, did not (of 10 expanded clones, 3 randomly selected ones were transplanted in three to five mice each; data not shown). This suggests that most of the expanded cells are qualitatively short-term repopulating cells.
Because constitutive HOXB4 expression does not alter the growth or differentiation properties of mouse ES cells significantly, manipulation of the hematopoietic compartment could already be performed at this level (19). Thus, subsequent hematopoietic differentiation and transplantation of molecularly defined ES-cell-derived clones could ensure safe genetic manipulation of the hematopoietic compartment.
Conclusion
We have shown that ES-HCs can fully recapitulate adult bone marrow function in vivo when HOXB4 is ectopically expressed. This observation included its dosage-dependent influence on lineage differentiation down to the side effect of myeloid proliferation. We therefore postulate that enforced HOXB4 expression leads to similar fates of ES-HCs and BM-HCs. Thus, ES cells should be considered a realistic promising alternative to bone marrow stem cells for efficient and safe somatic gene therapy. However, the route of advanced cell therapy will certainly require expression of HOXB4 in a regulated manner to avoid potential unwanted effects.
Supplementary Material
Acknowledgments
We thank S. Knoess, G. Arman-Kalcek, M. Id, and T. Neumann for excellent technical support and Z. Li, H. Grundtke, R. Baumann, and B. Polifka for support during irradiation and transplantation of experimental animals. For critical comments and discussions, we thank S. Heinrichs, H. Auer, and A. Palmetshofer. This work was supported by the German Research Foundation (DFG, KL1311/2-3) Deutsche Krebshilfe (10-1763-OS5), the Austrian Industrial Research Promotion Fund (FFF, 808714), and the Austrian Science Fund (FWF, SFB 006).
Author contributions: S.P., S.C., B.S., E.W., and H.K. designed research; S.P., S.C., B.S., A.M., E.W., U.M., and H.K. performed research; P.S. contributed new reagents/analytic tools; S.P., S.C., B.S., K.K., E.W., U.M., P.S., W.O., C.B., H.B., and H.K. analyzed data; and H.K. wrote the paper.
Abbreviations: HSC, hematopoietic stem cell; ES-HC, ES cell-derived hematopoietic cell; BM-HC, bone marrow hematopoietic cells; EB, embryoid bodies; wPRE, posttranscriptional regulatory element of woodchuck hepatitis virus; MSCV, murine stem cell virus.
References
- 1.Cheshier, S. H., Morrison, S. J., Liao, X. & Weissman, I. L. (1999) Proc. Natl. Acad. Sci. USA 96, 3120–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glimm, H., Oh, I. H. & Eaves, C. J. (2000) Blood 96, 4185–4193. [PubMed] [Google Scholar]
- 3.Kondo, M., Wagers, A. J., Manz, M. G., Prohaska, S. S., Scherer, D. C., Beilhack, G. F., Shizuru, J. A. & Weissman, I. L. (2003) Annu. Rev. Immunol. 21, 759–806. [DOI] [PubMed] [Google Scholar]
- 4.Just, U., Stocking, C., Spooncer, E., Dexter, T. M. & Ostertag, W. (1991) Cell 64, 1163–1173. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, J., Niu, C., Ye, L., Huang, H., He, X., Tong, W. G., Ross, J., Haug, J., Johnson, T., Feng, J. Q., et al. (2003) Nature 425, 836–841. [DOI] [PubMed] [Google Scholar]
- 6.Calvi, L. M., Adams, G. B., Weibrecht, K. W., Weber, J. M., Olson, D. P., Knight, M. C., Martin, R. P., Schipani, E., Divieti, P., Bringhurst, F. R., et al. (2003) Nature 425, 841–846. [DOI] [PubMed] [Google Scholar]
- 7.Thorsteinsdottir, U., Mamo, A., Kroon, E., Jerome, L., Bijl, J., Lawrence, H. J., Humphries, K. & Sauvageau, G. (2002) Blood 99, 121–129. [DOI] [PubMed] [Google Scholar]
- 8.Varnum-Finney, B., Xu, L., Brashem-Stein, C., Nourigat, C., Flowers, D., Bakkour, S., Pear, W. S. & Bernstein, I. D. (2000) Nat. Med. 6, 1278–1281. [DOI] [PubMed] [Google Scholar]
- 9.Reya, T., Duncan, A. W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423, 409–414. [DOI] [PubMed] [Google Scholar]
- 10.Sauvageau, G., Thorsteinsdottir, U., Eaves, C. J., Lawrence, H. J., Largman, C., Lansdorp, P. M. & Humphries, R. K. (1995) Genes Dev. 9, 1753–1765. [DOI] [PubMed] [Google Scholar]
- 11.Antonchuk, J., Sauvageau, G. & Humphries, R. K. (2001) Exp. Hematol. 29, 1125–1134. [DOI] [PubMed] [Google Scholar]
- 12.Antonchuk, J., Sauvageau, G. & Humphries, R. K. (2002) Cell 109, 39–45. [DOI] [PubMed] [Google Scholar]
- 13.Klump, H., Schiedlmeier, B., Vogt, B., Ryan, M., Ostertag, W. & Baum, C. (2001) Gene Ther. 8, 811–817. [DOI] [PubMed] [Google Scholar]
- 14.Schiedlmeier, B., Klump, H., Will, E., Arman-Kalcek, G., Li, Z., Wang, Z., Rimek, A., Friel, J., Baum, C. & Ostertag, W. (2003) Blood 101, 1759–1768. [DOI] [PubMed] [Google Scholar]
- 15.Krosl, J., Beslu, N., Mayotte, N., Humphries, R. K. & Sauvageau, G. (2003) Immunity 18, 561–571. [DOI] [PubMed] [Google Scholar]
- 16.Krosl, J., Austin, P., Beslu, N., Kroon, E., Humphries, R. K. & Sauvageau, G. (2003) Nat. Med. 9, 1428–1432. [DOI] [PubMed] [Google Scholar]
- 17.Buske, C., Feuring-Buske, M., Abramovich, C., Spiekermann, K., Eaves, C. J., Coulombel, L., Sauvageau, G., Hogge, D. E. & Humphries, R. K. (2002) Blood 100, 862–868. [DOI] [PubMed] [Google Scholar]
- 18.Kyba, M., Perlingeiro, R. C. & Daley, G. Q. (2002) Cell 109, 29–37. [DOI] [PubMed] [Google Scholar]
- 19.Rideout, W. M., 3rd, Hochedlinger, K., Kyba, M., Daley, G. Q. & Jaenisch, R. (2002) Cell 109, 17–27. [DOI] [PubMed] [Google Scholar]
- 20.Li, Z., Dullmann, J., Schiedlmeier, B., Schmidt, M., von Kalle, C., Meyer, J., Forster, M., Stocking, C., Wahlers, A., Frank, O., et al. (2002) Science 296, 497. [DOI] [PubMed] [Google Scholar]
- 21.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415–419. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, H. J. & Largman, C. (1992) Blood 80, 2445–2453. [PubMed] [Google Scholar]
- 23.Sauvageau, G., Lansdorp, P. M., Eaves, C. J., Hogge, D. E., Dragowska, W. H., Reid, D. S., Largman, C., Lawrence, H. J. & Humphries, R. K. (1994) Proc. Natl. Acad. Sci. USA 91, 12223–12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pineault, N., Helgason, C. D., Lawrence, H. J. & Humphries, R. K. (2002) Exp. Hematol. 30, 49–57. [DOI] [PubMed] [Google Scholar]
- 25.Abate-Shen, C. (2002) Nat. Rev. Cancer 2, 777–785. [DOI] [PubMed] [Google Scholar]
- 26.Brun, A. C., Fan, X., Bjornsson, J. M., Humphries, R. K. & Karlsson, S. (2003) Mol. Ther. 8, 618–628. [DOI] [PubMed] [Google Scholar]
- 27.Wiles, M. V. & Keller, G. (1991) Development (Cambridge, U.K.) 111, 259–267. [DOI] [PubMed] [Google Scholar]
- 28.Carotta, S., Pilat, S., Mairhofer, A., Schmidt, U., Dolznig, H., Steinlein, P. & Beug, H. (2004) Blood 104, 1873–1880. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, M., Donelly, M., Lewis, A., Mehrotra, A.P., Wilkie, J. & Gani, D. (1999) Bioorg. Chem. 27, 55–79. [Google Scholar]
- 30.Hawley, R. G., Fong, A. Z., Burns, B. F. & Hawley, T. S. (1992) J. Exp. Med. 176, 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grez, M., Akgun, E., Hilberg, F. & Ostertag, W. (1990) Proc. Natl. Acad. Sci. USA 87, 9202–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hildinger, M., Eckert, H. G., Schilz, A. J., John, J., Ostertag, W. & Baum, C. (1998) Gene Ther. 5, 1575–1579. [DOI] [PubMed] [Google Scholar]
- 33.Cherry, S. R., Biniszkiewicz, D., van Parijs, L., Baltimore, D. & Jaenisch, R. (2000) Mol. Cell. Biol. 20, 7419–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho, S. K., Bourdeau, A., Letarte, M. & Zuniga-Pflucker, J. C. (2001) Blood 98, 3635–3642. [DOI] [PubMed] [Google Scholar]
- 35.Chen, C. Z., Li, M., de Graaf, D., Monti, S., Gottgens, B., Sanchez, M. J., Lander, E. S., Golub, T. R., Green, A. R., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 15468–15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, C. Z., Li, L., Li, M. & Lodish, H. F. (2003) Immunity 19, 525–533. [DOI] [PubMed] [Google Scholar]
- 37.von Lindern, M., Deiner, E. M., Dolznig, H., Parren-Van Amelsvoort, M., Hayman, M. J., Mullner, E. W. & Beug, H. (2001) Oncogene 20, 3651–3664. [DOI] [PubMed] [Google Scholar]
- 38.Mazurier, F., Fontanellas, A., Salesse, S., Taine, L., Landriau, S., Moreau-Gaudry, F., Reiffers, J., Peault, B., Di Santo, J. P. & de Verneuil, H. (1999) J. Interferon Cytokine Res. 19, 533–541. [DOI] [PubMed] [Google Scholar]
- 39.Audet, J., Miller, C. L., Rose-John, S., Piret, J. M. & Eaves, C. J. (2001) Proc. Natl. Acad. Sci. USA 98, 1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kogan, S. C., Ward, J. M., Anver, M. R., Berman, J. J., Brayton, C., Cardiff, R. D., Carter, J. S., de Coronado, S., Downing, J. R., Fredrickson, T. N., et al. (2002) Blood 100, 238–245. [DOI] [PubMed] [Google Scholar]
- 41.Kyba, M., Perlingeiro, R. C. & Daley, G. Q. (2003) Methods Enzymol. 365, 114–129. [DOI] [PubMed] [Google Scholar]
- 42.Baum, C. & Fehse, B. (2003) Curr. Opin. Mol. Ther. 5, 458–462. [PubMed] [Google Scholar]
- 43.Williams, D. & Baum, C. (2004) Nature 429, 129. [DOI] [PubMed] [Google Scholar]
- 44.Hildinger, M., Abel, K. L., Ostertag, W. & Baum, C. (1999) J. Virol. 73, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zufferey, R., Donello, J. E., Trono, D. & Hope, T. J. (1999) J. Virol. 73, 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.