Abstract
Cadmium-induced glycine-rich protein (cdiGRP) is a cell wall-associated factor that increases callose levels in plant vasculature. To better understand the cdiGRP/callose regulation system, we identified a tobacco protein, GrIP (cdiGRP-interacting protein, GrIP), that associates with cdiGRP and localizes at the plant cell wall. Constitutive overexpression of GrIP enhanced the accumulation of the cdiGRP protein and callose in vasculature-associated cells with or without treatment with cadmium ions. That GrIP gene expression was not affected by cadmium ions indicated that GrIP does not directly modulate the callose levels induced by the treatment. Instead, GrIP most likely functions by further elevating the accumulated amount of cdiGRP, the expression of which is up-regulated by the cadmium ions. Interestingly, the levels of cdiGRP mRNA were not affected by constitutive expression of GrIP, demonstrating that the enhancement in cdiGRP protein accumulation by GrIP overexpression occurs posttranslationally. Collectively, these observations suggest that GrIP interacts with cdiGRP and increases its level of accumulation; in turn, the elevated amounts of cdiGRP induce callose deposits in the plant cell walls. Therefore, GrIP and cdiGRP represent sequentially acting factors in a biochemical pathway that regulates callose accumulation in the plant vasculature.
Keywords: cell wall, plasmodesmata, protein–protein interaction
The plasmodesma is a unique channel structure that spans the cell wall and connects neighboring cells to enable cytoplasmic exchange in plants. Accumulating evidence shows that many endogenous macromolecules are trafficked through this channel, enabling plant cells to communicate with each other (reviewed in refs. 1–5). Although the importance of symplastic molecular exchange through the plasmodesmata is widely accepted, its regulatory mechanism is not well characterized. One of the important parameters describing the molecular permeability of plasmodesmata is the size exclusion limit, which sets the largest size of the molecules that can traffic through the plasmodesmata through passive transport. Plant cells in different tissues at different developmental stages are known to possess plasmodesmata with different size exclusion limit (6).
Viruses are known to use the plasmodesmal channels to translocate cell-to-cell in plants by using their movement factors to interact with and modify the plasmodesmata (reviewed in refs. 7–10). Detailed studies on plant virus movement have revealed that size exclusion limit is not the sole determinant for molecular translocation through plasmodesmata; rather, the channel must possess an active regulatory machinery that may differ between the plasmodesmata at different tissue interfaces (reviewed in ref. 7). For example, coat protein-deficient tobacco mosaic virus can accumulate in the vascular parenchyma but not in the companion cells in Nicotiana tabacum cv. Xanthi-nn, suggesting that the plasmodesmata at the vascular parenchyma/companion cell boundary in the host do not permit movement of the mutant virus (11). The interface between bundle sheath and phloem cells can also serve as a barrier for bromoviruses in some hosts (12–14), suggesting that plasmodesmata at this boundary are more restrictive for bromovirus movement. Furthermore, the tobacco etch virus HAT strain can move cell-to-cell and load into the vascular tissue but cannot unload from it in systemic leaves in the restrictive host N. tabacum V20 (15). Similarly, when wild-type tobamoviruses, such as tobacco mosaic virus or turnip vein clearing virus, are inoculated onto tobacco plants treated with subtoxic concentrations of cadmium ions, the inoculated tobamovirus cannot unload from the vasculature in systemic leaves, whereas it is able to spread from cell to cell and load into the vasculature (16–18). These observations suggest that plasmodesmata at the interfaces between different types of vascular tissue or at the boundaries between vascular and nonvascular tissue possess characteristic features and are able to restrict traffic of some viruses unidirectionally.
Although it is clear that the plasmodesmata are equipped with molecular devices that can strictly control the traffic through these channels, the mechanism of this control remains unclear. One of the possible molecular machineries for plasmodesmal regulation may be the callose synthesis/degradation system at the plasmodesmata. Callose is a 1,3-β-d-glucan (19), synthesized by callose synthase and degraded by 1,3-β-d-glucanase (20, 21), which has been shown to accumulate around the neck region of the plasmodesmata (22). Because several studies have suggested that callose accumulation around the plasmodesmata restricts virus translocation through that channel (16, 22–27), callose could be a major restrictive factor for transport through the plasmodesmata.
Previously, we identified a tobacco cell-wall protein, cadmium-induced glycine-rich protein (cdiGRP), as one of the regulatory factors for callose accumulation in N. tabacum vascular-associated cells (16). We demonstrated that cdiGRP is specifically induced by subtoxic levels of cadmium ions and is accumulated in the cell walls of plant vascular tissues (16). Importantly, constitutive cdiGRP expression inhibited systemic transport of turnip vein clearing virus, whereas antisense suppression of cdiGRP production allowed turnip vein clearing virus movement in the presence of cadmium ions (16). Because overexpression of cdiGRP induced callose accumulation in vascular tissue (16), and the callose localized to the plasmodesmata in that tissue (unpublished data), cdiGRP most likely exerted its inhibitory effect on turnip vein clearing virus transport by enhancing callose deposits at the plasmodesmata in the vasculature (16).
To better understand the cdiGRP/callose regulation system, we set out to identify its additional molecular participants. Here, we identified a tobacco protein, cdiGRP-interacting protein (GrIP), that associates with cdiGRP, increases cdiGRP accumulation levels, and induces callose deposition in the plant vasculature.
Materials and Methods
Yeast Two-Hybrid Assay. Saccharomyces cerevisiae strain L40 (MATa his3del200 trp1–901 leu2–3 112 ade2 lys2–80lam URA3::(lexAop)-lacZ LYS2::(lexAop)4-HIS3) (28) was grown in yeast extract/peptone/dextrose or the appropriate selective minimal medium under standard conditions (29). Plasmids were introduced into yeast cells by using a standard lithium acetate protocol (28). The N. tabacum cv. Turk cDNA library in pGAD424 (LEU2+, Clontech) was screened with cdiGRP in pSTT91 (TRP1+; ref. 30) as bait as described in refs. 28 and 31, and positive clones were selected on a histidine-deficient selective medium and confirmed by β-galactosidase assay (32). False positives were eliminated by using the pBTM116 vector (TRP1+; ref. 28), expressing either human lamin C or topoisomerase I, known to function as nonspecific activators in the two-hybrid system (28, 33, 34).
Transgenic Plants. The GrIP cDNA was first inserted in sense orientation as a PCR-amplified PstI fragment into a plant expression vector, pCd, containing the 35S promoter of cauliflower mosaic virus, tobacco mosaic virus translational enhancer, and the nopaline synthase polyA signal (35). The entire expression cassette was subcloned as a BamHI-XbaI fragment into the binary vector pBIN19 (GenBank accession no. U09365.1), carrying a kanamycin resistance selection marker, and introduced into the disarmed Agrobacterium strain C1C58, which was then used to transform tobacco plants as described in ref. 36. The resulting transgenic plants were selected on a kanamycin-containing medium and maintained for 1 month under sterile conditions on a MS basal medium with no exogenous growth regulators (37). Plants were then transferred to soil in a greenhouse, allowed to set seed, and the transgenic progeny were selected by germinating the seeds on MS agar in the presence of kanamycin. Kanamycin-resistant seedlings were maintained in tissue culture for 2 weeks, transferred to soil, and grown to the four- to six-leaf stage for use in the experiments. For CdCl2 treatment, plants were grown and maintained under conditions described in refs. 16–18.
Coimmunoprecipitation and Western Blotting. Antibody was raised in mouse against a chemically synthesized peptide sequence derived from the GrIP amino acid sequence (residues 52–71, Covance Research Products, Denver, PA). The anti-cdiGRP rabbit antiserum was described in ref. 16. Surgically isolated young tobacco leaf mid-ribs (500 mg) were ground in 2.5 ml of extraction buffer [1% (vol/vol) Igeopal CM-630/5 mM DTT/1 mM PMSF in 10 mM sodium phosphate buffer, pH 7.2]. Although some glycine-rich proteins may be difficult to solubilize under mild conditions (38), we detected cdiGRP in tissue extract prepared with this relatively mild extraction buffer. The mixture was clarified by centrifugation at 10,000 × g for 2 min, and the extract was aliquoted into microfuge tubes. These 500-μl aliquots were incubated with 2.5 μl of anti-cdiGRP rabbit antiserum or preimmune mouse serum at 4°C. After 2 h of incubation, Protein G-conjugated agarose beads (25 μl, Sigma-Aldrich) were added to the mixture, which was then gently rocked at 4°C overnight. After three extensive washes in the extraction buffer, the captured immunocomplexes were released by mixing in 50 μl of SDS/PAGE loading buffer (39) and boiling for 5 min. The supernatant was collected after a brief centrifugation and subjected to Western blotting. Briefly, the samples were electrophoresed on a 12% SDS polyacrylamide gel under reducing conditions and resolved proteins were electrophoretically transferred to poly(vinylidene difluoride) membrane. The blot was incubated for 1 h in blocking solution [4% skim milk in Tris-buffered saline Tween 20 (TBST) (10 mM Tris·HCl/150 mM NaCl, pH 8.0 containing 0.05% (vol/vol) Tween 20)] and probed with anti-cdiGRP rabbit antiserum, anti-GrIP peptide mouse antiserum, and rabbit or mouse preimmune serum (diluted 1:250 in 1% (vol/vol) skim milk in TBST) for 1 h. After extensive washing with TBST, the membrane was incubated with a 1:4,000 diluted solution of horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit antibody (Jackson ImmunoResearch). After additional washing, proteins recognized by the antibodies were visualized by chemiluminescence with the ECL system (Amersham Pharmacia).
Immunohistochemistry and in Situ Hybridization. For immunoelectron microscopy, mid-rib tissues harvested from young wild-type tobacco leaves (≈4 cm in length) were processed for use with the Durcupan ACM embedding protocol (40). Ultra-thin sections (70–80 nm) were reacted with anti-GrIP antiserum, followed by anti-mouse IgG conjugated to 10-nm colloidal gold as described in refs. 16 and 40, and examined under a JEOL 100C transmission electron microscope.
For fluorescent microscopy, plant tissues were harvested, fixed, and embedded in Paraplast Plus as described in refs. 16 and 41. The samples were sectioned and processed for immunofluorescence or in situ hybridization. For immunofluorescent microscopy, 10-μm transverse sections were dewaxed, rehydrated, blocked, and reacted with a mixture of anti-cdiGRP rabbit polyclonal antiserum and anti-callose-specific mouse monoclonal antibody (Biosupplies, Parkville, Australia) as described in refs. 16 and 41. For controls, the sections were reacted with rabbit preimmune antiserum. The sections were further probed with goat Cy5-conjugated anti-rabbit IgG secondary antibody and Alexa Fluor 488-conjugated anti-mouse IgG+M secondary antibody (Jackson ImmunoResearch). This analysis of double-stained sections allowed simultaneous detection of qualitative changes in the levels of cdiGRP and callose in situ, within plant tissues, but it was not suitable for precise quantification of the immunocomplexes.
For in situ hybridization, 20-μm transverse sections were dewaxed, treated with proteinase K, dehydrated again, and hybridized with digoxigenin (DIG)-labeled probes according to the manufacturer's protocols. For the negative control, the probe was omitted from the hybridization solution. The samples were then reacted with Cy5-conjugated anti-DIG antibody (Jackson ImmunoResearch). All fluorescence microscopy was performed by using a Zeiss LSM 5 Pascal confocal laser scanning microscope.
Results and Discussion
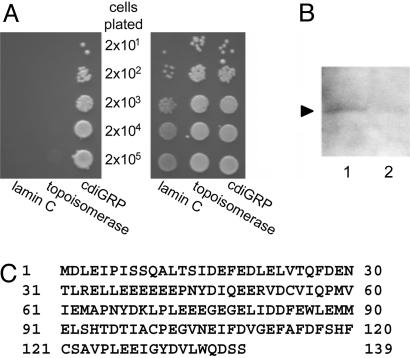
Identification of GrIP. To isolate potential interactors of cdiGRP, we used a two-hybrid screen (28, 42) with a N. tabacum cv. Turk cDNA library and the cdiGRP protein as bait. Screening of ≈1 × 106 transformants resulted in the identification and isolation of four independent cDNA clones producing cdiGRP interactors. One of the clones was designated GrIP and characterized in detail. Coexpression of GrIP and cdiGRP activated the HIS3 reporter gene and enabled yeast transformants to grow on a histidine dropout medium, whereas yeast cells cotransformed with cdiGRP and DNA topoisomerase I or lamin C as negative controls standard for the two-hybrid interactions (33, 34), were unable to survive in the absence histidine (Fig. 1A Left). Cells expressing all three combinations of the proteins grew to the same extent in the presence of histidine (Fig. 1 A Right), indicating that the tested proteins did not adversely and nonspecifically affect yeast cell physiology. Collectively, the results demonstrated the specificity of the interaction between GrIP and cdiGRP in the two-hybrid system. GrIP also interacted with cdiGRP, lacking its N-terminal secretion signal sequence (data not shown), suggesting that GrIP is able to recognize the processed and secreted form of cdiGRP.
Fig. 1.
Identification of GrIP. (A) Specific GrIP–cdiGRP interaction in the two-hybrid system. The indicated cell inocula were plated on growth media without leucine, tryptophan, and histidine (Left) or without leucine and tryptophan (Right). Growth on histidine-deficient medium represents selective conditions for protein–protein interaction. (B) Detection of in vivo interaction between GrIP and cdiGRP by coimmunoprecipitation with anti-cdiGRP antiserum (lane 1) or with preimmune serum (lane 2). Arrow indicates a ≈16 kDa protein species detected in the immunocomplexes by using Western blotting with anti-GrIP antiserum. (C) Deduced amino acid sequence of GrIP (GenBank accession no. DQ007343).
Next, we carried out coimmunoprecipitation of cdiGRP followed by Western blot analysis of the immunocomplexes by using anti-GrIP antiserum. Fig. 1B shows that a protein with a relative electrophoretic mobility of ≈16 kDa, consistent with the calculated molecular mass of GrIP (16.1 kDa, see Fig. 1C), was recognized by anti-GrIP antiserum in the immunocomplexes precipitated with anti-cdiGRP antiserum, whereas this protein was not detected after coimmunoprecipitation by using control, preimmune serum. These results support the notion of a direct interaction between GrIP and cdiGRP in vivo.
Sequence analysis of the GrIP cDNA revealed a single ORF encoding a protein of 139 amino acid residues (Fig. 1C). Interestingly, GrIP has a high content of acidic residues (20% glutamic acid and 10% aspartic acid). Based on the available databases [e.g., TAIR (www.arabidopsis.org), MOTIF (http://motif.genome.jp), and InterPro (www.ebi.ac.uk/interpro)], the GrIP amino acid sequence did not contain any known functional domains or targeting motifs. The GrIP cDNA displayed modest homology to a partial mRNA sequence from Atropa belladonna (GenBank accession no. AJ309388), the function of which is unknown. No significant homologies to the GrIP amino acid sequence were found in any other organisms.
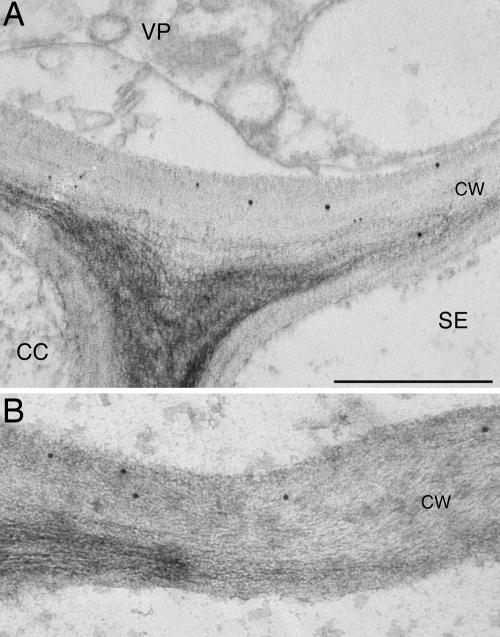
Subcellular Localization. To obtain initial insight into the biological function of GrIP, we determined its subcellular localization in planta. Vascular tissue from a wild-type plant was analyzed by immunoelectron microscopy with anti-GrIP antiserum. GrIP-specific antibodies decorated the cell wall, but not other cellular compartments and organelles (Fig. 2 and data not shown). No cell wall staining was observed in control experiments without primary antiserum (data not shown). This cell wall-specific localization of GrIP is similar to the cell wall association described for cdiGRP (16).
Fig. 2.
Immunohistochemical detection of GrIP in cell walls of the tobacco leaf phloem. CC, companion cells; SE, sieve elements; VP, vascular parenchyma. (A and B) Illustrated are cell walls (CW) in different cells associated with vasculature. (Scale bar: 0.5 μm.)
GrIP Overexpression Enhances Accumulation of cdiGRP and Callose. Ideally, the phenotype of plant mutants that do not express GrIP would help determine the function of this protein. However, antisense and RNA interference approaches did not yield plant lines with detectably suppressed GrIP expression (data not shown), suggesting that GrIP may be essential for the plant's survival. We therefore took the reverse approach, generating transgenic tobacco lines that overexpress GrIP. Seven independent lines were generated, and two of them (GrIP-S1 and GrIP-S2), which exhibited the highest degree of GrIP expression and, thus, most likely contained multiple copies of the GrIP transgene (data not shown), were analyzed for the levels of cdiGRP and callose in the presence or absence of subtoxic concentrations of cadmium ions.
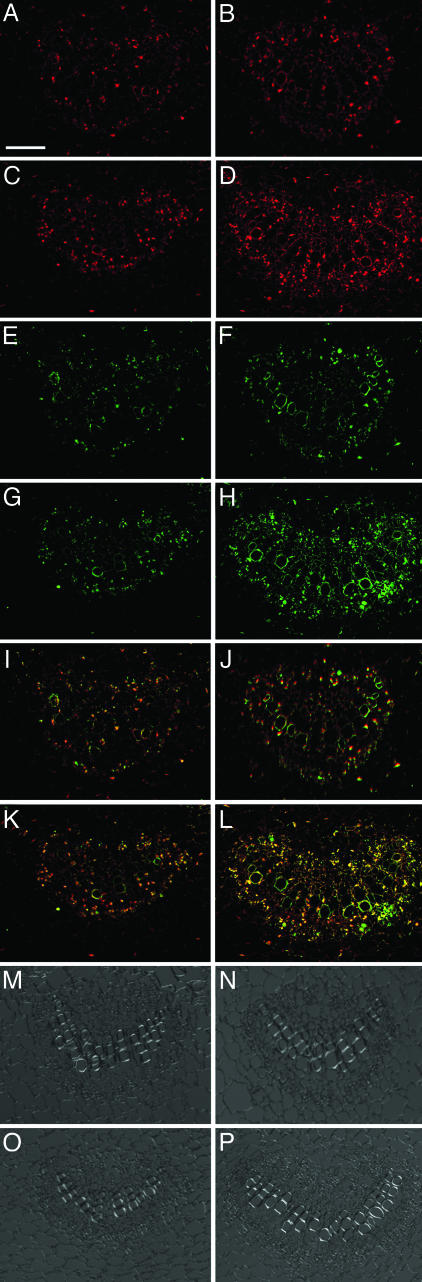
Previously, we found the optimal concentration of cadmium ions for maximum induction of cdiGRP and callose to be between 10 and 20 μM (unpublished data). Here, we used the suboptimal concentration of 5 μM to better observe the potential changes and differences in cdiGRP and callose accumulation levels. To detect cdiGRP and callose in the same tissue sections, they were immunostained with both anti-cdiGRP and anti-callose antibodies simultaneously. Essentially identical results were obtained with plant lines GrIP-S1 and GrIP-S2, and they are shown for the GrIP-S1 line. Fig. 3 demonstrates that cdiGRP levels were significantly higher in GrIP-S1 plants (Fig. 3 C and D) than in the wild-type plants (Fig. 3 A and B), with or without treatment (Fig. 3 B and D and Fig. 3 A and C, respectively) with 5 μM CdCl2. Moreover, induction of cdiGRP by cadmium ions was more dramatic in GrIP-overexpressing plants than in the wild-type plants; the level of cdiGRP accumulation in GrIP-S1 plants was significantly enhanced by 5 μM CdCl2 (compare Fig. 3 C to D), whereas it was only weakly induced by the same treatment in wild-type plants (compare Fig. 3 A to B).
Fig. 3.
Elevated levels of cdiGRP and callose in GrIP-S1 plants. (A–D) cdiGRP in untreated wild-type plants, wild-type plants treated with 5 μM CdCl2, untreated GrIP-S1 plants, and GrIP-S1 plants treated with 5 μM CdCl2, respectively. (E–H) Callose in sections shown in A–D, respectively. (I–L) Merged images from A and E, B and F, C and G, and D and H, respectively. cdiGRP- and callose-specific signals are red and green, respectively; overlapping cdiGRP- and callose-specific signals are yellow. (M–P) Phase contrast images of sections shown in A, E, and I; B, F, and J; C, G, and K; and D, H, and L, respectively. Vascular tissue is seen as smaller cells arranged in bundles and nonvascular tissue is represented by larger cells surrounding the vascular bundles. (Scale bar: 50 μm.)
Accumulated callose levels correlated closely with the level of cdiGRP in both GrIP-overexpressing and wild-type plants. As seen in Fig. 3, higher callose accumulations were observed in GrIP-S1 plants (Fig. 3 G and H) compared to wild-type plants (Fig. 3 E and F) both with and without exposure to CdCl2 (Fig. 3 H and F and G and E, respectively). The effect of CdCl2 on callose accumulation was also enhanced in GrIP-overexpressing plants (compare differences between Fig. 3 F and E to those between Fig. 3 H and G), suggesting that higher levels of GrIP expression up-regulate callose accumulation in vascular tissues.
Both cdiGRP and callose accumulated in the vascular tissues, predominantly in phloem-associated cells, in both wild-type and GrIP-overexpressing plants. Furthermore, merged images in Fig. 3 I–L show that cdiGRP and callose accumulations (red and green signals, respectively) are enhanced at the overlapping cellular sites (yellow signal) within vascular bundles that are clearly distinguished in phase contrast images of the same sections (Fig. 3 M–P). Because overexpression of GrIP promotes accumulation of both cdiGRP and callose (see Fig. 3), whereas overexpression of cdiGRP induces accumulation of callose (16) but not GrIP (data not shown), GrIP may regulate callose levels indirectly by altering the amount of cdiGRP in the plant vasculature.
Although GrIP overexpression caused a significant increase in the levels of cdiGRP and callose in the absence of cadmium ions or in the presence of suboptimal cadmium concentrations (i.e., 5 μM, see Fig. 3), this increase was insufficient to block movement of tobamoviruses, such as tobacco mosaic virus (data not shown).
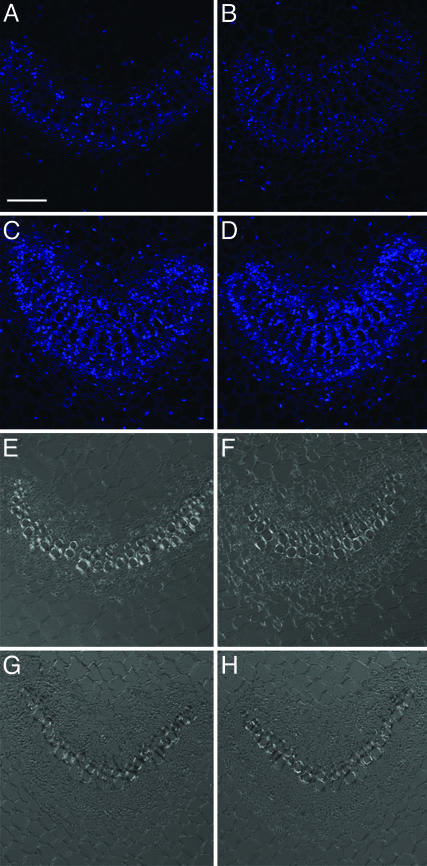
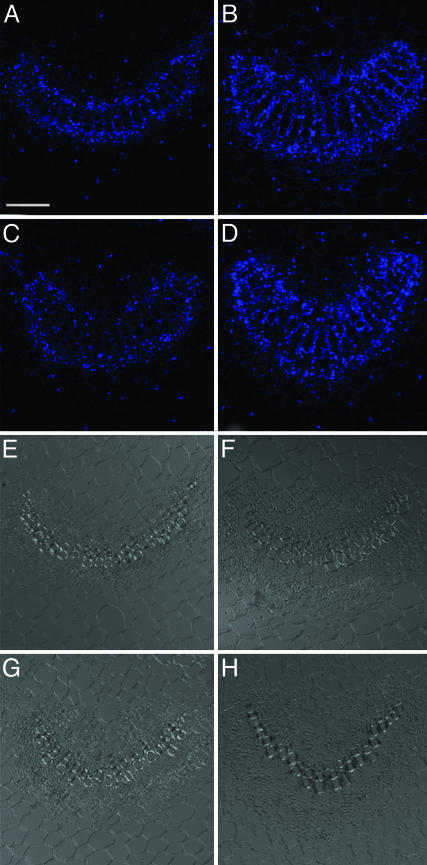
GrIP Modulates cdiGRP Accumulation Levels Posttranslationally. In situ hybridization experiments revealed GrIP mRNA predominantly in the vascular-associated cells of wild-type plants, without (Fig. 4 A and E) or with (Fig. 4 B and F) 5 μM CdCl2 treatment. This tissue-specific expression of GrIP parallels that of cdiGRP, which is also expressed in the vasculature (see Fig. 3 and ref. 16). Moreover, GrIP mRNA expression level was unaffected by cadmium ions, showing that, unlike cdiGRP, the GrIP gene expression is independent of the presence of this heavy metal. As expected, when overexpressed from a general promoter in GrIP-S1 transgenic plants, GrIP mRNA was observed in both vascular and nonvascular tissues (Fig. 4 C and G and Fig. 4 D and H, respectively).
Fig. 4.
GrIP transcripts in wild-type and GrIP-S1 plants. (A and E) Untreated wild-type plants. (B and F) Wild-type plants treated with 5 μM CdCl2. (C and G) Untreated GrIP-S1 plants. (D and H) GrIP-S1 plants treated with 5 μM CdCl2. (A–D) Cy5 confocal images are represented. (E–H) Phase contrast images of the same sections are shown. Vascular tissue is seen as smaller cells arranged in bundles, and nonvascular tissue is represented by larger cells surrounding the vascular bundles. (Scale bar: 50 μm.)
Similarly to the wild-type plants (see Fig. 4 A, B, E, and F), the GrIP expression level in GrIP-S1 plants was not enhanced by CdCl2 treatment (Fig. 4 C, D, G, and H), indicating that the elevated levels of cdiGRP and callose in GrIP-overexpressing plants treated with cadmium ions (see Fig. 3) are not due to induction of the expression of the GrIP transgene after the treatment. Moreover, in GrIP-S1 plants, both cdiGRP and callose accumulated only in the vasculature, whereas GrIP mRNA was also expressed in nonvascular tissues (Fig. 3). This discrepancy between levels of GrIP mRNA and cdiGRP protein is presumably because the cdiGRP gene expression is limited to the vasculature (see Figs. 3 and 5), and, thus, the cdiGRP protein is not present in significant amounts in other tissues even when they accumulate GrIP in GrIP-overexpressing lines. This result suggests that overexpression of GrIP itself is not sufficient to induce callose accumulation, which also requires the presence of cdiGRP. Thus, cdiGRP most likely regulates callose deposition by affecting the callose synthesis/degradation machinery, whereas GrIP modulates this process by altering the cdiGRP levels.
Fig. 5.
cdiGRP transcripts in wild-type and GrIP-S1 plants. (A and E) Untreated wild-type plants. (B and F) Wild-type plants treated with 5 μM CdCl2. (C and G) Untreated GrIP-S1 plants. (D and H) GrIP-S1 plants treated with 5 μM CdCl2. (A–D) Cy5 confocal images are represented. (E–H) Phase contrast images of the same sections are shown. Vascular tissue is seen as smaller cells arranged in bundles, and nonvascular tissue is represented by larger cells surrounding the vascular bundles. (Scale bar: 50 μm.)
The elevated levels of cdiGRP obser ved in GrIP-overexpressing plants may be due to posttranslational events, i.e., GrIP may interact with cdiGRP, promoting its stabilization and further accumulation. Alternatively, GrIP may simply enhance the cdiGRP gene expression. To distinguish between these possibilities, we compared the level and expression pattern of cdiGRP mRNA in wild-type and GrIP-overexpressing plants. Cadmium ion treatment induced the accumulation of cdiGRP mRNA to a similar extent in both the wild-type (Fig. 5 A, B, E, and F) and GrIP-S1 plants (Fig. 5 C, D, G, and H). Importantly, GrIP expression did not affect the level of cdiGRP mRNA, because the accumulation levels of the latter in wild-type plants were the same as in GrIP-overexpressing plants when treated with the same concentrations of cadmium ions (compare B to D in Fig. 5). These results demonstrate that the elevated levels of the cdiGRP protein in GrIP-overexpressing plants are not due to transcriptional activation of the cdiGRP gene but occur posttranslationally.
How does GrIP promote accumulation of the cdiGRP protein in vascular tissues? We suggest that direct interaction of GrIP with cdiGRP stabilizes cdiGRP and allows it to accumulate to higher levels within the plant cell walls. Protein–protein interactions that result in stabilization and higher accumulation levels of one of the interacting proteins are known in mammalian systems; for example, p57Kip2 binds to MyoD to stabilize it, thereby increasing the half-life of the protein (43), whereas ZBP-89 binds to p53, retaining it in the cell nucleus and protecting it from degradation (44). Because GrIP is not an abundant cellular protein and its levels are not affected by cadmium treatment and/or presence of cdiGRP (data not shown), it may act as a limiting factor for accumulation of cdiGRP and, consequently, deposition of callose, which are significantly increased after overexpression of GrIP. Thus, GrIP may represent one of the key components of the cdiGRP-mediated callose regulation system in tobacco vasculature.
Acknowledgments
We thank Sue Vanhorn for her technical help with the immunoelectron microscopy and Camille Vainstein for her proofreading of the manuscript. The work in our laboratory is supported by grants from the National Institutes of Health, the National Science Foundation, the U.S. Department of Agriculture, the U.S.–Israel Binational Agricultural Research and Development Fund, and the U.S.–Israel Binational Science Foundation (to V.C.).
Author contributions: S.U. and V.C. designed research; S.U. performed research; and S.U. and V.C. wrote the paper.
Abbreviations: cdiGRP, cadmium-induced glycine-rich protein; GrIP, cdiGRP-interacting protein.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ007343).
References
- 1.Oparka, K. J. (2004) Trends Plant Sci. 9, 33–41. [DOI] [PubMed] [Google Scholar]
- 2.Zambryski, P. C. (2004) J. Cell Biol. 164, 165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas, W. J. & Lee, J. Y. (2004) Nat. Rev. Mol. Cell. Biol. 5, 712–726. [DOI] [PubMed] [Google Scholar]
- 4.Heinlein, M. & Epel, B. L. (2004) Int. Rev. Cytol. 235, 93–164. [DOI] [PubMed] [Google Scholar]
- 5.Ueki, S. & Citovsky, V. (2005) Proc. Natl. Acad. Sci. USA 102, 1817–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim, I., Cho, E., Crawford, K. M., Hempel, F. D. & Zambryski, P. C. (2005) Proc. Natl. Acad. Sci. USA 102, 2227–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waigmann, E., Ueki, S., Trutnyeva, K. & Citovsky, V. (2004) Crit. Rev. Plant Sci. 23, 195–250. [Google Scholar]
- 8.Leisner, S. M. & Howell, S. H. (1993) Trends Microbiol. 1, 314–317. [DOI] [PubMed] [Google Scholar]
- 9.Oparka, K. J. & Santa Cruz, S. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 323–347. [DOI] [PubMed] [Google Scholar]
- 10.Ghoshroy, S., Lartey, R., Sheng, J. & Citovsky, V. (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 27–49. [DOI] [PubMed] [Google Scholar]
- 11.Wolf, S., Deom, C. M., Beachy, R. N. & Lucas, W. J. (1989) Science 246, 377–379. [DOI] [PubMed] [Google Scholar]
- 12.Thompson, J. R. & Garcia-Arenal, F. G. (1998) Mol. Plant--Microbe Interact. 11, 109–114. [Google Scholar]
- 13.Wintermantel, W. M., Banerjee, N., Oliver, J. C., Paolillo, D. J. & Zaitlin, M. (1997) Virology 231, 248–257. [DOI] [PubMed] [Google Scholar]
- 14.Goodrick, B. J., Kuhn, C. W. & Hussey, R. S. (1991) Phytopathology 81, 1426–1431. [Google Scholar]
- 15.Schaad, M. C. & Carrington, J. C. (1996) J. Virol. 70, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueki, S. & Citovsky, V. (2002) Nat. Cell Biol. 4, 478–485. [DOI] [PubMed] [Google Scholar]
- 17.Citovsky, V., Ghoshroy, S., Tsui, F. & Klessig, D. F. (1998) Plant J. 16, 13–20. [DOI] [PubMed] [Google Scholar]
- 18.Ghoshroy, S., Freedman, K., Lartey, R. & Citovsky, V. (1998) Plant J. 13, 591–602. [DOI] [PubMed] [Google Scholar]
- 19.Stone, B. A. & Clarke, A. E. (1992) Chemistry and Biology of 1,3-β-glucans (La Trobe Univ. Press, Victoria, Australia).
- 20.Kauss, H. (1996) in Membranes: Specialized Functions in Plants, eds. Smallwood, M., Knox, J. P. & Bowles, D. J. (BIOS Scientific Publishers, Oxford), pp. 77–92.
- 21.Kauss, H. (1985) J. Cell Sci. Suppl. 2, 89–103. [DOI] [PubMed] [Google Scholar]
- 22.Northcote, D. H., Davey, R. & Lay, J. (1989) Planta 178, 353–366. [DOI] [PubMed] [Google Scholar]
- 23.Botha, C. E. & Cross, R. H. (2000) Micron 31, 713–721. [DOI] [PubMed] [Google Scholar]
- 24.Bucher, G. L., Tarina, C., Heinlein, M., Di Serio, F., Meins, F., Jr. & Iglesias, V. A. (2001) Plant J. 28, 361–369. [DOI] [PubMed] [Google Scholar]
- 25.Iglesias, V. A. & Meins, F., Jr. (2000) Plant J. 21, 157–166. [DOI] [PubMed] [Google Scholar]
- 26.Beffa, R. & Meins, F., Jr. (1996) Gene 179, 97–103. [DOI] [PubMed] [Google Scholar]
- 27.Beffa, R. S., Hofer, R.-M., Thomas, M. & Meins, F., Jr. (1996) Plant Cell 8, 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollenberg, S. M., Sternglanz, R., Cheng, P. F. & Weintraub, H. (1995) Mol. Cell. Biol. 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser, C., Michaelis, S. & Mitchell, A. (1994) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY)
- 30.Sutton, A., Heller, R. C., Landry, J., Choy, J. S., Sirko, A. & Sternglanz, R. (2001) Mol. Cell. Biol. 21, 3514–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballas, N. & Citovsky, V. (1997) Proc. Natl. Acad. Sci. USA 94, 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durfee, T., Becherer, K., Chen, P.-L., Yeh, S.-H., Yang, Y., Kilburn, A. E., Lee, W.-H. & Elledge, S. J. (1993) Genes Dev. 7, 555–569. [DOI] [PubMed] [Google Scholar]
- 33.Bartel, P., Chien, C. T., Sternglanz, R. & Fields, S. (1993) in Cellular Interactions in Development: A Practical Approach, ed. Hartley, D. A. (IRL, New York), pp. 153–179.
- 34.Park, H. & Sternglanz, R. (1998) Chromosoma 107, 211–215. [DOI] [PubMed] [Google Scholar]
- 35.Tzfira, T., Vaidya, M. & Citovsky, V. (2001) EMBO J. 20, 3596–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horsch, R. B., Fry, J. E., Hoffman, N. L., Eichholtz, D., Rogers, S. G. & Fraley, R. T. (1985) Science 227, 1229–1231. [Google Scholar]
- 37.Murashige, T. & Skoog, F. (1962) Physiol. Plant. 15, 473–497. [Google Scholar]
- 38.Domingo, C., Sauri, A., Mansilla, E., Conejero, V. & Vera, P. (1999) Plant J. 20, 563–570. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli, U. K. (1970) Nature 277, 680–685. [DOI] [PubMed] [Google Scholar]
- 40.Ghoshroy, S. & Citovsky, V. (1998) J. Virol. Methods 74, 223–229. [DOI] [PubMed] [Google Scholar]
- 41.Dixon, D. C. & Klessig, D. F. (1995) in Methods in Plant Molecular Biology, eds. Maliga, P., Klessig, D. F., Cashmore, A. R., Gruissem, W. & Varner, J. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 101–110.
- 42.Fields, S. & Song, O. (1989) Nature 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 43.Reynaud, E. G., Leibovitch, M. P., Tintignac, L. A., Pelpel, K., Guillier, M. & Leibovitch, S. A. (2000) J. Biol. Chem. 275, 18767–18776. [DOI] [PubMed] [Google Scholar]
- 44.Bai, L. & Merchant, J. L. (2001) Mol. Cell. Biol. 21, 4670–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]