Abstract
Accurate procentriole formation is critical for centriole duplication. However, the holistic transcriptional regulatory mechanisms underlying this process remain elusive. Here, we show that KAT7 crotonylation, facilitated by the crotonyltransferase hMOF, competes against its acetylation regulated by the deacetylase HDAC2 at the K432 residue upon DNA damage stimulation. This competition diminishes its histone acetyltransferase activity, leading to the inhibition of procentriole formation in colorectal cancer cells. Mechanistically, the reduction of KAT7 histone acetyltransferase activity by the antagonistic effect of KAT7 crotonylation against its acetylation decreases the gene expression associated with procentriole formation by modulating the enrichment of H3K14ac at their promoters and plays an important role in colorectal tumorigenesis. Furthermore, KAT7 crotonylation and acetylation are associated with the prognosis in colorectal cancer patients. Collectively, our findings uncover a previously unidentified role of KAT7 in the regulation of procentriole formation and colorectal tumorigenesis via competitive antagonism of its crotonylation against acetylation.
Subject terms: Acetylation, Colon cancer, Transcriptional regulatory elements
Here, the authors suggest a role of the lysine acetyltransferase KAT7 in inhibiting procentriole formation and colorectal tumorigenesis via competitive antagonism of its crotonylation against acetylation.
Introduction
Lysine acetyltransferase KAT7 (also known as HBO1 or MYST2) is a member of the MYST family of acetyltransferases which is widely distributed and highly conserved in various organisms from yeast to humans1,2. KAT7 has a typical histone acetyltransferase (HAT) catalytic activity zone that enables it to act as the unique acetyltransferase of HBO1 complex1,3. KAT7 exhibits acetyltransferase activity towards both histone H3 (K14) and H4 (K5, K8, K12), which exerts diverse functions in various biological processes2–5. In recent years, numerous studies have reported that KAT7 is upregulated in multiple malignancies and participates in tumor cell proliferation, cell cycle distribution, drug resistance, and tumor stem cell maintenances2–8. Furthermore, abnormal expression of KAT7 is associated with clinical features, diagnosis, therapy and prognosis of patients6,9,10. In addition, post-transcriptional modifications (PTMs) of KAT7 are involved in tumor development11,12. For example, KAT7 is identified as a substrate for cyclin E/CDK2 and undergoes phosphorylation at residue T88 to promote the enrichment of cancer stem-like cells in breast cancer11. Song et al. reported that Plk1-mediates phosphorylation of KAT7 transcriptionally increases c-Fos expression and consequently elevates the expression of its downstream multidrug resistance 1 (MDR1), which contributes to gemcitabine resistance in pancreatic cancer12. These findings provide valuable insights into the key role played by KAT7 or its PTMs in various types of tumor development. Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world with increasing incidence and mortality rates globally13. It is a metabolically active tumor with crucial genetic and epigenetic alterations. However, the direct role of KAT7 on CRC progression remains poorly understood. It is also imperative to elucidate the potential impact of KAT7 PTMs in CRC occurrence and development.
PTMs are at the center of many cellular signaling events and play an important role in the pathogenesis of many diseases, including colorectal cancer14–17. In our previous study, we reported that KAT8 acetylation inhibits the recruitment of RNA pol II to the promoter of the lipolysis-related genes, and mediates the invasive and migratory potential of CRC cells18. Recently, HIF-2α has been found to induce ATX expression by recruiting p300/CBP, thereby promoting crotonylation of histone H3 in the ATX promoter region during hypoxia in SW480 colon cancer cells16. These findings reveal the critical role of acetylation or crotonylation in CRC. Despite the phosphorylation and ubiquitination of KAT7 have been reported, the functional implications of other PTMs on KAT7 remain elusive11,12,19. By scanning the data of proteomics of KAT7 PTMs, KAT7 is found to be acetylated or crotonylated in mammalian cells20,21. For example, quantitative acetylome analysis reveals a two-fold increase in the acetylation levels of KAT7 following treatment with HDACs inhibitor suberoylanilide hydroxamic acid (SAHA) and MS-27520. Furthermore, crotonylome profile demonstrates that Lysine 537 site of KAT7 is crotonylated in CDYL KO cells21. These proteomics findings provide evidence for the existence of KAT7 acetylation and crotonylation. However, their functional elucidation in various biological processes remains unknown. Does KAT7 acetylation or crotonylation contribute to the progression of CRC? To address this question, we investigate whether KAT7 acetylation or crotonylation exists in CRC and what is the critical role of its acetylation or crotonylation in CRC development.
Centriole duplication is a complex process and requires precise regulation to ensure that each centriole replicates only once in a cell cycle22,23. Centriole duplication begins by forming the procentriole. Precise procentriole formation is important for centriole duplication, bipolar spindle assembly and genome integrity maintenance24,25. A small set of core components fundamental for procentriole formation has been identified in humans, including CEP192, CEP152, Polo-like kinase 4 (PLK4), SCL/TAL1 interrupting locus (STIL) and Spindle assembly abnormal protein 6 homolog (SAS6)24–26. Following binding to its centriole receptors CEP152 and CEP192, a ring-to-dot transformation of PLK4 focuses on the wall of the parent centriole, marking the site of procentriole formation27,28. Subsequently, PLK4 phosphorylates STIL to promote the recruitment and binding of SAS6 to organize the central cartwheel29,30. Once the cartwheel is assembled, the centriole protein CEP135 interacts with CPAP to recruit multiple centriole proteins to format and stabilize triplet microtubule blades that induce centriole elongation24,25,31. Among the basic components of the procentriole, PLK4 has been identified as the trigger of centriole biogenesis and plays a critical role in the initiating centriole duplication26,32. Overexpression of PLK4 induces the formation of multiple daughter centrioles and leads to abnormal centrosome amplification26,33. In addition, stabilizing PLK4 by CEP131 leads to centrosome amplification and CRC progression34. In contrast, depletion or inhibition of PLK4 kinase activity results in diminished centriole replication26,32,35. Although significant progress has been made in unraveling the molecular constituents of centrioles over the past two decades, our understanding of holistic transcriptional regulation governing procentriole formation remains limited and warrants further investigation.
In this study, we find a positive correlation between higher levels of KAT7 crotonylation and lower levels of KAT7 acetylation with good prognosis in CRC patients. Further mechanistic studies reveal that competitive antagonism of KAT7 crotonylation against acetylation decreases its HAT activity, thereby reducing the enrichment of H3K14ac at the promoter of genes associated with procentriole formation to diminish centriole duplication. Collectively, our findings uncover an unidentified regulatory role of KAT7 in procentriole formation and colorectal tumorigenesis through competitive antagonism of its crotonylation against acetylation. It may serve as a potential target for the treatment of colorectal cancer.
Results
KAT7 crotonylation and acetylation correlate with the prognosis of CRC patients
KAT7 is a crucial molecule that controls tumor development2. However, the direct effect of KAT7 on CRC progression is unidentified. To investigate the role of KAT7 in colorectal tumorigenesis, we first analyzed the expression of KAT7 in different tumors based on The Cancer Genome Atlas (TCGA) database. We found that KAT7 expression exhibited significant differences between the tumor and adjacent normal tissues in various types of cancers (Fig. 1a). We observed the opposite effects of KAT7 on the overall survival in different cancer patients, indicating the complex and multifaceted roles of KAT7 in cancer progression (Fig. 1b, c). It was noteworthy that there is no significant difference observed in KAT7 expression in colorectal cancer tissues and adjacent normal tissues, and its expression levels did not exhibit any correlation with the prognosis of CRC patients (Fig. 1a, d). To validate the findings from the TCGA database analysis, we collected colon tumor tissues and matched adjacent noncancerous colon tissues from 153 CRC patients. The basic characteristics of the 153 patients were provided in Supplementary Table 1. The KAT7 protein levels in these tissues were detected by western blotting and semi-quantified by scanning the intensity of each band, and the representative bands are shown in Fig. 1e. We found that KAT7 expression was higher in CRC tumor tissue compared to matched adjacent normal tissue (Fig. 1f). However, Kaplan-Meier analysis showed no correlation between the KAT7 expression and overall survival among CRC patients (Fig. 1g).
Fig. 1. KAT7 crotonylation and acetylation correlate with the prognosis of CRC patients.
a Graphs are drawn using Sanger Box based on TCGA database. The expression levels of KAT7 were analyzed in different types of tumor tissues (T, T=tumor sample numbers) and their adjacent normal tissues (N, N=normal sample numbers) in the TCGA cohort. Two-tailed unpaired student’s t-test was used. P-values represented in the figure. Data represent mean ± SD of samples. n = 1 technical replicate. b–d Kaplan-Meier survival analysis was used to plot survival curves in human tumor specimens with high or low KAT7 expression based on the TCGA Human Protein Atlas database. n= tumor sample numbers. P-value based on two-tailed log-rank test represented in figure. (b) Liver Hepatocellular Carcinoma (c) Kidney Renal Clear Cell Carcinoma (d) Colon Adenocarcinoma. e The representative western blotting images of KAT7 in 153 pairs of CRC tissues and matched adjacent noncancerous colon tissues. n = 1 biological replicate due to the precious and limited tissue sample. Semi-quantification of proteins by scanning grayscale values. f Relative expression of KAT7 in 153 pairs of CRC tissues compared to normal adjacent tissues. KAT7 expression levels in normal adjacent tissues were set as 1, and KAT7 expression levels in CRC tissues were set as the gray value ratio of CRC tissue to normal tissue. Two-tailed paired student’s t-test was used. P < 0.0001. g Survival analysis of CRC patients according to KAT7 expression. Kaplan-Meier survival analysis was used to plot survival curves in CRC samples with high or low KAT7 expression. n = tumor sample numbers. P-value based on the two-tailed log-rank test represented in figure. h Definition of the samples used for KAT7 post-transcriptional modification study. i–k Kaplan-Meier survival analysis was used to plot survival curves in CRC samples with the crotonylation or acetylation levels of KAT7. n = tumor sample numbers. (i) crotonylation (j) acetylation (k) crotonylation and acetylation. The levels of KAT7 crotonylation and acetylation are relative. The ratio of the gray value of crotonylation or acetylation to the KAT7 gray value was defined as the relative gray value of the modification. A high level of modification was defined when the relative gray value of modification in CRC tissue exceeded than that in adjacent normal tissue, and vice versa. P-value based on the log-rank test is represented in the figure. P(i): crotonylation high vs crotonylation low; P(j): acetylation high vs acetylation low; P(k): crotonylation high and acetylation low vs crotonylation low and acetylation high. Source data are provided as a Source Data file.
Considering epigenetic alterations have vital roles in colorectal carcinogenesis36, we next investigate whether PTMs of KAT7 are involved in the prognosis of CRC patients. To conduct a more focused analysis, we selected 74 pairs of samples in which the difference in KAT7 expression between malignant CRC tissues and matched adjacent normal tissues was less than ±1.5-fold (Fig. 1h). The basic characteristics of the study population were provided in Supplementary Table 2. According to the proteomics data of KAT7 PTMs, we detected the levels of KAT7 crotonylation and acetylation in these samples by immunoprecipitation (representative bands were shown in Supplementary Fig. 1a, b). The association between KAT7 crotonylation or acetylation and the clinicopathological parameters were summarized in Supplementary Tables 3 and 4. In terms of the overall survival analysis, we found that the increase in KAT7 crotonylation levels was associated with longer survival in colon cancer patients (Fig. 1i). On the contrary, higher levels of KAT7 acetylation were linked to a poor prognosis in CRC patients (Fig. 1j). Notably, patients with higher KAT7 crotonylation and lower KAT7 acetylation levels exhibited substantially higher overall survival rates compared to patients with lower KAT7 crotonylation and higher KAT7 acetylation levels (Fig. 1k). These findings suggest that KAT7 crotonylation and acetylation are associated with the prognosis of CRC patients.
There are dynamic changes in KAT7 crotonylation and acetylation after DNA damage stimulation in CRC cells
Given the correlation between KAT7 crotonylation and acetylation with the prognosis of CRC patients, we wondered what the mechanism underlying this observation was. Firstly, we sought the effective stimuli to change the levels of KAT7 crotonylation and acetylation. It has been reported that DNA damage stimulations can cause an increase in non-histone protein crotonylation21. Therefore, we tried to detect the change of KAT7 crotonylation following various DNA damage drug treatments. We found that DNA damage drugs, especially etoposide, effectively increased KAT7 crotonylation in different CRC cells (Fig. 2a–c). Meanwhile, no significant changes in KAT7 mRNA and protein levels were detected after treatment with various doses or durations of etoposide in HCT116 cells were detected, establishing that etoposide is an effective stimulus for investigating the function of KAT7 crotonylation in CRC cells (Fig. 2d–g). KAT7 crotonylation increased significantly in a dose- and time-dependent manner following etoposide treatment (Fig. 2h, i). It is noteworthy that the elevation of KAT7 crotonylation was always accompanied by a decrease in its acetylation, whatever with different times or doses of etoposide treatment (Fig. 2h, i). Previous studies have shown that NaCr can substantially elevate intracellular levels of crotonyl-CoA and increase the level of protein crotonylation37. Furthermore, we selected NaCr as a stimulus to elevate the levels of KAT7 crotonylation. Surprisingly, the dynamic changes in KAT7 crotonylation and acetylation were observed after NaCr treatment (Fig. 2j). These results implicate that there is a dynamic balance between KAT7 crotonylation and acetylation in response to DNA damage stress in CRC cells.
Fig. 2. There are dynamic changes in KAT7 crotonylation and acetylation after etoposide treatment in CRC cells.
a–c KAT7 crotonylation was detected by immunoprecipitation assay in different colon cancer cell lines upon DNA damage stimulation. (a) HCT116 cell line (b) HT29 cell line (c) sw1116 cell line. d, e HCT116 cells were treated with etoposide (0, 20 μM and 40 μM) for 8 h (d) or 40 μM etoposide for 0, 4, 8 h (e). KAT7 mRNA levels were analyzed by real-time PCR. One-way ANOVA was used. Exact P-values represented in the figure. Error bars represent mean ± SD. f, g HCT116 cells were stimulated under the same condition above. KAT7 protein levels were detected by western blotting. α-tubulin was used as a loading control. h, i KAT7 crotonylation and acetylation were detected in HCT116 cells by immunoprecipitation assay with gradient doses (h) or times (i) of etoposide treatment. j KAT7 crotonylation and acetylation were detected in HCT116 cells by immunoprecipitation assay with NaCr treatment (20 mM, 12 h). The levels of crotonylation or acetylation are relative. The relative grayscale value of the band in the first lane (control group) was set as 1, and the ratio of the grayscale value of the other bands to the grayscale value of the first band was set as the relative grayscale value of the band. Each experiment was three independent biological replicates. Source data are provided as a Source Data file.
Competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue induced by DNA damage stimulation attenuates its HAT activity
Recently, the dynamic regulation of protein PTMs in colorectal tumorigenesis has been demonstrated38. Based on the aforementioned data, we postulated that there is a competitive relationship between crotonylation and acetylation at some site of KAT7. To validate our hypothesis, we first identified the specific crotonylation and acetylation sites of KAT7 upon DNA damage stimulation. Recombinant KAT7 plasmid was transfected into HCT116 cells, and mass spectrometry (MS) was performed. Conservation analysis revealed that four lysine sites (K401, K415, K432 and K462) among the acetylation sites of KAT7 identified by MS are highly conserved from Xenopus to Homo sapiens, suggesting that they may have important, evolutionarily conserved functions (Fig. 3a, b and Supplementary Fig. 2a, b). To investigate whether these four acetylation sites participate in this process, we individually mutated lysine to arginine of these four acetylation sites and then transfected Myc-KAT7 and these four mutations into HCT116 cells. The immunoprecipitation assay showed that the level of KAT7 acetylation reduced after transfection of cells with four KAT7 mutations in cells (Fig. 3c). However, no significant changes in KAT7 crotonylation were observed in these cells (Fig. 3d). These findings suggest that, under physiological conditions, KAT7 acetylation occurs predominantly at these specific sites. Interestingly, KAT7 crotonylation was significantly increased in the Myc-KAT7, Myc-KAT7-K401R, Myc-KAT7-K415R and Myc-KAT7-K462R transfected cells after etoposide treatment, whereas the levels of KAT7 crotonylation was almost unchanged in the K432R transfected cells, indicating that K432 might be the major crotonylation site of KAT7 induced by DNA damage stimulation. Additionally, KAT7 acetylation levels did not further decrease in the K432R mutant group, but not in the other three mutant groups after etoposide treatment, suggesting that KAT7 deacetylation after etoposide treatment may occur at the K432 residue (Fig. 3e). These investigations suggest that K432 may serve as a potential competitive site for KAT7 crotonylation against its acetylation in response to etoposide treatment.
Fig. 3. Competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue induced by DNA damage stimulation attenuates its HAT activity.
a HCT116 cells were transfected with Flag-KAT7 plasmid and the acetylation sites of KAT7 were identified by MS. n = 1 biological replicate. b Alignment of MS-characterized putative KAT7 acetylation residues among different species. c, d HCT116 cells were transfected with Myc-KAT7, Myc-KAT7-K401R, Myc-KAT7-K415R, Myc-KAT7-K432R and Myc-KAT7-K462R plasmids for immunoprecipitation assay to detect the acetylation (c) and crotonylation (d) levels of KAT7. e HCT116 cells were transfected with the plasmids mentioned above, and then treated with etoposide (40 μM, 8 h). An immunoprecipitation assay was performed to detect the crotonylation or acetylation levels of KAT7. f HCT116 cells were treated with etoposide (40 μM, 8 h), and then the acetylation changes of histone substrate of KAT7 were detected to examine the HAT activity of KAT7. g, h In vitro HAT activity assay was performed to detect the HAT activity of KAT7 purified from HCT116 cells with (g) etoposide (40 μM, 8 h) or (h) NaCr (20 mM, 12 h) stimulation. i In vitro HAT activity assay was performed to detect the HAT activity of KAT7 purified from HCT116 cells transfected with Myc-KAT7-WT or Myc-KAT7-K432R plasmids. j, k In vitro HAT activity assay was performed to detect the HAT activity of KAT7 purified from HCT116 cells transfected with Myc-KAT7-WT or Myc-KAT7-K432R plasmids after etoposide (40 μM, 8 h) (j) or NaCr (20 mM, 12 h) (k) stimulation. The levels of crotonylation or acetylation are relative. The relative grayscale value of the band in the first lane (control group) was set as 1, and the ratio of the grayscale value of the other bands to the grayscale value of the first band was set as the relative grayscale value of the band. The experiments in Fig. 3c–k were three independent biological replicates. Source data are provided as a Source Data file.
We subsequently explored the function of competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue upon DNA damage stimulation. It has been reported that K274 of hMOF is an important site to mediate its acetyltransferase activity39. KAT7 K432 is highly sequence homologous with hMOF K27440. Therefore, we speculated that competitive antagonism of KAT7 crotonylation against acetylation at residue K432 may diminish the catalytic HAT activity of KAT7. We initially assessed the acetylation levels of H3K14 and H4K5/8/12, substrates of KAT7, following etoposide treatment41,42. We observed that H3K14 and H4K5/8/12 acetylation were significantly reduced after etoposide stimulation, indicating that etoposide treatment may attenuate the HAT activity of KAT7 (Fig. 3f). To further confirm the direct effect of KAT7 crotonylation and acetylation in this process, HCT116 cells were treated with etoposide or NaCr, and KAT7 protein was purified by immunoprecipitation for in vitro HAT assay. Acetylation of H3K14 and H4K5/8 were significantly reduced in both etoposide- and NaCr- stimulated cells, indicating that competitive antagonism of KAT7 crotonylation against acetylation effectively attenuates the HAT activity of KAT7 (Fig. 3g, h and Supplementary Fig 2c, d).
Next, we explored the role of the KAT7 K432 residue in the regulation of KAT7 HAT activity by in vitro HAT assay. We found that H3K14 and H4K5/8 acetylation was significantly reduced in the Myc-KAT7-K432R transfected group compared with the Myc-KAT7-WT transfected group in vitro, indicating the pivotal role of the K432 residue in medicating KAT7 HAT activity (Fig. 3i and Supplementary Fig 2e). Moreover, we observed that H3K14 and H4K5/8 acetylation is not reduced following etoposide or NaCr stimulation in the Myc-KAT7-K432R transfected group compared to the Myc-KAT7-WT transfected group by in vitro HAT assay, suggesting that the K432 residue is a key site for medicating KAT7 HAT activity through its antagonism of KAT7 crotonylation against acetylation (Fig. 3j, k). Altogether, these observations indicate that competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue induced by DNA damage stimulation attenuates its HAT activity.
hMOF is the predominant crotonyltransferase of KAT7 upon DNA damage stimulation
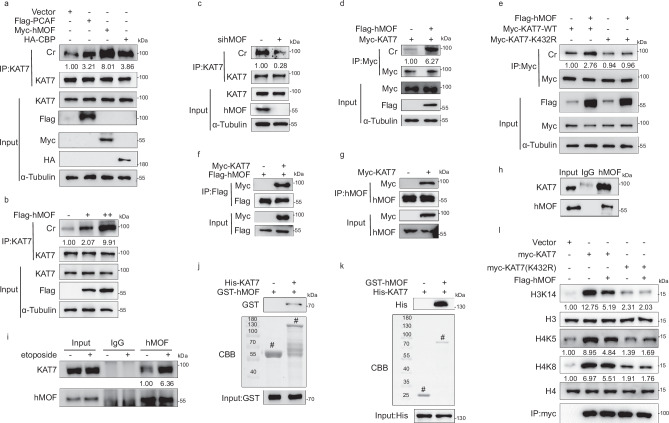
Having known the critical function of the antagonistic effect of KAT7 crotonylation against its acetylation upon DNA damage stress, we next attempted to identify the specific histone crotonyltransferase (HCT) of KAT7 crotonylation in this process. Due to the poor antigenic epitope, we failed to generate a site-specific antibody against KAT7 acetylation and crotonylation at residue K432. This made it difficult to establish a specific link to KAT7 K432 crotonylation and acetylation. Consequently, we had to resort to using a pan-crotonylation antibody for the detection of KAT7 crotonylation. We transfected HCT116 cells with different HCT plasmids PCAF, hMOF and CBP. Overexpression of these three plasmids resulted in varying degrees of increase in KAT7 crotonylation, with the most significant enhancement after hMOF overexpression (Fig.4a). RNAi experiments also demonstrated the predominant role of hMOF in regulating KAT7 crotonylation (Supplementary Fig. 3a). Meanwhile, we have observed that the acetyltransferase activity of hMOF is inhibited upon etoposide treatment, thereby excluding the impact of hMOF acting as an acetyltransferase on KAT7 acetylation (Supplementary Fig. 3b–d). Both overexpression of Flag-tagged hMOF and knockdown of hMOF using siRNA confirmed the effect of hMOF on KAT7 crotonylation (Fig. 4b, c). The in vitro crotonylation assay also showed that hMOF can catalyze KAT7 crotonylation (Fig. 4d). When hMOF was overexpressed in KAT7-WT and KAT7-K432R transfected cells, we found that KAT7 crotonylation did not increase in the KAT7-K432R group compared to the KAT7-WT group, showing that hMOF can crontonylate KAT7 at the K432 residue (Fig. 4e).
Fig. 4. hMOF is the predominant crotonyltransferase of KAT7 upon DNA damage stimulation.
a HCT116 cells were transfected with plasmids as indicated, and harvested to identify the specific HCTs for KAT7 crotonylation by immunoprecipitation. b Flag-tagged hMOF were transfected with dose gradient into HCT116 cells to confirm the effect of hMOF on KAT7 crotonylation by immunoprecipitation. c HCT116 cells were transfected with hMOF siRNA to detect KAT7 crotonylation by immunoprecipitation. d HCT116 cells were transfected with Myc-KAT7 or Flag-hMOF, and then the Myc-KAT7 and Flag-hMOF proteins were purified for in vitro crotonylation assay. The levels of KAT7 crotonylation was detected by immunoprecipitation. e HCT116 cells were transfected with Myc-KAT7-WT or Myc-KAT7-K432R with or without hMOF overexpression. KAT7 crotonylation was detected by immunoprecipitation assay. f–h Exogenous, semi-exogenous and endogenous Co-IP were performed to detect the interaction between KAT7 and hMOF in HCT116 cells. i Endogenous Co-IP was performed to detect the interaction between KAT7 and hMOF in response to etoposide (40 μM, 8 h) treatment in HCT116 cells. j, k GST-hMOF FL and His-KAT7 FL were purified, and then western blotting was performed to detect the direct binding of KAT7 and hMOF in vitro. The bands were stained with Coomassie brilliant blue staining. # indicates the specific bands. l HCT116 cells were transfected with Myc-KAT7-WT or Myc-KAT7-K432R with or without hMOF overexpression and then KAT7 was purified by Myc-tag antibody for in vitro HAT activity assay. The relative grayscale value of the band in the first lane (control group) was set as 1, and the ratio of the grayscale value of the other bands to the grayscale value of the first band was set as the relative grayscale value of the band. Each experiment was three independent biological replicates. Source data are provided as a Source Data file.
Subsequently, the exogenous, semi-exogenous and endogenous Co-IP assays showed that KAT7 can interact with hMOF (Fig. 4f–h). Additionally, we observed that the interaction between KAT7 and hMOF significantly increased after etoposide stimulation, showing that hMOF can interact with and crotonylate KAT7 upon etoposide stimulation in vivo (Fig. 4i). Furthermore, the direct binding of KAT7 and hMOF was proven by GST pull-down assay (Fig. 4j, k). The domain mapping experiments of hMOF and KAT7 were shown in Supplementary Fig. 3j–m. These findings support that hMOF interacts with KAT7 and crontonylates KAT7 in vivo and in vitro.
Finally, we overexpressed hMOF in KAT7-WT and KAT7-K432R transfected cells and purified KAT7 protein for in vitro HAT assay. As shown in Fig. 4l, overexpression of hMOF significantly attenuates the HAT activity of KAT7 in the KAT7-WT group. However, there is no significant reduction in the HAT activity of KAT7 after overexpression of hMOF in the KAT7 K432R group, indicating that KAT7 crotonylation at the K432 residue by hMOF effectively attenuates the HAT activity of KAT7. Collectively, these findings demonstrate that hMOF is the predominant crotonyltransferase responsible for KAT7 crotonylation.
HDAC2 is the major deacetylase of KAT7 upon DNA damage stimulation
Having identified the predominant crotonyltransferase of KAT7, we next sought to identify the deacetylase responsible for KAT7 deacetylation. We treated HCT116 cells with either the class I and II HDAC inhibitor trichostatin A (TSA) or the class III HDAC inhibitor nicotinamide (NAM), and detected KAT7 acetylation changes using an immunoprecipitation assay. Our results demonstrated that exposure to TSA, but not NAM, leads to a marked elevation in KAT7 acetylation, suggesting that class I or II HDACs may mediate KAT7 deacetylation (Fig. 5a). To further elucidate the specific histone deacetylase involved in KAT7 deacetylation, we purified proteins associated with KAT7 following etoposide treatment using tandem affinity purification (TAP) and subjected them to MS analysis. Among the proteins interacting with KAT7 identified by MS, HDAC2 was implicated as a potential candidate responsible for the deacetylation of KAT7 (Fig. 5b, Supplementary Data File 1). Both overexpression of Flag-tagged HDAC2 and knockdown of HDAC2 using shRNA confirmed the effect of HDAC2 on KAT7 deacetylation (Fig. 5c, d). Additionally, an in vitro deacetylation assay verified the direct role of HDAC2 in KAT7 deacetylation, showing its ability to deacetylate KAT7 in vitro (Fig. 5e). These findings showed that HDAC2 may be the major deacetylase of KAT7.
Fig. 5. HDAC2 is the major deacetylase of KAT7 upon DNA damage stimulation.
a HCT116 cells were treated with TSA (3 μM, 12 h) and NAM (5 mM, 12 h). Cells were harvested to detect the acetylation levels of KAT7 by immunoprecipitation. b HCT116 cells were transfected with Flag-KAT7 plasmid and then stimulated with 40 μM etoposide for 8 h. KAT7-associated proteins were identified by MS. n = 1 biological replicate. The table lists the selected proteins identified by MS. The full protein list is provided in Supplementary Data File 1. c Flag-tagged HDAC2 were transfected in dose-dependent manner into HCT116 cells to confirm the effect of HDAC2 on KAT7 acetylation by immunoprecipitation. d Detection of KAT7 acetylation levels in HDAC2 stable knockdown HCT116 cells by immunoprecipitation. e HCT116 cells were transfected with Myc-KAT7 or Flag-HDAC2, and then the Myc-KAT7 and Flag-HDAC2 proteins were purified for in vitro deacetylation assay. KAT7 acetylation was detected by immunoprecipitation. f–h Exogenous, semi-exogenous and endogenous Co-IP were performed to detect the interaction between KAT7 and HDAC2 in HCT116 cells. i Endogenous Co-IP was performed to detect the interaction between KAT7 and HDAC2 in response to etoposide (40 μM, 8 h) treatment in HCT116 cells. j, k GST-HDAC2 FL and His-KAT7 FL were purified, and then western blotting was performed to detect the direct binding of KAT7 and HDAC2 in vitro. The bands were stained with Coomassie brilliant blue staining. # indicates the specific bands. The relative grayscale value of the band in the first lane (control group) was set as 1, and the ratio of the grayscale value of the other bands to the grayscale value of the first band was set as the relative grayscale value of the band. The experiments in Fig. 5a, c-k were three independent biological replicates. Source data are provided as a Source Data file.
Furthermore, a Co-IP assay was performed to explore the potential molecular interaction between HDAC2 and KAT7. Exogenous, semi-exogenous and endogenous Co-IP assays showed that KAT7 and HDAC2 can interact with each other (Fig. 5f–h). Additionally, we observed an enhanced interaction between KAT7 and HDAC2 upon etoposide treatment (Fig. 5i). These data suggest that HDAC2 can interact with and deacetylate KAT7 upon etoposide treatment in vivo. Moreover, the direct binding of KAT7 and HDAC2 was detected by GST pull-down assay (Fig. 5j, k). The domain mapping experiments of HDAC2 and KAT7 were shown in Supplementary Fig. 4d–g. Taken together, these data suggest that HDAC2 can interact with KAT7 and deacetylate KAT7 in vivo and in vitro.
Reduction of KAT7 HAT activity inhibits the expression of genes associated with procentriole formation
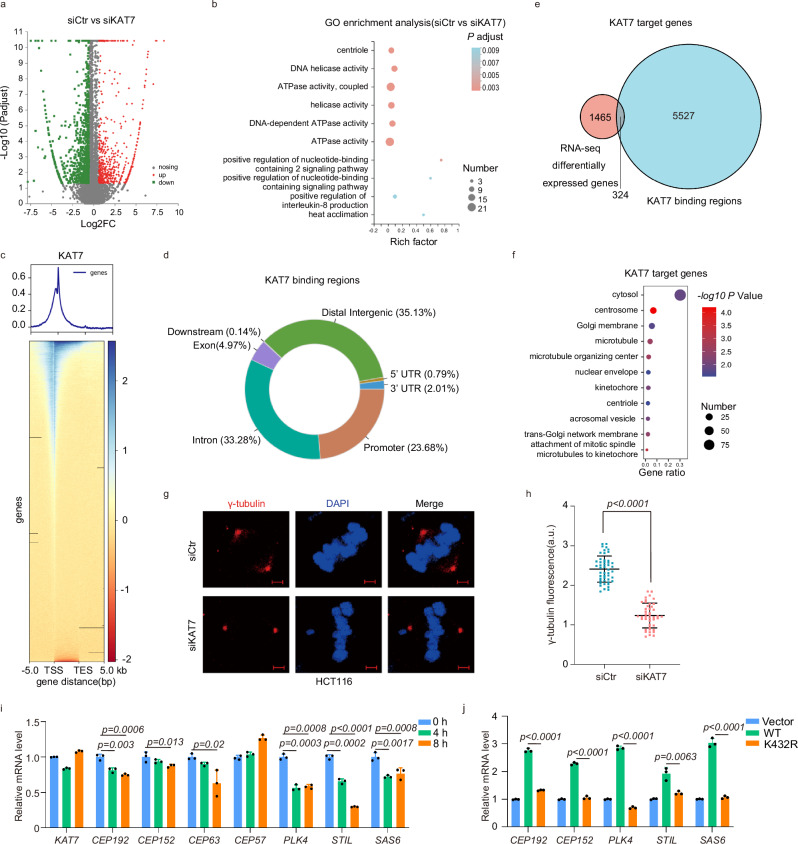
Our data thus far suggest that the competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue effectively inhibits KAT7 HAT activity. However, it remains unclear how the reduction of KAT7 HAT activity by the competition influences CRC progression. To further elucidate the impact of reduced KAT7 HAT activity, we first explored the cellular functions of KAT7 using RNA-sequencing analysis with KAT7 knockdown cells. The knockdown efficiency of KAT7 was detected by western blotting (Supplementary Fig. 5a). Volcano plot analyses revealed that a total of 632 upregulated and 1157 downregulated differentially expressed genes (changes ≥1.5- fold) between KAT7 siRNA-transfected cells and non-specific siRNA-transfected cells (Fig. 6a, Supplementary Data File 2). Gene Ontology (GO) analysis revealed that these differentially expressed genes involve in centriole-related biological processes (Fig. 6b). To confirm the direct effect of KAT7 on these differentially expressed genes, we conducted a comprehensive search in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/)43 and found the KAT7 ChIP-seq data [GSE33007] in the RKO CRC cell line44. Through the analysis of ChIP-seq data, 5851 KAT7-binding regions of genes were detected (Fig. 6c). As shown in Fig. 6d, 23.68%, 33.28% and 35.13% of the KAT7-binding regions are located in gene promoters, intronic and intergenic regions, respectively. By analysing the KAT7 ChIP-seq and RNA-seq data, the 324 genes were found to contain KAT7 binding regions among the differentially expressed genes (Fig. 6e). GO analysis of these overlapping genes demonstrated their involvement in centrosome processes (Fig. 6f). These results suggest that KAT7 may directly mediate the expression of centrosome related genes.
Fig. 6. Reduction of KAT7 HAT activity inhibits the expression of genes associated with procentriole formation.
a Volcano plot showing the mRNA changes upon the depletion of KAT7 in HCT116 cells. Downregulated (green) and upregulated (red) genes (fold change ≥ 1.5) are marked in blue or red, respectively. Adjusted P-value based on benjamini-Hochberg method and fold change was used to draw volcano plot. n = 1 biological replicate. b GO enrichment analysis of the molecular functions and ontology of biological processes with changed genes in (a). Adjusted P-value in GO enrichment analysis based on fisher’s exact test. The color of the circle indicates the adjusted P value. The circle size indicates the number of differentially expressed genes (DEGs). c Heatmaps showing the occupancy of KAT7 in RKO cells centered on peak summits ± 5 kb. The ChIP-seq signal heatmap using a 10 kb window was centered on peak regions. d Genomic distribution of KAT7 binding regions in RKO cells. e Venn diagram showing overlap of KAT7-binding regions and the differentially expressed genes of RNA-seq in KAT7 siRNA cells. f Gene ontology enrichment of overlapped genes in (e) operated by Database for Annotation, Visualization and Integrated Discovery (DAVID). The circle size represents the number of KAT7 target genes associated with each pathway. The circle color gradient represents the P value. P-value based on fisher’s exact test. g HCT116 cells were transfected with non-specific siRNA or KAT7 siRNA, and then were stained with γ-tubulin (red) and DAPI (blue) for immunofluorescence. Scale bar, 2 μm. h The fluorescence intensity of γ-tubulin foci was measured by Image J as the volume of centrosome. Dot plots depict the volume of centrosomes. Centrosome volumes were averaged across the two centrosomes in each cell (control group: n = 49 cells; KAT7 knockdown group: n = 44 cells). Two-tailed unpaired student’s t-test was used. P-values represented in the figure. Error bars represent mean ± SD. i HCT116 cells were treated with etoposide (0, 4 or 8 h, 40 μM), and then cells were harvested to examine the expression of genes encoding procentriole formation proteins by real-time PCR. n = 3 independent biological replicates. One-way ANOVA was used. P-values represented in the figure. Error bars represent mean ± SD. j HCT116 cells were transfected with Myc-Vector, Myc-KAT7-WT or Myc-KAT7-K432R plasmids, and then cells were harvested to examine the expression of genes encoding procentriole formation proteins by real-time PCR. n = 3 independent biological replicates. One-way ANOVA was used. P-values represented in the figure. Error bars represent mean ± SD. Source data are provided as a Source Data file.
To further validate the regulatory effect of KAT7 in centrosome, we detected the expression of several genes encoding centrosome-associated proteins using the qPCR assay. A large number of centrosome-associated genes are downregulated in KAT7 knockdown cells compared with control cells (Supplementary Fig. 5b). We also found that the centrosome volumes, defined by gamma-tubulin staining, significantly decrease in KAT7 knockdown HCT116 cells (Fig. 6g, h). These data indicate that KAT7 may mediate the expression of genes associated with centrosome.
Having established the regulatory role of KAT7 on gene expression related to centrosome, we next explored the role of reduced KAT7 HAT activity by competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue in this process. We detected these gene expressions after etoposide treatment and found that most of the core genes encoding procentriole formation proteins including CEP192, CEP152, PLK4, STIL and SAS6 decrease significantly among the changed genes, suggesting that reduction of KAT7 HAT activity may be involved in the process of procentriole formation (Fig. 6i and Supplementary Fig. 5c). Considering the consistent effect of crotonylation increased and acetylation decreased of KAT7 on its HAT activity and the pivotal role of the K432 residue in medicating KAT7 HAT activity, we employed the K432R mutation to represent the status of low KAT7 HAT activity. When transfected KAT7-WT (normal HAT activity) and KAT7-K432R mutant (low HAT activity) plasmids, we found that the relative mRNA levels of CEP192, CEP152, PLK4, STIL and SAS6 increase significantly in KAT7-WT transfected cells and this effect is reversed in KAT7-K432R transfected cells (Fig. 6j). Remarkably, a significant reduction in PLK4 (the trigger of centriole biogenesis) protein levels was observed in KAT7 knockdown cells (Supplementary Fig. 5d). We also observed that the increased protein levels of PLK4 in KAT7-WT transfected cells were sharply decreased in KAT7-K432R transfected HCT116 cells (Supplementary Fig. 5e). The alterations in PLK4 protein levels provide supporting evidence for the regulatory role of KAT7 on genes involved in procentriole formation. In summary, our data conclude that reduction of KAT7 HAT activity diminishes the gene expression associated with procentriole formation.
Reduction of KAT7 HAT activity inhibits centriole duplication by impairing the enrichment of H3K14ac at the promoter of genes associated with procentriole formation
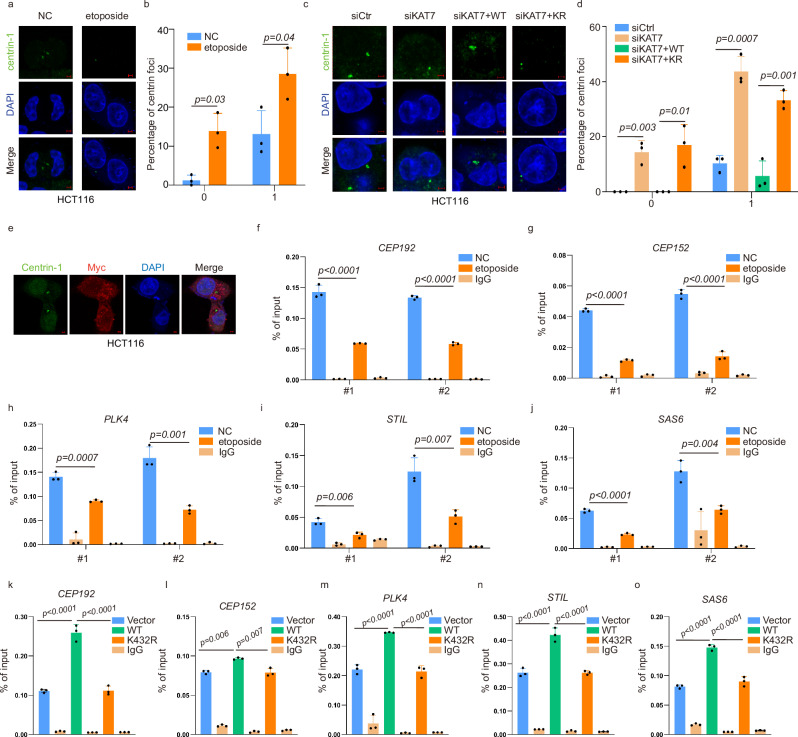
Given that the reduced HAT activity of KAT7 inhibited the expression of genes associated with procentriole formation, we conducted an immunofluorescence assay to further investigate its impact on centriole duplication. As shown in Fig. 7a, etoposide treatment led to a reduction of centriole number, manifested by the acquisition of less than two centrin-1 foci. We observed that approximately 42.1% of HCT116 cells treated with etoposide exhibited either none or only one centriole (Fig. 7b). We next investigated the effect of KAT7 depletion on centriole duplication. Immunofluorescence and microscopy analysis demonstrated that KAT7 knockdown resulted in a significant reduction of centriole number, with 58.1% of KAT7 knockout cells showing just less than two centrioles (Fig. 7c, d). Remarkably, overexpressing KAT7-WT, but not KAT7-K432R, was able to rescue the phenotype induced by KAT7 knockdown (Fig. 7c, d). These findings suggest that inhibition of KAT7 HAT activity can effectively suppress centriole duplication in CRC cells.
Fig. 7. Reduction of KAT7 HAT activity inhibits centriole duplication by impairing the enrichment of H3K14ac at the promoter of genes associated with procentriole formation.
a HCT116 cells were treated with or without etoposide (40 μM, 8 h), and then were stained with centrin-1 (green) and DAPI (blue) for immunofluorescence. A total of 300 cells were captured and representative images are shown. n = 3 independent biological replicates. Scale bar, 2 μm. b The proportion of cells with the indicated numbers of foci were counted. Two-tailed unpaired student’s t-test was used. Exact P-values represented in the figure. Each bar represents mean ± SD. c HCT116 siRNA cells were transfected with Myc-vector, Myc-KAT7-WT or Myc-KAT7-K432R plasmids, and then were stained with centrin-1 (green) and DAPI (blue) for immunofluorescence. A total of 300 cells were captured and representative images are shown. n = 3 independent biological replicates. Scale bar, 2 μm. d The proportion of cells with the indicated numbers of foci were counted. One-way ANOVA was used. Exact P-values represented in the figure. Each bar represents mean ± SD. e HCT116 cells were transfected with Myc-KAT7, and then were stained with centrin-1 (green), Myc (red) and DAPI (blue) for immunofluorescence. Representative confocal images are provided. n = 5 randomly captured images. Scale bar, 2 μm. f–j HCT116 cells were treated with or without etoposide (40 μM, 8 h). A ChIP assay was performed to detect the enrichment of H3K14ac at the promoter regions of genes associated with procentriole formation. n = 3 independent biological replicates. One-way ANOVA was used. P-values represented in the figure. Each bar represents mean ± SD for biological triplicate experiments. f CEP192, g CEP152, h PLK4, i STIL and j SAS6. k–o HCT116 cells were transfected with Myc-vector, Myc-KAT7-WT or Myc-KAT7-K432R plasmids and a ChIP assay was performed to detect the enrichment of H3K14ac at the gene promoter regions of genes associated with procentriole formation. n = 3 independent biological replicates. One-way ANOVA was used and multiple comparisons were applied in these groups. P-values represented in the figure. Each bar represents mean ± SD. (k) CEP192, (l) CEP152, (m) PLK4, (n) STIL and (o) SAS6. Source data are provided as a Source Data file.
Having known the key role of KAT7 HAT activity reduction in centriole duplication, we further explored the mechanism underlying this phenomenon. The co-localization of KAT7 with centrioles was first detected by immunofluorescence assay. As shown in Fig. 7e, KAT7 is localized within the nucleus and cytoplasm without any detectable co-localization with centrioles, indicating that KAT7 may not regulate centriole replication by acting directly with centrioles. The analysis of gene tracks showed that KAT7 locates on the promoter regions of genes associated with procentriole formation (Supplementary Fig. 6). Considering the effect of inhibition of KAT7 HAT activity following etoposide treatment on H3K14 and H4K5/8 acetylation, we speculated that KAT7 might regulate the transcriptional activity of genes associated with procentriole formation by influencing the enrichment of H3K14ac or H4K5/8ac at their promoter regions. To verify this, we selected two fragments of approximately 300 bp in size within the 1200 bp region upstream of the transcription start site (TSS) of these genes and designed specific primers for chromatin immunoprecipitation quantitative PCR (ChIP-qPCR). The ChIP-qPCR assay showed a reduction in H3K14ac enrichment at the promoter regions of CEP192, CEP152, PLK4, STIL and SAS6 after etoposide treatment (Fig. 7f–j). Meanwhile, we observed a significant increase in H3K14ac enrichment at these genes’ promoters upon transfection with KAT7-WT plasmid, which was reversed by overexpression of KAT7-K432R (Fig. 7k–o). Collectively, these data demonstrate that reduction of KAT7 HAT activity inhibits procentriole formation by decreasing H3K14ac enrichment at the promoter regions of these genes, thereby impeding centriole duplication.
Reduction of KAT7 HAT activity inhibits tumor cell proliferation and colorectal tumorigenesis
Thus far, having clarified the underlying mechanism by which reduced HAT activity of KAT7 mediates procentriole formation, we have now validated its pivotal role in colorectal cancer cell proliferation and tumorigenesis. We found that knockdown of KAT7 can effectively inhibit the proliferation of CRC cells (Fig. 8a). The colony formation assay also showed that the colony forming efficiency decreased by approximately 18.0% in KAT7 knockdown cells compared with control cells (Fig. 8b, c). Consistently, we found that cell proliferation is inhibited in KAT7-K432R transfected cells compared with KAT7-WT transfected cells (Fig. 8d). Additionally, consistent results were obtained from the colony formation assay (Fig. 8e, f). Moreover, both the proliferation and the colony forming efficiency of CRC cells were decreased after treatment with the KAT7 HAT activity inhibitor WM-3835 (Fig. 8g–i). These data demonstrate that reduction of KAT7 HAT activity can effectively inhibit colorectal cancer cell proliferation.
Fig. 8. Reduction of KAT7 HAT activity inhibits tumor cell proliferation and colorectal tumorigenesis.
a HCT116 cells were transfected with non-specific siRNA or KAT7 siRNA, and cell proliferation was assessed by CCK-8 assay. n = 3 independent biological replicates. Two-tailed unpaired student’s t-test was used. Exact P-values represented in the figure. Error bars represent mean ± SD. b, c Clonogenic growth of non-specific siRNA or KAT7siRNA transfected HCT116 cells was observed (b), and the statistical chart was shown in (c). n = 3 independent biological replicates. Two-tailed unpaired student’s t-test was used. Exact P-values represented in the figure. Each bar represents mean ± SD. d HCT116 cells were transfected with Myc-vector, Myc-KAT7-WT or Myc-KAT7-K432R plasmids, and cell proliferation of each group was assessed by CCK-8 assay. n = 3 independent biological replicates. One-way ANOVA was used. Exact P-values represented in the figure. Error bars represent mean ± SD. e, f Clonogenic growth of each group was observed (e), and the statistical chart was shown in (f). n = 3 independent biological replicates. One-way ANOVA was used. Exact P-values represented in the figure. Error bars represent mean ± SD. g HCT116 cells were treated with KAT7 HAT activity inhibitor WM-3835 (0, 50 μM and 100 μM) for 24 h, and cell proliferation was assessed by CCK-8 assay. n = 3 independent biological replicates. One-way ANOVA was used. Exact P-values represented in the figure. Error bars represent mean ± SD. h, i Clonogenic growth of the cells treated with WM−3835 was observed (h), and the statistical chart was shown in (i). n = 3 independent biological replicates. One-way ANOVA was used. Exact P-values represented in the figure. Error bars represent mean ± SD. j Xenograft experiment was performed in BALB/c nude mice injected with stably transfected HCT116-KAT7-WT and HCT116-KAT7-K432R cells. Measurement of tumor diameters was carried out every 3 days one week after injection, and tumor volumes were calculated. n = 9 mice for each group. Two-tailed paired student’s t-test was used. Exact P-values represented in the figure. Error bars represent mean ± SD. k, l Mice were executed 19 days after treatment (k), and the tumors were dissected and photographed (l). m, n Measurement of weight (m) and volume (n) of tumors. Dots represent the volume and weight of KAT7-WT tumors (n = 9 mice) or KAT7 K432R tumors (n = 9 mice). Two-tailed paired student’s t-test was used. Exact P-values represented in the figure. Error bars represent mean ± SD. o A working model outlining the proposed mechanism by which competitive antagonism of KAT7 crotonylation against acetylation regulates centriole duplication and colorectal tumorigenesis. Source data are provided as a Source Data file.
Finally, we utilized cell line-derived xenograft models to investigate the role of reduced KAT7 HAT activity reduction in tumorigenesis. KAT7-WT or KAT7-K432R stably transfected HCT116 cells were established and injected subcutaneously into BALB/c nude mice. One week later, the length and width of the tumors were measured every 3 days. Remarkably, KAT7-K432R significantly inhibited tumor growth in the xenograft mice (Fig. 8j–l). The tumor weight and volume exhibited a significant reduction in the KAT7-K432R group compared to the KAT7-WT group (Fig. 8m, n). Together, our findings further confirm the inhibitory effect of KAT7 HAT activity reduction on tumor cell proliferation and colorectal tumorigenesis.
Discussion
In this study, we aim to investigate the function of KAT7 PTMs on centriole duplication and its contribution to CRC progression. We observed that the levels of KAT7 crotonylation and acetylation were correlated with the prognosis of CRC patients (Fig. 1i–k). Further mechanistic studies revealed that the antagonistic effect of KAT7 crotonylation against acetylation at K432 residue attenuated its HAT activity upon DNA damage stimulation. The decreased HAT activity of KAT7 resulted in a diminished enrichment of H3K14ac at the promoter regions of genes related to procentriole formation, ultimately inhibiting centriole duplication (Fig. 8o). Overall, our findings uncover a previously unidentified role of KAT7 in regulating centriole duplication and colorectal tumorigenesis through competitive antagonism of KAT7 crotonylation against acetylation.
Initiation and progression of CRC are induced by the cumulation of a variety of genetic and epigenetic aberrations in colonic epithelial cells. Apart from DNA methylation, chromatin remodeling and non-coding RNAs, PTMs also play important role in the development of CRC15,45–47. For example, PHF5A acetylation-induced alternative splicing is shown to stabilize KDM3A mRNA and contribute to colon carcinogenesis by promoting the capacity of cancer cells for stress resistance15. In addition to individual modification, the dynamic balance between modifications has garnered increasing attention. Zhu et al. reported that dephosphorylation of malic enzyme 1 (ME1) at residue S336, which is regulated by PGAM5, enables increased ACAT1-mediated acetylation at K337. The dynamic balance between ME1 phosphorylation and acetylation affects NADPH generation, lipid metabolism and susceptibility to colorectal tumorigenesis38. Qian Z et al. found that AKT1 decrotonylation facilitates AKT1 phosphorylation and activation to promote myogenic differentiation48. In this study, we found that competitive antagonism of KAT7 crotonylation against acetylation at the K432 residue induced by DNA damage drug etoposide stimulation attenuates KAT7 HAT activity and inhibits CRC tumor cell proliferation and tumorigenesis. As we know, excessive proliferation of tumor cells can result in replication stress and excessive spontaneous DNA damage. It is reasonable to propose that replication stress and excessive spontaneous DNA damage can induce KAT7 K432 crotonylation, which may play important role in the proliferation and development of tumor cells. While it is challenging to directly link KAT7 K432 crotonylation or acetylation with clinical samples due to the lack of a site-specific antibody targeting the K432, the observed changes in KAT7 modifications detected by pan-crotonylation or -acetylation antibodies in colorectal cancer patient samples are consistent with those found in CRC cells. We observed that patients with higher levels of KAT7 crotonylation and lower levels of KAT7 acetylation exhibited significantly increased overall survival rates compared to patients with lower levels of KAT7 crotonylation and higher levels of KAT7 acetylation (Fig. 1i–k). Our findings shed light on the antagonistic effect of KAT7 crotonylation against its acetylation in mediating KAT7 HAT activity, providing valuable insights into the role of KAT7 in CRC progression.
Therefore, what is the primary underlying factor driving the upregulation of KAT7 crotonylation and concurrent downregulation of its acetylation? Previous studies have indicated that levels of histone crotonylation increase while levels of acetylation decrease in response to elevated crotonyl-CoA, and vice versa49,50. Notably, compared to acetyl-CoA which is abundantly present within cells, the intracellular abundance of crotonyl-CoA is approximately 1000-fold lower. The relatively low intracellular abundance of crotonyl-CoA makes it more susceptible to change and more likely to impact cellular crotonylation levels51. Here, we observed a significant upregulation expression of several genes associated with crotonyl-CoA production following etoposide treatment (Supplementary Fig. 7a). Additionally, we measured the concentration of crotonyl-CoA by ELISA and found that it increased about 2.1-fold after etoposide treatment (Supplementary Fig. 7b). These findings suggest that the increase in crotonyl-CoA generation may lead to an accelerated accumulation of KAT7 crotonylation up etoposide stimulation.
The mechanism by which acyltransferases or deacylases selectively catalyze certain protein acylation/deacylation in vivo remains elusive. It is an open question in the field of post-translational modifications of proteins. In our study, we found that hMOF and HDAC2 are involved in the dynamic balance between KAT7 crotonylation and acetylation. We found that hMOF may function as an HAT for KAT7 (Supplementary Fig. 3b). However, we observed that the acetylation level of H4K16, the main histone target of hMOF, decreased after etoposide treatment. These findings suggest that the acetyltransferase activity of hMOF may be inhibited following etoposide treatment. In further experiments, we showed that etoposide can inhibit the acetylation level of hMOF K274, a site is reported to be required for hMOF HAT activity52,53, indicating that etoposide may inhibit the HAT activity of hMOF (Supplementary Fig. 3c, d). Meanwhile, we obtained the crystal structure of WT-hMOF, K274 mutant hMOF and crotonyl-CoA structure from the PDB website and PubChem website. Then, we calculated the binding energy between hMOF (WT and K274 mutant) with crotonyl-CoA, respectively. There was no significant difference in the binding energy between hMOF (WT and K274 mutant) and crotonyl-CoA, indicating that the K274 mutation of hMOF does not affect the affinity between hMOF and crotonyl-CoA (Supplementary Fig. 3e–i). The results suggest that the K274 mutation of hMOF may not affect the HCT activity of hMOF. From these data, we speculated that hMOF might mainly play its HCT activity on KAT7 crotonylation instead of its HAT activity on KAT7 acetylation after etoposide treatment, due to the inhibitory effect of etoposide on its HAT activity. As we know, the concentration of acetyl-CoA is much higher than that of crotonyl-CoA in cells. Although we found that several genes associated with crotonyl-CoA production are upregulated and that the level of crotonyl-CoA increases after etoposide treatment (Supplementary Fig. 7a, b), the mechanism by which hMOF exerts its HCT activity under the condition of losing its HAT activity after etoposide treatment is still not fully elucidated in our study. This is a weak point of our model and needs to be further investigated.
HDAC2 was reported to regulate both histone acetylation and crotonylation54. We also identified the deacetylase and decrotonylase activity of HDAC2 on KAT7 deacetylation and decrotonylation following etoposide treatment (Supplementary Fig. 4a–c). According to our data, we have proposed a model to demonstrate the regulatory role of hMOF and HDAC2 in this process. Under physiological conditions, the K432 site of KAT7 is predominantly acetylated (Fig. 3c, d). After etoposide treatment, HDAC2 primarily shows a preference for deacetylating the K432 residue of KAT7 after binding to the substrate. Meanwhile, the accelerated accumulation of KAT7 crotonylation by hMOF occurs due to the increased levels of crotonyl-CoA (Supplementary Fig. 7b). Upon continuous exposure to etoposide, there is an increase in KAT7 crotonylation. At the same time, HDAC2 gradually exerts its deacetylase and decrotonylase activity (Supplementary Fig. 4a–c). However, as a result of the accelerated accumulation of KAT7 crotonylation, there is an overall increase in KAT7 crotonylation and a decrease in acetylation levels. Due to the limitations in detecting KAT7 decrotonylation in cells, we can only investigate the role of HDAC2 in KAT7 decrotonylation after etoposide treatment in vitro. Our model demonstrates a potential mechanism by which hMOF and HDAC2 mediate the acetylation and decrotonylation of KAT7 following etoposide treatment, although it has limitations. It will be very useful for us to understand the regulatory mechanism of the dynamic balance between crotonylation and acetylation in cells.
As a result of decreasing KAT7 HAT activity by the antagonistic effect of KAT7 crotonylation against its acetylation, the inhibition of centriole duplication was observed in colorectal cancer HCT116 cells (Fig. 7a–d). Surprisingly, we found that reduction of KAT7 HAT activity regulates the expression of genes involved in procentriole formation (Fig. 6i, j). Procentriole formation is important for centrosome duplication. However, the mechanisms governing this process remain largely unknown. PLK4, STIL and SAS-6 are key factors in the initiation of centriole duplication and their assembly on the wall of the mother centriole appears to mark the main steps of procentriole formation55–57. Here, we found that the holistic gene expression associated with procentriole formation, including CEP192, CEP152, PLK4, STIL and SAS6, decreased both in KAT7 knockdown cells and in etoposide-stimulation cells (Supplementary Fig. 5b, c and Fig. 6i). Mechanistic studies indicated that attenuating KAT7 HAT activity effectively reduced H3K14ac enrichment at the promoter regions of these related genes (Fig. 7f–o). Our study presents a mechanism by which competitive antagonism of KAT7 crotonylation against acetylation reduces the HAT activity of KAT7, resulting in the comprehensive transcriptional inhibition of genes related to procentriole formation.
Actually, reduction of KAT7 HAT activity not only regulates the expression of genes associated with procentriole formation but also controls the expression of other centrosome-associated genes (Supplementary Fig. 8a). Here, we found that the transcriptional activity of AURKA, CEP215, CEP55 and CEP128 was reduced following etoposide treatment (Supplementary Fig. 8b–e). Further ChIP-qPCR assay showed that reduction of KAT7 HAT activity regulates the expression of AURKA, CEP215, CEP55 and CEP128 by decreasing the enrichment of H4K5ac or H4K8ac at their promoters (Supplementary Fig. 8f–i). Proteins encoded by these four genes are also important for centrosome, from centriole engagement to spindle organization58–61. In the current study, the increase of multipolar spindles was observed in KAT7 knockdown cells or KAT7-K432R transfected cells compared with control cells (Supplementary Fig. 8j, k). These findings reveal the important regulatory function of KAT7 on centrosome. Additionally, reduction of KAT7 HAT activity was observed to effectively suppress the invasion of CRC cells, hinting its multiple functions on tumorigenesis (Supplementary Fig. 9).
In conclusion, our data reveal an unidentified biological function of KAT7 in centriole duplication. Reduction of KAT7 HAT activity by competitive antagonism of KAT7 crotonylation against acetylation decreases the global transcriptional activity of genes related to procentriole formation through reducing the enrichment of H3K14ac at their promoters, thereby impeding centriole duplication. These findings contribute to our understanding of the epigenetic impact of KAT7 on CRC cell proliferation and CRC progression. KAT7 crotonylation and acetylation may serve as a potential target for CRC therapy.
Methods
Ethical statement and confirmation
This study was in compliance with all relevant ethical requirements. The protocols for animal experiments were rigorously reviewed and approved by the Institutional Animal Care and Use Committees (IACUC) of Peking University (LA2022008). Informed consent was obtained from each patient, and the study protocol was approved by the Clinical Research Ethics Committee of Peking University Cancer Hospital & Institute (2021KT13).
Cell culture
Human colon cancer HCT116 cells were purchased from the Cell Resource Center, Peking Union Medical College (the Headquarter of National Infrastructure of Cell Line Resource, NSTI) and cultured in McCoy’s 5 A. HT-29, SW116 and HeLa were cultured in DMEM supplemented with 10% heat-inactivated FBS (Gemini, South America). All cells were cultured at 37 °C under 5% CO2.with penicillin/Streptomycin.
Mouse tumor models
Animal experiments were proven by the Institutional Animal Care and Use Committee of Peking University Health Science Center. The four-week-old male BALB/c nude mice (n = 9 mice) were obtained from the Department of Laboratory Animal Science of Peking University Health Science Center, Beijing. Mice were housed in a well-equipped animal facility, where humidity is controlled between 30% and 70%. Mice are kept on a constant 12-h dark-light cycle and maintained at 20–25 °C. Mice were allowed food and water ad libitum. BALB/c nude mice were implanted with 1 × 106 cells subcutaneously into the shoulders that stably overexpressed KAT7-WT and KAT7-K432R in HCT116-KO cells. One week later, tumor length and width were measured every 3 days by calipers, and tumor volume was calculated using the formula length × (width)2. The mice were sacrificed and the tumors dissected, weighed and photographed on day 19, when a remarkable difference between the two groups appeared. All animal used was approved by the Institutional Animal Care and Use Committee (IACUC) of Peking University, China. 2000 mm3 was the maximum tumor size allowed, and this was not exceeded in any of the groups.
Plasmid construction and transfection
KAT7, hMOF and HDAC2 cDNA were cloned into 3× Flag CMV10, pCMV-Myc or TF-pCold-His. All indicated mutations of KAT7 were generated using a site-directed mutagenesis kit (FM111-01, transgene, China). Transient plasmid transfection was performed by using Lipofectamine 2000 (Invitrogen).
RNA interference (RNAi)
Cells were harvested for western blotting, qRT-PCR or ChIP assay after transfection with siRNA oligonucleotides using Lipofectamine 2000 (Invitrogen)62. The siRNA sequences (5′-3′) used in this study are as follows: siKAT7: CCCUUCCUGUUCUAUGUUATT; sihMOF: GAAAGAGAUCUACCGCAATT; siCBP-1: GAUGCUGCUUCCAAACAUAAATT; siCBP-2: GAGCCAUCUAGUGCAUAAATT; siPCAF: GCAGATACCAAACAAGTTT; shHDAC2: CAGTCTCACCAATTTCAGAAA; shhMOF: GCAAGATCACTCGCAACCAAA.
LC-MS/MS analysis
For LC-MS/MS analysis, recombinant KAT7 plasmid was transfected into HCT116 cells, and then mass spectrometry (MS) was performed. The sample (n = 1) was dissolved in solvent A (0.1% formic acid in 2% acetonitrile) and separated using the EASY-nLC 1000 UPLC system. The gradient consisted of increasing mobile solvent B (0.1% formic acid in 90% acetonitrile) from 7% to 25% over 40 min, 25% to 35% over 12 min, rising to 80% over 4 min and then holding at 80% for the last 4 min, all at a constant flow rate of 500 nL/min. The peptides were subjected to NSI source followed by MS/MS on Q Exactive (Thermo) coupled online to the UPLC system. Secondary mass spectral data were retrieved using MaxQuant (v1.5.2.8). Tandem mass spectra were searched against the KAT7 (Homo sapiens) database. Trypsin/P was specified as the cleavage enzyme, allowing up to four missing cleavages for KAT7, when searching the Kac and Kcr databases. The mass error was set to 10 p.p.m. for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys was designated as fixed modification. Acetylation (Lys) and crotonylation (Lys) were designated as variable modifications. Protein identification and false-discovery rate (FDR) identified by PSM were set at 1%.
Protein extraction and western blotting
Protein expressions were detected by western blotting63. Equal amounts of proteins were prepared and were run on 9 to 15% SDS-PAGE gels. Anti-KAT7 (ab70183, abcam, Cambridge, MA, USA), anti-crotonylation (PTM-501, PTM, China), anti-acetylation (PTM-105, PTM, China), anti-hMOF (ab200660, abcam, USA), anti-HDAC2 (ab7029, abcam, USA), anti-Flag (F3165, Sigma, St Louis, MO, USA), anti-HA (bs-0966 R, Bioss, China), anti-GST (sc-138, Santa Cruz, CA, USA), anti-His (PM032, MBL, Naka-ku, Nagoya, Japan), anti-MYC (M047-3, MBL, Naka-ku, Nagoya, Japan), anti-H3K14ac (ab52946, abcam, USA), anti-H4K5ac (ab51997, abcam, USA), anti-H4K8ac (ab45166, abcam, USA), anti-H4K12ac (ab177793, abcam, USA), anti-H4K16ac (ab109463, abcam, USA), anti-H3 (ab1791, abcam, USA), anti-H4 (ab10158, abcam, USA), anti-centrin1 (12794-1-AP, Proteintech, China), anti-γ-tubulin (66320-1-Ig, Proteintech, China), anti-PLK4 (SAB1408790, Sigma, USA), anti-CEP215 (A15376, ABclonal, China), anti-GAPDH (ab8245, Abcam, USA), anti-β-actin (C1313, Applygen, China) and anti-α-tubulin (BE0031, EASYBIO, China) antibodies were used. The blots were exposed to an enhanced chemiluminescence kit (Amersham Corp.). All the uncropped and unprocessed scans of blots in this study can be found in the Source Data.
Co-Immunoprecipitation (Co-IP)
Cells were lysed and then incubated with antibodies for 2 h on ice64. The immunoprecipitants were pulled down with protein A- or G-agarose beads (GE Healthcare, Little Chalfont, UK). The samples were mixed and rotated slowly at 4 °C for 4 h. After three washes with lysis buffer, the pellets were resuspended in 2× SDS loading buffer and then analyzed by western blotting.
GST pull-down assay
His-tagged proteins and GST or GST fusion proteins were purified from bacteria. Purified recombinant proteins were incubated in TEN buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA) for 4 h at 4 °C. After three washes with TEN buffer, the beads were boiled with 2× SDS loading buffer and analyzed by western blotting or Coomassie brilliant blue (CBB) staining.
In vitro crotonylation assay
For the in vitro crotonylation reaction, recombinant Flag-hMOF, pCMV-Myc and Myc-KAT7 proteins were purified from HCT116 cells. The Myc-hMOF recombinant proteins (0.2 μg) and crotonyl-CoA (10 μM) were incubated with 30 μl reaction buffer (50 mM Tris-HCl, pH 8.0, 10% glycerol, 100 nM TSA, 5 mM nicotinamide, 0.1 mM EDTA, 1 mM DTT and 1× proteinase inhibitor cocktail) at 30 °C for 1 h. The mixture was analyzed by western blotting using a pan anti-crotonylation antibody after stop with 2× SDS loading buffer.
In vitro deacetylation and decrotonylation assay
For the in vitro deacetylation reaction of Fig. 5, recombinant Flag-HDAC2, pCMV-Myc and Myc-KAT7 proteins were purified from HCT116 cells. Flag-HDAC2 recombinant proteins (0.2 μg) were incubated with purified pCMV-Myc or Myc-KAT7 recombinant proteins in 30 μl deacetylation reaction buffer at 37 °C for 1 h. The mixture was analyzed by western blotting using a pan anti-acetylation antibody after stop with 2× SDS loading buffer.
For the in vitro deacetylation and decrotonylation reaction of Supplementary Fig. 4, HCT116 cells treated with or without etoposide (40 μM, 4 h), and then the KAT7 and HDAC2 proteins were purified in 30 μl deacetylation or decrotonylation reaction buffer. The reaction was performed at 37 °C for 1 h and analyzed by western blotting using a pan anti-acetylation or anti-crotonylation antibody after stop with 2× SDS loading buffer.
Crotonyl-CoA concentration measurement by ELISA
HCT116 cells were plated in 10cm-wells and cultured with either etoposide (40 μM) or control diluent. After 8 h, cells were lysed and the concentration of Crotonyl-CoA was measured by ELISA kit (MM-51450H2, MEIMIAN, China).
RNA extraction and qRT-PCR
Total RNA was extracted by TRIzol agent (Applygen, China). Reverse transcription was performed with a Quantscript RT Kit (Promega, WI, USA. qRT-PCR assays were performed with SYBR PCR mix agent (Vazyme, China). The primers are listed in Supplementary Data File 3.
RNA-sequencing assay
High quality RNA was extracted from control or KAT7 knockdown cells using TRIzol reagent. RNA sequencing was performed by Majorbio Bio-pharm Technology (China). A fold change ≥ 1.5 was used to define differentially expressed genes (DEGs) and p < 0.05 corrected for false discovery rate (FDR). Genes were analyzed for Gene Ontology (GO) enrichment using the Bioinformatics resources database DAVID to determine the significant molecular functions and biological process of DEGs.
Chromatin immunoprecipitation sequencing (ChIP-seq) analysis
The KAT7 ChIP-seq data [GSE33007] were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). ChIP-seq peaks were called by the peak-finding algorithm MACS2 v2.2.7.1. Bigwig files were generated using deepTools v3.4.3. Annotation of peaks was performed using HOMER.
Chromatin immunoprecipitation (ChIP) assay
HCT116 cells were harvested and lysed in lysis buffer for ChIP assay after treatment64. After reverse cross-linking (65 °C overnight), the DNA was purified and analyzed by qPCR. The primer sequences for all ChIP-qPCR are listed in Supplementary Data File 4.
Immunofluorescence
Cells were fixed in cold 4% paraformaldehyde, permeabilized in ice methanol and then blocked with 2% BSA. The indicated primary antibodies were added and then incubated overnight at 4 °C, followed by secondary antibodies (conjugated to FITC/TRITC) after washing with PBS for 2 h at room temperature avoiding light. Finally, the cells were stained with DAPI for 5 min, and images were captured using an Olympus FV1000-IX81 confocal microscope.
Centriole duplication assay
HCT116 cells were grown on coverslips housed in 35-mm dishes until 40% confluence and were treated with different stimuli. The cells were then stained with DAPI or centrin-1 antibodies for the nucleus or centriole visualization, respectively. Images were captured using an Olympus FV1000-IX81 confocal microscope, and the numbers of the centrioles in each cell were counted.
Cell proliferation and Colony formation assay
For the cell proliferation assay, cells were treated with stimulation needed and then harvested. Cell viability was evaluated by a cell counting kit 8 (Yeasen, China).
For the colony formation assay, cells were transfected with plasmids needed and then cultured for 14 days. Cells were fixed and then stained with 0.25% Coomassie brilliant blue. All the experiments were performed in triplicate.
Transwell assay
HCT116 cells were plated into the upper Transwell chambers and cultured in serum-free DMEM. After 48 h of culture, the cells in the lower chamber were fixed and stained with 0.1% crystal violet, and were quantified in three randomly selected areas using Image J software. All the experiments were performed in triplicate. The quantified results are presented as mean ± SD.
Molecular docking
The wild-type hMOF (PDB ID: 5WCI) and K274 mutant hMOF (PDB ID: 5J8F) protein crystal structures were obtained from the PDB website. Pymol (version 2.5) and autodock (version 1.57) software were used to process the protein molecules, remove all water molecules from the structure, and hydrogenate the protein. Molecular docking was performed using Autodock software with the center of the grid box located at the active site of the protein. The binding energy of wild-type and K274 mutant hMOF to Cr-CoA (PubChem ID: 5497143) was analyzed. The key hMOF amino acid sites involved in the binding were identified.
Patients and clinical specimens
Matched primary CRC tissues and matched adjacent normal tissues were obtained from surgical specimens of CRC patients at Peking University Cancer Hospital & Institute and were frozen in liquid nitrogen. These cases were selected on the basis of a clear pathological diagnosis, excluding patients who had received any form of preoperative anticancer treatment. Overall survival was calculated from the start of the surgery until death. Informed consent was obtained from each patient, and the study protocol was approved by the Clinical Research Ethics Committee of Peking University Cancer Hospital & Institute. The baseline characteristics of the patients were shown in Supplementary Table 1.
Statistics and reproducibility
The results of all the experiments were repeated at least three times as indicated in the figure legend, except for mass spectrometry and RNA sequencing. Immunogel electrophoresis of clinical samples was performed only once due to the precious and limited tissue sample. All data represent mean ± SD for at least three independent experiments. A two-tailed unpaired Student’s t-test was used to assess the significance of differences between two selected groups. One-way analysis of variance (ANOVA) was performed to compare data among multiple groups by using PRISM statistical analysis software (GraphPad Software, Inc., San Diego, CA). Survival analysis was performed using Kaplan-Meier analysis and curves were compared using the log-rank test. p value < 0.05 was considered statistically significant, and the statistical details are provided in the figures and figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank Prof. Chuanmao Zhang (Peking University, China) for sharing PLK4 and Centrin-1 antibodies. The authors also thank Prof. Jiemin Wong (East China Normal University, China) for sharing the HDAC plasmids, Prof. Jianping Ding (Chinese Academy of Sciences, China) for sharing the Flag-KAT7 plasmid, and Prof. Jun Chen (Peking University, China) for sharing the Myc-KAT7 and His-KAT7 plasmids. We appreciate Prof. Jiadong Wang (Peking University, China) for his supervision in this study. This research was supported by Beijing Natural Science Foundation (L244065 to Y.Y.), the National Natural Science Foundation of China (82472744 to Y.Y.), and the Beijing Natural Science Foundation (7222103 to Y.Y.).
Author contributions
Y.Y. conceived, supervised the project, and designed the experiments. M.W. and G.M. analyzed the data and performed most of the experiments. B.Q., S.W., J.L., Y.M., X.Z. and K.C. performed some of the experiments and helped in the animal experiments. C.T. and B.Q. analyzed the profiling results. Z.L. obtained the clinical specimens. W.W. and E.Y. supervised part of the project. Y.Y., G.M. and M.W. wrote the manuscript. Y.Y., M.W., and B.Q. revised the manuscript. All authors discussed the results and commented on the manuscript.
Peer review
Peer review information
Nature Communications thanks Saadi Khochbin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that all data of this study are available within the article and the Supplementary information and Source data files. The data supporting the findings of an association between KAT7 protein expression and tumor survival were obtained from UCSC Xena (https://xenabrowser.net/datapages/). The public dataset supporting the KAT7 ChIPseq in this study is available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33007(reuse). The raw RNA-seq data in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE236701. The mass spectrometry proteomics data on the identification of KAT7 acetylation and KAT7-interaction proteins have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD043707 and PXD043664, respectively. The data of the protein expression level of KAT7 and its crotonylation and acetylation are available on Figshare (10.6084/m9.figshare.28227701). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Meng Wang, Guanqun Mu, Bingquan Qiu.
Contributor Information
Ence Yang, Email: yangence@bjmu.edu.cn.
Yang Yang, Email: yangsh@bjmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-57546-7.
References
- 1.Iizuka, M. & Stillman, B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem.274, 23027–23034 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Lan, R. F. & Wang, Q. Q. Deciphering structure, function and mechanism of lysine acetyltransferase HBO1 in protein acetylation, transcription regulation, DNA replication and its oncogenic properties in cancer. Cell. Mol. Life Sci.77, 637–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iizuka, M. et al. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene436, 108–114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacPherson, L. et al. HBO1 is required for the maintenance of leukaemia stem cells. Nature577, 266–270 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Yang, Y. Q. et al. The histone lysine acetyltransferase HBO1 (KAT7) regulates hematopoietic stem cell quiescence and self-renewal. Blood139, 845–858 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Zhong, W. H., Liu, H. P., Deng, L., Chen, G. H. & Liu, Y. B. HBO1 overexpression is important for hepatocellular carcinoma cell growth. Cell Death Dis.12, 549 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mi, Y. Y. et al. A first-in-class HBO1 inhibitor WM-3835 inhibits castration-resistant prostate cancer cell growth in vitro and in vivo. Cell Death Dis.14, 67 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma, Y. et al. KAT7 promotes radioresistance through upregulating PI3K/AKT signaling in breast cancer. J. Radiat. Res64, 448–456 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Z. et al. HBO1 promotes cell proliferation in bladder cancer via activation of Wnt/β-catenin signaling. Mol. carcinogenesis57, 12–21 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Wang, Y. et al. High-expression HBO1 predicts poor prognosis in gastric cancer. Am. J. Clin. Pathol.152, 517–526 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Duong, M. T. et al. Hbo1 is a cyclin E/CDK2 substrate that enriches breast cancer stem-like cells. Cancer Res73, 5556–5568 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song, B. et al. Plk1 phosphorylation of orc2 and hbo1 contributes to gemcitabine resistance in pancreatic cancer. Mol. Cancer Ther.12, 58–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Li, W., Li, F. F., Zhang, X., Lin, H. K. & Xu, C. Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment. Signal Transduct. Tar.6, 422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, Z. et al. Acetylation of PHF5A Modulates Stress Responses and Colorectal Carcinogenesis through Alternative Splicing-Mediated Upregulation of KDM3A. Mol. cell74, 1250–1263.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Qu, M. et al. Hypoxia increases ATX expression by histone crotonylation in a HIF-2α-dependent manner. Int J. Mol. Sci.24, 7031 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, S. et al. The function and related diseases of protein crotonylation. Int J. Biol. Sci.17, 3441–3455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu, B. et al. KAT8 acetylation-controlled lipolysis affects the invasive and migratory potential of colorectal cancer cells. Cell Death Dis.14, 164 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, Y. et al. Protein kinase D1 phosphorylation of KAT7 enhances its protein stability and promotes replication licensing and cell proliferation. Cell Death Discov.6, 89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science325, 834–840 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Yu, H. et al. Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair. Sci. Adv.6, eaay4697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loncarek, J. & Bettencourt-Dias, M. Building the right centriole for each cell type. J. Cell Biol.217, 823–835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nigg, E. A. & Holland, A. J. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Bio19, 297–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banterle, N. & Gonczy, P. Centriole biogenesis: from identifying the characters to understanding the plot. Annu Rev. Cell Dev. Bi33, 23–49 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Strnad, P. & Gonczy, P. Mechanisms of procentriole formation. Trends Cell Biol.18, 389–396 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Habedanck, R., Stierhof, Y. D., Wilkinson, C. J. & Nigg, E. A. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol.7, 1140–1146 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Sonnen, K. F., Schermelleh, L., Leonhardt, H. & Nigg, E. A. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open1, 965–976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, J. E. et al. Autophosphorylation-induced self-assembly and STIL-dependent reinforcement underlie Plk4’s ring-to-dot localization conversion around a human centriole. Cell Cycle19, 3419–3436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzhindzhev, N. S. et al. Plk4 phosphorylates Ana2 to trigger sas6 recruitment and procentriole formation. Curr. Biol.24, 2526–2532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moyer, T. C. & Holland, A. J. PLK4 promotes centriole duplication by phosphorylating STIL to link the procentriole cartwheel to the microtubule wall. Elife8, e46054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, Y. C. et al. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. Embo J.32, 1141–1154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bettencourt-Dias, M. et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol.15, 2199–2207 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Peel, N., Stevens, N. R., Basto, R. & Raff, J. W. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol.17, 834–843 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, D. H. et al. Cep131 overexpression promotes centrosome amplification and colon cancer progression by regulating Plk4 stability. Cell Death Dis.10, 570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong, Y. L. et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science348, 1155–1160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung, G., Hernandez-Illan, E., Moreira, L., Balaguer, F. & Goel, A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat. Rev. Gastro Hepat.17, 111–130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao, M. et al. LINC00922 decoys SIRT3 to facilitate the metastasis of colorectal cancer through up-regulation the H3K27 crotonylation of ETS1 promoter. Mol. Cancer22, 163 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, Y. H. et al. Dynamic regulation of ME1 phosphorylation and acetylation affects lipid metabolism and colorectal tumorigenesis. Mol. cell77, 138–149.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Sun, B. et al. Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274. Cell Res.21, 1262–1266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, H. et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. Embo J.31, 58–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kueh, A. J., Dixon, M. P., Voss, A. K. & Thomas, T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol.31, 845–860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doyon, Y. et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. cell21, 51–64 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Barrett, T. et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res37, D885–D890 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avvakumov, N. et al. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol. Cell Biol.32, 689–703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su, R., Wu, X., Tao, L. & Wang, C. The role of epigenetic modifications in colorectal cancer metastasis. Clin. Exp. Metastasis39, 521–539 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaiopoulos, A. G., Athanasoula, K. & Papavassiliou, A. G. Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim Biophys. Acta1842, 971–980 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y. Q. et al. SATB2-AS1 suppresses colorectal carcinoma aggressiveness by inhibiting SATB2-dependent snail transcription and epithelial-mesenchymal transition. Cancer Res.79, 3542–3556 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Qian, Z. et al. Decrotonylation of AKT1 promotes AKT1 phosphorylation and activation during myogenic differentiation. J. Adv. Res.50, 117–133 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, G. R. & Hirschey, M. D. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. cell54, 5–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, S. M. et al. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol. cell67, 853–866.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Sabari, B. R. et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. cell58, 203–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han, Z. et al. Revealing the protein propionylation activity of the histone acetyltransferase MOF (males absent on the first). J. Biol. Chem.293, 3410–3420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu, L. et al. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res21, 1182–1195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly, R. D. W. et al. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci. Rep.8, 14690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arquint, C. & Nigg, E. A. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem Soc. T44, 1253–1263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopes, C. A. M. et al. PLK4 trans-Autoactivation Controls Centriole Biogenesis in Space. Dev. Cell35, 222–235 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Zitouni, S., Nabais, C., Jana, S. C., Guerrero, A. & Bettencourt-Dias, M. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Bio15, 433–452 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Wang, X. T., Baumann, C., De La Fuente, R. & Viveiros, M. M. CEP215 and AURKA regulate spindle pole focusing and aMTOC organization in mouse oocytes. Reproduction159, 261–274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kashihara, H. et al. Cep128 associates with Odf2 to form the subdistal appendage of the centriole. Genes Cells24, 231–243 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Xu, Z. Y. et al. Cep55 regulates spindle organization and cell cycle progression in meiotic oocyte. Sci. Rep.-Uk5, 16978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrera, J. A. et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev. Cell18, 913–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou, T. Y. et al. Cytoplasmic SIRT6-mediated ACSL5 deacetylation impedes nonalcoholic fatty liver disease by facilitating hepatic fatty acid oxidation. Mol. cell82, 4099–4115.e9 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Gao, T. et al. PKCζ Phosphorylates SIRT6 to Mediate Fatty Acid β-Oxidation in Colon Cancer Cells. Neoplasia21, 61–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li, M. T. et al. p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death Dis.9, 941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that all data of this study are available within the article and the Supplementary information and Source data files. The data supporting the findings of an association between KAT7 protein expression and tumor survival were obtained from UCSC Xena (https://xenabrowser.net/datapages/). The public dataset supporting the KAT7 ChIPseq in this study is available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33007(reuse). The raw RNA-seq data in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE236701. The mass spectrometry proteomics data on the identification of KAT7 acetylation and KAT7-interaction proteins have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD043707 and PXD043664, respectively. The data of the protein expression level of KAT7 and its crotonylation and acetylation are available on Figshare (10.6084/m9.figshare.28227701). Source data are provided with this paper.