Abstract
This study was to explore the regulatory effect of long non‐coding RNA LINC01559 on Docetaxel resistance in breast carcinoma (BCa) and its underlying mechanism. In the present study, we found that LINC01559 expression was elevated and LINC01559 overexpression facilitated docetaxel resistance in BCa cells. Moreover, it was revealed that the upregulation of LINC01559 in BCa cells was induced by FTO‐mediated demethylation in an m6A‐YTHDF2‐dependent manner. Additionally, Dual‐luciferase reporter assay confirmed the binding ability between LINC01559 and miR‐1343‐3p, and Pearson correlation analysis showed a negative correlation between them. Particularly, miR‐1343‐3p inhibition partly abolished the suppression on docetaxel resistance in BCa cells caused by LINC01559 knockdown. To sum up, FTO‐mediated epigenetic upregulation of LINC01559 promoted cell resistance to Docetaxel in BCa by negatively regulating miR‐1343‐3p.
Keywords: breast carcinoma, docetaxel resistance, FTO, LINC01559, N6‐methyladenosine
1. INTRODUCTION
Breast carcinoma (BCa) is one of the most diagnosed and life‐threatening malignant tumors in females around the world. 1 It is also the second leading cause of cancer‐related deaths among females worldwide. 2 Although considerable improvements have been made in the diagnosis and therapeutic strategies of BCa, the prognosis of patients with BCa remains unsatisfactory, which was mainly due to frequent metastasis and chemoresistance. 3 Chemotherapy is a very important and effective therapeutic method for breast carcinoma treatment, 4 and taxanes are often applied for chemotherapy. 5 As a second‐generation taxane, Docetaxel (DTX) has been approved as an anticancer drug for the treatment of multiple human cancers, including breast cancer. 6 At present, DTX is still an anticancer drug for first‐line chemotherapy in breast carcinoma. 7 Although DTX can effectively block its target, it could also lead to drug resistance and even tumor recurrence. 8 Therefore, further investigation is still needed for the research on DTX resistance in BCa.
Long non‐coding RNAs (LncRNAs) are RNA transcripts with more than 200 nucleotides in length without protein‐coding ability. 9 It has been widely recognized that lncRNAs are abnormally expressed in many cancers and participate in a variety of biological processes, including cell proliferation and apoptosis. For example, upregulated lncRNA ZFAS1 promotes cell proliferation and induces apoptosis in esophageal squamous cell carcinoma by upregulating STAT3 and downregulating miR‐124. 10 Wang et al. reported that lncRNA‐ATB was upregulated in non‐small cell lung cancer and induced cell apoptosis by miR‐200a/β‐catenin pathway. 11 A recent study also revealed that lncRNA WEE2‐AS1 contributed to triple negative breast cancer through the miR‐32‐5p/TOB1 axis. 12 Additionally, numerous studies have demonstrated that lncRNAs induce chemoresistance in multiple human cancers, such as hepatocellular carcinoma, 13 colorectal cancer, 14 and breast carcinoma. 15 Hu et al. reported that LINC01559 was significantly associated with poor prognosis in invasive breast carcinoma according to bioinformatics analysis. 16 Nevertheless, the relationship between the level of LINC01559 expression in BCa and DTX resistance is still unknown. Hence, a scientific exploration of the potential mechanism of LINC01559 in regulating BCa cell resistance to DTX needs to be performed.
Fat mass and obesity‐associated protein (FTO) is an RNA demethylase that removes the m6A modification from mRNA. 17 More recently, several studies have shown that FTO expression is increased in various types of cancer, including leukemia, glioblastoma, and bladder cancer. 18 , 19 , 20 In breast cancer, FTO has been indicated to promote cancer cell proliferation, migration, and invasion, through its effects on mRNA stability and translation. 21 Nevertheless, the role of FTO in docetaxel‐resistance of breast cancer has never been investigated.
In this study, the biological effect of LINC01559 on DTX resistance as well as the specific mechanism by which LINC01559 affect DTX sensitivity in BCa was explored through in vitro BCa model. Therefore, our findings provide a new chemotherapy target for BCa treatment.
2. MATERIALS AND METHODS
2.1. Cell culture
Immortalized nonmalignant human mammary epithelial cell line (MTSV1‐7) and human breast tumor cell lines (T‐47D, BT‐474, MAD‐MB‐468, and MDA‐MB‐231) purchased from BeNa Culture Collection (Beijing, China) were cultured in RPMI‐1640 medium with 10% FBS at 37°C with 5% CO2.
2.2. Establishment of DTX‐resistant BCa cells
To develop DTX‐resistant BCa cells, T‐47D and BT‐474 cells were continuously exposed to increasing concentrations of DTX, starting from 10 nM terminating with 1 μg/mL over a period of 8 months till T‐47D and BT‐474 cells had become resistant to DTX. To maintain the DTX‐resistance characterization, DTX‐resistant T‐47D (T‐47D/DR) cells and DTX‐resistant BT‐474 (BT‐474/DR) cells were incubated in medium containing 10 nM DTX.
2.3. Cell transfection
siRNA against LINC01559 (si‐LINC01559#1: GCACCCAACAUGUUGGAUACG; si‐LINC01559#2: GCCCUAAAUGUGGUUGGAUCA), FTO (si‐FTO: GGACCUGGUUAGGAUCCAA), YTHDF2 (si‐YTHDF2: GACCAAGAAUGGCAUUGCA), siRNA Negative Control (si‐NC: GCCUUCCGCAAAGUGUUCUAG), miR‐1343‐3p mimics (CUCCUGGGGCCCGCACUCUCGC), miR‐1343‐3p inhibitor (GCGAGAGUGCGGGCCCCAGGAG), and their relative negative controls (NC mimics: UUGUACUACACAAAAGUACUG; NC inhibitor: CAGUACUUUUGUGUAGUACAA) were obtained from GenePharma (Shanghai, China). For overexpression, the full length of LINC01559, FTO, or YTHDF2 was sub‐cloned into the pcDNA3.1 vector (Shanghai GenePharma Co., Ltd). The final plasmids were referred to as pcDNA3.1/LINC01559, pcDNA3.1/FTO, or pcDNA3.1/YTHDF2 with empty pcDNA3.1 as control. Cells were transfected with these vectors using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Subsequent experiments were performed 48 h after transfection.
2.4. RT‐qPCR assay
Total RNA was isolated from BCa cells with TRIzol (Invitrogen) and reversely transcribed into cDNA via PrimeScript RT Reagent Kit (Takara, Japan). Then, RT‐qPCR assay was carried out with SYBR Green Master Mix (Takara) on Bio‐Rad system. Afterwards, relative LINC01559 and miR‐1343‐3p expressions were calculated by the 2−ΔΔCt method, respectively. All the primers used are listed: GAPDH forward: 5′‐GGAGCGAGATCCCTCCAAAAT‐3′, reverse: 5′‐GGCTGTTGTCATACTTCTCATGG‐3′; U6 forward: 5′‐TGCGGGTGCTCGCTTCGGC‐3′, reverse: 5′‐CCAGTGCAGGGTCCGAGGT‐3′; LINC01559 forward: 5′‐GTCCTGCAGAACTCCCTCTT 3′, reverse: 5′‐AGTCCTGGAGCTGCAGAAAT‐3′; miR‐1343‐3p forward: 5′‐CTAGTGCAGTTGTGACTCTACCCAGGAAA‐3′, reverse: 5′‐AGCTTTTCCTGGGTAGAGTCACAACTGCA‐3′.
2.5. Western blot
Total proteins were isolated using the RIPA lysis buffer (Beyotime, China) and subjected to 10% polyacrylamide gel electrophoresis. After being transferred to PVDF membrane, blocked with 5% skim milk, incubated with primary antibodies (anti‐GAPDH, anti‐FTO, and anti‐YTHDF2) and secondary antibodies, the proteins signals were finally detected by ECL kit (EpiZyme, China).
2.6. CCK‐8 assay
To assess DTX‐induced cytotoxicity in BCa cells, CCK‐8 assay was adopted to determine the half‐maximal inhibitory concentration (IC50) values. BCa cells were seeded in 96‐well plates and incubated in medium with DTX at different concentrations (0–160 nM) for 48 h.
To detect cell viability, the transfected BCa cells were seeded into 96‐well plates and cultured in medium supplemented with DTX (10 nM) for 24, 48, or 72 h. Subsequently, 10 μL CCK‐8 reagent (Beyotime, China) was added to each well. Then, the cells were incubated for 2 h. The absorbance at 450 nm was measured under a microplate reader (Bio‐Rad, USA).
2.7. Colony formation assay
The transfected BCa cells were seeded into six‐well plates (500 cells/well) and treated with 10 nM DTX. After 2 weeks, the cells were washed with PBS twice, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet. The number of colonies (>50 cells) was counted with a light microscope (Olympus, Japan).
2.8. Flow cytometry assay
Annexin V‐fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Yeasen, China) was adopted to measure apoptotic cells. Briefly, cells were collected and washed three times with PBS. Then, cells were cultured in binding buffer with FITC and propidium iodide (Solarbio) for 15 min. At last, cell apoptosis was assessed with a flow cytometer (Agilent, China).
2.9. Dual‐luciferase reporter assay
The wild type and mutant type of LINC01559 (LINC01559‐WT/LINC01559‐MUT) were cloned into pGL3 vectors (Promega), and then co‐transfected into BCa cells with miR‐1343‐3p mimics or NC mimics. The luciferase activity was determined by the dual‐luciferase assay system (Promega) according to the manufacturer's instructions.
2.10. Methylated RNA immune precipitation (MeRIP) assay
MeRIP assay was performed using MeRIP m6A Kit (Merck Millipore, USA) following the manufacturer's instructions. Briefly, total RNAs were extracted, and incubated with m6A antibody‐conjugated protein A/G magnetic beads. After washing with IP buffer, methylated RNA was eluted followed by ethanol precipitation. The enrichment level of m6A containing mRNA was analyzed using RT‐qPCR.
2.11. RNA stability assay
Transfected cells were treated with Actinomycin d (Act D, 5 μg/mL) for indicated time points. Total RNAs were isolated and subjected to RT‐qPCR analysis.
2.12. Statistical analysis
All statistical analyses were performed by GraphPad Prism version 6.0 software. A Student's t‐test and ANOVA analysis were used to analyze the difference between groups. The association between LINC01559 and miR‐1343‐3p expressions was analyzed by Pearson's correlation analysis. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. LINC01559 is upregulated in DTX‐resistant BCa cell lines
To explore the role of LINC01559 in DTX resistance in BCa, LINC01559 expression in normal human mammary epithelial cell line (MTSV1‐7) and BCa cell lines (T‐47D, BT‐474, MAD‐MB‐468, and MDA‐MB‐231) was measured by RT‐qPCR. The data obtained indicated that LINC01559 level was markedly upregulated in BCa cell lines, especially in T‐47D and BT‐474 cell lines, compared with normal human mammary epithelial cell line (Figure 1A). Therefore, T‐47D and BT‐474 cell lines were used for subsequent experiments. To further illustrate the relationship between LINC01559 and Docetaxel resistance in BCa cells, DTX‐resistant BCa cells (T‐47D/DR and BT‐474/DR) were established. As shown in Figure 1B, the IC50 of DTX was remarkably increased in DTX‐resistant BCa cells (T‐47D/DR and BT‐474/DR) compared with their corresponding parental cells. In addition, RT‐qPCR assay also disclosed that LINC01559 expression was significantly enhanced in T‐47D/DR and BT‐474/DR cells, compared with original T‐47D and BT‐474 cells (Figure 1C). These results suggested that LINC01559 was highly expressed in DTX‐resistant BCa cells.
FIGURE 1.

LINC01559 is upregulated in DTX‐resistant cell lines. (A) RT‐qPCR analysis was applied to evaluate LINC01559 expression in normal human mammary epithelial cell line (MTSV1‐7) and BCa cell lines (T‐47D, BT‐474, MAD‐MB‐468, and MDA‐MB‐231) (n = 3). (B) CCK‐8 assay detected the IC50 of T‐47D, T‐47D/DR, BT‐474, and BT‐474/DR treated with DTX at 0, 5, 10, 20, 40, 80, and 160 nM (n = 3). (C) The LINC01559 expression were assessed in DTX‐resistant BCa cell lines (T‐47D/DR and BT‐474/DR), compared with that in the corresponding parental cell lines (T‐47D and BT‐474) (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001.
3.2. LINC01559 promotes docetaxel resistance in DTX‐resistant BCa cells
To further explored the effect of LINC01559 on Docetaxel resistance in vitro, LINC01559 overexpression and knockdown were performed, respectively. The LINC01559 level was apparently upregulated after transfection with LINC01559 overexpression plasmid and downregulated after transfection with si‐LINC01559#1 and si‐LINC01559#2 (Figure 2A,B). CCK‐8 and colony formation assays were performed to evaluate the impact of LINC01559 on cell viability. The results showed that DTX‐resistant BCa cell growth was improved after LINC01559 overexpression; however, LINC01559 knockdown impaired cell viability of DTX‐resistant BCa cells (Figure 2C–F). Besides, CCK‐8 assay manifested that LINC01559 upregulation attenuated the sensitivity of T‐47D/DR and BT‐474/DR cells to DTX and increased the IC50 value of DTX in T‐47D/DR and BT‐474/DR cells, while LINC01559 downregulation enhanced the sensitivity of T‐47D/DR and BT‐474/DR cells to DTX and decreased the IC50 value of DTX in T‐47D/DR and BT‐474/DR cells (Figure 2G,H). Moreover, the apoptotic rate of T‐47D/DR and BT‐474/DR cells was declined after LINC01559 overexpression, but increased after LINC01559 inhibition (Figure 2I). Taken together, LINC01559 facilitated the resistance of T‐47D/DR and BT‐474/DR cells to DTX.
FIGURE 2.

LINC01559 promotes Docetaxel resistance in DTX‐resistant BCa cells. (A and B) LINC01559 overexpression plasmid (pcDNA3.1/LINC01559) and two siRNAs for LINC01559 (50 nM si‐LINC01559#1 and 50 nM si‐LINC01559#2) were transfected into T‐47D/DR and BT‐474/DR cells. RT‐qPCR assays were used to confirm the efficiency of LINC01559 overexpression and knockdown (n = 3). (C and D) CCK‐8 assay evaluated the viability of T‐47D/DR and BT‐474/DR cells after designated treatments at 0, 24, 48, and 72 h (n = 3). (E and F) The colony formation ability of treated cells at day 0 and day 10 was evaluated, respectively. (G and H) IC50 value of DTX in the transfected T‐47D/DR and BT‐474/DR cells treated with DTX at 0, 5, 10, 20, 40, 80, and 160 nM was determined (n = 3). (I) The effect of LINC01559 overexpression and knockdown on cell apoptosis in T‐47D/DR and BT‐474/DR cells was detected by flow cytometry (n = 3). *p < 0.05; **p < 0.01.
3.3. LINC01559 targets miR‐1343‐3p and negatively regulates its expression in DTX‐resistant BCa cells
To further explore the underlying molecular mechanisms of LINC01559, we searched the downstream targets of LINC01559 on the StarBase website. MiR‐1343‐3p and miR‐6783‐3p were predicted as direct targets of LINC01559 (Figure 3A). Then dual‐luciferase assay was performed to verify the binding ability between LINC01559 and miR‐1343‐3p or miR‐6783‐3p and only miR‐1343‐3p affected the luciferase activity of LINC01559‐WT (Figure 3B; Figure S1A). In addition, it was found that miR‐1343‐3p expression in T‐47D/DR and BT‐474/DR cells was apparently increased when LINC01559 expression was downregulated and decreased when LINC01559 expression was upregulated (Figure 3C,D). Hence, it was suggested that LINC01559 was a sponge for miR‐1343‐3p and inversely regulated miR‐1343‐3p expression in DTX‐resistant BCa cells.
FIGURE 3.

LINC01559 targets miR‐1343‐3p and negatively regulates its expression in DTX‐resistant BCa cells. (A) StarBase website predicted the binding site between LINC01559 and miR‐1343‐3p or miR‐6783‐3p. (B and C) Dual luciferase reporter assay was performed to evaluated the luciferase activity of LINC01559‐WT and LINC01559‐Mut reporters in T‐47D/DR and BT‐474/DR cells transfected with NC mimics (50 nM) or miR‐1343‐3p mimics (50 nM) (n = 3). (D and E) The effect of LINC01559 overexpression or knockdown on miR‐1343‐3p expression was detected in T‐47D/DR and BT‐474/DR cells by RT‐qPCR, respectively (n = 3). **p < 0.01.
3.4. MiR‐1343‐3p partly reverses the effect of LINC01559 on Docetaxel resistance in DTX‐resistant BCa cells
Since LINC01559 interacted with miR‐1343‐3p in DTX‐resistant BCa cells, further investigation was carried out to explore whether LINC01559 regulated DTX resistance in BCa via miR‐1343‐3p. First, miR‐1343‐3p mimic or inhibitor was transfected in T‐47D/DR and BT‐474/DR cells (Figure 4A). It was observed that the repressive effect of LINC01559 downregulation on the viability and colony formation ability of T‐47D/DR and BT‐474/DR cells were reversed by miR‐1343‐3p inhibitor (Figure 4B–E). Moreover, CCK‐8 assay indicated that LINC01559 knockdown‐induced increase in DTX susceptibility and decline in IC50 value of T‐47D/DR and BT‐474/DR cells were partially abrogated by miR‐1343‐3p suppression (Figure 4F,G). Additionally, decreased miR‐1343‐3p expression also eliminated the stimulation on T‐47D/DR and BT‐474/DR cell apoptosis caused by LINC01559 silencing (Figure 4H). Oppositely, miR‐1343‐3p mimics partially abrogated the effects of LINC01559 abundance on T‐47D/DR and BT‐474/DR cells (Figure S2A–D). Therefore, the above results implied that LINC01559 participated in resistance to DTX of BCa cells by sponging miR‐1343‐3p.
FIGURE 4.

MiR‐1343‐3p partly reverses the effect of LINC01559 on Docetaxel resistance in DTX‐resistant BCa cells. (A) RT‐qPCR detected the expression level of miR‐1343‐3p in T‐47D/DR and BT‐474/DR cells transfected with 50 nM NC inhibitor, 50 nM miR‐1343‐3p inhibitor, 50 nM NC mimics, or 50 nM miR‐1343‐3p mimics (n = 3). (B–E) CCK‐8 and colony formation assays were used to detect cell proliferation of T‐47D/DR and BT‐474/DR cells (n = 3). (F and G) CCK‐8 assay was employed to evaluate DTX sensitivity of T‐47D/DR and BT‐474/DR cells by calculating IC50 value (n = 3). (H) Flow cytometry analysis was used to assess apoptosis of T‐47D/DR and BT‐474/DR cells (n = 3). *p < 0.05; **p < 0.01.
3.5. LINC01559 is upregulated by FTO‐mediated m6A demethylation
The m6A modification at the post‐transcriptional level has been demonstrated to play pivotal roles in RNA metabolism and function in cancers. 22 We hypothesized that m6A modification might cause the upregulation of LINC01559 in BCa. The SRAMP m6A analysis website predicted two m6A modification sites of very high confidence in the promoter region of LINC01559 (Figure 5A). Subsequently, RNA m6A quantification assay uncovered that the overall m6A enrichment of RNA was lower in BCa cells (T‐47D, BT‐474, MAD‐MB‐468, and MDA‐MB‐231) than in MTSV1‐7 cells (Figure 5B). Next, the m6A2Target website (http://rm2target.canceromics.org) was applied, which forecasted that FTO may regulate the m6A modification of LINC01559 (Figure 5C). Meanwhile, FTO was confirmed to be highly expressed in BCa cells (Figure 5D). MeRIP‐qPCR revealed that the overexpression of FTO decreased the m6A modification of LINC01559 mRNAs in BCa cells, while the downregulation of FTO increased the level (Figure 5E). Moreover, FTO overexpression resulted in a significant elevation of LINC01559 expression and FTO knockdown contributed to the decrease of LINC01559 expression in T‐47D and BT‐474 cells (Figure 5F). These results indicated that FTO regulated the m6A demethylation of LINC01559 and promoted LINC01559 expression.
FIGURE 5.
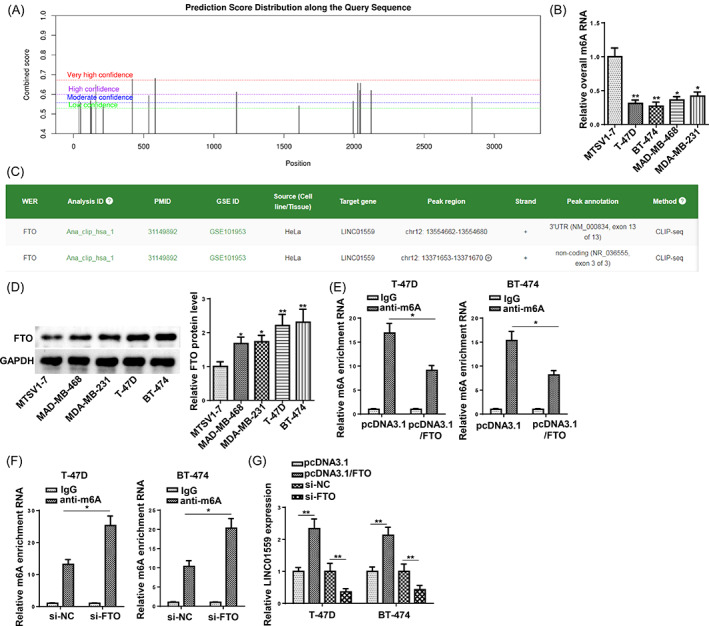
FTO mediates m6A demethylation of LINC01559 and promotes its upregulation. (A) The putative m6A modification sites of LINC01559 RNA were analyzed by SRAMP. (B) The overall m6A enrichment of RNA in BCa cells (T‐47D, BT‐474, MAD‐MB‐468, and MDA‐MB‐231) and MTSV1‐7 cells were examined (n = 3). (C) The possible m6A RNA modification regulator for LINC01559 was predicted by m6A2Target website. (D) FTO protein level in BCa and MTSV1‐7 cells was detected by western blot (n = 3). (E) The m6A enrichment of LINC01559 in T‐47D and BT‐474 cells overexpressed or depleted with FTO (n = 3). (F) The expression of LINC01559 in T‐47D and BT‐474 cells transfected with pcDNA3.1, pcDNA3.1/FTO, 50 nM si‐NC, or 50 nM si‐FTO (n = 3). *p < 0.05; **p < 0.01.
3.6. FTO promotes LINC01559 up‐regulation in an YTHDF2‐dependent manner
The m6A reader, YTHDF2, has been proved to destabilize and facilitate the degradation of m6A‐modified RNA molecules. 23 To verify whether YTHDF2 regulated the m6A‐modified LINC01559 transcript in BCa cells, the protein expression of YTHDF2 was compared between BCa cells and MTSV1‐7 cells. Western blot showed that YTHDF2 was remarkably down‐regulated in BCa cells (Figure 6A). Then, YTHDF2 was silenced or overexpressed in T‐47D and BT‐474 cells (Figure 6B). LINC01559 expression was remarkably upregulated following YTHDF2 knockdown while ectopic YTHDF2 expression caused a drastic suppression on LINC01559 level (Figure 6C). More importantly, YTHDF2 deficiency retarded the decay rate of LINC01559 in BCa cells whereas the abundance of YTHDF2 contributed to the degradation of LINC01559 transcript in BCa cells treated with Act D (an RNA synthesis inhibitor) (Figure 6D,E). Furthermore, the ablation of YTHDF2 rescued the decrease of LINC01559 level induced by FTO depletion (Figure 6F). Overexpressed YTHDF2 abated the elevation of LINC01559 level caused by FTO abundance (Figure 6G). Furthermore, the protein expression of YTHDF2 was suppressed in T‐47D/DR and BT‐474/DR in comparison with that in T‐47D and BT‐474 cells (Figure S3A). Besides, IC50 of T‐47D and BT‐474 cells transfected with si‐YTHDF2 or pcDNA3.1/YTHDF2 was evaluated. The results indicated that YTHDF2 depletion increased the IC50 value while YTHDF2 supplementation decreased it (Figure S3B). In sum, FTO facilitated LINC01559 up‐regulation in BCa cells in an m6A‐YTHDF2‐dependent manner.
FIGURE 6.

FTO promotes LINC01559 up‐regulation in an YTHDF2‐dependent manner. (A) YTHDF2 protein level in BCa and MTSV1‐7 cells was compared (n = 3). (B) The protein expression of YTHDF2 in T‐47D and BT‐474 cells transfected with 50 nM si‐NC, 50 nM si‐YTHDF2, pcDNA3.1, or pcDNA3.1/YTHDF2 (n = 3). (C) LINC01559 expression in YTHDF2‐silenced or overexpressed T‐47D and BT‐474 cells (n = 3). (D and E) The effect of knockdown or over‐expression of YTHDF2 on the stability of LINC01559 transcript in BCa cells in the presence of ActD (5 μg/mL) was tested by qRT‐PCR (n = 3). (F) The expression of LINC01559 in BCa cells transfected with 50 nM si‐NC, 50 nM si‐FTO, or 50 nM si‐FTO + 50 nM si‐YTHDF2 (n = 3). (G) The expression of LINC01559 in BCa cells transfected with pcDNA3.1, pcDNA3.1/FTO, orpcDNA3.1/FTO + pcDNA3.1/YTHDF2 (n = 3). *p < 0.05.
4. DISCUSSION
Drug resistance is still a challenge for antitumor chemotherapy today. There are numerous studies focusing on the functions of lncRNAs in the chemoresistance during the chemotherapy for human cancers, including BCa. 24 Emerging studies revealed that lncRNAs could regulate chemoresistance in BCa. Dong et al. reported that lncRNA TINCR promoted EMT process and induced trastuzumab resistance in BCa. 25 Shi et al. revealed that lncRNA DILA1 conferred resistance to tamoxifen in BCa by upregulating the level of Cyclin D1 protein. 26 Additionally, Zhang et al. elaborated that LINC00461 contributed to BCa progression and conferred Docetaxel resistance by serving as a sponge for miR‐411‐5p. 27 LINC01559 has been recognized as an oncogene and is highly expressed in several human cancers. To cite an instance, exosome‐transferred LINC01559 contributes to cell proliferation, migration, and stemness in gastric cancer through decreasing PTEN expression and increasing PGK1. 28 A report from Lou et al. indicated that LINC01559 was highly expressed in pancreatic cancer and promoted its progression through LINC01559/miR‐607/YAP pathway. 29 The above study implied that LINC01559 was an oncogene and exerted a vital role in the drug resistance of BCa.
In this study, we established two DTX‐resistant cells models (T‐47D/DR and BT‐474/DR) to explore the DTX‐resistance mechanism of LINC01559 in BCa. By RT‐qPCR analysis, we discovered that LINC01559 was clearly upregulated in DTX‐resistant BCa cells, suggesting LINC01559 was involved in DTX resistance in BCa. Besides, functional analysis exhibited that LINC01559 knockdown repressed cell proliferation and attenuated DTX resistance of T‐47D/DR and BT‐474/DR cells. LINC01559 silencing also induced apoptosis of T‐47D/DR and BT‐474/DR cells under DTX treatment. The above data indicated that LINC01559 functioned as an oncogene and promoted DTX resistance in BCa in vitro.
It has been widely recognized that lncRNAs participated in the chemoresistance of tumors via interacting with miRNAs. For example, lncRNA TTN‐AS1 sponges miR‐134‐5p and upregulates MBTD1 to promote drug resistance and tumor growth in osteosarcoma. 30 Li et al. reported that UCA1 induced cisplatin resistance in ovarian cancer by suppressing miR‐143 expression and increasing FOSL2 expression. 31 LncRNA NR2F1‐AS1 was demonstrated to accelerate oxaliplatin resistance in hepatocellular carcinoma by regulating ABCC1 as a sponge of miR‐363. 32 In this study, we discovered miR‐1343‐3p as a downstream target of LINC01559 through bioinformatic analysis. As reported in recent studies, miR‐1343‐3p served as an oncogene or tumor suppressor in different cancers. For instance, miR‐1343‐3p was obviously upregulated in papillary thyroid carcinoma (PTC) cell lines and facilitated PTC progression by downregulating ATG7 protein. 33 However, Qi et al. stated that miR‐1343‐3p induced apoptosis and slowed down glioma progression by downregulating ANXA11 expression. 34 Moreover, Li et al. discovered that miR‐1343‐3p was lowly expressed in ovarian cancer (OC) and repressed OC cell growth and migration. 35 In the current study, miR‐1343‐3p expression was suppressed by LINC01559, which was consistent with a previous study. 36 Furthermore, the results of rescue experiments showed that miR‐1343‐3p depletion reversed the impact on DTX susceptibility of T‐47D/DR and BT‐474/DR cells induced by LINC01559 silencing. Consequently, it was concluded that LINC01559 regulated the sensitivity of BCa to DTX by interacting with miR‐1343‐3p in vitro.
The upstream regulatory mechanisms of LINC01559 in BCa was also investigated. Mounting evidence demonstrates that aberrant methylations play essential roles in the expression of lncRNAs. Herein, we demonstrated that FTO suppressed the enrichment of m6A‐modified LINC01559 mRNAs, leading to the upregulation of LINC01559 in BCa. Studies have shown that FTO and YTHDF2 can act in opposition to regulate the fate of m6A‐modified mRNA. 37 More importantly, FTO has been identified to interact with YTHDF2 during the development of human cancers. For example, in an m6A‐YTHDF2‐dependent way, FTO facilitates the advancement of multiple myeloma by promoting the posttranscriptional activation of HSF1. 38 FTO knockdown inhibits melanoma tumorigenicity and sensitizes melanoma to anti‐PD‐1 treatment through the mediation of YTHDF2. 39 FTO inhibits LINC00022 decay via the m6A reader YTHDF2 to promote tumorigenesis in esophageal squamous cell carcinoma. 40 Therefore, we explored whether YTHDF2 was involved in the FTO‐mediated upregulation of LINC01559 in BCa cells. Mechanistically, YTHDF2 degraded the LINC01559 mRNAs containing m6A and suppressed LINC01559 mRNA translation. Additionally, FTO regulated LINC01559 activity in BCa cells in a YTHDF2‐dependent manner.
In summary, it was demonstrated that FTO‐mediated m6A demethylation of LINC01559 could impair DTX sensitivity of BCa cells by sponging miR‐1343‐3p, indicating LINC01559 is a potential biomarker and therapeutic target for BCa.
CONFLICT OF INTEREST STATEMENT
All authors declare no conflict of interest.
Supporting information
FIGURE S1: (A) The luciferase activity of LINC01559‐WT and LINC01559‐Mut reporters in T‐47D/DR and BT‐474/DR cells transfected with miR‐1343‐3p mimics or NC mimics.
FIGURE S2: T‐47D/DR and BT‐474/DR cells were transfected with pcDNA3.1, pcDNA3.1/LINC01559, or pcDNA3.1/LINC01559 + miR‐1343‐3p mimics. (A, B) The proliferation of the treated cells was evaluated using CCK‐8 assay and colony formation assay (n = 3). (C) The IC50 value of T‐47D/DR and BT‐474/DR cells exposed to DTX (n = 3). (D) Cell apoptosis was assessed using flow cytometry assay (n = 3). *p < 0.05; **p < 0.01.
FIGURE S3: (A) The protein expression of YTHDF2 in T‐47D, BT‐474, T‐47D/DR, and BT‐474/DR cells was detected using western blot. (B) IC50 of T‐47D and BT‐474 cells transfected with si‐NC, si‐YTHDF2, or pcDNA3.1, pcDNA3.1/YTHDF2. **p < 0.01; ***p < 0.001.
Lin W, Mo C‐Q, Kong L‐J, Chen L, Wu K‐L, Wu X. FTO‐mediated epigenetic upregulation of LINC01559 confers cell resistance to docetaxel in breast carcinoma by suppressing miR‐1343‐3p. Kaohsiung J Med Sci. 2023;39(9):873–882. 10.1002/kjm2.12728
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13(11):1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones SE. Metastatic breast cancer: the treatment challenge. Clin Breast Cancer. 2008;8(3):224–33. [DOI] [PubMed] [Google Scholar]
- 4. Wawruszak A, Luszczki JJ, Grabarska A, Gumbarewicz E, Dmoszynska‐Graniczka M, Polberg K, et al. Assessment of interactions between cisplatin and two histone deacetylase inhibitors in MCF7, T47D and MDA‐MB‐231 human breast cancer cell lines—an Isobolographic analysis. PLoS One. 2015;10(11):e0143013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J, Zheng R, Wang Z, Yang Y, Wang M, Zou W. Efficacy and safety of vinorelbine plus cisplatin vs. gemcitabine plus cisplatin for treatment of metastatic triple‐negative breast cancer after failure with anthracyclines and Taxanes. Med Sci Monit. 2017;23:4657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith IC, Heys SD, Hutcheon AW, Miller ID, Payne S, Gilbert FJ, et al. Neoadjuvant chemotherapy in breast cancer: significantly enhanced response with docetaxel. J Clin Oncol. 2002;20(6):1456–66. [DOI] [PubMed] [Google Scholar]
- 7. Hsu SK, Chang WT. The role of necroptosis in ROS‐mediated cancer therapies and its promising applications. Cancer. 2020;12(8):2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H, Vo T, Hajar A, Li S, Chen X, Parissenti AM, et al. Multiple mechanisms underlying acquired resistance to taxanes in selected docetaxel‐resistant MCF‐7 breast cancer cells. BMC Cancer. 2014;14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA‐124/STAT3 axis. J Exp Clin Cancer Res. 2019;38(1):477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang T, Tang X, Liu Y. LncRNA‐ATB promotes apoptosis of non‐small cell lung cancer cells through MiR‐200a/beta‐catenin. J BUON. 2019;24(6):2280–6. [PubMed] [Google Scholar]
- 12. Wang R, Huang Z, Qian C, Wang M, Zheng Y, Jiang R, et al. LncRNA WEE2‐AS1 promotes proliferation and inhibits apoptosis in triple negative breast cancer cells via regulating miR‐32‐5p/TOB1 axis. Biochem Biophys Res Commun. 2020;526(4):1005–12. [DOI] [PubMed] [Google Scholar]
- 13. Li W, Dong X, He C, Tan G, Li Z, Zhai B, et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR‐21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2019;38(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xian Z, Hu B, Wang T, Zeng J, Cai J, Zou Q, et al. lncRNA UCA1 contributes to 5‐fluorouracil resistance of colorectal cancer cells through miR‐23b‐3p/ZNF281 Axis. Onco Targets Ther. 2020;13:7571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J, Xie S, Yang J, Xiong H, Jia Y, Zhou Y, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu Y, Gu X, Duan Y, Shen Y, Xie X. Bioinformatics analysis of prognosis‐related long non‐coding RNAs in invasive breast carcinoma. Oncol Lett. 2020;20(1):113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li E, Xia M, Du Y. METTL3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the EGF/RAD51 axis. elife. 2022;11:e75231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)‐Methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huff S, Tiwari SK, Gonzalez GM, Wang Y, Rana TM. m(6)A‐RNA demethylase FTO inhibitors impair self‐renewal in glioblastoma stem cells. ACS Chem Biol. 2021;16(2):324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tao L, Mu X, Chen H, Jin D, Zhang R, Zhao Y, et al. FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin Transl Med. 2021;11(2):e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6‐methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishigami Y, Ohira T, Isokawa Y, Suzuki Y. A single m(6)A modification in U6 snRNA diversifies exon sequence at the 5′ splice site. Nat Commun. 2021;12(1):3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Zhang N, Jiao X, Wang C, Sun W, He Y, et al. Downregulation of microRNA‐6125 promotes colorectal cancer growth through YTHDF2‐dependent recognition of N6‐methyladenosine‐modified GSK3β. Clin Transl Med. 2021;11(10):e602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ayers D, Vandesompele J. Influence of microRNAs and long non‐coding RNAs in cancer chemoresistance. Genes (Basel). 2017;8(3):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial‐mesenchymal transition by targeting MicroRNA‐125b in breast cancer. Mol Cancer. 2019;18(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Shi Q, Li Y, Li S, Jin L, Lai H, Wu Y, et al. LncRNA DILA1 inhibits cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat Commun. 2020;11(1):5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang C, Wang J, Zhang J, Qu H, Tang X. LINC00461 overexpression can induce docetaxel resistance in breast cancer by interacting with miR‐411‐5p. Onco Targets Ther. 2020;13:5551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang L, Bo X, Yi X, Xiao X, Zheng Q, Ma L, et al. Exosome‐transferred LINC01559 promotes the progression of gastric cancer via PI3K/AKT signaling pathway. Cell Death Dis. 2020;11(9):723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lou C, Zhao J, Gu Y, Li Q, Tang S, Wu Y, et al. LINC01559 accelerates pancreatic cancer cell proliferation and migration through YAP‐mediated pathway. J Cell Physiol. 2020;235(4):3928–38. [DOI] [PubMed] [Google Scholar]
- 30. Fu D, Lu C, Qu X, Li P, Chen K, Shan L, et al. LncRNA TTN‐AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR‐134‐5p/MBTD1 axis. Aging. 2019;11(19):8374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Niu H, Qin Q, Yang S, Wang Q, Yu C, et al. lncRNA UCA1 mediates resistance to cisplatin by regulating the miR‐143/FOSL2‐signaling pathway in ovarian cancer. Mol Ther Nucleic Acids. 2019;17:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J. LncRNA NR2F1‐AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR‐363. J Cell Mol Med. 2018;22(6):3238–45. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Qin Y, Sun W, Wang Z, Dong W, He L, Zhang T, et al. ATF2‐induced lncRNA GAS8‐AS1 promotes autophagy of thyroid cancer cells by targeting the miR‐187‐3p/ATG5 and miR‐1343‐3p/ATG7 axes. Mol Ther Nucleic Acids. 2020;22:584–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi J, Wang Z, Zhao Z, Liu L. EIF3J‐AS1 promotes glioma cell growth via up‐regulating ANXA11 through sponging miR‐1343‐3p. Cancer Cell Int. 2020;20:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y, Zhao Z, Sun D, Li Y. Novel long noncoding RNA LINC02323 promotes cell growth and migration of ovarian cancer via TGF‐beta receptor 1 by miR‐1343‐3p. J Clin Lab Anal. 2021;35(2):e23651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X, Wang J, Xie F, Mou T, Zhong P, Hua H, et al. Long noncoding RNA LINC01559 promotes pancreatic cancer progression by acting as a competing endogenous RNA of miR‐1343‐3p to upregulate RAF1 expression. Aging. 2020;12(14):14452–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu B, Liu J, Zhang J, Mu T, Feng X, Ma R, et al. Regulatory role of RNA N(6)‐methyladenosine modifications during skeletal muscle development. Front Cell Dev Biol. 2022;10:929183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu A, Zhang J, Zuo L, Yan H, Chen L, Zhao F, et al. FTO promotes multiple myeloma progression by posttranscriptional activation of HSF1 in an m(6)A‐YTHDF2‐dependent manner. Mol Ther. 2022;30(3):1104–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang S, Wei J, Cui YH, Park G, Shah P. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti‐PD‐1 blockade. Nat Commun. 2019;10(1):2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, et al. RNA m6A demethylase FTO‐mediated epigenetic up‐regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40(1):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1: (A) The luciferase activity of LINC01559‐WT and LINC01559‐Mut reporters in T‐47D/DR and BT‐474/DR cells transfected with miR‐1343‐3p mimics or NC mimics.
FIGURE S2: T‐47D/DR and BT‐474/DR cells were transfected with pcDNA3.1, pcDNA3.1/LINC01559, or pcDNA3.1/LINC01559 + miR‐1343‐3p mimics. (A, B) The proliferation of the treated cells was evaluated using CCK‐8 assay and colony formation assay (n = 3). (C) The IC50 value of T‐47D/DR and BT‐474/DR cells exposed to DTX (n = 3). (D) Cell apoptosis was assessed using flow cytometry assay (n = 3). *p < 0.05; **p < 0.01.
FIGURE S3: (A) The protein expression of YTHDF2 in T‐47D, BT‐474, T‐47D/DR, and BT‐474/DR cells was detected using western blot. (B) IC50 of T‐47D and BT‐474 cells transfected with si‐NC, si‐YTHDF2, or pcDNA3.1, pcDNA3.1/YTHDF2. **p < 0.01; ***p < 0.001.


