Abstract
The epithelial–mesenchymal transition (EMT) is closely associated with Crohn's disease (CD) related intestinal fibrosis, a condition whose prevalence is increasing annually among children. Recently, the CD marker gene microarray screening revealed an upregulation of circ_0001666 in the colon tissues of CD patients, but its underlying mechanisms remain unclear. In this study, we explored the molecular mechanism of circ_0001666 in regulating EMT‐mediated fibrosis in CD in vitro. The levels of circ_0001666 and EMT‐associated proteins were assessed in CD clinical samples, and a CD cell model was established using TGF‐β1 to induce human intestinal epithelial cells (HIECs). Additionally, the expression levels of genes and proteins related to EMT and fibrosis were analyzed by quantitative real‐time PCR and western blot, cell migration, and invasion were assessed via wound healing assay and transwell, respectively, and RNA pull‐down and RNA immunoprecipitation assays were performed to verify the relationship between SRSF1 and BMP7 or circ_0001666. Circ_0001666 was overexpressed in the intestinal mucosal tissues of CD patients and was positively correlated with EMT. Silencing circ_0001666 inhibited the migration, invasion, EMT, and fibrosis of HIECs induced by TGF‐β1. Mechanistically, circ_0001666 regulated BMP7 expression by interacting with SRSF1. Furthermore, the effects of inhibiting circ_0001666 on HIECs could be partially reversed by overexpressing SRSF1 or silencing BMP7. Collectively, circ_0001666 regulates TGF‐β1‐induced HIEC migration, invasion, EMT, and fibrosis. Circ_0001666 also promoted EMT‐mediated fibrosis by interacting with SRSF1 to accelerate BMP7 mRNA decay. These findings provide new insights into the pathogenesis of CD and suggest that circ_0001666 might be a potential therapeutic target for CD.
Keywords: BMP7, circ_0001666, Crohn's disease, epithelial–mesenchymal transition, fibrosis
Abbreviations
- BMP7
bone morphogenetic protein 7
- CD
Crohn's disease
- circRNA
circular RNA
- ECM
extracellular matrix
- EMT
epithelial–mesenchymal transition
- HIEC
human intestinal epithelial cell
- IBD
inflammatory bowel disease
- qPCR
quantitative real‐time PCR
- RBP
RNA binding protein
- SRSF1
serine/arginine‐rich splicing factor 1
- TGF‐β
transforming growth factor‐β
- TNF
tumor necrosis factor
- UC
ulcerative colitis
1. INTRODUCTION
Inflammatory bowel disease (IBD) is mainly divided into Crohn's disease (CD) and ulcerative colitis (UC) and has a reported incidence of approximately 25% in children. 1 Globally, the incidence of CD in pediatric patients has been increasing annually, 2 and studies have shown that more than 30% of children have intestinal complications 5 years after suffering from IBD. 3 CD is a chronic intestinal inflammatory disease resulting from changes in the intestinal mucosal immunity due to a large number of cytokines and white blood cell infiltration in the intestinal walls. Long‐term repeated intestinal inflammation and tissue remodeling may induce the deposition of intestinal wall collagen and intestinal fibrosis. 4 Intestinal fibrosis is one of the most common and threatening complications of CD. 5 A study on chronic intestinal inflammation in animal models revealed that CD‐related intestinal fibrosis was initiated by epithelial–mesenchymal transition (EMT). 6 , 7 EMT is mainly characterized by the loss of the polarized phenotype of epithelial cells and the transformation into mesenchymal‐like myofibroblasts, which is mainly manifested by the co‐expression of epithelial markers (E‐cadherin and cytokeratin) and mesenchymal markers (α‐SMA and Vimentin) in EMT cells. 8 Existing literature showed that EMT is highly correlated with the formation of CD intestinal fibrosis, and transforming growth factor‐β (TGF‐β) is the most powerful medium of EMT in vitro and in vivo. 9 , 10 , 11 EMT induced by TGF‐β1 can promote the growth of intestinal epithelial cells. 12 Therefore, TGF‐β1 is mainly used as a common molecule to induce fibrosis models in vitro and in vivo. 13 It is of certain significance to explore the pathogenesis of CD by verifying the specific molecular mechanism of EMT on intestinal fibrosis.
Circular RNAs (circRNAs) represent a type of noncoding RNA with a closed‐loop covalent structure. Due to their unique circular structure, circRNAs have a more stable structure than other types of RNAs. 14 CircRNAs are specifically expressed in various cells and tissues and participate in the regulation of diseases, including IBD. 15 Analysis of colon tissues of CD patients by circRNA microarray revealed a total of 218 circRNAs with varying degrees of expression changes. 16 These studies suggest that circRNA might be involved in the regulation of CD. Recent studies have shown that noncoding RNAs (especially circRNAs) are widely involved in intestinal fibrosis. 17 CircRNA CDR1 regulates fibrosis in patients with pulmonary silicosis through the EMT. 18 Circ_0114428 was also confirmed to regulate renal fibrosis through EMT in renal fibrosis. 19 Recently, microarray screening of CD marker genes revealed an upregulation of circ_0001666 in the colon tissues of CD patients. 20 It is speculated that circ_0001666 may be a potential novel marker for CD. However, the specific regulatory mechanism of circ_0001666 remains to be clarified.
Bone morphogenetic protein 7 (BMP7), a member of the TGF‐β superfamily, has been shown to have different regulatory roles in different diseases. 21 In renal fibrosis, BMP7 prevents renal tubular fibrosis by inhibiting the deposition of the extracellular matrix (ECM). 22 Similarly, BMP7 can regulate EMT and fibrosis of intestinal cells. 6 In this present study, we verified the regulatory relationship of circ_0001666 and BMP7 in children with CD. Using molecular biotechnology at the cellular level, we found that circ_0001666 promoted intestinal epithelial cell EMT in pediatric patients with CD by influencing the BMP7 expression in combination with Serine/arginine‐rich splicing factor 1 (SRSF1).
2. MATERIALS AND METHODS
2.1. Clinical tissue collection
The colon mucosal tissues of 18 children with clinically active CD (average age 12 years) were obtained perioperatively during surgical resection, immediately placed in liquid nitrogen, and stored at −80°C for further analysis. In addition, colonic mucosal biopsy tissues were obtained from 18 healthy controls (HCs), with roughly matching age (average age 8 years) and sex, during colonoscopy at our hospital. The diagnosis of CD was based on the Lennard‐Jones criteria. 23 Children with the following conditions were excluded from this study: autoimmune diseases, a history of malignant tumors or complications, severe renal and/or renal dysfunction, and a past history of surgery or severe infection. The specific characteristic parameters of patients with CD and controls are shown in Table 1. All patients provided written informed consent, and all experiments were approved by the Ethics Committee of Hunan Children's Hospital.
TABLE 1.
Clinical characteristics of patients.
| HC | CD | |
|---|---|---|
| n | 18 | 18 |
| Age (years) | 8.55 ± 2.65 | 12.5 ± 1.2 |
| Sex (male/female) | 6/12 | 7/11 |
| BMI (kg/m2) | 18.03 ± 0.82 | 17.64 ± 0.7 |
| PCDAI | 0 | 24.45 ± 3.62* |
| Iron (μmol/L) | 15.43 ± 1.73 | 5.1 ± 1.35* |
| TIBC (μmol/L) | 60.45 ± 2.31 | 49.78 ± 4.74 |
| Albumin (g/L) | 46.58 ± 1.37 | 43.4 ± 5.08 |
| Hemoglobin (g/L) | 128.6 ± 3.82 | 113 ± 4.57 |
| Hematocrit (%) | 0.36 ± 0.02 | 0.35 ± 0.02 |
| Platelet count (Giga/L) | 375.8 ± 28.39 | 436.9 ± 35.42 |
| Treatment (stopped before surgery) | ||
| 5‐ASA (n) | N/A | 4 |
| Steroid (n) | N/A | 2 |
| Azathioprine (n) | N/A | 4 |
| Anti‐TNF (n) | N/A | 3 |
Abbreviations: BMI, body mass index; CD, Crohn's disease; HC, healthy control; N/A, not applicable; PCDAI, pediatric Crohn's disease activity index; TIBC, total iron binding capacity; TNF, tumor necrosis factor.
p < 0.05 versus HC.
2.2. Cell culture and induction
Human intestinal epithelial cells (HIECs) were purchased from American Type Culture Collection. The cells were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 1% antibiotics (100 U/mL penicillin and streptomycin) at 37°C and 5% CO2. The cells were grown to about 80% confluence and were induced with 10 ng/mL TGF‐β1 (Okaybio) for 72 h.
2.3. Cell transfection
The SRSF1 sequence was inserted into pcDNA3.1 (Thermo Fisher Scientific) to construct an overexpression vector (ov‐SRSF1), and the empty vector was used as a negative control (ov‐NC). Small interfering RNA (siRNA) targeting circ_0001666 (si‐circ1, si‐circ2), BMP7 (si‐BMP7), and si‐NC were designed and synthesized by GenePharma. Then, the cells were transfected with Liposome 3000 transfection reagent (Beyotime) for 48 h.
2.4. RNA extraction and quantitative real‐time PCR
First, the cells or tissues to be examined were collected, and total RNA was extracted using the TRIzol reagent (Invitrogen). Then, RNA concentration was quantified and stored at −80°C. The PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa, 6210A) was used for cDNA synthesis. SYBR (Vazyme) was used to prepare a quantitative real‐time PCR (qPCR) reaction system for qPCR system operation. The expression levels of RNAs were normalized to GAPDH. The primers used in this study are listed in Table 2.
TABLE 2.
Primers used in this study for qPCR assay.
| Primer | Forward (5′‐3′) | Reverse (5′‐3′) |
|---|---|---|
| hsa_circ_0001666 | CTGCCTAGCTGTCAAGGAGTGG | TCCGGGAAAGGATCTGGAATG |
| SRSF1 | TATCCGCGACATCGACCTCAAG | AAACTCCACCCGCAGACGGTAC |
| N‐cadherin | AGGGGACCTTTTCCTCAAGA | TCAAATGAAACCGGGCTATC |
| E‐cadherin | TCACATCCTACACTGCCCAG | AGTGTCCCTGTTCCAGTAGC |
| Snail | TGCCCTCAAGATGCACATCCGA | GGGACAGGAGAAGGGCTTCTC |
| α‐SMA | TATCCCCGGGACTAAGACGGG | CAGAGCCCAGAGCCATTGTC |
| Collagen III | GGAGAGTCCATGGATGGTGG | TTTGCTCCATTCCCCAGTGT |
| Fibronectin | CAGTGGGAGACCTCGAGAAG | CACTGTGACAGCAGGAGCAT |
| MMP9 | TTCCAAACCTTTGAGGGCGA | CTGTACACGCGAGTGAAGGT |
| BMP7 | GCAGCACAATTTG GGAA | ACAGGTGTTTCGAGAACTGGC |
| GAPDH | CCAGGTGGTCTCCTCTGA | GCTGTAGCCAAATCGTTGT |
2.5. Western blot
HIECs were lysed with RIPA Lysis Buffer (Beyotime) on ice. Total protein was quantified using a bicinchoninic acid detection kit (Beyotime). The proteins were separated using 10% SDS‐PAGE, transferred onto a PVDF membrane, and 5% skimmed milk was used to block non‐specific sites. Then, primary antibodies were added and incubated at 4°C overnight, following which a secondary antibody (dilution, 1:5000) was added to the membrane and incubated for 1 h. The protein bands were observed using an enhanced chemiluminescence kit, and the intensity of the bands was quantified using the Image‐pro Plus v6.0 analysis software. The primary antibodies used in this study were purchased from Abcam and comprised anti‐E‐cadherin (ab40772, 1:10,000), anti‐N‐cadherin (ab76011, 1:5000), anti‐Vimentin (ab92547, 1:1000), anti‐Snail (ab216347, 1:1000), anti‐α‐SMA (124964, 1:10,000), anti‐Collagen III (ab7778, 1:5000), anti‐Fibronectin (ab2413, 1:1000), anti‐MMP9 (ab76003, 1:5000), anti‐SRSF1 (ab238523, 1:1000), anti‐BMP7 (ab129156, 1:5000), and anti‐GAPDH (ab9485, 1:2500).
2.6. Cell migration measurement
Cell migration was assessed by wound healing assay. Briefly, HIECs (3.5 × 105 cells/well) were inoculated in a 12‐well plate for 12 h. Then, a small duct nozzle was used to form a wound on the plate, and the width of the scratch was recorded at 0 and 24 h using an inverted phase‐contrast microscope (Olympus) to quantify the cells.
2.7. Cell invasion detection
The invasion ability of HIECs was analyzed by transwell chambers (Costar). HIECs (4 × 104) were resuspended in a serum‐free medium and inoculated onto Matrigel in the upper chamber. At the same time, 500 μL of medium containing 10% serum was added to the lower chamber. After incubation, the transmembrane cells were fixed with methanol (Aladdin), stained with 0.1% crystal violet (Sigma‐Aldrich), and observed under a microscope (Olympus).
2.8. Subcellular fractionation location
The cells were digested from the culture flask and collected. They were resuspended in PBS, following which the Ambion® PARIS™ Kit (AM1921, Thermo Fisher Scientific) and Cytoplasmic & Nuclear RNA Purification Kit (NGB‐21000, Norgen Biotek) were used to extract and isolate RNA from the cytoplasm and nucleus. Quantitative analysis was performed separately by qPCR.
2.9. RNA pull‐down assay
The samples were tested using the Pierce™ Magnetic RNA‐Protein Pull‐Down Kit (Thermo Fisher Scientific). Briefly, circ_0001666 or BMP7 and their respective antisense RNAs were transcribed and biotin‐labeled, after which the biotinylated RNAs were incubated with cell lysates prepared using an immunoprecipitation buffer. Following a 30‐min incubation, streptavidin magnetic beads (Thermo Fisher Scientific) were used to isolate RNA‐protein complexes, which were incubated for an additional 30 min. Finally, western blot was used to quantify the retrieved proteins from RNA‐binding protein complexes.
2.10. RNA immunoprecipitation assay
The interaction between SRSF1 and circ_0001666 or BMP7 was detected using the EZ‐Magna RNA Immunoprecipitation (RIP) Kit (Millipore). Briefly, the cells (5 × 106) were lysed for 30 min at 4°C and incubated with RIP buffer containing magnetic beads conjugated to antibodies against Ago2 (Millipore), anti‐SRSF1 antibody or immunoglobulin (Ig) G (Abcam) for 2 h. Then, the samples were eluted thrice, and co‐precipitated RNAs associated with SRSF1 were extracted for qPCR analysis and subsequent quantification. IgG was used as the negative RIP control.
2.11. RNA stability
For BMP7 mRNA stability determination, 5 μg/mL actinomycin D (Sigma‐Aldrich) was added to the cell culture medium to be tested and cultured for 0, 6, and 12 h. Then, the total RNA in the cells was extracted for qPCR quantification.
2.12. Statistical analysis
All data were analyzed using the GraphPad Prism v5.0 software. The data are expressed as mean ± standard deviation (SD). The difference between multiple groups was analyzed by one‐way analysis of variance with Tukey's post hoc test, and the difference between two groups was analyzed using Student's t test. p Values <0.05 were considered statistically significant.
3. RESULTS
3.1. Circ_0001666 is overexpressed in children with CD
First, we collected the colon mucosal tissues from 18 children with CD and HCs. As shown in Figure 1A, circ_0001666 was overexpressed in the colonic mucosa of the CD patients. The expression of EMT markers, mainly include E‐cadherin, N‐cadherin, and Snail, is closely related to fibrosis phenotype. 24 E‐cadherin was significantly downregulated in the colonic mucosa of patients with CD, while N‐cadherin and Snail were upregulated (Figure 1B–D). Correlation analysis indicated that circ_0001666 expression in CD was negatively correlated with E‐cadherin and positively correlated with N‐cadherin and Snail (Figure 1E–G). Then, we constructed a CD cell model using TGF‐β1‐induced HIECs. The results showed that circ_0001666 was increased in intestinal epithelial cells induced by TGF‐β1 (Figure 1H). In summary, these findings confirm the overexpression of circ_0001666 in children with CD and indicate its involvement in regulating EMT.
FIGURE 1.

Circ_0001666 is overexpressed in children with Crohn's disease. (A) qPCR detected circ_0001666 expression in the colonic mucosa of patients. (B–D) E‐cadherin, N‐cadherin, and Snail expressions in the colonic mucosa of patients were detected by qPCR. (E) Statistical analysis of the correlation between circ_0001666 and E‐cadherin expression. (F) Statistical analysis of the correlation between circ_0001666 and N‐cadherin expression. (G) Statistical analysis of the correlation between circ_0001666 and Snail expression. n = 18. The intestinal epithelial cells HIECs were treated with 10 ng/mL TGF‐β1 for 72 h. (H) Circ_0001666 expression in the intestinal epithelial cells induced by TGF‐β1 was analyzed by qPCR. EMT, epithelial–mesenchymal transition; HIEC, human intestinal epithelial cell; qPCR, quantitative real‐time PCR. n = 3. **p < 0.01, ***p < 0.001.
3.2. Circ_0001666 regulates intestinal epithelial cell migration, invasion and EMT
Here, we investigated the regulatory effect of circ_0001666 in the constructed CD cell model. HIECs were transfected with circ_0001666 specific siRNA (si‐circ1 and si‐circ2) to silence circ_0001666 expression in the cells. Our results revealed that both siRNAs effectively silenced circ_0001666 expression in HIECs, with si‐circ2 exhibiting a more significant knockdown effect, which was then used in subsequent experiments (Figure 2A). We then examined the effect of circ_0001666 downregulation on the migration and invasion of HIECs induced by TGF‐β1. Our findings demonstrate that the inhibition of circ_0001666 significantly suppressed the migration and invasion of HIECs induced by TGF‐β1 (Figure 2B,C). Moreover, analysis of EMT marker proteins (E‐cadherin, N‐cadherin, Vimentin, and Snail) demonstrated that the inhibition of circ_0001666 expression increased the protein levels of E‐cadherin but decreased those of N‐cadherin, Vimentin, and Snail (Figure 2D). Collectively, these results indicated that circ_0001666 suppression inhibited the migration, invasion, and EMT of intestinal epithelial cells induced by TGF‐β1.
FIGURE 2.

Circ_0001666 regulates intestinal epithelial cell migration, invasion, and EMT. HIECs were transfected with si‐NC or si‐circ, followed by 10 ng/mL TGF‐β1 treatment for 72 h. (A) Circ_0001666 expression was analyzed by qPCR (HIECs without TGF‐β1 treatment). (B) Wound healing assay was performed to assess cell migration. (C) Cell invasion was measured by transwell assay. (D) The levels of EMT marker proteins were analyzed by western blot. EMT, epithelial–mesenchymal transition; HIEC, human intestinal epithelial cell; qPCR, quantitative real‐time PCR. n = 3. **p < 0.01, ***p < 0.001.
3.3. Circ_0001666 regulated the expression of fibrosis markers
Here, we analyzed the relationship between circ_0001666 and cell fibrosis. We found that the silencing of circ_0001666 significantly suppressed the expressions of fibrosis‐related genes (α‐SMA, Collagen III, Fibronectin, and MMP9) in TGF‐β1‐treated HIECs (Figure 3A–D). Consistent with the mRNA expression results, inhibiting circ_0001666 reduced fibrosis‐related protein levels in TGF‐β1‐treated HIECs (Figure 3E). Taken together, these results indicate that silencing circ_0001666 could inhibit the fibrosis of HIECs induced by TGF‐β1.
FIGURE 3.
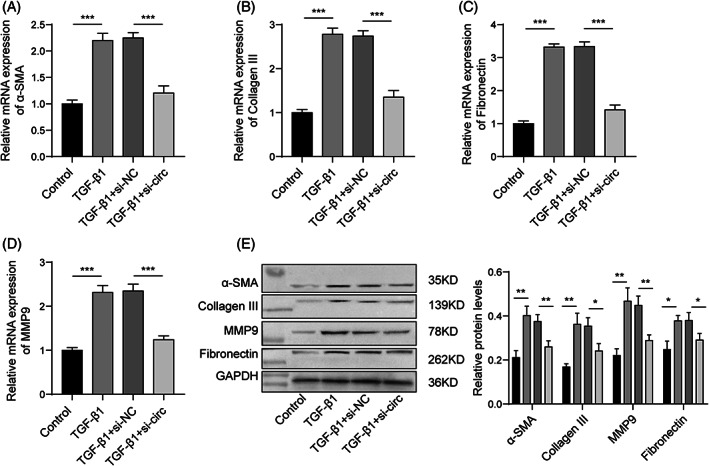
Circ_0001666 regulates the expression of fibrosis markers. HIECs were transfected with si‐NC or si‐circ, followed by 10 ng/mL TGF‐β1 treatment for 72 h. (A–D) The expressions of genes related to fibrosis were detected by qPCR. (E) Western blotting measured fibrosis‐related protein levels. HIEC, human intestinal epithelial cell; qPCR, quantitative real‐time PCR. n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
3.4. Circ_0001666 regulates BMP7 expression by interacting with SRSF1
We further investigated the possible downstream regulatory mechanism of circ_0001666 and its potential association with BMP7 in intestinal epithelial cells. BMP7 is known to play a role in the regulation of EMT and fibrosis in intestinal cells. 6 In CD patients, BMP7 expression was decreased and negatively correlated with circ_0001666 expression (Figure 4A). Inhibition of circ_0001666 in HIECs upregulated BMP7 expression and protein level (Figure 4B,C). Consistent with the distribution of GAPDH in the control group, we found that circ_0001666 was mainly distributed in the cytoplasm, while U6 was mainly located in the nucleus (Figure 4D). The suppression of circ_0001666 significantly increased the stability of BMP7 mRNA (Figure 4E). Thus, circ_0001666 might affect BMP7 expression by regulating the stability of BMP7 mRNA. CircRNA can be used as a dynamic scaffold molecule to regulate protein‐gene interactions. 25 Through the prediction of bioinformatics (RBPsuite: http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/), it is found that SRSF1 is associated with circ_0001666 and BMP7. To explore whether circ_0001666 could regulate BMP7 by binding to RNA binding protein (RBP), we verified whether SRSF1 binds to circ_0001666 and BMP7 by RNA pull‐down and RIP. We found that SRSF1 was upregulated in CD patients and positively correlated with circ_0001666 expression (Figure 4F). Additionally, as shown in Figure 4G,H, SRSF1 had a direct binding effect with circ_0001666 and BMP7. Next, we overexpressed SRSF1 and found that BMP7 expression was reduced, while silencing circ_0001666 could partially reverse the regulatory effect of SRSF1 (Figure 4I,J). These results indicate that circ_0001666 can target the expression of BMP7 by binding to SRSF1. Lastly, we investigated the stability of BMP7 mRNA and found that the upregulation of SRSF1 promoted the decay of BMP7 mRNA, but the knockdown of circ_0001666 reversed the regulation of SRSF1 on BMP7 mRNA decay (Figure 4K). Collectively, circ_0001666 promoted BMP7 mRNA decay to inhibit its expression by interacting with SRSF1.
FIGURE 4.

Circ_0001666 regulates BMP7 expression by interacting with SRSF1. (A) BMP7 expression in the colonic mucosa of patients was detected by qPCR, and statistical analysis of the correlation between circ_0001666 and BMP7 was performed. (B, C) The expressions of BMP7 after the knockdown of circ_0001666 were detected by qPCR and western blot. (D) The distribution of circ_0001666 in cells was determined by qPCR. (E) The knockdown effects of circ_0001666 on the stability of BMP7 mRNA were analyzed by qPCR. (F) SRSF1 expression in the colonic mucosa of patients was detected by qPCR, and statistical analysis of the correlation between circ_0001666 and SRSF1 was performed. (G, H) The relationship between SRSF1 and circ_0001666 or BMP7 was verified by RNA pull‐down and RIP. HIECs were transfected with ov‐NC, ov‐SRSF1, ov‐SRSF1 and si‐NC, and ov‐SRSF1 and si‐circ. (I) SRSF1 expression was detected by qPCR. (J) BMP7 expression was detected by qPCR. (K) The stability of BMP7 mRNA was analyzed by qPCR. EMT, epithelial–mesenchymal transition; HIEC, human intestinal epithelial cell; qPCR, quantitative real‐time PCR. n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
3.5. Circ_0001666/SRSF1/BMP7 axis regulates intestinal epithelial cell migration, invasion, and EMT
First, we suppressed BMP7 expression in the cells by transfecting si‐BMP7 (Figure 5A). We found that circ_0001666 silencing reduced TGF‐β1‐induced cell migration and invasion while overexpressing SRSF1 or silencing BMP7 could partially reverse this trend (Figure 5B,C). Additionally, the knocking down of circ_0001666 inhibited TGF‐β1‐induced cell EMT while upregulating SRSF1 or inhibiting BMP7 reversed this effect (Figure 5D). Taken together, circ_0001666 promoted intestinal epithelial cell migration, invasion, and EMT through the SRSF1/BMP7 axis.
FIGURE 5.

The circ_0001666/SRSF1/BMP7 axis regulates intestinal epithelial cell migration, invasion, and EMT. HIECs were transfected with si‐NC and si‐BMP7. (A) BMP7 expression in cells transfected with si‐BMP7 was determined by qPCR (HIECs without TGF‐β1 treatment). HIECs were transfected with si‐NC, si‐circ, si‐circ and ov‐SRSF1, and si‐circ and si‐BMP7, followed by treatment with 10 ng/mL TGF‐β1 for 72 h. (B) Wound healing assay was performed to analyze cell migration. (C) Cell invasion was measured by transwell. (D) Western blot was conducted to determine the expressions of EMT marker proteins. EMT, epithelial–mesenchymal transition; HIEC, human intestinal epithelial cell; qPCR, quantitative real‐time PCR. n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
3.6. Circ_0001666/SRSF1/BMP7 axis regulates the expression of fibrosis markers
Here, we verified the effects of the circ_0001666/SRSF1/BMP7 axis on cell fibrosis. The results showed that the knockdown of circ_0001666 inhibited the expressions of fibrosis‐related genes (α‐SMA, Collagen III, Fibronectin, and MMP9) in cells induced by TGF‐β1, while the overexpression of SRSF1 or the inhibition of BMP7 restored the inhibitory effects of si‐circ on fibrosis‐related genes (Figure 6A–D). Additionally, the results of protein expressions were consistent with those obtained with gene expressions. Silencing circ_0001666 inhibited α‐SMA, Collagen III, Fibronectin, and MMP9 protein levels in TGF‐β1‐induced cells, while simultaneously upregulating SRSF1 or silencing BMP7 partially reversed the effects of si‐circ (Figure 6E). Thus, the circ_0001666/SRSF1/BMP7 axis might mediate fibrosis.
FIGURE 6.

The circ_0001666/SRSF1/BMP7 axis regulates the expression of fibrosis markers. HIECs were transfected with si‐NC, si‐circ, si‐circ and ov‐SRSF1, and si‐circ and si‐BMP7, followed by treatment with 10 ng/mL TGF‐β1 for 72 h. (A–D) The expressions of fibrosis‐related genes were analyzed by qPCR. (E) Fibrosis‐related protein levels were detected by western blot. (F) The schematic diagram demonstrates the role of circ_0001666 on EMT‐mediated fibrosis in intestinal epithelial cells. EMT, epithelial–mesenchymal transition; HIEC, human intestinal epithelial cell; qPCR, quantitative real‐time PCR. n = 3. *p < 0.05, **p < 0.01, ***p < 0.001.
4. DISCUSSION
CD is an uncurable chronic gastrointestinal inflammation. At present, the treatment of CD is mainly performed via drugs and surgical treatment to achieve remission. 26 Anti‐tumor necrosis factor (TNF) antibody therapy, which began in 1998, changed the treatment mode of CD and improved the remission rate of patients. However, at present, about one third of CD patients treated with anti‐TNF antibodies may not respond to the treatment due to immunogenicity or insufficient serum antibodies, and it is impossible to benefit from another TNF antagonist. 27 We found that circ_0001666 expression was upregulated in CD children's colonic mucosal tissues. Further, the establishment of the CD cell model using TGF‐β1‐treated HIECs confirmed that circ_0001666 promoted cell migration, invasion, EMT, and fibrosis by interacting with SRSF1 to accelerate BMP7 mRNA decay (Figure 6F).
Approximately 33% of CD patients develop complications due to intestinal fibrosis and require surgery. 8 In CD mouse model exploration, it was shown that intestinal fibrosis was caused by EMT. 6 A sustained inflammatory response promotes EMT, mainly via the decrease of E‐cadherin (adhesion molecule) expression in epithelial cells, which weakens the cell–cell connection. At the same time, the expression of N‐cadherin, a marker of interstitial cells, increased, and the expression of TGF‐β, which promotes cell motility and fibrosis, increased by activating the common Snail1 pathway of EMT. 28 , 29 Yang et al. found that total flavone of Abelmoschus manihot (TFA) promoted the migration and invasion of intestinal epithelial cells induced by TGF‐β1 and inhibited EMT levels through the Smad and MAPK signaling pathways. 30 Our results also revealed changes in the expression of EMT marker proteins in the colonic mucosal tissues of CD patients, suggesting that the development of CD might be related to EMT. Further, EMT was also identified in TGF‐β1‐treated HIECs, which might be related to the abnormal expression of circRNA. Recently, microarray analysis showed an uncontrolled expression of a large number of circRNA in CD. 31 CircRNA_103765 regulates DLL4 through the sponge adsorption of miR‐30 and inhibits the apoptosis of HIECs induced by TNF‐α. 31 The upregulation of hsa_circRNA_102610 promotes intestinal epithelial cell proliferation and EMT by targeting miR‐130a‐3p. 14 Consistent with previous reports, our study found that circ_0001666 was overexpressed in children with CD and regulated intestinal epithelial cell migration, invasion, EMT, and fibrosis.
One type of circRNA regulatory mechanism is interaction with RBP. 32 In liver cancer, circZKSCAN1 was shown to regulate the cell cycle and apoptosis of liver cancer cells by competitively binding FMRP to regulate the Wnt/β‐catenin pathway. 33 SRSF1 is a kind of RBP that often combines with RNA to play a regulatory role. For instance, circRPAP2 can bind to the oncoprotein SRSF1, and SRSF1 decreases the stability of PTK2 mRNA and promotes the progression of breast cancer. 34 In lung cancer, the combination of SRSF1 and circCDR1 regulates malignant behavior and apoptosis. 35 Furthermore, SRSF1 is reported to be highly expressed during TGF‐β1 induction and promote fibrosis progression. 36 , 37 We found in HIECs that circ_0001666 regulated the expression of BMP7 by interacting with SRSF1. Studies have found that overexpression of BMP7 reduces the EMT and migration of retinal pigment epithelial cells induced by TGF‐β2. 38 BMP7 was also reported to participate in intestinal inflammation, EMT, and fibrosis, and BMP7 could prevent this process. 6 , 11 However, Burke et al. found that BMP7 is increased in CD intestinal tissues, 39 which is contrary to our results. The patient's tissues we took were the colonic mucosa of children with active CD (average age 12 years old), while Burke et al.'s research samples came from the transmural biopsy of macro‐narrow small intestine of CD patients (average age 22.3 years old). 39 By analyzing patient clinical information, the differences in results may be related to factors such as the location of the tissue taken and the patient's age, which needs to be further explored in subsequent research. Our additional investigations also confirmed that knocking down circ_0001666 significantly inhibited HIECs cell migration, invasion, EMT, and fibrosis, while SRSF1 overexpression or BMP7 silencing partially reversed this trend, indicating that circ_0001666 could influence the EMT and fibrosis of HIECs by regulating the expression of BMP7 mediated by SRSF1. When circRNA binds to RBP to regulate downstream genes, the increase in RBP level may be due to increased RBP biogenesis efficiency or increased RBP mRNA stability. 40 Since circ_0001666 is mainly distributed in the cytoplasm, circ_0001666 may affect the expression of SRSF1 by affecting the stability of SRSF1 mRNA. Nevertheless, we cannot rule out the possibility that a small amount of circ_0001666 observed in the nucleus may affect SRSF1 biogenesis. Therefore, it is necessary to further study the regulation of circ_0001666 on SRSF1.
Despite these interesting observations, this present research also has some limitations. We aim to conduct further experimental verification in CD animal models. At the same time, we also aim to perform in‐depth explorations on the regulatory role and related mechanisms of circ_0001666 in intestinal fibroblasts.
In conclusion, circ_0001666 modulates the stability of BMP7 mRNA by interacting with SRSF1 to regulate cell migration, invasion, EMT, and fibrosis in a CD model in vitro. Collectively, our research provides a new direction for the study of the molecular mechanism of CD and a new target for the exploration of targeted therapy that could be used as a reference to improve the outcomes of pediatric CD patients.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
The authors thank the anonymous reviewers who helped to improve the paper.
Li J, Xu J‐Z, Dou B, Huang T‐F, Chen J, Wang T‐M, et al. Circ_0001666 upregulation promotes intestinal epithelial cell fibrosis in pediatric Crohn's disease via the SRSF1/BMP7 axis. Kaohsiung J Med Sci. 2023;39(10):966–977. 10.1002/kjm2.12734
REFERENCES
- 1. Yu YR, Rodriguez JR. Clinical presentation of Crohn's, ulcerative colitis, and indeterminate colitis: symptoms, extraintestinal manifestations, and disease phenotypes. Semin Pediatr Surg. 2017;26(6):349–55. [DOI] [PubMed] [Google Scholar]
- 2. Mutanen A, Pakarinen MP. Perianal Crohn's disease in children and adolescents. Eur J Pediatr Surg. 2020;30(5):395–400. [DOI] [PubMed] [Google Scholar]
- 3. Duricova D, Fumery M, Annese V, Lakatos PL, Peyrin‐Biroulet L, Gower‐Rousseau C. The natural history of Crohn's disease in children: a review of population‐based studies. Eur J Gastroenterol Hepatol. 2017;29(2):125–34. [DOI] [PubMed] [Google Scholar]
- 4. Quencer KB, Nimkin K, Mino‐Kenudson M, Gee MS. Detecting active inflammation and fibrosis in pediatric Crohn's disease: prospective evaluation of MR‐E and CT‐E. Abdom Imaging. 2013;38(4):705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santacroce G, Lenti MV, Di Sabatino A. Therapeutic targeting of intestinal fibrosis in Crohn's disease. Cell. 2022;11(3):429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flier SN, Tanjore H, Kokkotou EG, Sugimoto H, Zeisberg M, Kalluri R. Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J Biol Chem. 2010;285(26):20202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grgic I, Duffield JS, Humphreys BD. The origin of interstitial myofibroblasts in chronic kidney disease. Pediatr Nephrol. 2012;27(2):183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scharl M, Huber N, Lang S, Furst A, Jehle E, Rogler G. Hallmarks of epithelial to mesenchymal transition are detectable in Crohn's disease associated intestinal fibrosis. Clin Transl Med. 2015;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mehta SJ, Lewis A, Nijhuis A, Jeffery R, Biancheri P, Di Sabatino A, et al. Epithelial down‐regulation of the miR‐200 family in fibrostenosing Crohn's disease is associated with features of epithelial to mesenchymal transition. J Cell Mol Med. 2018;22(11):5617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macias‐Ceja DC, Coll S, Bauset C, Seco‐Cervera M, Gisbert‐Ferrandiz L, Navarro F, et al. IFNgamma‐treated macrophages induce EMT through the WNT pathway: relevance in Crohn's disease. Biomedicine. 2022;10(5):1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wenxiu J, Mingyue Y, Fei H, Yuxin L, Mengyao W, Chenyang L, et al. Effect and mechanism of TL1A expression on epithelial‐mesenchymal transition during chronic colitis‐related intestinal fibrosis. Mediators Inflamm. 2021;2021:5927064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Xiao Y, Ge W, Zhou K, Wen J, Yan W, et al. miR‐200b inhibits TGF‐beta1‐induced epithelial‐mesenchymal transition and promotes growth of intestinal epithelial cells. Cell Death Dis. 2013;4(3):e541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai Y, Wang W, Yin P, Gao J, Na L, Sun Y, et al. Ruxolitinib alleviates renal interstitial fibrosis in UUO mice. Int J Biol Sci. 2020;16(2):194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin J, Ye YL, Hu T, Xu LJ, Zhang LP, Ji RN, et al. Hsa_circRNA_102610 upregulation in Crohn's disease promotes transforming growth factor‐beta1‐induced epithelial‐mesenchymal transition via sponging of hsa‐miR‐130a‐3p. World J Gastroenterol. 2020;26(22):3034–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin L, Zhou G, Chen P, Wang Y, Han J, Chen M, et al. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020;11(6):456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qiao YQ, Cai CW, Shen J, Zheng Q, Ran ZH. Circular RNA expression alterations in colon tissues of Crohn's disease patients. Mol Med Rep. 2019;19(5):4500–6. [DOI] [PubMed] [Google Scholar]
- 17. Zhou LY, Lin SN, Rieder F, Chen MH, Zhang SH, Mao R. Noncoding RNAs as promising diagnostic biomarkers and therapeutic targets in intestinal fibrosis of Crohn's disease: the path from bench to bedside. Inflamm Bowel Dis. 2021;27(7):971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yao W, Li Y, Han L, Ji X, Pan H, Liu Y, et al. The CDR1as/miR‐7/TGFBR2 axis modulates EMT in silica‐induced pulmonary fibrosis. Toxicol Sci. 2018;166(2):465–78. [DOI] [PubMed] [Google Scholar]
- 19. Li B, Sun G, Yu H, Meng J, Wei F. Circ_0114428 promotes proliferation, fibrosis and EMT process of high glucose‐induced glomerular mesangial cells through regulating the miR‐185‐5p/SMAD3 axis. Autoimmunity. 2022;55(7):462–72. [DOI] [PubMed] [Google Scholar]
- 20. Hu YA, Zhu Y, Liu G, Yao X, Yan X, Yang Y, et al. Expression profiles of circular RNAs in colon biopsies from Crohn's disease patients by microarray analysis. J Clin Lab Anal. 2021;35(6):e23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun R, Guan H, Liu W, Liang J, Wang F, Li C. Expression of BMP7 in cervical cancer and inhibition of epithelialmesenchymal transition by BMP7 knockdown in HeLa cells. Int J Mol Med. 2020;45(5):1417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu L, Wang Y, Yan R, Liang L, Zhou X, Liu H, et al. BMP‐7 inhibits renal fibrosis in diabetic nephropathy via miR‐21 downregulation. Life Sci. 2019;238:116957. [DOI] [PubMed] [Google Scholar]
- 23. Sun CM, Wu J, Zhang H, Shi G, Chen ZT. Circulating miR‐125a but not miR‐125b is decreased in active disease status and negatively correlates with disease severity as well as inflammatory cytokines in patients with Crohn's disease. World J Gastroenterol. 2017;23(44):7888–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lupinacci S, Perri A, Toteda G, Vizza D, Puoci F, Parisi OI, et al. Olive leaf extract counteracts epithelial to mesenchymal transition process induced by peritoneal dialysis, through the inhibition of TGFbeta1 signaling. Cell Biol Toxicol. 2019;35(2):95–109. [DOI] [PubMed] [Google Scholar]
- 25. Wang Z, Lei X. Matrix factorization with neural network for predicting circRNA‐RBP interactions. BMC Bioinform. 2020;21(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen T, Duerksen DR. Enteral nutrition in the management of pediatric and adult Crohn's disease. Nutrients. 2018;10(5):537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmitt H, Billmeier U, Dieterich W, Rath T, Sonnewald S, Reid S, et al. Expansion of IL‐23 receptor bearing TNFR2+ T cells is associated with molecular resistance to anti‐TNF therapy in Crohn's disease. Gut. 2019;68(5):814–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ortiz‐Masia D, Gisbert‐Ferrandiz L, Bauset C, Coll S, Mamie C, Scharl M, et al. Succinate activates EMT in intestinal epithelial cells through SUCNR1: a novel protagonist in fistula development. Cell. 2020;9(5):1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang H, Shen J, Ran Z. Epithelial‐mesenchymal transition in Crohn's disease. Mucosal Immunol. 2018;11(2):294–303. [DOI] [PubMed] [Google Scholar]
- 30. Yang BL, Zhu P, Li YR, Xu MM, Wang H, Qiao LC, et al. Total flavone of Abelmoschus manihot suppresses epithelial‐mesenchymal transition via interfering transforming growth factor‐beta1 signaling in Crohn's disease intestinal fibrosis. World J Gastroenterol. 2018;24(30):3414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ye Y, Zhang L, Hu T, Yin J, Xu L, Pang Z, et al. CircRNA_103765 acts as a proinflammatory factor via sponging miR‐30 family in Crohn's disease. Sci Rep. 2021;11(1):565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang M, Wang T, Xiao G, Xie Y. Large‐scale profiling of RBP‐circRNA interactions from public CLIP‐seq datasets. Genes. 2020;11(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9(12):3526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu Y, Fang L. CircRPAP2 regulates the alternative splicing of PTK2 by binding to SRSF1 in breast cancer. Cell Death Discov. 2022;8(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu J, Huang L, Bao T, Duan K, Cheng Y, Zhang H, et al. CircCDR1as mediates PM(2.5)‐induced lung cancer progression by binding to SRSF1. Ecotoxicol Environ Saf. 2023;249:114367. [DOI] [PubMed] [Google Scholar]
- 36. Sun J, Jin T, Niu Z, Guo J, Guo Y, Yang R, et al. LncRNA DACH1 protects against pulmonary fibrosis by binding to SRSF1 to suppress CTNNB1 accumulation. Acta Pharm Sin B. 2022;12(9):3602–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun J, Jin T, Su W, Guo Y, Niu Z, Guo J, et al. The long non‐coding RNA PFI protects against pulmonary fibrosis by interacting with splicing regulator SRSF1. Cell Death Differ. 2021;28(10):2916–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao H, Ge T, Zhang Y, Li M, Yang S, Li H, et al. BMP7 antagonizes proliferative vitreoretinopathy through retinal pigment epithelial fibrosis in vivo and in vitro. FASEB J. 2019;33(3):3212–24. [DOI] [PubMed] [Google Scholar]
- 39. Burke JP, Ferrante M, Dejaegher K, Watson RW, Docherty NG, De Hertogh G, et al. Transcriptomic analysis of intestinal fibrosis‐associated gene expression in response to medical therapy in Crohn's disease. Inflamm Bowel Dis. 2008;14(9):1197–204. [DOI] [PubMed] [Google Scholar]
- 40. Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF‐A axis. Nat Commun. 2019;10(1):4317. [DOI] [PMC free article] [PubMed] [Google Scholar]


