Abstract
Cells contain a plethora of structurally diverse lipid species, which are unevenly distributed across the different cellular membrane compartments. Some of these lipid species require vesicular trafficking to reach their subcellular destinations. Here, we review recent advances made in the field that contribute to understanding lipid sorting during endomembrane trafficking.
Keywords: membrane nanodomains, endocytosis, membrane trafficking, membrane curvature, membrane rafts, complex sphingolipids
1 Introduction
The plasma- and endo-membranes of eukaryotic cells are two-dimensionally diffusing fluids comprised of myriads of lipid species. They form a barrier, separating compartments into selective reaction spaces, and embed the trans- and membrane proteins of the cell. It is estimated that each cell consists of tens of thousands of lipid species (Gerl et al., 2012) with each cellular membrane compartment or organelle having its unique and distinct lipid composition and thus membrane identity (Harayama and Riezman, 2018; van Meer et al., 2008). The different membrane and organelle compartments, however, are interconnected through vesicular transport and thus in constant exchange. For the plasma membrane (PM), for instance, it is estimated that the equivalent of its total surface area is turned over every 15 min (Koval and Pagano, 1991; Mayor et al., 1993). Given this continuous vesicular endomembrane flux along the endocytic and secretory systems, concomitant lipid and protein sorting are thus key cellular processes necessary to enable eukaryotic cells to maintain membrane homeostasis among organelles (van Meer et al., 2008).
There is an increasing body of research demonstrating how individual or bulk lipids can be supplied to different organelles through either lipid transfer proteins specific for individual lipids or through membrane contact sites that connect different organelles. Both pathways bypass vesicular trafficking (interested readers might be referred to the following references: Anders and Mattjus, 2021; Khaddaj and Kukulski, 2023; Melia and Reinisch, 2022; Samaha et al., 2019; Wong et al., 2019).
However, these lipid transport conduits are not available for all lipid species. Due to their large hydrophilic headgroup, complex sphingolipids (cSLs), sphingomyelin and especially the glycosphingolipids (GSLs), are trapped in the outer membrane leaflet and cannot rely on lipid transfer proteins for sorting (Sokoya et al., 2022; van Meer et al., 2008; Young et al., 1992). Rather, complex SLs depend heavily on vesicular trafficking to reach their subcellular destinations. The mechanisms by which cells preferentially sort these lipids during trafficking to their respective compartments and organelles, and the underlying biophysical driving forces, remain open questions.
This is the topic of this review: to collect our current understanding about the mechanisms by which cells can sort their plethora of different SL species that rely on vesicular trafficking to maintain compartment and organelle homeostasis.
2 Lipid self-organization and membrane nanodomains
How membrane lipids interact with each other is critical for vesicular-based lipid sorting. Membrane lipids, including SLs, are structurally extremely diverse. This diversity stems from different headgroups, e.g., for complex sphingolipids, this can be a choline (sphingomyelin), or a diversity of different sugar headgroups for the glycosphingolipids. Phospholipids occur as phosphatidylcholine (PtdCho), -ethanolamine (PtdEtn) or -serine (PtdSer), but all lipids also differ in their acyl chain structures. The acyl chains of lipids can vary in their hydrocarbon chain length and in their degree of saturation. Most phospholipids are ‘hybrid’ lipids and contain one saturated acyl chain at the sn1 position and one cis mono- or polyunsaturated fatty acid at their sn2 position. Phospholipids with two saturated or two unsaturated acyl chains are relatively scarce. Sphingolipids on the other hand, are anchored in the membrane through a ceramide portion, characterized by a long chain sphingoid base connected to an acyl chain of varying length, between 14 and typically 24 hydrocarbons. The sphingoid base is usually 18 or 20 hydrocarbons in length and comprises a trans carbon double bond at C4. The acyl chain, however, is predominantly saturated or contains one cis carbon double bond (Figure 1; Hannun and Obeid, 2018; Merrill, 2011).
FIGURE 1.
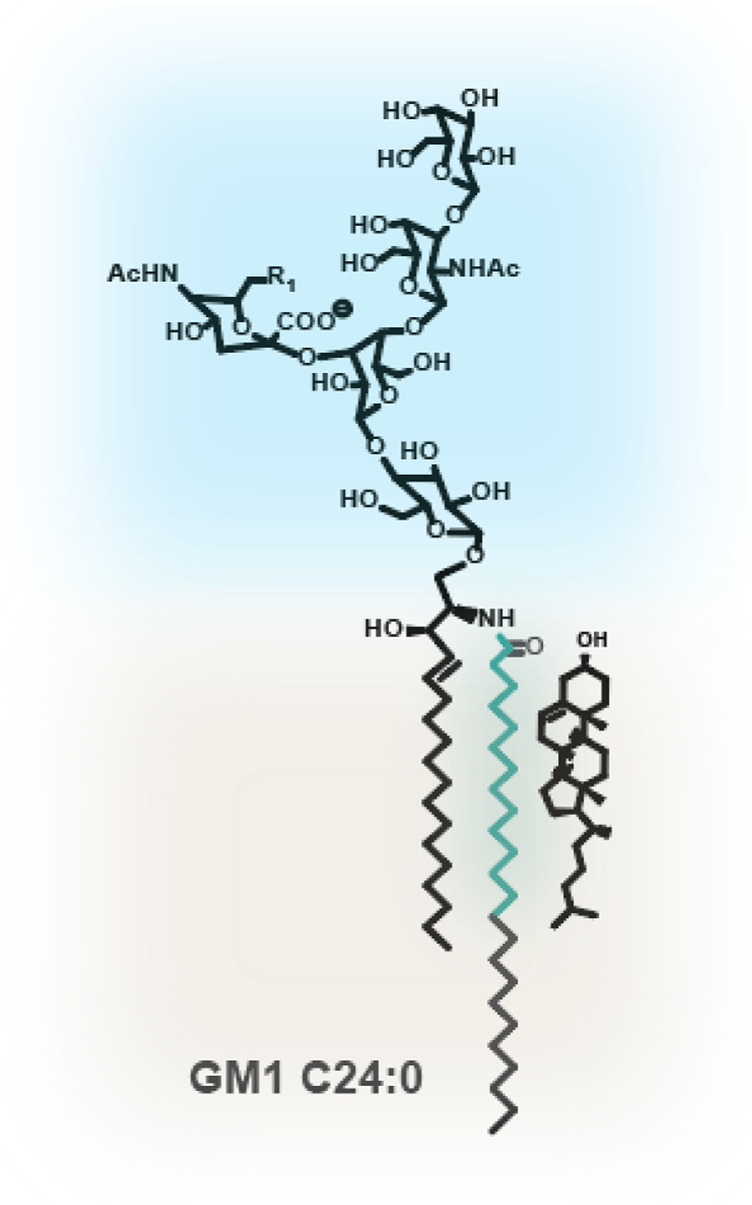
Glycosphingolipid GM1. GSLs such as GM1 contain a large and hydrophilic oligosaccharyl headgroup, protruding into the extracellular space. The ceramide is composed of a C18 or C20 sphingoid base, containing a trans double carbon bond at C4. The acyl chains can vary dramatically in length and degree of unsaturation. Depicted is a C24 fully saturated acyl chain. In turquoise is the C14* motif. 14 fully saturated hydrocarbons from the amide bond at the water-bilayer interface are required for assembly with cholesterol.
There are two ways in which lipids can organize in the membrane, first by shaping the membrane physically or by creating lateral heterogeneity. The individual attributes of a lipid, the ratio between the acyl chain structures, and the size of a lipid’s headgroup give lipids an intrinsic shape or geometry (Pinot et al., 2014; Luzzati et al., 1966; Luzzati et al., 1968; Florence et al., 2004; Vanni et al., 2014; Maggio et al., 1978; Cebecauer et al., 2018; Frolov et al., 2011). Each cis carbon double bond induces a kink in the acyl chain tail, thus requiring more physical space than its saturated counterpart and therefore reducing the ability of the lipid to pack side-by-side (Chiantia et al., 2006; Dietrich et al., 2001; Hitchcock et al., 1974; McIntosh and Simon, 1986; Olbrich et al., 2000). Additionally, the two acyl chains can vary in hydrocarbon chain length, for some requiring interdigitation into the opposing leaflet. Apart from the acyl chain structure, the size of the head group affects the lipid’s overall shape (Rawicz et al., 2000). Phosphatidylcholine (PtdCho), phosphatidylserine (PtdSer) and the sphingolipid, sphingomyelin are cylindrical lipids, while lipids such as phosphatidylethanolamine (PtdEtn), phosphatic acid, and diacylglycerol (DAG) or cholesterol, have a smaller polar headgroup, and thus adopt a conical shape. GSLs, with their large oligosaccharyl headgroup adopt an inverse conical shape, with the headgroup requiring more space than the ceramide (Hitchcock et al., 1974). A lipid’s structural features (both the acyl chain structures and size of the headgroup) also critically influence its ability to interact with other lipids and pack side-by-side (see below for the concept of membrane nanodomains, rafts and the liquid-ordered phase). They give rise to small lateral inhomogeneities within the membrane space, where lipids are not uniformly distributed or ‘mixed’, but instead can be highly organized and form membrane domains with distinct lipid compositions (Figure 2; Heberle and Feigenson, 2011; Almeida, 2009).
FIGURE 2.
Membrane nanodomains or lipid rafts. Saturated phospholipids (blue), sphingolipids (red) and cholesterol (yellow) assemble into membrane nanodomains or lipid rafts. These phase separate from the phospholipids with predominantly unsaturated acyl chains (black).
One example of how lipids can self-associate is the concept of membrane nanodomains and lipid rafts (Simons and Ikonen, 1997). These nanodomains or rafts are membrane areas enriched in cholesterol, saturated lipids, and especially sphingo- and glycosphingolipids, and are thought to organize the plasma membrane into heterogeneous sub-domains, to compartmentalize cellular functions (Brown and London, 1998a; Pike, 2006), e.g., immune signaling (Dinic et al., 2015; Field et al., 1995; Gupta and DeFranco, 2007), endocytosis (Kim et al., 2017), host-viral/toxin interaction processes (Chinnapen et al., 2007; Dick et al., 2012; Johannes, 2017; Wang et al., 2008), and protein clustering (Arumugam et al., 2021). Importantly, another physiological role for nanodomains is believed to be the sorting and trafficking platform for membrane components between subcellular organelles (Diaz-Rohrer et al., 2014; Schuck and Simons, 2004; Smart et al., 1996).
The lipid raft hypothesis was originally conceived to explain differences in membrane sorting between the apical and basolateral membranes of polarized epithelial cells (Simons and Ikonen, 1997), but the overall concept was already suggested earlier by Stier and Sackmann (1973). It assumes that rafts or nanodomains have different physical properties by creating a highly packed and ordered lipid environment. This leads to an altered membrane miscibility, with increased membrane thickness and rigidity and a reduced diffusiveness of its components (Lingwood and Simons, 2010; Simons and Sampaio, 2011; Sezgin et al., 2017; Simons and Toomre, 2000; Cebecauer et al., 2018). The nanodomain or raft concept is supported by a wealth of in vitro studies on model membrane systems, where SLs, phospholipids with saturated acyl chains, and cholesterol phase-segregate into liquid-ordered (Lo) regions, with tight lipid-lipid packing, due to their preferred interactions. Similar to the liquid-disordered (Ld) phase, the Lo phase is still fluid, allowing molecular motion of the individual components, albeit at reduced diffusiveness. The Ld phase is characterized by weak lipid-lipid packing, higher permeability, and low membrane rigidity. The distinctiveness between these two phases allows them to coexist over a large compositional spectrum (Bacia et al., 2005; Brown and London, 1998b; Feigenson, 2009; Hjort Ipsen et al., 1987; Sezgin et al., 2012; Simons and Vaz, 2004; Veatch and Keller, 2005; Wesołowska et al., 2009).
In addition, the concept is supported by many atomistic simulations characterizing cholesterol interactions in membrane bilayers, with favorable packing between cholesterol and saturated lipid acyl chains (Martinez-Seara et al., 2010; Róg et al., 2007).
While we have a good understanding of the physicochemical principles that drive phase separation and raft formation in artificial membrane systems, their existence, relevance, and locations in live cells remain controversial to this day. Evidence for macroscopic phase separated Lo domains in live cells comes predominantly from work on the vacuole of the budding yeast Saccharomyces cerevisiae. Here, the vacuolar membrane and membrane associated proteins start to phase separate when the yeast is entering the stationary growth phase. These vacuolar Lo domains show similar characteristics than what is observed in GUVs (Leveille et al., 2022; Moeller and Thomson, 1979; Moeller et al., 1981; Toulmay and Prinz, 2013). Interestingly, in a recent study, Kim et al. could demonstrate that this vacuolar phase separation is driven by a change in lipid trafficking and thus resulting redistribution of cellular complex SL into the vacuole (Kim and Budin, 2024).
Instead of large macroscopic phase separated domains, mammalian live cell plasma and endomembranes are thought to contain small membrane nanodomains, which are highly dynamic and typically less than 20 nm in size (Kenworthy and Edidin, 1998; Lagerholm et al., 2005; Lingwood and Simons, 2010; Nichols, 2003; Sharma et al., 2004; Varma and Mayor, 1998; Veatch and Keller, 2003; Hancock, 2006; Levental et al., 2020). Direct evidence for their existence comes from studies investigating the differential behavior and dynamics of, e.g., fluorescently labeled lipids or GPI-anchored proteins (Eggeling et al., 2009; Kinoshita et al., 2017; Komura et al., 2016; Mueller et al., 2011; Saha et al., 2016; Stone et al., 2017; Honigmann et al., 2014). This includes our own work, where, using a GSL library with varied ceramide structures in live cells, we found evidence that incorporation of GSLs into membrane nanodomains requires a specific number of saturated carbon atoms. We termed this motif within the acyl chain the “C14* motif” (Figure 1). This stretch of 14+ saturated hydrocarbons from the amide bond at the water-bilayer interface most likely represents the minimal motif within an acyl chain to associate and accommodate cholesterol packing (Arumugam et al., 2021; Schmieder et al., 2022). While many studies, including our own, have demonstrated a necessity for cholesterol in nanodomain formation, the Kraft group, interestingly, using a technique called NanoSIMS, could not detect such cholesterol-sphingolipid domains in the PM; instead, they observed local sphingolipid-exclusive enrichments (Yeager et al., 2016).
Additionally, a restriction for macroscopic phase separation in the PM of live cells, is presumably the dynamic cortical actin cytoskeleton, which likely affects the location, size, and timing of nanodomain domain formation (Badizadegan et al., 2000; Kraft, 2016; Kusumi et al., 2005a; Liu and Fletcher, 2006). Specifically, nanodomains in the outer membrane leaflet are coupled to the preexisting actin-myosin networks inside the cell, which are mediated by the inner leaflet lipid PS (Raghupathy et al., 2015). Molecular simulations, e.g., demonstrated that filamentous supports, modeling the cortical actin cytoskeleton, coupled to lipids, have the potential to segregate membranes into corrals and stabilize domain formation, even at relatively low connectivity to the membrane, supporting the picket-fence model of membrane organization through the actin cytoskeleton (Levental et al., 2020; Tsai et al., 2024; Kusumi et al., 2005b; Kusumi et al., 2004 Kusumi et al., 1999) (Figure 3). Furthermore, extracellular binding to and cross-linking of nanodomain components by, e.g., endogenous lectins or exogenous toxins, which bind and crosslink the extracellular headgroup of especially GSLs, can result in stabilization and/or coalescence of membrane nanodomains and thereby affect their lifetime and function (Roemer et al., 2007; Szklarczyk et al., 2013; Hammond et al., 2005; Arumugam et al., 2021; Koyama-Honda et al., 2020; Day et al., 2015; Garner and Baum, 2008; Johannes et al., 2018; Raghunathan et al., 2016).
FIGURE 3.
Membrane nanodomains in live cells. The actin cytoskeleton, as well as membrane associated proteins are thought to stabilize and affect nanodomain size in live cell membranes. Membrane proteins which bind to PtdSer or PtdIns crosslink the membrane to the cortical actin cytoskeleton.
3 Lipid landscape of a cell
The lipid landscape of a cell, meaning the distinct membrane compositions of cellular organelles, has been described by the concept of evolutionarily conserved ‘lipid territories,” delineating the organelle and vesicular intermediates as two ends of distinct lipidome spectrums, with a “PM territory” on the one side and an “ER territory” on the other (Bigay and Antonny, 2012; Kim and Burd, 2023). The “plasma membrane (PM) territory” comprises the plasma membrane itself, the trans Golgi network (TGN), as well as the secretory and endolysosomal networks, while the endoplasmic reticulum (ER), the cis, and medial cisternae of the Golgi apparatus belong to the “ER territory.” The distinction between these two membrane territories is based on differences in a) lipid compositions, especially SL and cholesterol, leading to differences in b) membrane order, while also differing in c) the net charge of the leaflets and d) the degree of lipid species asymmetry between the bilayer leaflets (Holthuis and Menon, 2014; Kim and Burd, 2023). The membranes of the “ER territory” are characterized by a low membrane order due to a relative absence of nanodomain forming lipids, specifically in SL but also cholesterol (van Meer et al., 2008). The “PM territory,” on the other hand, arises from the synthesis of nanodomain-forming SLs and a consequent sequestration and enrichment of cholesterol within the late Golgi compartments due to their preferential interactions (Orci et al., 1981; Hanada et al., 2003; Sharpe et al., 2011; Slotte, 2013). SLs and cholesterol enrich gradually in the outer membrane leaflets of the secretory pathway leading to the plasma membrane. Their assembly into nanodomains leads to a high degree of packing order characterizing this membrane territory. Sequestration of cholesterol and SL in the outer membrane leaflet and the presence of PtdSer and phosphatidyl inositol (PtdIns) species on the cytoplasmic leaflet give rise to a highly asymmetric membrane. The increased order and associated increase in membrane thickness allow for the required barrier function in the PM. The gradual increase in membrane thickness through the synthesis of SLs within the Golgi and TGN and the following increase in cholesterol have been hypothesized to be a means to sort PM proteins in the Golgi for PM delivery by hydrophobic mismatch (Bigay and Antonny, 2012; Kim and Burd, 2023) of the transmembrane domain with membrane thickness. Indeed, the transmembrane domains of PM-resident transmembrane proteins contain slightly longer TMs and generally sort into Lo domains (Lorent et al., 2017; Munro, 1995; Quiroga et al., 2013).
The differences in SL and GSL composition between the two membrane territories are believed to rely on the differential SL trafficking and sorting between them. Importantly, the physico-chemical features of these territories seem to be conserved throughout eukaryotic evolution (Bigay and Antonny, 2012; Kim and Burd, 2023).
The importance of the SL gradient in organelle identity and its maintenance was recently illustrated by a study by Sokoya et al. (2022). Here, the sphingomyelin gradient in the secretory pathway was disrupted by the mislocalization of sphingomyelin synthase to the ER due to a pathogenic mutation. The subsequent synthesis of sphingomyelin in the ER and lack thereof in the TGN and PM resulted in manifold changes in the overall amounts of many different lipid species, with altered overall membrane lipid packing within the secretory pathway, and aberrant cholesterol accumulation in cytoplasmic vesicles, leading to osteoporosis and skeletal dysplasia in the patients (Pekkinen et al., 2019; Sokoya et al., 2022).
4 Lipid sorting principles
In the previous section, we provided an overview of the SL and GSL distributions across the two different cellular lipid territories. Without lipid sorting, however, vesicular trafficking, interconnecting the different territories, would quickly erode this gradient. How can cells maintain SL and GSLs compositions across their organelles without re-distribution by lipid transfer proteins or through membrane contact sites? There is compelling evidence that lipids are sorted differentially into transport carriers in live cells for both the secretory and endocytic pathways (Figure 4).
FIGURE 4.

Endocytic membrane trafficking pathways within cell. After plasma membrane (PM) cargo is endocytosed, cargo is sorted within the sorting endosome (SE). Pathway specific tubules are pulled from the SE, serving the recycling (back to the PM), retrograde (PM to Golgi to ER) or in polarized epithelial cells the transcytotic (linking apical and basolateral membranes) pathways. Cargo destined for degradation remains in the vesicular part of the endosome.
While SL synthesis starts with ceramide production in the ER, ceramide itself is trafficked to the Golgi by both vesicular trafficking and ceramide-specific transport proteins (Funato and Riezman, 2001). Only the addition of headgroup in the Golgi lumens, especially the large and hydrophilic oligosaccharide of GSLs destines SL and GSLs to vesicular trafficking for sorting. Recent studies in both polarized epithelial cells and nonpolarized cells show sphingomyelin sorting and enrichment into specific TGN-derived vesicles, thus supporting the evidence of Golgi-to-PM lipid sorting for SL and cholesterol (Deng et al., 2016; Klemm et al., 2009; Meer and Sprong, 2004; Wakana et al., 2020).
Early studies showed that, within the endosomal system, saturated and thus nanodomain-forming lipids were depleted from endosomal recycling tubules compared to unsaturated lipids (Gruenberg, 2003; Maxfield and McGraw, 2004; Mukherjee et al., 1999; Mukherjee and Maxfield, 2000). And within the retrograde pathways (trafficking from the PM to the Golgi and back to the ER), COPI-coated vesicles were found to be depleted of SLs (Brügger et al., 2000; Manneville et al., 2008). These results are in line with our own observations utilizing a GSL library of different ceramide structures. GSL species lacking a C14* motif and thus unable to form membrane nanodomains (Figure 1) were found in endosomal sorting tubules of the recycling, the retrograde, as well as in polarized epithelial cells transcytotic pathways (Schmieder et al., 2022; te Welscher et al., 2014; Chinnapen et al., 2012; Chinnapen et al., 2012). However, GSL species containing a C14* motif, which enables incorporation of the GSL into membrane nanodomains, were instead significantly depleted from these pathways and were sorted instead into the degradative pathway. Additionally, we could identify distinct lipid domains within enlarged endocytic carriers, where C14* motif containing GM1 species were segregated from transferrin receptor positive and C14* motif-lacking GM1 species (Schmieder et al., 2022).
4.1 Curvature based lipid sorting
Apart from lipid composition, membrane curvature also changes throughout the endomembrane compartments, suggesting a role for curvature as a means of sorting lipids (Black et al., 2013). Membrane vesicles, which facilitate inter-organelle traffic, are produced by budding and fission of the membrane from a donor organelle. This induction of highly curved membranes is thought to facilitate sorting of lipids.
Over the years, there has been a wealth of in vitro evidence to support how individual lipid species can be preferentially sorted through a curvature-based sorting mechanism. Initially, lipid shape was thought to be a prime candidate for how lipids might be sorted across membrane curvature. This was based on the idea that lipids might distribute spontaneously to differentially curved membrane regions according to the intrinsic geometrical shape of the lipid (see concept above, C. Black et al., 2014; Cheney et al., 2017; Hatzakis et al., 2009; Lodish, 2008). The molecular basis of this argument was that specific lipid species are not cylindrical-shaped but are conical and/or inverse-conical, and therefore would preferentially sort into membrane areas with curvature that accommodates and complements their shape. For instance, lipids with an inverse conical shape, comprising lipids with a large headgroup to acyl chain ratio, e.g., lysoPC with a single tail and a large headgroup, PtdIns or the GSLs, favor membrane regions of positive curvature bending the monolayer away from their large headgroups (reviewed in: Chernomordik and Kozlov, 2003; Di Paolo and De Camilli, 2006; Zimmerberg and Kozlov, 2006). On the other hand, lipids with both acyl chains being unsaturated and a small headgroup would sort to negative curvature (Kamal et al., 2009). In line with this, several studies found an enrichment of phosphatic acid and other inverse conical shaped lipids at the neck of highly negatively curved membranes (Crowley et al., 2024; Putta et al., 2016; Zhukovsky et al., 2019; see also above references). Supplementing these in vitro studies, molecular dynamics simulations also show that lipids have the propensity to sort to a membrane region based on their intrinsic shape, sensing the spontaneous curvature of the membrane (Baoukina et al., 2018; Beltrán-Heredia et al., 2019; Koenig et al., 2023). Despite the wealth of studies in this area, the consensus is that lipid shape alone, while important, does not completely account for the measurable amount of lipid sorting required in live cells and that lipid-lipid or lipid-protein based interactions are necessary to amplify curvature-based sorting (Callan-Jones et al., 2011; Cooke and Deserno, 2006). This is also supported by our own work in live cells, where we find that, rather than the size of the SL headgroup, it is the structure of the ceramide domain with the presence or absence of the C14* motif that determines intracellular trafficking (Duclos et al., 2020; Garcia-Castillo et al., 2018; Schmieder et al., 2022; Chinnapen et al., 2012).
4.2 Nanodomain based lipid sorting
An alternative model or complementary concept to the above-presented idea of curvature-induced lipid sorting, is that curvature preference, or indeed, induction, could arise not due to the physical properties of singular lipid species but as an emergent behavior of lipid organization in the membrane. Under this umbrella, segregated membrane nanodomains would provide an explanation for the differential lipid distributions and consequently lipid territories observed throughout the cell. Rather than an individual lipid molecule sensing membrane curvature, membrane nanodomains, with their unique physical properties, e.g., their low bending modulus, would detect and/or induce curvature preferences (C. Black et al., 2014; Huttner and Zimmerberg, 2001). Small and local inhomogeneities in membrane composition within an organelle could thus give rise to vesicles with different lipid compositions. In addition, lipid inhomogeneity might minimize the energy costs of bending the membrane (Mukherjee et al., 1999; Mukherjee and Maxfield, 2000; van Meer and Sprong, 2008; Maxfield and McGraw, 2004; van Meer and Sprong, 2008). This interplay between curvature and phase separation has been demonstrated in vitro for membranes close to phase separation or demixing (Baumgart et al., 2003; 2005; Jülicher and Lipowsky, 1996; Lipowsky, 1993; Ogunyankin et al., 2013; Ogunyankin and Longo, 2013; Parthasarathy et al., 2006; Sorre et al., 2009; Veatch and Keller, 2003; Woodward et al., 2023). Roux et al. (2005), Ikonen (2008) for instance, were able to demonstrate that lipid tubes pulled from Lo-Ld phase separated vesicles were almost exclusively in the Ld phase, implicating that tightly packed SL, or PtdCho with fully saturated acyl chains, disfavor curvature. These results are consistent with those by Mukherjee et al. (1999) and our own work. We observed GSLs species with the C14* motif within endosomal recycling tubules - in the absence of cholesterol, implicating that rather than lipid shape, incorporation of GSLs with C14* motif into membrane nanodomains is the driver for the observed differences in GSL sorting (Schmieder et al., 2022).
Interestingly, work by the Lippincott-Schwartz group showed in an elegant study phase separated domains within the endosomal network by hypotonic swelling and cooling, implicating that small diffraction limited nanodomains could also exist within the endosomal network, not only the PM (King et al., 2020). This is in agreement with, as mentioned above, our results where we also observed segregated domains within endosomal vesicles (Schmieder et al., 2022).
4.3 Alternatives: proteins associated with lipid sorting
An important caveat in many experiments investigating lipid sorting is, that they are often conducted in cell-free, and thus, protein-free systems. However, it is unlikely that curvature or nanodomain-based lipid sorting are the sole driving forces for lipid sorting within a cell and that this process would occur without protein assistance. There are few examples of such protein-assisted lipid sorting. Convincing evidence comes from studies on caveolin and the transport of cholesterol to the PM (E. J. Smart et al., 1996). Such selective transport of cholesterol to the PM by caveolin would most likely affect the concomitant transport of cSL as well. This is in agreement with work by the Nichols group, which suggests that caveolin, apart from transporting cholesterol and SM to the PM, is also required for endocytic trafficking of excess SL to the lysosome (Shvets et al., 2015).
Apart from caveolin, this idea is supported specifically for cSL-binding toxins such as Shiga toxin or Cholera toxin or cSL-binding galectins (Chinnapen et al., 2012; Römer and Elling, 2011; Safouane et al., 2010, p. 2010; Sorre et al., 2009; Tian and Baumgart, 2009). Here, the geometry of multivalent binding of nanodomain cSL by the proteins induces lipid compression and membrane bending (Arumugam et al., 2021; Ewers et al., 2010; Kabbani et al., 2020; Koyama-Honda et al., 2020; Roemer et al., 2007; Watkins et al., 2019). This process facilitates uptake of the membrane-protein complexes through recruitment of cellular trafficking machinery and forms the basis of the glycolipid-lectin driven endocytosis (Simunovic et al., 2017; Day and Kenworthy, 2015; Rydell et al., 2013; Johannes et al., 2015; Simunovic et al., 2017). Intriguingly, cSL lipid structures required for these processes to occur, differ between the different clustering proteins. Simian virus 40 requires cSL with C14* motif to induce membrane invaginations and endocytosis (Ewers et al., 2010), Shiga toxin and Cholera toxin however require cSL that do not contain C14* motif (Roemer et al., 2007; Chinnapen et al., 2012).
A concept for all the cSL sorting events mentioned in this review could be envisioned, where different proteins might immobilize and stabilize particular cSL distributions in the membrane and thus sort them in the process (Figure 5).
FIGURE 5.
cSL sorting models. In cellular membranes curvature, nanodomain formation and cellular proteins recognizing local cSL heterogeneities contribute to their differential, subcellular sorting.
5 Open questions
We hypothesize that, most likely, SL sorting within the endomembrane system is a synergy of all three sorting mechanisms presented here. SL species that are structurally able to incorporate into membrane nanodomains are sorted as such, most likely aided by the cellular protein machinery. Conversely, SL that are unable to assemble into membrane nanodomains might experience sole curvature-based sorting mechanisms more strongly.
Most interesting is the recent discovery of bulk lipid exchange at membrane contact sites, which virtually interconnect all organelles. We envision that such bulk lipid exchange or the specific depletion/supplementation with certain lipid species could rapidly change membrane composition and fluidities in small organelles and thus drive demixing or curvature generation. An example is VPS 13C, mediating lipid exchange between the ER with the endosome, potentially supplying phospholipids to the endosome to ensure tubule formation (Suzuki et al., 2024).
Given the recent advances in the development of better lipid probes and membrane sensors, in combination with advancements in molecular dynamics simulations of more realistic membrane compositions and over longer time scales, it will be intriguing to see which endocytic and secretory proteins might function in the specific delivery of SLs to certain compartments.
Acknowledgments
We thank Krishnan Raghunathan for helpful discussions and corrections during the preparation of this review.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Charles A. King Trust Postdoctoral Research Fellowship and BCH OFD/BTREC/CTREC Faculty Career Development grant (SS), the National Institutes of Health R37 DK048106 (WL) and National Institutes of Health RO1 DK104868 (WL).
Author contributions
VS: Visualization, Writing–original draft, Writing–review and editing. KE: Writing–original draft, Writing–review and editing. WL: Funding acquisition, Supervision, Writing–original draft, Writing–review and editing. SS: Conceptualization, Funding acquisition, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Conflict of interest
WL is scientific founder and board member of Transcera Inc that is seeking to translate discoveries on GSL endosome sorting to clinical practice.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Almeida P. F. F. (2009). Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta 1788, 72–85. 10.1016/j.bbamem.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Anders P. E. B., Mattjus P. (2021). Who moves the sphinx? An overview of intracellular sphingolipid transport. Biochimica Biophysica Acta - Mol. Cell. Biol. Lipids 1866 (11), 1388–1981. 10.1016/j.bbalip.2021.159021 [DOI] [PubMed] [Google Scholar]
- Arumugam S., Schmieder S., Pezeshkian W., Becken U., Wunder C., Chinnapen D., et al. (2021). Ceramide structure dictates glycosphingolipid nanodomain assembly and function. Nat. Commun. 12, 3675. 10.1038/s41467-021-23961-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K., Schwille P., Kurzchalia T. (2005). Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc. Natl. Acad. Sci. U. S. A. 102, 3272–3277. 10.1073/pnas.0408215102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badizadegan K., Dickinson B. L., Wheeler H. E., Blumberg R. S., Holmes R. K., Lencer W. I. (2000). Heterogeneity of detergent-insoluble membranes from human intestine containing caveolin-1 and ganglioside GM1. Am. J. Physiol. Gastrointest. Liver Physiol. 278, G895–G904. 10.1152/ajpgi.2000.278.6.G895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baoukina S., Ingolfsson H. I., Marrink S. J., Tieleman D. P. (2018). Curvature-induced sorting of lipids in plasma membrane tethers. Adv. theory simulations 1. 10.1002/adts.201800034 [DOI] [Google Scholar]
- Baumgart T., Das S., Webb W. W., Jenkins J. T. (2005). Membrane elasticity in giant vesicles with fluid phase coexistence. Biophysical J. 89, 1067–1080. 10.1529/biophysj.104.049692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T., Hess S. T., Webb W. W. (2003). Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824. 10.1038/nature02013 [DOI] [PubMed] [Google Scholar]
- Beltrán-Heredia E., Tsai F.-C., Salinas-Almaguer S., Cao F. J., Bassereau P., Monroy F. (2019). Membrane curvature induces cardiolipin sorting. Commun. Biol. 2, 225–227. 10.1038/s42003-019-0471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J., Antonny B. (2012). Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 23, 886–895. 10.1016/j.devcel.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Black B. A., Sun C., Zhao Y. Y., Gänzle M. G., Curtis J. M. (2013). Antifungal lipids produced by lactobacilli and their structural identification by normal phase LC/atmospheric pressure photoionization–MS/MS. J. Agric. Food Chem. 61, 5338–5346. 10.1021/jf400932g [DOI] [PubMed] [Google Scholar]
- Black J. C., Cheney P. P., Campbell T., Knowles M. K. (2014). Membrane curvature based lipid sorting using a nanoparticle patterned substrate. Soft Matter 10, 2016–2023. 10.1039/C3SM52522H [DOI] [PubMed] [Google Scholar]
- Brown D. A., London E. (1998a). Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164, 103–114. 10.1007/s002329900397 [DOI] [PubMed] [Google Scholar]
- Brown D. A., London E. (1998b). Functions of lipid rafts in biological membranes. Annu. Rev. Cell. Dev. Biol. 14, 111–136. 10.1146/annurev.cellbio.14.1.111 [DOI] [PubMed] [Google Scholar]
- Brügger B., Sandhoff R., Wegehingel S., Gorgas K., Malsam J., Helms J. B., et al. (2000). Evidence for segregation of sphingomyelin and cholesterol during formation of copi-coated vesicles. J. Cell. Biol. 151, 507–518. 10.1083/jcb.151.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan-Jones A., Sorre B., Bassereau P. (2011). Curvature-driven lipid sorting in biomembranes. Cold Spring Harb Perspect Biol. 3 (2), a004648. 10.1101/cshperspect.a004648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebecauer M., Amaro M., Jurkiewicz P., Sarmento M. J., Šachl R., Cwiklik L., et al. (2018). Membrane lipid nanodomains. Chem. Rev. 118 (23), 11259–11297. 10.1021/acs.chemrev.8b00322 [DOI] [PubMed] [Google Scholar]
- Cheney P. P., Weisgerber A. W., Feuerbach A. M., Knowles M. K. (2017). Single lipid molecule dynamics on supported lipid bilayers with membrane curvature. Membranes 7, 15. 10.3390/membranes7010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L. V., Kozlov M. M. (2003). Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72, 175–207. 10.1146/annurev.biochem.72.121801.161504 [DOI] [PubMed] [Google Scholar]
- Chiantia S., Ries J., Kahya N., Schwille P. (2006). Combined AFM and two-focus SFCS study of raft-exhibiting model membranes. ChemPhysChem 7, 2409–2418. 10.1002/cphc.200600464 [DOI] [PubMed] [Google Scholar]
- Chinnapen D.J.-F., Chinnapen H., Saslowsky D., Lencer W. I. (2007). Rafting with cholera toxin: endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol. Lett. 266, 129–137. 10.1111/j.1574-6968.2006.00545.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnapen D. J.-F., Hsieh W.-T., te Welscher Y. M., Saslowsky D. E., Kaoutzani L., Brandsma E., et al. (2012). Lipid sorting by ceramide structure from plasma membrane to ER for the cholera toxin receptor ganglioside GM1. Dev. Cell. 23, 573–586. 10.1016/j.devcel.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke I. R., Deserno M. (2006). Coupling between lipid shape and membrane curvature. Biophysical J. 91, 487–495. 10.1529/biophysj.105.078683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J., Hilpert C., Monticelli L. (2024). “Chapter Eight - predicting lipid sorting in curved membranes,” in Methods in enzymology, biophysical approaches for the study of membrane structure—Part B: theory and simulations. Editors Deserno, M., Baumgart T. (Academic Press; ), 287–307. 10.1016/bs.mie.2024.03.022 [DOI] [PubMed] [Google Scholar]
- Day C. A., Baetz N. W., Copeland C. A., Kraft L. J., Han B., Tiwari A., et al. (2015). Microtubule motors power plasma membrane tubulation in clathrin-independent endocytosis. Traffic 16, 572–590. 10.1111/tra.12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C. A., Kenworthy A. K. (2015). Functions of cholera toxin B-subunit as a raft cross-linker. Essays Biochem. 57, 135–145. 10.1042/bse0570135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Rivera-Molina F. E., Toomre D. K., Burd C. G. (2016). Sphingomyelin is sorted at the trans Golgi network into a distinct class of secretory vesicle. Proc. Natl. Acad. Sci. 113, 6677–6682. 10.1073/pnas.1602875113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rohrer B., Levental K. R., Levental I. (2014). Rafting through traffic: membrane domains in cellular logistics. Biochimica Biophysica Acta (BBA) - Biomembr. 1838, 3003–3013. 10.1016/j.bbamem.2014.07.029 [DOI] [PubMed] [Google Scholar]
- Dick R. A., Goh S. L., Feigenson G. W., Vogt V. M. (2012). HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. U. S. A. 109, 18761–18766. 10.1073/pnas.1209408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C., Bagatolli L. A., Volovyk Z. N., Thompson N. L., Levi M., Jacobson K., et al. (2001). Lipid rafts reconstituted in model membranes. Biophys. J. 80, 1417–1428. 10.1016/S0006-3495(01)76114-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinic J., Riehl A., Adler J., Parmryd I. (2015). The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Sci. Rep. 5, 10082. 10.1038/srep10082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Duclos R. I., Blue K. D., Rufo M. J., Chen X., Guo J. J., Ma X., et al. (2020). Conjugation of peptides to short-acyl-chain ceramides for delivery across mucosal cell barriers. Bioorg Med. Chem. Lett. 30, 127014. 10.1016/j.bmcl.2020.127014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C., Ringemann C., Medda R., Schwarzmann G., Sandhoff K., Polyakova S., et al. (2009). Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162. 10.1038/nature07596 [DOI] [PubMed] [Google Scholar]
- Ewers H., Roemer W., Smith A., Bacia K., Dmitrieff S., Chai W., et al. (2010). GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell. Biol. 12, 11–18. 10.1038/ncb1999 [DOI] [PubMed] [Google Scholar]
- Feigenson G. W. (2009). Phase diagrams and lipid domains in multicomponent lipid bilayer mixtures. Biochim. Biophys. Acta 1788, 47–52. 10.1016/j.bbamem.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field K. A., Holowka D., Baird B. (1995). Fc epsilon RI-mediated recruitment of p53/56lyn to detergent-resistant membrane domains accompanies cellular signaling. Proc. Natl. Acad. Sci. U. S. A. 92, 9201–9205. 10.1073/pnas.92.20.9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence A. T., Nasseri B., Arunothyanun P. (2004). “Does shape matter? Spherical, polyhedral, and tubular vesicles,” in Carrier-based drug delivery (Washington: American Chemical Society; ), 75–84. [Google Scholar]
- Frolov V. A., Shnyrova A. V., Zimmerberg J. (2011). Lipid polymorphisms and membrane shape. Cold Spring Harb. Perspect. Biol. 3 (11), a004747. PMID: 21646378; PMCID: PMC3220359. 10.1101/cshperspect.a004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K., Riezman H. (2001). Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell. Biol. 155, 949–959. 10.1083/jcb.200105033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castillo M. D., Chinnapen D.J.-F., te Welscher Y. M., Gonzalez R. J., Softic S., Pacheco M., et al. (2018). Mucosal absorption of therapeutic peptides by harnessing the endogenous sorting of glycosphingolipids. eLife 7, e34469. 10.7554/eLife.34469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner O. B., Baum L. G. (2008). Galectin-glycan lattices regulate cell-surface glycoprotein organization and signalling. Biochem. Soc. Trans. 36 (Pt 6), 1472–1477. 10.1042/BST0361472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerl M. J., Sampaio J. L., Urban S., Kalvodova L., Verbavatz J.-M., Binnington B., et al. (2012). Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell. Biol. 196, 213–221. 10.1083/jcb.201108175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. (2003). Lipids in endocytic membrane transport and sorting. Curr. Opin. Cell. Biol. 15, 382–388. 10.1016/S0955-0674(03)00078-4 [DOI] [PubMed] [Google Scholar]
- Gupta N., DeFranco A. L. (2007). Lipid rafts and B cell signaling. Seminars Cell. and Dev. Biol. Membr. Lipid Microdomains Roles Signal. Dis. 3D Chromatin 18, 616–626. 10.1016/j.semcdb.2007.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. T., Heberle F. A., Baumgart T., Holowka D., Baird B., Feigenson G. W. (2005). Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 102 (18), 6320–6325. 10.1073/pnas.0405654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., et al. (2003). Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809. 10.1038/nature02188 [DOI] [PubMed] [Google Scholar]
- Hancock J. F. (2006). Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell. Biol. 7 (6), 456–462. PMID: 16625153; PMCID: PMC2782566. 10.1038/nrm1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun Y. A., Obeid L. M. (2018). Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell. Biol. 19, 175–191. 10.1038/nrm.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama T., Riezman H. (2018). Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell. Biol. 19, 281–296. 10.1038/nrm.2017.138 [DOI] [PubMed] [Google Scholar]
- Hatzakis N. S., Bhatia V. K., Larsen J., Madsen K. L., Bolinger P.-Y., Kunding A. H., et al. (2009). How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 5, 835–841. 10.1038/nchembio.213 [DOI] [PubMed] [Google Scholar]
- Heberle F. A., Feigenson G. W. (2011). Phase separation in lipid membranes. Cold Spring Harb. Perspect. Biol. 3, a004630. 10.1101/cshperspect.a004630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. B., Mason R., Thomas K. M., Shipley G. G. (1974). Structural chemistry of 1,2 dilauroyl-DL-phosphatidylethanolamine: molecular conformation and intermolecular packing of phospholipids. Proc. Natl. Acad. Sci. U.S.A. 71 (8), 3036–3040. 10.1073/pnas.71.8.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjort Ipsen J., Karlström G., Mourtisen O. G., Wennerström H., Zuckermann M. J. (1987). Phase equilibria in the phosphatidylcholine-cholesterol system. Biochimica Biophysica Acta (BBA) - Biomembr. 905, 162–172. 10.1016/0005-2736(87)90020-4 [DOI] [PubMed] [Google Scholar]
- Holthuis J. C. M., Menon A. K. (2014). Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57. 10.1038/nature13474 [DOI] [PubMed] [Google Scholar]
- Honigmann A., Mueller V., Ta H., Schoenle A., Sezgin E., Hell S. W., et al. (2014). Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat. Commun. 5, 5412. 10.1038/ncomms6412 [DOI] [PubMed] [Google Scholar]
- Huttner W. B., Zimmerberg J. (2001). Implications of lipid microdomains for membrane curvature, budding and fission: commentary. Curr. Opin. Cell. Biol. 13, 478–484. 10.1016/S0955-0674(00)00239-8 [DOI] [PubMed] [Google Scholar]
- Ikonen E. (2008). Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell. Biol. 9, 125–138. 10.1038/nrm2336 [DOI] [PubMed] [Google Scholar]
- Johannes L. (2017). Shiga toxin—a model for glycolipid-dependent and lectin-driven endocytosis. Toxins (Basel) 9, 340. 10.3390/toxins9110340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L., Jacob R., Leffler H. (2018). Galectins at a glance. J. Cell. Sci. 131 (9), jcs208884. 10.1242/jcs.208884 [DOI] [PubMed] [Google Scholar]
- Johannes L., Parton R. G., Bassereau P., Mayor S. (2015). Building endocytic pits without clathrin. Nat. Rev. Mol. Cell. Biol. 16 (5), 311–321. Epub 2015 Apr 10. PMID: 25857812. 10.1038/nrm3968 [DOI] [PubMed] [Google Scholar]
- Jülicher F., Lipowsky R. (1996). Shape transformations of vesicles with intramembrane domains. Phys. Rev. E 53, 2670–2683. 10.1103/PhysRevE.53.2670 [DOI] [PubMed] [Google Scholar]
- Kabbani A. M., Raghunathan K., Lencer W. I., Kenworthy A. K., Kelly C. V. (2020). Structured clustering of the glycosphingolipid GM1 is required for membrane curvature induced by cholera toxin. Proc. Natl. Acad. Sci. U.S.A. 117 (26), 14978–14986. 10.1073/pnas.2001119117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal M. M., Mills D., Grzybek M., Howard J. (2009). Measurement of the membrane curvature preference of phospholipids reveals only weak coupling between lipid shape and leaflet curvature. Proc. Natl. Acad. Sci. 106, 22245–22250. 10.1073/pnas.0907354106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy A. K., Edidin M. (1998). Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J. Cell. Biol. 142, 69–84. 10.1083/jcb.142.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaddaj R., Kukulski W. (2023). Piecing together the structural organisation of lipid exchange at membrane contact sites. Curr. Opin. Cell. Biol. 83, 102212. 10.1016/j.ceb.2023.102212 [DOI] [PubMed] [Google Scholar]
- Kim H., Budin I. (2024). Intracellular sphingolipid sorting drives membrane phase separation in the yeast vacuole. J. Biol. Chem. 300 (1), 105496. 10.1016/j.jbc.2023.105496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Singh A., Del Poeta M., Brown D. A., London E. (2017). The effect of sterol structure upon clathrin-mediated and clathrin-independent endocytosis. J. Cell. Sci. 130, 2682–2695. 10.1242/jcs.201731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Burd C. G. (2023). Lipid sorting and organelle identity. Cold Spring Harb. Perspect. Biol. 15, a041397. 10.1101/cshperspect.a041397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C., Sengupta P., Seo A. Y., Lippincott-Schwartz J. (2020). ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. 117, 7225–7235. 10.1073/pnas.1910854117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Suzuki K. G. N., Matsumori N., Takada M., Ano H., Morigaki K., et al. (2017). Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. J. Cell. Biol. 216, 1183–1204. 10.1083/jcb.201607086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R. W., Ejsing C. S., Surma M. A., Kaiser H.-J., Gerl M. J., Sampaio J. L., et al. (2009). Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell. Biol. 185, 601–612. 10.1083/jcb.200901145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig M., De Vries R., Grünewald F., Marrink S. J., Pezeshkian W. (2023). Curvature-induced lipid sorting beyond the critical packing parameter. 10.1101/2023.12.15.571845 [DOI] [Google Scholar]
- Komura N., Suzuki K. G. N., Ando H., Konishi M., Koikeda M., Imamura A., et al. (2016). Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat. Chem. Biol. 12, 402–410. 10.1038/nchembio.2059 [DOI] [PubMed] [Google Scholar]
- Koval M., Pagano R. E. (1991). Intracellular transport and metabolism of sphingomyelin. Biochimica Biophysica Acta (BBA) - Lipids Lipid Metabolism 1082, 113–125. 10.1016/0005-2760(91)90184-J [DOI] [PubMed] [Google Scholar]
- Koyama-Honda I., Fujiwara T. K., Kasai R. S., Suzuki K. G. N., Kajikawa E., Tsuboi H., et al. (2020). High-speed single-molecule imaging reveals signal transduction by induced transbilayer raft phases. J. Cell. Biol. 219 (12), e202006125. PMID: 33053147; PMCID: PMC7563750. 10.1083/jcb.202006125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft M. L. (2016). Sphingolipid organization in the plasma membrane and the mechanisms that influence it. Front. Cell. Dev. Biol. 4, 154. 10.3389/fcell.2016.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Koyama-Honda I., Suzuki K. (2004). Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic 5 (4), 213–230. PMID: 15030563. 10.1111/j.1600-0854.2004.0178.x [DOI] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., et al. (2005a). High-Speed single-molecule tracking of membrane molecules. [DOI] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., et al. (2005b). Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophysics 34, 351–378. 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Suzuki K., Koyasako K. (1999). Mobility and cytoskeletal interactions of cell adhesion receptors. Curr. Opin. Cell. Biol. 11 (5), 582–590. PMID: 10508652. 10.1016/s0955-0674(99)00020-4 [DOI] [PubMed] [Google Scholar]
- Lagerholm B. C., Weinreb G. E., Jacobson K., Thompson N. L. (2005). Detecting microdomains in intact cell membranes. Annu. Rev. Phys. Chem. 56, 309–336. 10.1146/annurev.physchem.56.092503.141211 [DOI] [PubMed] [Google Scholar]
- Leveille C. L., Cornell C. E., Merz A. J., Keller S. L. (2022). Yeast cells actively tune their membranes to phase separate at temperatures that scale with growth temperatures. Proc. Natl. Acad. Sci. U. S. A. 119 (4), e2116007119. PMID: 35046036; PMCID: PMC8795566. 10.1073/pnas.2116007119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Levental K. R., Heberle F. A. (2020). Lipid rafts: controversies resolved, mysteries remain. Trends Cell. Biol. 30, 341–353. 10.1016/j.tcb.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Lipowsky R. (1993). Domain-induced budding of fluid membranes. Biophysical J. 64, 1133–1138. 10.1016/S0006-3495(93)81479-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. P., Fletcher D. A. (2006). Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys. J. 91, 4064–4070. 10.1529/biophysj.106.090852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F. (2008). Molecular cell biology. Macmillan. [Google Scholar]
- Lorent J. H., Diaz-Rohrer B., Lin X., Spring K., Gorfe A. A., Levental K. R., et al. (2017). Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 8, 1219. 10.1038/s41467-017-01328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzati V., Reiss-Husson F., Rivas E., Gulik-Krzywicki T. (1966). Structure and polymorphism in lipid-water systems, and their possible biological implications. Ann. N. Y. Acad. Sci. 137, 409–413. 10.1111/j.1749-6632.1966.tb50172.x [DOI] [PubMed] [Google Scholar]
- Luzzati V., Tardieu A., Gulik-Krzywicki T. (1968). Polymorphism of lipids. Nature 217, 1028–1030. 10.1038/2171028a0 [DOI] [PubMed] [Google Scholar]
- Maggio B., Cumar F. A., Caputto R. (1978). Interactions of gangliosides with phospholipids and glycosphingolipids in mixed monolayers. Biochem. J. 175, 1113–1118. 10.1042/bj1751113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manneville J.-B., Casella J.-F., Ambroggio E., Gounon P., Bertherat J., Bassereau P., et al. (2008). COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc. Natl. Acad. Sci. 105, 16946–16951. 10.1073/pnas.0807102105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Seara H., Róg T., Karttunen M., Vattulainen I., Reigada R. (2010). Cholesterol induces specific spatial and orientational order in cholesterol/phospholipid membranes. PLoS One 5, e11162. 10.1371/journal.pone.0011162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F. R., McGraw T. E. (2004). Endocytic recycling. Nat. Rev. Mol. Cell. Biol. 5, 121–132. 10.1038/nrm1315 [DOI] [PubMed] [Google Scholar]
- Mayor S., Presley J. F., Maxfield F. R. (1993). Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J. Cell. Biol. 121, 1257–1269. 10.1083/jcb.121.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh T. J., Simon S. A. (1986). Area per molecule and distribution of water in fully hydrated dilauroylphosphatidylethanolamine bilayers. Biochemistry 25 (17), 4948–4952. 10.1021/bi00365a034 [DOI] [PubMed] [Google Scholar]
- Meer G., Sprong H. (2004). Membrane lipids and vesicular traffic. Curr. Opin. Cell. Biol. 16, 373–378. 10.1016/j.ceb.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Melia T. J., Reinisch K. M. (2022). A possible role for VPS13-family proteins in bulk lipid transfer, membrane expansion and organelle biogenesis. J. Cell. Sci. 135, jcs259357. 10.1242/jcs.259357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill A. H. (2011). Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111, 6387–6422. 10.1021/cr2002917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller C. H., Mudd J. B., Thomson W. W. (1981). Lipid phase separations and intramembranous particle movements in the yeast tonoplast. Biochim. Biophys. Acta 643 (2), 376–386. PMID: 7013807. 10.1016/0005-2736(81)90082-1 [DOI] [PubMed] [Google Scholar]
- Moeller C. H., Thomson W. W. (1979). An ultrastructural study of the yeast tomoplast during the shift from exponential to stationary phase. J. Ultrastruct. Res. 68 (Issue 1), 28–37. ISSN 0022-5320. 10.1016/S0022-5320(79)90139-4 [DOI] [PubMed] [Google Scholar]
- Mueller V., Ringemann C., Honigmann A., Schwarzmann G., Medda R., Leutenegger M., et al. (2011). STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys. J. 101, 1651–1660. 10.1016/j.bpj.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Maxfield F. R. (2000). Role of membrane organization and membrane domains in endocytic lipid trafficking. Traffic 1, 203–211. 10.1034/j.1600-0854.2000.010302.x [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Soe T. T., Maxfield F. R. (1999). Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J. Cell. Biol. 144, 1271–1284. 10.1083/jcb.144.6.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. (1995). An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14, 4695–4704. 10.1002/j.1460-2075.1995.tb00151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. J. (2003). GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 13, 686–690. 10.1016/s0960-9822(03)00209-4 [DOI] [PubMed] [Google Scholar]
- Ogunyankin M. O., Huber D. L., Sasaki D. Y., Longo M. L. (2013). Nanoscale patterning of membrane-bound proteins formed through curvature-induced partitioning of phase-specific receptor lipids. Langmuir 29, 6109–6115. 10.1021/la401011d [DOI] [PubMed] [Google Scholar]
- Ogunyankin M. O., Longo M. L. (2013). Metastability in pixelation patterns of coexisting fluid lipid bilayer phases imposed by e-beam patterned substrates. Soft Matter 9, 2037–2046. 10.1039/C2SM27027G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich K., Rawicz W., Needham D., Evans E. (2000). Water permeability and mechanical strength of polyunsaturated lipid bilayers. Biophys. J. 79 (1), 321–327. 10.1016/S0006-3495(00)76294-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Montesano R., Meda P., Malaisse-Lagae F., Brown D., Perrelet A., et al. (1981). Heterogeneous distribution of filipin--cholesterol complexes across the cisternae of the Golgi apparatus. Proc. Natl. Acad. Sci. U. S. A. 78, 293–297. 10.1073/pnas.78.1.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R., Yu C., Groves J. T. (2006). Curvature-Modulated phase separation in lipid bilayer membranes. Langmuir 22, 5095–5099. 10.1021/la060390o [DOI] [PubMed] [Google Scholar]
- Pekkinen M., Terhal P. A., Botto L. D., Henning P., Mäkitie R. E., Roschger P., et al. (2019). Osteoporosis and skeletal dysplasia caused by pathogenic variants in SGMS2 . JCI Insight 4, e126180. 10.1172/jci.insight.126180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike L. J. (2006). Rafts defined: a report on the Keystone symposium on lipid rafts and cell function. J. Lipid Res. 47, 1597–1598. 10.1194/jlr.E600002-JLR200 [DOI] [PubMed] [Google Scholar]
- Pinot M., Vanni S., Pagnotta S., Lacas-Gervais S., Payet L.-A., Ferreira T., et al. (2014). Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science 345, 693–697. 10.1126/science.1255288 [DOI] [PubMed] [Google Scholar]
- Putta P., Rankenberg J., Korver R. A., van Wijk R., Munnik T., Testerink C., et al. (2016). Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochimica Biophysica Acta (BBA) - Biomembr. 1858, 2709–2716. 10.1016/j.bbamem.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Quiroga R., Trenchi A., González Montoro A., Valdez Taubas J., Maccioni H. J. F. (2013). Short transmembrane domains with high-volume exoplasmic halves determine retention of Type II membrane proteins in the Golgi complex. J. Cell. Sci. 126, 5344–5349. 10.1242/jcs.130658 [DOI] [PubMed] [Google Scholar]
- Raghunathan K., Wong T. H., Chinnapen D. J., Lencer W. I., Jobling M. G., Kenworthy A. K. (2016). Glycolipid crosslinking is required for cholera toxin to partition into and stabilize ordered domains. Biophysical J. 111 (12), 2547–2550. 10.1016/j.bpj.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy R., Anilkumar A. A., Polley A., Singh P. P., Yadav M., Johnson C., et al. (2015). Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 161, 581–594. 10.1016/j.cell.2015.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawicz W., Olbrich K. C., McIntosh T., Needham D., Evans E. (2000). Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophysical J. 79 (1), 328–339. 10.1016/S0006-3495(00)76295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., et al. (2007). Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675. 10.1038/nature05996 [DOI] [PubMed] [Google Scholar]
- Römer C. E., Elling L. (2011). Galectins: structures, binding properties and function in cell adhesion, in: biomaterials - physics and chemistry. IntechOpen. 10.5772/24647 [DOI] [Google Scholar]
- Róg T., Pasenkiewicz-Gierula M., Vattulainen I., Karttunen M. (2007). What happens if cholesterol is made smoother: importance of methyl substituents in cholesterol ring structure on phosphatidylcholine-sterol interaction. Biophys. J. 92, 3346–3357. 10.1529/biophysj.106.095497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A., Cuvelier D., Nassoy P., Prost J., Bassereau P., Goud B. (2005). Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 24, 1537–1545. 10.1038/sj.emboj.7600631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydell G. E., Svensson L., Larson G., Johannes L., Roemer W. (2013). Human GII.4 norovirus VLP induces membrane invaginations on giant unilamellar vesicles containing secretor gene dependent α1,2-fucosylated glycosphingolipids. Biochimica Biophysica Acta (BBA) - Biomembr. 1828 (8), 1840–1845. ISSN 0005-2736. 10.1016/j.bbamem.2013.03.016 [DOI] [PubMed] [Google Scholar]
- Safouane M., Berland L., Callan-Jones A., Sorre B., Römer W., Johannes L., et al. (2010). Lipid cosorting mediated by Shiga toxin induced tubulation. Traffic 11, 1519–1529. 10.1111/j.1600-0854.2010.01116.x [DOI] [PubMed] [Google Scholar]
- Saha S., Anilkumar A. A., Mayor S. (2016). GPI-anchored protein organization and dynamics at the cell surface. J. Lipid Res. 57, 159–175. 10.1194/jlr.R062885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha D., Hamdo H. H., Wilde M., Prause K., Arenz C. (2019). Sphingolipid-transporting proteins as cancer therapeutic targets. Int. J. Mol. Sci. 20 (14), 3554. 10.3390/ijms20143554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder S. S., Tatituri R., Anderson M., Kelly K., Lencer W. I. (2022). Structural basis for acyl chain control over glycosphingolipid sorting and vesicular trafficking. Cell. Rep. 40, 111063. 10.1016/j.celrep.2022.111063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Simons K. (2004). Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell. Sci. 117, 5955–5964. 10.1242/jcs.01596 [DOI] [PubMed] [Google Scholar]
- Sezgin E., Kaiser H.-J., Baumgart T., Schwille P., Simons K., Levental I. (2012). Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051. 10.1038/nprot.2012.059 [DOI] [PubMed] [Google Scholar]
- Sezgin E., Levental I., Mayor S., Eggeling C. (2017). The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell. Biol. 18, 361–374. 10.1038/nrm.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Varma R., Sarasij R. C., Ira N., Gousset K., Krishnamoorthy G., et al. (2004). Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 116, 577–589. 10.1016/s0092-8674(04)00167-9 [DOI] [PubMed] [Google Scholar]
- Sharpe L. J., Luu W., Brown A. J. (2011). Akt phosphorylates Sec24: new clues into the regulation of ER-to-golgi trafficking. Traffic 12, 19–27. 10.1111/j.1600-0854.2010.01133.x [DOI] [PubMed] [Google Scholar]
- Shvets E., Bitsikas V., Howard G., Hansen C. G., Nichols B. J. (2015). Dynamic caveolae exclude bulk membrane proteins and are required for sorting of excess glycosphingolipids. Nat. Commun. 6, 6867. 10.1038/ncomms7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Ikonen E. (1997). Functional rafts in cell membranes. Nature 387, 569–572. 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- Simons K., Sampaio J. L. (2011). Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697. 10.1101/cshperspect.a004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Toomre D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 1, 31–39. 10.1038/35036052 [DOI] [PubMed] [Google Scholar]
- Simons K., Vaz W. L. C. (2004). Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33, 269–295. 10.1146/annurev.biophys.32.110601.141803 [DOI] [PubMed] [Google Scholar]
- Simunovic M., Manneville J. B., Renard H. F., Evergren E., Raghunathan K., Bhatia D., et al. (2017). Friction mediates scission of tubular membranes scaffolded by BAR proteins. Cell. 170 (1), 172–184.e11. 10.1016/j.cell.2017.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte J. P. (2013). Biological functions of sphingomyelins. Prog. Lipid Res. 52, 424–437. 10.1016/j.plipres.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Smart E. J., Ying Y., Donzell W. C., Anderson R. G. W. (1996). A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J. Biol. Chem. 271, 29427–29435. 10.1074/jbc.271.46.29427 [DOI] [PubMed] [Google Scholar]
- Sokoya T., Parolek J., Foged M. M., Danylchuk D. I., Bozan M., Sarkar B., et al. (2022). Pathogenic variants of sphingomyelin synthase SMS2 disrupt lipid landscapes in the secretory pathway. eLife 11, e79278. 10.7554/eLife.79278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B., Callan-Jones A., Manneville J.-B., Nassoy P., Joanny J.-F., Prost J., et al. (2009). Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc. Natl. Acad. Sci. 106, 5622–5626. 10.1073/pnas.0811243106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stier A., Sackmann E. (1973). Spin labels as enzyme substrates Heterogeneous lipid distribution in liver microsomal membranes. Biochimica Biophysica Acta (BBA) - Biomembr. 311, 400–408. 10.1016/0005-2736(73)90320-9 [DOI] [PubMed] [Google Scholar]
- Stone M. B., Shelby S. A., Núñez M. F., Wisser K., Veatch S. L. (2017). Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife 6, e19891. 10.7554/eLife.19891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S. W., West M., Zhang Y., Fan J. S., Roberts R. T., Odorizzi G., et al. (2024). A role for Vps13-mediated lipid transfer at the ER–endosome contact site in ESCRT-mediated sorting. J. Cell. Biol. 223, e202307094. 10.1083/jcb.202307094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk O. M., González-Segredo N., Kukura P., Oppenheim A., Choquet D., Sandoghdar V., et al. (2013). Receptor concentration and diffusivity control multivalent binding of Sv40 to membrane bilayers. PLOS Comput. Biol. 9 (11), e1003310. 10.1371/journal.pcbi.1003310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher Y. M., Chinnapen D.J.-F., Kaoutzani L., Mrsny R. J., Lencer W. I. (2014). Unsaturated glycoceramides as molecular carriers for mucosal drug delivery of GLP-1. J. Control. Release 175, 72–78. 10.1016/j.jconrel.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A., Baumgart T. (2009). Sorting of lipids and proteins in membrane curvature gradients. Biophysical J. 96, 2676–2688. 10.1016/j.bpj.2008.11.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., Prinz W. A. (2013). Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell. Biol. 202 (1), 35–44. 10.1083/jcb.201301039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.-C., Guérin G., Pernier J., Bassereau P. (2024). Actin-membrane linkers: insights from synthetic reconstituted systems. Eur. J. Cell. Biol. 103, 151402. 10.1016/j.ejcb.2024.151402 [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D. R., Feigenson G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 9, 112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni S., Hirose H., Barelli H., Antonny B., Gautier R. (2014). A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat. Commun. 5, 4916. 10.1038/ncomms5916 [DOI] [PubMed] [Google Scholar]
- Varma R., Mayor S. (1998). GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801. 10.1038/29563 [DOI] [PubMed] [Google Scholar]
- Veatch S. L., Keller S. L. (2003). Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophysical J. 85, 3074–3083. 10.1016/S0006-3495(03)74726-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch S. L., Keller S. L. (2005). Seeing spots: complex phase behavior in simple membranes. Biochimica Biophysica Acta (BBA) - Mol. Cell. Res. Lipid Rafts Model. Membr. Cells 1746, 172–185. 10.1016/j.bbamcr.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Wakana Y., Hayashi K., Nemoto T., Watanabe C., Taoka M., Angulo-Capel J., et al. (2020). The ER cholesterol sensor SCAP promotes CARTS biogenesis at ER–Golgi membrane contact sites. J. Cell. Biol. 220, e202002150. 10.1083/jcb.202002150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., et al. (2008). SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell. Res. 18, 290–301. 10.1038/cr.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins E., Majewski J., Chi E. Y., Gao H., Florent J.-C., Johannes L. (2019). Shiga toxin induces lipid compression: a mechanism for generating membrane curvature. Nano Lett. 19 (10), 7365–7369. 10.1021/acs.nanolett.9b03001 [DOI] [PubMed] [Google Scholar]
- Wesołowska O., Michalak K., Maniewska J., Hendrich A. B. (2009). Giant unilamellar vesicles - a perfect tool to visualize phase separation and lipid rafts in model systems. Acta Biochim. Pol. 56, 33–39. 10.18388/abp.2009_2514 [DOI] [PubMed] [Google Scholar]
- Wong L. H., Gatta A. T., Levine T. P. (2019). Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell. Biol. 20, 85–101. 10.1038/s41580-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Woodward X., Javanainen M., Fábián B., Kelly C. V. (2023). Nanoscale membrane curvature sorts lipid phases and alters lipid diffusion. Biophysical J. 122, 2203–2215. 10.1016/j.bpj.2023.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager A. N., Weber P. K., Kraft M. L. (2016). Three-dimensional imaging of cholesterol and sphingolipids within a Madin-Darby canine kidney cell. Biointerphases 11, 02A309. 10.1116/1.4939681 [DOI] [PubMed] [Google Scholar]
- Young W. W., Lutz M. S., Blackburn W. A. (1992). Endogenous glycosphingolipids move to the cell surface at a rate consistent with bulk flow estimates. J. Biol. Chem. 267, 12011–12015. 10.1016/S0021-9258(19)49798-6 [DOI] [PubMed] [Google Scholar]
- Zhukovsky M. A., Filograna A., Luini A., Corda D., Valente C. (2019). Phosphatidic acid in membrane rearrangements. FEBS Lett. 593, 2428–2451. 10.1002/1873-3468.13563 [DOI] [PubMed] [Google Scholar]
- Zimmerberg J., Kozlov M. M. (2006). How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell. Biol. 7, 9–19. 10.1038/nrm1784 [DOI] [PubMed] [Google Scholar]





