Abstract
Advances in structure determination of the bacterial and eukaryotic transcription machinery have led to a dramatic increase in understanding the mechanism of transcription. Models for the specific assembly of the RNA Polymerase II transcription machinery at a promoter, conformational changes that occur during initiation of transcription, and the mechanism of initiation are discussed in light of recent developments.
Regulation of transcription, the synthesis of RNA from a DNA template, is one of the most important steps in control of cell growth and differentiation. Transcription is carried out by the enzyme RNA polymerase (Pol) along with other factors, termed general transcription factors. The general factors are involved in recognition of promoter sequences, the response to regulatory factors, and conformational changes essential to the activity of Pol during the transcription cycle1,2. Advances made over the past 11 years3–5 have revealed the structures of bacterial and eukaryotic Pols, several of the key general transcription factors, and most recently, structures and models for Pol II interacting with general transcription factors6–8. Combined with biochemical and genetic studies, these structures provide emerging views on the mechanism of the transcription machinery, the dynamic nature of protein-protein and protein-DNA interactions involved, and the mechanism of transcriptional regulation.
While the transcription machinery of eukaryotes is much more complex than that of prokaryotes or archaea, the general principals of transcription and its regulation are conserved. Bacteria and archaea have only one Pol, while eukaryotes utilize three nuclear enzymes, Pol I, II, and III, to synthesize different classes of RNA. The nuclear Pols share five common subunits, with the remainder showing strong similarity among the eukaryotic and archaeal enzymes2,9. Although these enzymes have many more subunits than bacterial Pol, subunits that comprise most of Pol II are homologous to subunits from all cellular Pols, implying that all these enzymes have the same basic structure and mechanism10. In bacteria, the sigma subunit is the sole general transcription factor-like polypeptide. Sigma recognizes promoter sequences, promotes conformational changes in the Pol-DNA complex upon initiation, and interacts directly with some transcription activators. In eukaryotes, sigma factor function has been replaced by a much larger set of polypeptides, with each of the three forms of Pol having their own set of associated general transcription factors2,11,12. The Pol II transcription machinery is the most complex, with a total of nearly 60 polypeptides (Table 1), only a few of which are required for transcription by the other nuclear Pols. In contrast, archaea utilize a simplified version of a Pol II/Pol III-like system, relying on only two essential general, factors, TBP (TATA binding protein), and TFB (related to the Pol II and Pol III general factors TFIIB and Brf1)9.
Table 1.
S. cerevisiae RNA Pol II general transcription factors and coactivators
| Factor | #Subunits | Function | ||
|---|---|---|---|---|
| TFIIA | 2 | Stabilizes TBP and TFIID-DNA binding. Blocks transcription inhibitors. Positive and negative gene regulation. | ||
| TFIIB | 1 | Binds TBP, Pol II, and promoter DNA. Helps fix transcription start site | ||
| TFIID | TBP | 1 | Binds TATA element and deforms promoter DNA. Platform for assembly of TFIIB, TFIIA and TAFs | |
| TAFs | 14 | Binds INR and DPE promoter elements. Target of regulatory factors. | ||
| Mediator | 24 | Binds cooperatively with Pol II. Kinase and acetyltransferase activity. Stimulates basal and activated transcription. Target of regulatory factors. | ||
| TFIIFa | 3 | Binds Pol II and is involved in Pol II recruitment to PIC and in Open Complex formation | ||
| TFIIE | 2 | Binds promoter near transcription start. May help open or stabilize the transcription bubble in the Open Complex. | ||
| TFIIH | 10 | Functions in transcription and DNA repair. Kinase and two helicase activities. Essential for Open Complex formation. Mutations in IIH can cause human disease. | ||
| SAGAb | TAFs | 5 | Function unknown | |
| Spts, Adas, Sgfs | 9 | SAGA structure. Interact with TBP, TFIIA, and Gcn5. | ||
| Gcn5 | 1 | Histone acetyltransferase. | ||
| Tra1 | 1 | Essential subunit, Activator target. Third largest yeast protein. | ||
| Ubp8 | 1 | Ubiquitin protease. |
Yeast has 1 extra non essential subunit compared to other organisms studied
Yeast also contain SLIK/SALSA, a closely related complex.
The RNA Pol II transcription cycle
Pol II transcription typically begins with the binding of gene-specific regulatory factors near the site of transcription initiation. These factors can act indirectly on the transcription machinery by recruiting factors that modify chromatin structure, or directly by interacting with components of the transcription machinery. In the simplest form of gene activation, both the direct and indirect mechanisms result in recruitment of the transcription machinery to a core promoter (the minimal DNA sequence needed to specify non-regulated or basal transcription; Fig. 1)13,14. The core promoter serves to position Pol II in a state termed the Preinitiation Complex (PIC) analogous to the bacterial Closed Complex. In this state, Pol II and the general factors are all bound to the promoter but are not in an active conformation to begin transcription. Next, a dramatic conformational change occurs in which 11–15 base pairs of DNA surrounding the transcription start site are melted and the template strand of the promoter is positioned within the active site cleft of Pol to form the Open Complex15. Initiation of transcription begins with synthesis of the first phosphodiester bond of RNA. In many systems, multiple short RNAs (3–10 bases), termed abortive products, are synthesized before Pol productively initiates synthesis of full length RNAs16,17. After synthesis of about 30 bases of RNA, Pol is thought to release its contacts with the core promoter and the rest of the transcription machinery and enter the stage of transcription elongation. Factors that promote productive RNA chain synthesis, RNA processing, RNA export, and chromatin modification can all be recruited to elongating Pol II18. After initiation of transcription by Pol II in vitro, many of the general transcription factors remain behind at the promoter in the Scaffold Complex19. This complex presumably marks genes that have been transcribed and enables the typically slow step of recruitment to be bypassed in subsequent rounds of transcription. Certain transcription activation domains can stabilize this Scaffold-promoter complex in vitro. The Scaffold Complex can then rapidly recruit the remaining general factors to promote transcription reinitiation.
Figure 1.
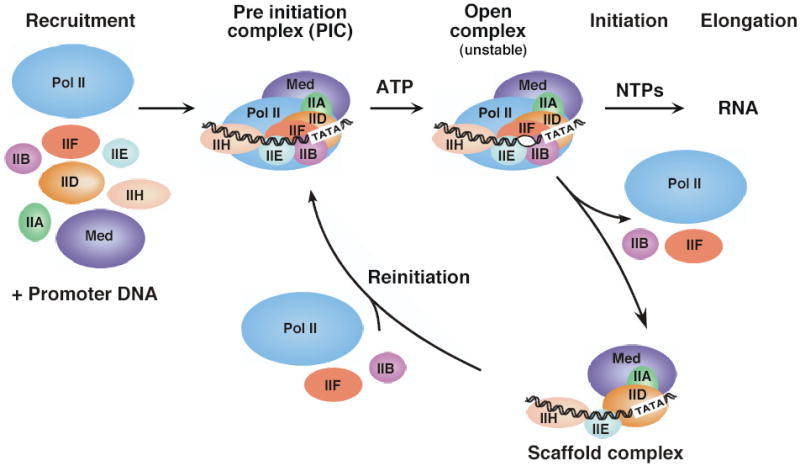
The pathway of transcription initiation and reinitiation for RNA Pol II.
Promoter recognition using multiple low affinity interactions
Recognition of the core promoter by the transcription machinery is essential for correct positioning and assembly of Pol II and the general factors. Sequence elements found in core promoters include the TATA element (TBP binding site), BRE (TFIIB recognition element), Inr (initiator element), and DPE (downstream promoter element)20. Most promoters contain one or more of these elements, but there is no one element that is absolutely essential for promoter function. The promoter elements are binding sites for subunits of the transcription machinery and serve to orient the transcription machinery at the promoter asymmetrically to direct unidirectional transcription.
The core domain of TBP consists of two imperfect repeats forming a saddle shaped molecule that binds the widened minor groove of an 8-bp TATA element, unwinding about a third of a helical turn and bending the DNA about 80 degrees toward the major groove21,22 ( Fig. 2a). At TATA containing promoters, formation of this protein-DNA complex is the initial step in assembly of the transcription machinery. While the TBP molecule is symmetrically shaped, the protein surface of the two repeats is very divergent, forming a large asymmetric protein-DNA interface, creating a platform for binding other components of the transcription machinery. Biochemical studies elegantly showed that TBP does not bind to TATA elements with high orientation specificity23, leading to the finding that other promoter elements in combination with TATA determine the orientation of transcription machinery assembly at a promoter. The BRE element was first recognized as a sequence contributing to high affinity binding of TFIIB and TFB to the human and archaeal TBP-DNA complex24,25. In archaea, where the DNA binding surface of the two TBP imperfect repeats is more symmetrical compared to eukaryotic TBPs, the BRE is the primary determinant of transcription orientation26,27.
Figure 2.
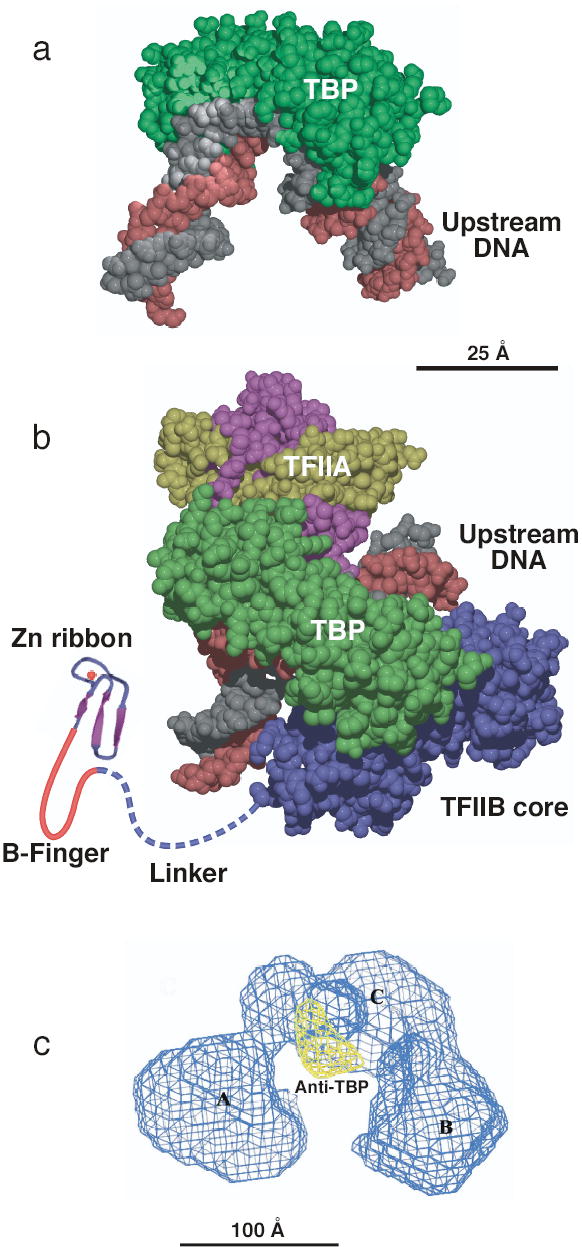
General transcription factor structures. (a) Structure of TBP (green) bound to TATA-DNA with B-form DNA (grey and red) modeled upstream and downstream of the TATA box21,22. (b) Structure model of the TBP-TFIIA-TFIIB-DNA complex. TBP (green) is shown from the top binding to the TFIIB core domain (TFIIBc, blue) and TFIIA (large subunit magenta, small subunit yellow). The zinc ribbon domain (shown as β strands with red Zn atom) connects to the B-finger domain is normally located in the PIC within the Pol II active site and is connected to the TFIIBc domain through a flexible linker. This model is a composite of the DNA-TBP-TFIIBc, DNA-TBP-TFIIA, and the TFIIB Zn ribbon NMR and crystal structures 8,41–43,50,125. (c) EM structure of human TFIID with density due to anti-TBP antibody shown in yellow and the three TFIID lobes labeled A, B, and C57. Figure 2c provided by E. Nogales.
The other two core promoter elements with proven function, Inr and DPE likely serve as binding sites for the TAF (TBP Associated Factor) subunits of the general factor TFIID. A combination of two TAFs (TAF1, 2) was found to specifically bind the Inr, and selection for an optimal TAF1-2 binding sequence led to identification of a sequence closely resembling the Inr element28. Additionally, UV crosslinking has shown that TAF1 and TAF2 are normally positioned close to the Inr29 and that TAF6 and TAF9 lie close to the DPE30. Proper function of a DPE containing promoter requires an Inr element, probably because these elements cooperatively promote the correct binding of TFIID30. In summary, specific binding of the transcription machinery at the core promoter derives from cooperative binding of two or more general transcription factor subunits to degenerate, low specificity promoter elements. The combination of these elements varies between promoters and, in some cases, the core promoter elements used determine activator and enhancer specificity31.
The role of TBP at TATA-less promoters.
It was initially believed that most Pol II promoters contained a TATA element, however, subsequent sequence analysis showed that only about 30% of mRNA genes analyzed in Drosophila contain a recognizable TATA32. Although TBP can recognize divergent AT-rich sequences because of its DNA binding mechanism (see below), there are a number of promoters which clearly do not have a TATA-like sequence about 30 bp upstream of the transcription start site that would be compatible with specific binding by TBP33. In Drosophila and human promoters, many of these non-TATA containing promoters contain some combination of Inr and DPE elements20.
Must TBP bind DNA in order to function? At promoters with a functional TATA, mutation of TATA away from the consensus severely decreases transcription34. From biochemical studies of the yeast HIS4 promoter, mutation of the TATA to a GC-rich sequence allowed recruitment of the transcription machinery to a promoter at a reduced level, but transcription initiation was completely abolished35. These results demonstrate that at one class of promoter, assembly of the transcription machinery into a productive complex requires that TBP bind the TATA element as seen in the crystal structure. Although TBP has tremendous flexibility in the ability to bind variants of the TATA sequence, not all sequences are compatible with TBP binding. For example, C or G in certain positions within TATA is incompatible with the DNA binding surface of TBP33. Since a large number of human and Drosophila promoters have no recognizable TATA element or even AT-rich regions upstream from the transcription start32, this suggests that if TBP interacts with DNA at these promoters, it must do so by a different mechanism from that seen at classical TATA elements. In support of this model, a mutation at the TBP binding surface that abolished detectable binding to a TATA element in vitro blocked transcription from a TATA-containing promoter but not from an Inr-containing promoter36. At promoters lacking TATA, TBP may nucleate protein-protein interactions among the general transcription factors and interact non-specifically with DNA, while DNA bending is facilitated by interaction of other factors such as TAFs with Inr and DPE elements.
Although there is only one gene encoding TBP in yeast and most archaea, higher eukaryotes have one or two copies of genes encoding TBP Related Factors (TRFs) in addition to TBP37,38. It is well established that TRFs promote transcription from a subset of Pol II genes in a cell-type specific fashion. Trf1, unique to insect cells, binds a TC-rich sequence rather than a TATA element and promotes transcription from a small subset of Pol II promoters as well as all Drosophila Pol III transcription39,40. Trf2, conserved between Drosophila, mouse, and human also directs transcription from a subset of promoters. Like TBP, Trf1 and Trf2 are both components of multi subunit complexes, although the identity of most of the Trf associated factors is not yet known.
General factors interacting directly with TBP
TFIIA and TFIIB are the two general factors that interact specifically and independently with TBP. The x-ray structure of these factors bound to the TBP-DNA complex has shown that both TFIIA and TFIIB recognize TBP as well as the DNA distorted by TBP binding41–43 ( Fig. 2b). Both factors recognize the DNA backbone and, as mentioned above, TFIIB can also make base specific contacts with the BRE. TFIIA is a heterodimer composed of two domains, the C-terminal domain contacting TBP-DNA and the N-terminal domain pointing directly away from TBP. TFIIA stabilizes TBP-DNA binding44 and strongly promotes binding of TFIID to DNA through an anti repression mechanism by competing with the TAF1 N-terminal domain (TAND) that occludes the DNA binding surface of TBP when TFIID is not bound to DNA45–47. This effect is particularly striking using human TFIID and certain transcription activators, where a dramatic change in DNA binding activity of TFIID is observed in the presence of TFIIA and activator48. TFIIA can also compete with the negative regulatory factors Mot1 and NC2 to promote TBP binding in vitro 2.
TFIIB contains two domains conserved in the Pol III and archaea factors Brf1 and TFB: an N-terminal zinc ribbon domain (ribbon) connected by a flexible linker to the C-terminal core domain (TFIIBc) that binds TBP-DNA ( Fig. 2b). Both the ribbon and core domains bind cooperatively to RNA Pol II; neither isolated TFIIB domain detectably interacts with Pol II6,49. The functional surface of the ribbon domain has been conserved in TFIIB, Brf1, and TFB and is essential for recruitment of Pol II to the PIC50. For RNA Pol III, the Brf1 ribbon domain is required for normal formation of the Open Complex, a function likely conserved in the Pol II system (see below). The linker connecting the ribbon and core domains contains a short conserved block of sequence that forms a loop termed the B-finger, which is positioned within the active site of Pol II8 where it functions in determining the transcription start site (see below).
TAFS are shared subunits of two large complexes
TFIID is a complex composed of TBP and about 14 TAFs (TBP associated factors), nearly all of which have been conserved through evolution51–53. The TAFs function in promoter recognition as well as in positive and negative regulation of transcription. Although yeast contains only one form of TFIID, at least 6 TAFs in mammalian and Drosophila TFIID have alternative subunits that change the subunit composition of TFIID in a cell type and developmental-specific fashion. TAFs have been implicated in gene regulation in both biochemical and genetic studies. In certain in vitro systems, TAFs can be functional targets for transcription activators54,55. A subset of TAFs has DNA binding activity and at least one TAF has the ability to bind acetylated nucleosomes, protein acetylase, and ubiquitylation activity56. Some TAFs are also subunits of complexes lacking TBP involved in covalent chromatin modification and transcriptional co activation such as yeast SAGA and SLIK/SALSA (Spt-Ada-Gcn5-Acetylase and SAGA-like complex), and the human complexes pCAF and STAGA (p300/CBP and Spt-TAF-Gcn5-Acetylase) 52. A core of 5 TAFs are found in both TFIID and the acetylase/coactivator complexes.
Multiple histone fold domain interactions shape TFIID structure.
The structure of human, Drosophila, and yeast TFIID has been determined at low resolution by electron microscopy57,58 ( Fig. 2c). TFIID contains 3 lobes (termed A, B, and C) arranged in a horseshoe shape, observed in both closed and open configurations. Immune localization of TBP within this complex showed that TBP lies in the center lobe on the inside of the horseshoe, presumably exposing its DNA binding surface57. Comparison of this structure to that of TFIID with either TFIIA or TFIIB localizes these two additional factors to either side of the TBP binding site in the center lobe. Depending on the experimental conditions used, TFIID protects 40 to 60 bp of DNA from DNAse I cleavage47,48. From the shape of TFIID, the two non-central lobes potentially provide a large surface for interaction with DNA. Again, further structural studies should clarify whether the known DNA interacting TAFs are on these surfaces.
The structures of several TAF subunits were solved by x-ray crystallography59,60. These studies, combined with sequence analysis, have shown that 9 out of 13 conserved TAF subunits have histone fold domains (HFDs)61. TAFs form at least 5 histone-like pairs essential for the function of TFIID (TAF pairs 4-12, 6-9, 3-10, 10-8, and 11-13). Mapping the location of the HFD TAFs to the TFIID structure using electron microscopy has given unexpected results62, the most surprising of which is that the TAFs shared between TFIID and SAGA are not found in a substructure of TFIID but are distributed among the three lobes. Although it is unknown whether the HFDs are involved in DNA binding, their potential DNA interaction must be different from those of the nucleosome, since many of the side chains in the HFDs that interact with DNA in the nucleosome are not conserved in the TAFs63. From structural studies of Pol II interactions with TFIIB and TFIIF, where these factors interact extensively with promoter DNA (see below), at least some of the extensive protein-DNA interactions between TAFs and promoter DNA likely change during PIC formation to allow access of Pol II and other general factors to the promoter.
TFIID is not universally required at all promoters.
Molecular genetic studies have shown that promoters vary widely in the requirement for TAFs to promote normal gene regulation8,64–66. Although most genes are dependent on at least some TAFs for normal regulation, an important class of promoters seems independent of any TAF. In yeast, these completely TAF-independent promoters recruit TBP but not the TAFs upon gene activation67,68. Since TFIID is a large complex protecting 40–60 bp of promoter DNA, it would be expected that significant structural differences exist between PICs formed with and without TAFs. However, at some TFIID-independent promoters, a TAF-containing complex such as SAGA may functionally replace the TFIID, consistent with results suggesting that TFIID and SAGA function overlap at many yeast genes69.
Pol II the center of attention
Pol II lies at the center of the transcription machinery, interacting with the general transcription factors in the PIC, breaking these interactions upon initiation and promoter clearance, and associating with another set of factors during elongation and termination. Nearly all Pol II subunits have clear counterparts in the other two nuclear Pols and in archaea. Pol II subunits can be classified into three overlapping categories: subunits of the core domain having homologous counterparts in bacterial Pol (Rpb1, 2, 3, and 11), subunits shared between all three nuclear polymerases (Rpb5, 6, 8, 10, and 12), and subunits specific to Pol II but not essential for transcription elongation (rpb4, 7, and 9).
Pol II structure.
A breakthrough in understanding the mechanism of transcription was achieved with the high-resolution structures of bacterial Pol and the Pol II enzyme70–72. Since the initial structural description of bacterial Pol, the structures of Pol in complex with the elongation factor GreB73, with Sigma factor (holoenzyme)74–76 as well as of holoenzyme in complex with a fork junction DNA77 (analogous to an intermediate in Open Complex formation) have been determined. The Pol II structure and models for protein interactions have been determined for the 10-subunit and 12 subunit enzymes without DNA78,79, the 10 subunit enzyme in two different transcribing complexes80,81 in complex with the general factor TFIIB6,8 and the elongation factor TFIIS82. Lower resolution EM structures have also been obtained for Pol II binding to the Mediator complex83 as well as with the general factor TFIIF7. The structures of these multi factor complexes are beginning to reveal the assembly mechanism for the general transcription machinery as well as identifying conformational changes in protein and DNA that must occur during transcription initiation.
The highest resolution Pol II structures have been those of the ten subunit enzyme lacking the Rpb4 and Rpb7 subunits72 (Fig. 3a,b). These two subunits are important for transcription initiation but not for elongation. Pol II is composed of four mobile elements termed Core, Clamp, Shelf, and Jaw Lobe that move relative to each other. The Core element (Rpb3, 10, 11, 12 and regions of Rpb1 and Rpb2 forming the active center)80 accounts for about half the mass of Pol II and is comprised mainly of subunits common to all cellularPols. At the center of the enzyme is a deep cleft where incoming DNA enters from one side and the active site is buried at the base. This cleft is formed by all four mobile elements and has been observed in both closed and open conformations in the 10 subunit enzyme. The Shelf and Jaw Lobe elements move relatively little and can rotate parallel to the active site cleft. The Clamp element, connected to the Core through a set of flexible switches, moves with a large swinging motion of up to 30Å to open and close the cleft. Recent work revealed the structure of the complete 12 subunit enzyme showing Rpb4/7 binding to a pocket formed by Rpb1, 2, and 6 at the base of the Clamp78,79. Rpb7 in this pocket acts as a wedge to lock the Clamp in the closed conformation. This striking finding has important implications for the mechanism of initiation and suggests that double stranded DNA never enters the active site cleft. Rather, it has been proposed that during Open Complex formation, the single stranded DNA template strand is inserted deep into the cleft to reach the active site. This mechanism is likely preserved in Pol I, Pol III, and archaeal Pol since these enzymes contain subunits homologous to Rpb4/7. In addition to locking the position of the Clamp, Rpb4/7 also provides a binding surface for other factors and possibly for RNA exiting the elongating Pol.
Figure 3.
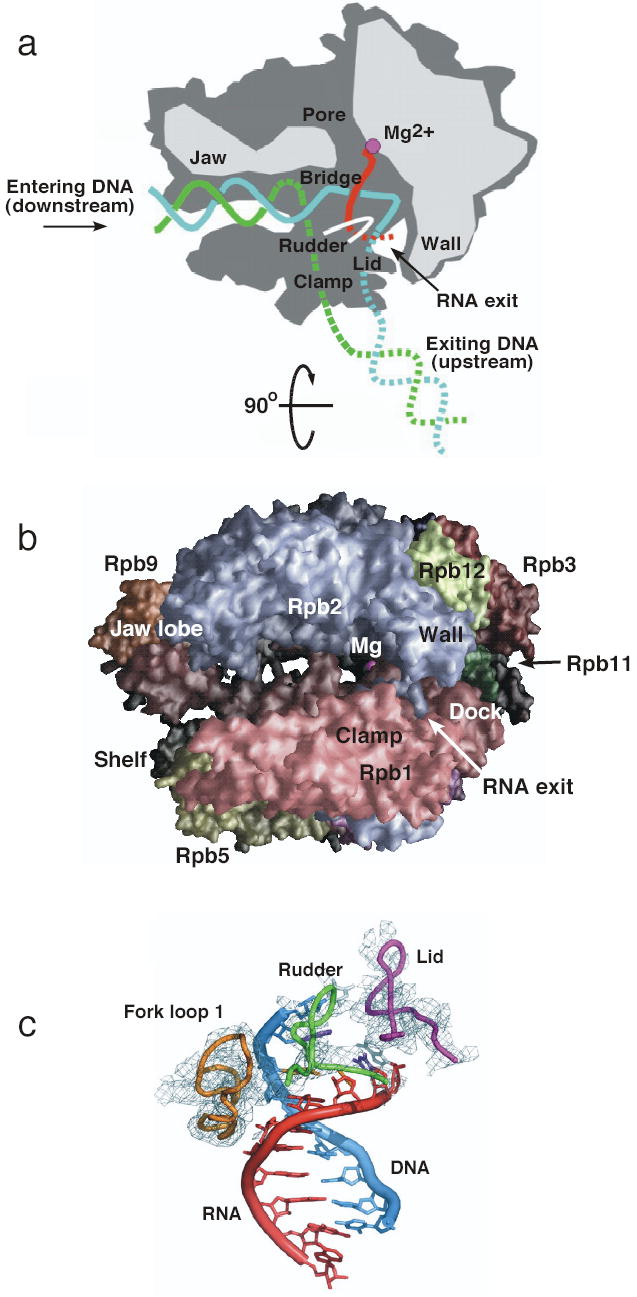
The ten subunit yeast RNA Pol II structure and structure of the elongation complex. (a) top view of RNA Pol II with the DNA template strand (blue) non-template strand (green), RNA (red) and active site Mg (magenta). Figure adapted from126 (b) Side view of Pol II looking into the active site cleft70– 72. Subunits are colored as follows: Rpb1, light red; Rpb2, light blue; Rpb12, light green; Rpb3, red; Rpb11, dark green; Rpb5, dark yellow; Rpb9, orange, active site Mg, magenta sphere. (c) details of the interactions between Pol II and the DNA-RNA hybrid in elongating Pol II. Figure from81.
The Pol II elongation complex structure.
Much insight on the mechanism of Pol II has been gained from structures of the elongating complex. The first structure determined was of Pol II transcribing a 3′ tailed template that had backtracked by one base with respect to the nucleotide addition site80. Several important details on the interaction of the enzyme with the nucleic acids were not visible, probably due to heterogeneity of this complex. In new work, the elongation complex was instead assembled from a 5′ tailed DNA oligonucleotide and a 9 base RNA complimentary to the 5′ tail, generating a complex in the post translocation state81. The structure of this complex was determined to 3.5 A resolution, revealing new important details on the mechanism of elongation. This structure clearly shows an 8 base pair RNA-DNA hybrid and interactions of the enzyme with both ends of the transcription bubble as well as with the RNA-DNA hybrid (Fig. 3c). The Pol II loop termed Lid appears to act as a wedge to drive apart the DNA and RNA strands at the upstream end of the transcription bubble and guide the RNA strand toward the RNA exit groove. The Rudder loop interacts with single stranded DNA after separation from the RNA strand, likely preventing reassociation with the exiting RNA. Finally, the newly revealed Fork Loop 1 interacts with the RNA-DNA hybrid, possibly stabilizing it. These three protein loops also interact with each other, forming a network of protein-protein and protein-nucleic acid interactions stabilizing the elongation complex.
The Pol II CTD molds itself to a binding partner.
Pol II undergoes regulatory phosphorylation and dephosphorylation as part of the transcription cycle with the Rpb1 C-terminal domain (CTD) the target of this modification18. The CTD, which is unique to Pol II, contains 25–52 repeats of the tandemly repeated heptad sequence YSPTSPS, with both Ser2 and Ser5 the sites of phosphorylation. The CTD acts as a platform for assembly of factors that regulate transcription initiation, elongation, termination, and mRNA processing. Pol II with a hypophosphorylated CTD is initially recruited to promoters during PIC formation and is phosphorylated at Ser5 during transcription initiation. Two cyclin dependent kinases, CDK7 and CDK8 are components of the PIC and target the CTD for phosphorylation2,84. Although previous work suggested that only CDK7 positively regulates transcription, new work in yeast indicates that both kinases can promote transcription in vivo and in vitro, as inhibition of both kinases together is required for maximal inhibition of transcription84. Phosphorylation of the CTD by these kinases also destabilizes the PIC leading to formation of the Scaffold complex. After initiation, other kinases such as CDK9/Ctk1 phosphorylate Ser2 resulting in recruitment of the RNA processing and polyadenylation/termination factors to elongating Pol II, allowing coupling of transcription and RNA processing18,85.
Until recently, the structural basis for CTD action was unclear as the many biochemically determined CTD binding partners had no obvious structural relationship. New structures of CTD interactions with two different binding partners have revealed that the CTD appears to mold itself to its binding partner, adopting different conformations. In one study, a complex of a single copy of the CTD with Ser2, 5-P bound to the Pin1 peptidylproline isomerase was solved86. This structure showed that the CTD bound as an extended coil, projecting every third residue onto one face of the coil. In another study, the structure of a four heptad Ser2-P CTD repeat was solved in complex with the guanylyltransferase Cgt187. In contrast to the CTD-Pin1 structure, 17 amino acids of the CTD repeats bound to an extended surface of Ctg1, anchored at both ends by electrostatic interactions with Ser5-P and with extensive hydrophobic CTD-Cgt1 interactions in between. This extensive surface contact between the CTD and Ctg1 suggested that mutation of any single residue would be unlikely to have a major effect on binding, a prediction borne out by mutagenesis studies87. The flexibility of the CTD, combined with covalent modification by phosphorylation, provides a way for the CTD to be interact with multiple structurally dissimilar partners, a paradigm that may hold true for some transcription activators and their targets. For example, many activation regions are very insensitive to mutagenesis and the strength of the activator often is dependent on the simple length of the activation region88–90, suggesting that some activators do not interact with their target as a folded globular domain. Interaction of these activators with an extended surface of their binding partners, similar to the interaction of the CTD with Ctg1, could explain these unusual properties of activators.
Structure of Mediator and connection to Pol II function.
Mediator is a large protein complex that interacts with Pol II, in part through the CTD, binding to the non phosphorylated form. Mediator was first identified in yeast, where it is composed of 24 subunits and is essential for both basal and activated transcription in unfractionated systems2. The yeast mediator binds cooperatively with Pol II and a subset of the general factors at an intermediate step in PIC formation35. Related Mediator complexes have been found in all eukaryotes examined, although the Mediator subunits are the least conserved of all the transcription machinery, consistent with the idea that many Mediator subunits serve as regulatory factor targets91,92. Biochemical fractionation has shown that about 40% of Mediator is in a stable complex with Pol II, consistent with studies showing that Mediator can be recruited to promoters independent of the rest of the transcription machinery93,94. Mediator consists of 3–4 domains or modules 95. In agreement with this, EM structures of the yeast Pol II-Med complex shows three domains (head, middle, and tail) with Mediator binding centered on the Rpb3 and Rpb11 subunits, on the opposite side from the active site cleft83 (Fig. 4). It is not yet known how the large Mediator complex fits into the context of the rest of the transcription machinery or how it transmits signals from regulatory factors to Pol II.
Figure 4.
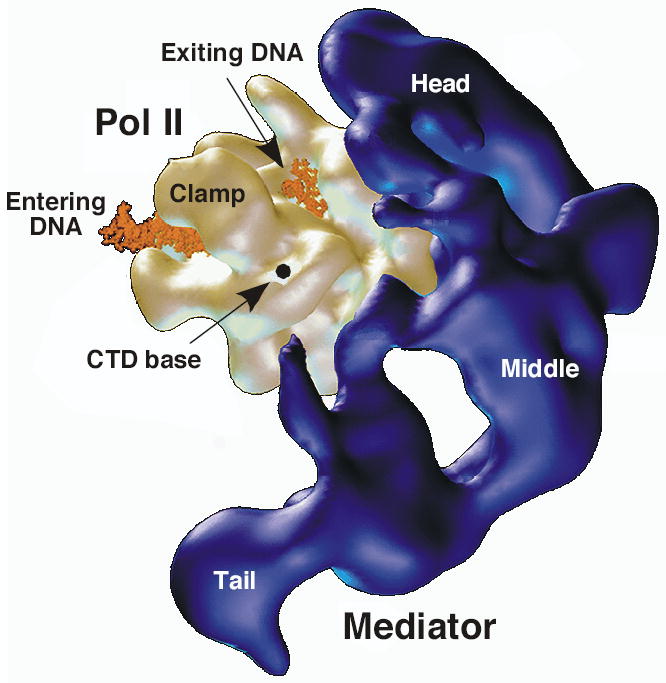
EM structure of the Pol II-Mediator complex. Mediator (dark blue) shown with head, middle, and tail domains. Pol II (gold) shown with modeled upstream and downstream DNA (orange). The dot represents the presumed beginning of the CTD. Figure from 83.
Assembly of Pol II with the transcription machinery
A major question remaining to be answered is how Pol assembles with the rest of the transcription machinery at a promoter. Important information on the architecture of the PIC came from photoreactive probes placed in promoter DNA and assembled into minimal PICs formed with TBP96–99 ( Fig. 5). These studies showed that the transcription machinery makes extensive interactions with promoter DNA between positions −43 and +24 with respect to the transcription start site. Two RNA Pol II subunits (Rpb1 and 2) make extensive DNA interactions over 60 bp. TFIIB and the small subunit of TFIIF (TFIIFβ) both interact with DNA on either side of the TATA and the large TFIIF subunit interacts with DNA downstream of TATA. TFIIE interacts with promoter DNA just upstream of the transcription start site while the TFIIH helicase subunit interacts downstream and possibly upstream of the transcription start. Any structural model for the PIC must account for these extensive Protein-DNA interactions.
Figure 5.
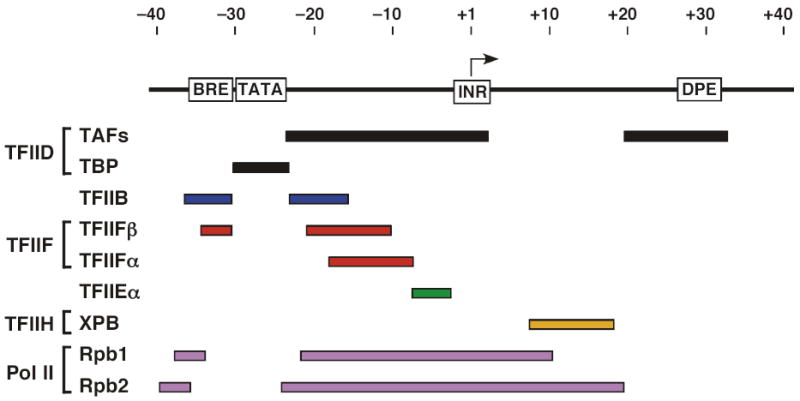
Summary of human general transcription factor protein-DNA crosslinks at a promoter. Top line represents promoter DNA with the position of functional elements indicated. Arrow represents the transcription start site. The bars below the line show the extent of protein-DNA crosslinking with the indicated general transcription factor using a crosslinker incorporated into the DNA backbone and crosslinked after treatment with Sarkosyl98. Use of a larger major groove crosslinking reagent without detergent challenge has shown even more extensive crosslinks across the promoter region99,127.
The Pol II-TFIIB interface.
Recently, two lines of evidence have shown how TFIIB interacts with Pol II, which in one case led to a model for a complex of Pol II with TBP, TFIIB and DNA. In the first set of experiments, photocrosslinking and hydroxyl radical generating probes were placed on TFIIB near the functional surface of the ribbon domain and assembled into PICs 6. Mapping the interaction of the ribbon domain with respect to the two largest Pol II subunits showed the ribbon domain fits into a pocket formed by the Wall, Dock, and Clamp domains near the RNA exit point. In this model, the functional surface of the ribbon interacts with the Dock domain, a region of Rpb1 best conserved in Pol II, Pol III, and archaea Pol, all of which utilize a TFIIB-like factor for initiation.
In new work from the Kornberg laboratory, the structure of a complex of TFIIB and the 10 subunit Pol II was determined79 ( Fig. 6a). In this structure, the position of the TFIIB ribbon domain agrees with the binding seen in the complete PIC as described above. Additionally, the conserved portion of the linker between the TFIIB ribbon and core domains enters the RNA exit channel and into the active site cleft analogous to the path of Sigma region 3.2. This linker sequence forms a hairpin-like structure termed the B-finger that is predicted to be located very near the upstream end of the RNA-DNA hybrid in elongating Pol II. In this location, the B finger would block productive elongation much like Sigma region 3.2 and may help stabilize or position the transcription bubble during open complex formation. The remainder of the TFIIB linker C-terminal to the B-finger was observed to exit back out though the RNA exit channel. The proposed location of the B-finger within the active site cleft is also consistent with recent results showing that in the Open Complex, archaeal TFB crosslinks to the template strand close to the transcription start site 100,101.
Figure 6.
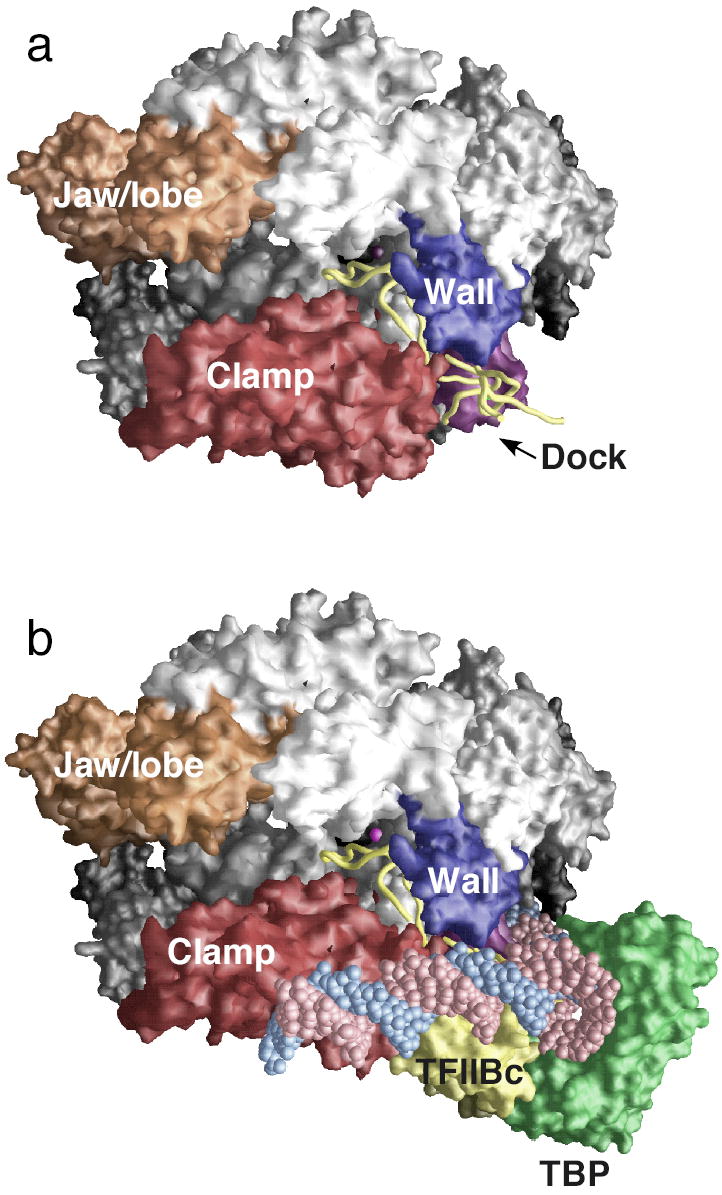
The Pol II-TFIIB complex and a model for the structure of the PIC. (a). X-ray structure of the Pol II-TFIIB complex with the TFIIB N-terminal domain backbone (yellow), and the clamp, wall, dock and jaw/lobe domains shown in red, dark blue, purple, and orange8. (b). Model for PIC structure based on the Pol II-TFIIB structure8. Colors the same as in panel (a) with TBP (green), the N-terminal TFIIB core domain (TFIIBc; yellow), and DNA (red and blue).
In the Pol II-TFIIB structure, electron density from the TFIIB core domain was located adjacent to the Pol Dock domain, on the opposite side of the ribbon domain interaction surface. Modeling the TFIIB-TBP-DNA structure to this location led to a model for a complex of Pol II with these factors and a predicted path of DNA in the preinitiation complex ( Fig. 6b). In this model, TBP mediates promoter DNA bending around Pol II and the DNA downstream from TBP runs along the outer edge of the Clamp element. This model will guide biochemical tests for interaction of the TFIIB core domain and promoter DNA with Pol II and will also serve as a template for modeling assembly of other general transcription factors. An unanswered question is whether the interaction of TFIIB differs between the Pol II-TFIIB complex and the PIC. Further structural and biochemical studies on higher order assemblies of Pol II and the general factors are needed to answer this question.
Pol II and TFIIF interact over an extended surface.
In other new work, cryo-EM was used to determine the structure of yeast Pol II in complex with the general factor TFIIF7 ( Fig. 7). TFIIF binds Pol II as a heteromer and contains two subunits that are conserved among human, insects and yeast, termed Tfg1 and Tfg2 in yeast (Rap74 and Rap30 in humans). The N-termini of both conserved subunits form a dimerization domain and the C-termini of both subunits are winged helix domains3. Biochemical and structural analysis has implicated regions of TFIIF involved in protein-protein interactions with Pol II, TFIIB, and the FCP1 phosphatase, as well as non specific protein-DNA interactions102–104. In the EM structure, TFIIF was observed to interact with a highly extended surface of Pol II along the edge of the Clamp element as well as with the Rpb4/7 subunits. The structure of Pol II with the Tfg2 subunit alone implicated Tfg2 binding to the extended clamp region and Tfg1 binding to Rpb4/7, although this experiment should be interpreted cautiously as the Tfg1/2 dimerization domain is unlikely to fold normally with only one subunit102.
Figure 7.
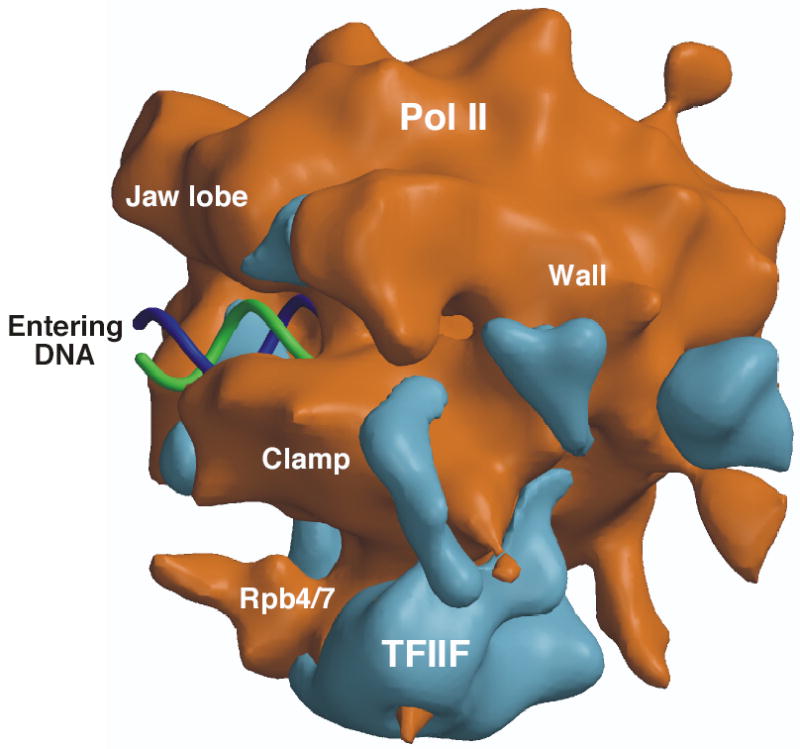
EM structure of the Pol II-TFIIF complex. Pol II is shown as a brown surface with TFIIF density in blue. The orientation shown is similar to that of Figs 3b and 6. Also shown is a model for downstream double stranded DNA entering the active site cleft (blue and green helix). Figure provided by F. Asturias.
Because the extended TFIIF domains seen along the Clamp element is in a similar general location to that seen with Sigma in the bacterial holoenzyme, the authors speculate that TFIIF is the structural homolog of Sigma factor. Previous sequence comparison suggested weak similarity between two regions of Tfg2/Rap30 and Sigma105,106. However, a more extensive comparison with many Sigma and Tfg2 family members does not reveal any striking similarity between these polypeptides (H-T Chen (Fred Hutchinson Cancer Res. Ctr.) and S. Hahn, unpublished). Nevertheless, it is possible that both factors play some of the same roles in the initiation process, such as helping to promote or stabilize opening of the DNA strands upon Open Complex formation.
Combining the TFIIB-Pol II structure model with the Pol II-TFIIF model, Asturias and coworkers propose a model for the structure of a minimal PIC in which TFIIF interacts with DNA downstream from the TATA and helps position DNA along the active site cleft of Pol II7. One complication in this proposal is that it does not seem to agree with protein-DNA crosslinking results that show a close overlap between promoter sequences upstream and downstream of the TATA contacted by both TFIIB and TFIIF96,97,99. This again suggests the possibility that the arrangement of the general factors on Pol II could change in higher order assemblies with DNA, a possibility that can be addressed by further structural and biochemical studies.
TFIIE and TFIIH function in steps after PIC formation.
The general factors TFIIE and TFIIH function primarily in steps after PIC formation and can be at least partially dispensable on promoters with a preformed transcription bubble 2. TFIIE binds independently to Pol II107 and is thought to stimulate both the kinase and helicase activities of TFIIH2,108. Biochemical analysis has suggested that TFIIE interacts with a number of other general transcription factors and may functionally interact with double stranded and single stranded promoter DNA. TFIIE is likely a heterodimer109 and electron crystallography studies suggest that TFIIE binds near the pol II cleft, consistent with the observed crosslinking of TFIIE to DNA immediately surrounding the transcription start site in the PIC110. The structure of the central core domain of the TFIIE β was determined to be a winged helix domain by NMR111. It has been proposed that this domain interacts with single stranded promoter DNA, but conclusive evidence for this has not been obtained.
TFIIH, which has a dual function in transcription and transcription-coupled DNA mismatch repair, is composed of two domains, a core domain containing two DNA helicase activities and a kinase domain termed CAK containing CDK7 112,113. Mutations in the human XPD helicase cause the diseases xeroderma pigmentosum and trichothiodystrophy. These mutations affect nucleotide excision repair and can also affect basal transcription as well as transcription activated by certain nuclear receptors114. The low resolution structure of the TFIIH core domain was determined by EM and was shown to be a ring shaped structure with the two helicase subunits located on either side of a prominent protrusion115,116. The center of the ring appeared to have dimensions sufficient to accommodate double stranded DNA, although it is not clear if DNA normally enters the ring. Models for how TFIIH fits into the PIC are speculative as there is no information available on the docking of TFIIH with any other factor. The XPB helicase is essential for Open Complex formation and is the only TFIIH subunit seen to crosslink to promoter DNA.
Open Complex formation and the transcription start site.
A major unanswered question about the mechanism of Pol II initiation is how melting of the DNA strands is initiated during Open Complex formation. Pol II is unique among cellular Pols in requiring the action of an ATP-dependent DNA helicase (XPB) for Open Complex formation. This requirement is puzzling since all cellular Pols have the same overall structure and catalytic mechanism. Aside from Pol II, archaeal and other eukaryotic Pols may use a mechanism similar to that seen in bacteria in which aromatic side chains from one of the general transcription factors act as a wedge to stabilize separation of the two DNA strands77.
Helicase requirement for opening the transcription bubble.
Although helicase activity is typically defined in vitro as the ability to processively remove a paired oligonucleotide from single stranded DNA, not all helicases act processively. Helicases act by destabilizing double stranded nucleic acid through the ATP hydrolysis-dependent motion of two separate domains that interact with single and double stranded nucleic acids117. Using this mechanism, the XPB helicase binding to promoter DNA as a subunit of TFIIH, likely initiates unwinding by introducing tortional strain in the DNA near the transcription start site. Because of uncertainty in the location of XPB-DNA interaction 3, the mechanism of helicase action is also uncertain. If the XPB helicase motifs bind at the site of single stranded bubble formation, XPB would directly initiate strand unwinding. If the helicase domain interacts only with downstream DNA, then this tortional strain would lead to initial DNA opening upstream from the point of destabilization. In either case, one or more of the general transcription factors likely acts like Sigma region 3.1 to trap the single stranded bubble and promote the insertion of this single stranded region into the active site of the enzyme. From the EM structure and protein-DNA crosslinking studies, TFIIE or the TFIIF subunit Tfg2 may be in a position to promote this reaction. Understanding this reaction will require more precise localization of the XPB helicase domain at the promoter and the identification of amino acids in general transcription factors located near the initial site of DNA melting. Also unexplained is how the related archaeal factors TBP and TFIIB promote open complex formation without the requirement for any other general factor.
Transcription start site selection.
Another major unsolved problem is understanding how the start site of transcription is selected. At TATA-containing promoters in vertebrates and Drosophila, the transcription start site is located about 30 bp downstream from the beginning of the TATA sequence. However, at S. cerevisiae TATA-containing promoters, the TATA appears to define a window of ∼40–120 bp in which transcription starts at preferred DNA sequences1. Transcription start sites in S. pombe are less heterogeneous, with initiation beginning 25–40 bp downstream from the TATA element118. A model to explain the transcription initiation site in higher eukaryotes would be that TFIIB binding to both Pol II and promoter DNA sets the distance needed for the DNA to travel from the TFIIB binding site on Pol II to the active site of the enzyme8. It is not obvious why this would be different in yeasts where the transcription machinery is largely conserved. Extensive genetic and biochemical studies in S. cerevisiae have identified mutations in Pol II subunits, TFIIB, and TFIIF altering the transcription start site119–123.
Based on the Pol II-TFIIB structure, it was proposed that the tip of the B-finger might play a role in recognition of the transcription start site8. This residue is not conserved between yeasts and human and it was postulated that, lacking a stable protein-DNA interaction, the promoter DNA would slip through the enzyme active site until a sequence that stably bound to the active site was located. This model is consistent with in vivo mapping experiments of S. cerevisiae promoters that found evidence of single stranded DNA over a wide region between the TATA and transcription start site124. Ultimately, determining the mechanism of start site selection will involve mapping the location of promoter DNA in the PIC both before and after ATP addition.
Conclusions
As exemplified by the similarities in cellular RNA polymerases, the general mechanism of transcription is similar in all cells. Despite this overall conservation, the transcription machinery is much more complex in eukaryotes, with the function of bacterial Sigma factor distributed among several general transcription factors. In eukaryotes, the transcription start site is determined in part by the precise binding of the transcription machinery to a promoter and this seems driven not by any one high affinity protein-DNA interaction, but rather by multiple low affinity and low specificity protein-DNA interactions. Since all transcription requires TBP but many promoters do not have a recognizable TATA element, the precise role of TBP in nucleating the assembly of the PIC at these non-TATA promoters is an open question. Although much progress has been made in structural analysis, an important challenge is to determine the structure of the PIC to test if structures and models of single general transcription factor-Pol II complexes reflect the structure of the much larger PIC. Also important, will be to determine the mechanism of the dramatic conformational changes which accompany transition to the Open Complex state. This mechanism, whereby single stranded DNA is positioned in the active site of Pol, is one of the most mysterious aspects of both bacterial and eukaryotic transcription. In future studies, integration of the structures discussed above with biochemical and structural work on activators and their targets will get at the heart of the mechanism of gene regulation and will examine in atomic detail how regulatory signals are transmitted to the transcription machinery.
Acknowledgments
I thank D. Bushnell, R. Kornberg, and F. Asturias for figures and for communication of results before publication, E. Nogales for figures, B. Moorefield and H-T. Chen for their comments on the manuscript, and H-T. Chen for discussions and help with figures. This work was supported by a grant from the NIH. S.H. is an associate investigator of the Howard Hughes Medical Institute.
Footnotes
The author declares that he has no competing financial interests.
References
- 1.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 3.Woychik NA, Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 4.Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol. 2003;6:93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 5.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen HT, Hahn S. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol Cell. 2003;12:437–447. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 7.Chung WH, et al. RNA Polymerase II/TFIIF Structure and Conserved Organization of the Initiation Complex. Mol Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- 8.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 9.Bell SD, Jackson SP. Transcription in Archaea. Cold Spring Harb Symp Quant Biol. 1998;63:41–51. doi: 10.1101/sqb.1998.63.41. [DOI] [PubMed] [Google Scholar]
- 10.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 11.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 12.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 13.Ptashne, M. & A., G. Genes and Signals, (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2002).
- 14.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Carey M, Gralla JD. Polymerase II Promoter Activation: Closed Complex Formation and ATP-Driven Start Site Opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 16.Luse DS, Jacob GA. Abortive initiation by RNA polymerase II in vitro at the adenovirus 2 major late promoter. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 17.Holstege FCP, Fiedler U, Timmers HTM. Three transitions in the RNA polymerase II transcription complex during initiation. Embo J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 19.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 20.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 22.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 23.Cox JM, et al. Bidirectional binding of the TATA box binding protein to the TATA box. Proc Natl Acad Sci U S A. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcrition: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qureshi SA, Jackson SP. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- 26.Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in archaea. Proc Natl Acad Sci U S A. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Littlefield O, Korkhin Y, Sigler PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci U S A. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250- TAF(II)150 complex recognizes the initiator. Embo J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oelgeschlager T, Chiang C-M, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 30.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans ind is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patikoglou GA, et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wobbe CR, Struhl K. Yeast and Human TATA-Binding Proteins Have Nearly Identical DNA Sequence Requirements for Transcription In Vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez E, et al. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci U S A. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 38.Davidson I. The genetics of TBP and TBP-related factors. Trends Biochem Sci. 2003;28:391–398. doi: 10.1016/S0968-0004(03)00117-8. [DOI] [PubMed] [Google Scholar]
- 39.Holmes MC, Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- 40.Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 41.Nikolov DB, et al. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 42.Geiger JH, Hahn S, Lee S, Sigler PB. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 43.Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 44.Weideman CA, et al. Dynamic interplay of TFIIA, TBP, and TATA DNA. J Mol Biol. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- 45.Kokubo T, Swanson MJ, Nishikawa JI, Hinnebusch AG, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu D, et al. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP [see comments] Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 47.Sanders SL, Garbett KA, Weil PA. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol Cell Biol. 2002;22:6000–6013. doi: 10.1128/MCB.22.16.6000-6013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 49.Pardee TS, Bangur CS, Ponticelli AS. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 50.Hahn S, Roberts S. The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev. 2000;14:719–730. [PMC free article] [PubMed] [Google Scholar]
- 51.Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 52.Green MR. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- 53.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 54.Chen J-L, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 55.Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 56.Wassarman DA, Sauer F. TAF(II)250: a transcription toolbox. J Cell Sci. 2001;114:2895–2902. doi: 10.1242/jcs.114.16.2895. [DOI] [PubMed] [Google Scholar]
- 57.Andel F, 3rd, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 58.Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286:2151–2153. doi: 10.1126/science.286.5447.2151. [DOI] [PubMed] [Google Scholar]
- 59.Xie X, et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 60.Werten S, et al. Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAF(II)135) and hTAF12 (hTAF(II)20) J Biol Chem. 2002;277:45502–45509. doi: 10.1074/jbc.M206587200. [DOI] [PubMed] [Google Scholar]
- 61.Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 62.Leurent C, et al. Mapping histone fold TAFs within yeast TFIID. Embo J. 2002;21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.9Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 64.Freiman RN, et al. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 65.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen WC, et al. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. Embo J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 68.Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 69.Lee TI, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 70.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 33 A resolution . Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 71.Cramer P, et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 72.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 73.Opalka N, et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 74.Mekler V, et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 75.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 76.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 77.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 78.Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc Natl Acad Sci U S A. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci U S A. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 81.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 82.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 83.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 84.Liu Y, et al. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the Scaffold Complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 86.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 87.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 88.Hope IA, Mahadevan S, Struhl K. Structural and Functional Characterization of the Short Acidic Transcriptional Activation Region of Yeast GCN4 Protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 89.Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 90.Jackson BM, Drysdale CM, Natarajan K, Hinnebusch AG. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 92.Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 93.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 94.Rani, G., Ranish, J.A. & Hahn, S. RNA polymerase II (Pol II)-TFIIF and Pol II-mediator complexes: the major stable Pol II complexes and their activity in transcription initiation and reinitiation. Mol. Cell. Biol.24(2004). [DOI] [PMC free article] [PubMed]
- 95.Kang JS, et al. The structural and functional organization of the yeast mediator complex. J Biol Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 96.Forget D, et al. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box . Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim T-K, et al. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 99.Forget D, Langelier M-F, Therien C, Trinh V, Coulombe B. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol Cell Biol. 2004;24:1122–1131. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bartlett MS, Thomm M, Geiduschek EP. Topography of the euryarchaeal transcription initiation complex. J Biol Chem. 2004;279:5894–5903. doi: 10.1074/jbc.M311429200. [DOI] [PubMed] [Google Scholar]
- 101.Renfrow MB, et al. Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex. J Biol Chem. 2004;279:2825–2831. doi: 10.1074/jbc.M311433200. [DOI] [PubMed] [Google Scholar]
- 102.Gaiser F, Tan S, Richmond TJ. Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 A resolution. J Mol Biol. 2000;302:1119–1127. doi: 10.1006/jmbi.2000.4110. [DOI] [PubMed] [Google Scholar]
- 103.Kamada K, Roeder RG, Burley SK. Molecular mechanism of recruitment of TFIIF- associating RNA polymerase C-terminal domain phosphatase (FCP1) by transcription factor IIF. Proc Natl Acad Sci U S A. 2003;100:2296–2299. doi: 10.1073/pnas.262798199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nguyen BD, et al. NMR structure of a complex containing the TFIIF subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1. Proc Natl Acad Sci U S A. 2003;100:5688–5693. doi: 10.1073/pnas.1031524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sopta M, Burton ZF, Greenblatt J. Structure and Associated DNA-Helicase Activity of a General Transcription Initiation Factor that Binds to RNA Polymerase II. Nature. 1989;341:410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- 106.Henry NL, et al. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 107.Bushnell DA, Bamdad C, Kornberg RD. A minimal set of RNA Pol II transcription protein interactions. J Biol Chem. 1996;271:20170–20174. doi: 10.1074/jbc.271.33.20170. [DOI] [PubMed] [Google Scholar]
- 108.Ohkuma Y. Multiple functions of general transcription factors TFIIE and TFIIH in transcription: possible points of regulation by trans-acting factors. J Biochem (Tokyo) 1997;122:481–489. doi: 10.1093/oxfordjournals.jbchem.a021777. [DOI] [PubMed] [Google Scholar]
- 109.Sayre MH, Tschochner H, Kornberg RD. Purification and properties of S. cerevisiae RNA polymerase II general initiation factor a. J Biol Chem. 1992;267:23383–23387. [PubMed] [Google Scholar]
- 110.Leuther KK, Bushnell DA, Kornberg RD. Two-dimensional crystallography of TFIIB- and IIE-RNA polymerase II complexes: implications for start site selection and initiation complex formation. Cell. 1996;85:773–779. doi: 10.1016/s0092-8674(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 111.Okuda M, et al. Structure of the central core domain of TFIIEbeta with a novel double- stranded DNA-binding surface. Embo J. 2000;19:1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coin F, Egly JM. Ten years of TFIIH. Cold Spring Harb Symp Quant Biol. 1998;63:105–110. doi: 10.1101/sqb.1998.63.105. [DOI] [PubMed] [Google Scholar]
- 113.Takagi, Y. et al. Revised subunit structure of yeast TFIIH and reconciliation with human TFIIH. J. Biol. Chem. (2003). [DOI] [PubMed]
- 114.Dubaele S, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 115.Schultz P, et al. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 116.Chang WH, Kornberg RD. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102:609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 117.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 118.Choi WS, Yan M, Nusinow D, Gralla JD. In vitro transcription and start site selection in Schizosaccharomyces pombe. J Mol Biol. 2002;319:1005–1013. doi: 10.1016/S0022-2836(02)00329-7. [DOI] [PubMed] [Google Scholar]
- 119.Hekmatpanah DS, Young RA. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berroteran RW, Ware DE, Hampsey M. The sua8 suppressors of S. Cerevisiae encode replacements of conserved residues within the largest subunit of RNA Polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutation. Mol Cell Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pinto I, Ware DE, Hampsey M. The Yeast SUA7 Gene Encodes a Homolog of Human Transcription Factor TFIIB and Is Required for Normal Start Site Selection In Vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 122.Faitar SL, Brodie SA, Ponticelli AS. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol Cell Biol. 2001;21:4427–4440. doi: 10.1128/MCB.21.14.4427-4440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun ZW, Tessmer A, Hampsey M. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2560–2566. doi: 10.1093/nar/24.13.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Giardina C, Lis JT. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 125.Chen HT, Legault P, Glushka J, Omichinski JG, Scott RA. Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription. Protein Sci. 2000;9:1743–1752. doi: 10.1110/ps.9.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci U S A. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Douziech M, et al. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol Cell Biol. 2000;20:8168–8177. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


