Abstract
Breast cancer is a disorder affecting women globally, and hence an early and precise classification is the best possible treatment to increase the survival rate. However, the breast cancer classification faced difficulties in scalability, fixed-size input images, and overfitting on limited datasets. To tackle these issues, this work proposes a Patho-Net model for breast cancer classification that overcomes the problems of scalability in color normalization, integrates the Gated Recurrent Unit (GRU) network with the U-Net architecture to process images without the need for resizing and computational efficiency, and addresses the overfitting problems. The proposed model collects and normalizes histopathology images using automated reference image selection with the Reinhard method for color standardization. Also, the Enhanced Adaptive Non-Local Means (EANLM) filtering is utilized for noise removal to preserve image features. These preprocessed images undergo semantic segmentation to isolate specific parts of an image, followed by feature extraction using an Improved Gray Level Co-occurrence Matrix (I-GLCM) to reveal fine patterns and textures in images. These features serve as input into the classification U-Net model integrated with GRU networks to improve the model performance. Finally, the classification result is expanded, and XAI is used for clear visual explanations of the model’s predictions. The proposed Patho-Net model, which uses the 100X BreakHis dataset, achieves an accuracy of 98.90% in the classification of breast cancer.
Keywords: Histopathology images, breast cancer, deep learning, gated recurrent unit, U-Net and classification
Introduction
Breast cancer increases the death rate of women worldwide, which spreads abnormal cell growth in the human body on a gradual basis [1,2]. Genetic factors and non-genetic characteristics like age, short breastfeeding periods, mammographic density, and drinking alcohol can contribute to the development of breast cancer. Common signs of breast tumors include swelling, breast lumps, tenderness, pain, and nipple discharge unrelated to breast milk [3]. The early diagnosis of breast cancer substantially reduces the death rate by enabling the timely selection of appropriate treatment. The imaging techniques of mammography, Breast Ultrasound (BUS), Computed Tomography (CT), Magnetic Resonance Imaging (MRI), and Positron Emission Tomography (PET) enhance the early identification of breast tumors [4,5]. It can be treated through surgery, radiation, chemo, and gene therapies, which produce several side effects [6]. To combat these side effects, researchers are exploring advanced AI techniques to enhance early diagnosis. Deep learning (DL) algorithms [7] can detect subtle patterns and structures that might escape human observation, contributing to the early detection of breast cancer.
Histopathological analysis plays a crucial part in the diagnosis of Breast cancer. Image processing techniques analyze the histopathological images through color normalization, preprocessing, segmentation, feature extraction, and classification [8]. Machine Learning (ML) encompasses a Support Vector Machine (SVM), XGBoost, decision trees, random forest, and logistic regression models to classify and predict breast cancer. It provides a reference for early diagnosis of breast cancer, focusing on recall as the primary evaluation index due to its importance in medical diagnosis for detecting malignant cells [9,10]. The DL framework detects breast cancer using Immunohistochemistry (IHC) images. The DL architecture recognizes breast mass tumors with an object detection algorithm and a Convolutional Neural Network (CNN). It examines the mammogram to pinpoint specific regions with high accuracy [11,12]. Automated detection of malignancy in breast tumors from mammography images using object detection models like YOLO and Mask R-CNN improves the accuracy of breast cancer detection by lowering false positive and false negative rates [13]. The DRD-UNet architecture detects and segments breast cancer tumors in histopathological images. It integrates the dilation, residual, and dense blocks for precise segmentation. The efficient UNet method integrating ResNet18, a channel attention mechanism, and deep supervision addresses the challenges in image analysis and enhances feature extraction capabilities [14,15]. Breast cancer detection still faces difficulties due to the lack of datasets and the significant diversity among tumors in terms of their shape, size, and location [16]. To overcome these problems a novel model has been proposed, and the contributions of the proposed work are as follows.
1. The proposed work utilizing the 100X BreakHis Dataset of histopathological images indicates multiple types of breast cancer in the development of DL models proposed for breast cancer diagnosis. 2. This paper could resolve the problem of scalability for big datasets with multiple classes by proposing a novel framework. This combines noise reduction using Enhanced Adaptive Non-Local Means (EANLM) technology for histopathology images to improve image quality, thereby obviating image enhancement processes. 3. This study of Gated Recurrent Unit (GRU) networks integrated with U-Net presents a solution for varying image sizes in imaging to make them flexible enough to record the intricate spatial features of tumors accurately. 4. This methodology focuses on Explainable Artificial Intelligence (XAI) technologies for creating visual explanations of classification conclusions, thus rendering the outputs more intelligible and corroborating the diagnoses based on obtained features.
This research is structured as follows: Section 2 reviews related work on breast cancer classification methods and models. Section 3 details the proposed Patho-Net to distinguish malignant and benign breast tumors. Section 4 presents the findings, showcasing the effectiveness of the proposed methodology. Finally, Section 5 offers conclusions.
Literature survey
The early research of the histopathological image classification of breast cancer using DL and ML has been listed below. Reshma et al. [17] created a classification strategy based on weighted feature selection, an improved genetic algorithm, and a CNN classifier to detect breast cancer. This model increased the accuracy of breast cancer classification by addressing the challenges of low-density regions. Nevertheless, the model categorization performance declined due to an insufficient initial training set. To address the problem of restricted data availability, Shah et al. [18] suggested a deep convolutional generative adversarial network. The synthetic mammograms generated by this approach faithfully replicate the inherent patterns found in the data. However, the model could benefit from additional datasets and more Generative Adversarial Network (GAN) architecture for further validation.
Mohanakurup et al. [19] suggested a Composite Dilated Backbone Network (CDBN) to identify breast cancer from histopathological images. Computer-aided design (CAD) phases improve the accuracy of early detection and decrease false diagnoses. However, the lack of label information in breast cancer histopathological images results in incorrect grouping. In the same way, Eltoukhy et al. [20] developed an automatic CAD system using Histopathology-RDNet, a self-training DL model, to classify multiclass histopathological breast images. The model demonstrated strong performance while dealing with the intricate nature of histopathological images. However, it was computationally expensive due to skip connections and numerous layers with different convolutional filter widths. For this, Sharma et al. [21] used a pre-trained Xception model to extract the distinguishable features from histopathological images for effective breast cancer treatment. The model preserved the spatial information to enhance the classification performance. While this model prevents the problems of overfitting, it does not address the issue of binary classification.
Lu et al. [22] proposed a paradigm for fine-grained lesion zone segmentation in high-resolution pathology using YOLOV4. The model effectively increased the segmented regions of interest’s average recognition precision. It could be further improved by reducing the false detection rate of the primary lesion region. Similarly, using the segmentation method, Tagnamas et al. [23] developed a Hybrid CNN transformer that integrates EfficientNetV2 and an Adaptive Vision Transformer (ViT) encoder to classify the breast tumor. The model successfully classified the segmented tumor regions as benign or malignant using a Multi-Layer Perceptron (MLP). However, it lacks contextual information about the tumor’s area. Hossain et al. [24] developed a fully automated RTC assessment method using the ViT to identify representative tumor regions and the Data-efficient Image Transformer (DeiT) for evaluating tumor cellularity in those regions. However, the model supports automatic tumor selection, it was only trained and tested for 20X magnified images.
Islam et al. [25] introduced an Ensemble deep CNN model that enhances breast cancer detection by incorporating the MobileNet, Xception models, and U-Net segmentation technique. The model utilized the GRAD-CAM XAI method to improve model interpretability and diagnosis accuracy. However, it could be further improved by integrating a CAD system for more effective detection of breast cancer. Correspondingly, Thakur et al. [26] developed a CNN model integrated with Early Stopping and ReduceLROnPlateau callbacks for breast cancer classification. The model effectively harnessed the power of advanced DL models, thereby revolutionizing the precision and reliability of breast cancer classification. Further inferential statistical tests were needed to improve the model’s predictive performance.
The existing models struggle with the lack of label information, generalization problems, unbalanced datasets, real-time detection, and computer resources. This paper uses the integration of Gated Recurrent Unit (GRU) networks with the U-Net architecture to avoid overfitting problems and improve generalizability and robustness. To avoid the scalability issue in color normalization, we used the Reinhard method to automatically select the reference image.
Proposed methodology
The proposed Patho-Net uses histopathology images that are taken from the Breakhis dataset to classify malignant and benign tumors. The process begins with the color normalization that selects the automatic reference image to improve the standardization and consistency of the color normalization process. Then the preprocessed images enhance the image quality using the ENALM technique. Next, the images are segmented to identify and perform to extract the regions of interest. From these regions, extract the features to train the U-net model to be classified as cancers (malignant) or non-cancers (benign). Finally, XAI techniques visually explain the model’s predictions. Figure 1 illustrates the architecture of the proposed framework.
Figure 1.
The framework for the proposed breast cancer classification.
Dataset
The 100X BreakHis Dataset contains a set of 2048 histopathology images for breast cancer classification. This dataset confronts clinicians with such dilemmas as they look for adenosis, ductal carcinoma, lobular carcinoma, and other types of pathologies. The set provides the integration of a DL model for the classification of breast cancer from histopathological images.
Color normalization
The color normalization method involves standardizing the color distribution of histopathology images to enhance the image analysis processes. In the normalization framework, the process used an automated reference image selection method, which was combined with the Reinhard method. It selects a reference image for every class and then identifies the different staining color characteristics within particular categories like benign or malignant. To achieve this, the following mathematical equation is used:
Where (μi ) is the mean color vector for class (C), (D) represents the number of images within the class (dataset), and (Vj,C ) is the color vector of the (jth) image in class (C). This equation (1) is used to calculate the mean color vector as it averages the color vectors of all the images in a class. To measure the variation in color distribution for the class, the Standard Deviation of the color vector for a given class (C) is calculated as:
Where, (σi,C ) denotes the standard deviation of the color vector for class (C). This equation (2) is utilized to find out the dispersion of the color vectors around the mean color vector for the class by taking the corresponding product of two matrices and the transposition of one of them to be measured.
Reference image selection
The reference image (jmin ) with the minimum Euclidean distance to the mean vector selected as the reference image for the class (C) and is calculated as:
Where (jmin) is the image index with the minimum Euclidean distance to the mean color vector (μi). Equation (3) will select the reference image via the detection of the image with the minimum Euclidean distance to the average color vector. After choosing the reference image, color normalization (equation 4) is performed for each image (I’) from the class (C) to get the result of the reference image.
Here, (μref ) and (σref ) are the mean and standard deviation of the color vector of the reference image, respectively. (I’) represents the normalized image, as shown in Figure 2.
Figure 2.
The left side shows the histopathological input images and the right side shows the color-normalized images using the Reinhard method.
Pre-processing
Following color normalization, the next step is to preprocess the data more efficiently by improving image quality and eliminating the noise using the EANLM technique. Non-local means noise reduction and is operated by image features like edges and details, identifying similar patches throughout the entire image and then calculating weights for each pixel based on the similarity of their surrounding patch. Still, the quality is due to the interference of noise, which might cause the blurring and loss of details. To address this, the work proposes EANLM for adjusting key parameters alteration of (h) (base filter), the setting of the template window size, and the setting of the search window size to get an optimal balance for effective noise reduction and maintain a clear image. Mathematically, this is expressed as:
Where, (I’) is a normalization constant for the image, v(Gm )-v(Gn ) measures the Euclidean distance between patches (Gm ) and (Gn ) with a Gaussian kernel parameterized by (σ), and (h) is the base filter strength for adjusting the sensitivity to pixel differences. It determines the weight, w(m,n) for each pixel (m,n) based on the similarity of its patch (Gm ) to neighboring patches (Gn ). A higher weight is assigned to pixels with similar neighboring patches, ensuring noise reduction while preserving important details.
This equation (6) computes the normalization constant (I’) to ensure that the weights sum up to one for each pixel for proper normalization. The following equation (7) calculates the filtered value EANLM(m) for each pixel. And, v(n) represents the intensity value of the pixel in the image.
These equations describe the process by which the EANLM reduces noise while retaining critical image features such as edges and textures for accurate analysis. The following equation (8) representing the noise reduction image using the EANLM method is:
Here, |P| denotes the set of all patches in the image, and {p1, p2, … p∞} represents the processed patches after noise reduction to preserve important details to improve overall image quality, as shown in Figure 3.
Figure 3.
Pre-processed patches after noise reduction using the ENALM technique.
Algorithm 1. Pseudocode for EANLM noise reduction method applied to histopathology images
| Objective: To remove the noise from the color-normalized image |
| Input: Histopathology Image (D) |
| Output: Pre-processed Image (P) |
| 1: for each pixel (m,n) in D do |
| 2: Compute normalization constant I’ using Eq. (6) |
| 3: Initialize EANLM(m) as the filtered value for pixel (m,n) |
| 4: for each patch (Gm ) in the template window around (m,n) do |
| 5: Calculate weight w(m,n) based on Eq. (5) |
| 6: Accumulate weighted intensity from patches (Gn ) in the search window around (m,n) using Eq. (7) |
| 7: Assign EANLM(m) to P(m,n) using Eq. (8) |
| 8: end for |
| 9: Return P as the pre-processed image after noise reduction |
Segmentation
In segmentation, an image is divided into different parts to isolate some objects of interest. In this particular instance, semantic segmentation sets a category for each pixel of an image, and every pixel is the provided information to which class it corresponds. It will isolate the malignant or benign regions to provide inputs for further feature extraction and classification. A set of mathematical formulas that are followed in semantic segmentation is:
This equation (9) represents the probability distribution (q(xt |P) of a patch (xt ) given to the processed image (P) after noise reduction. Here, (N) denotes a Gaussian distribution, (αt ) is a weight factor, and (1-αt ) represents the variance. It captures the structure of the image. To train the segmentation model, the following equation (10) defines the loss function (L) used by the model to segment the image.
This equation defines the loss function and the expectation (IE) over the distribution of patches (P), noise (∈), and time (t). The term (∈θ) denotes the predicted noise, and the loss function measures the difference between the predicted and actual noise and (αt). It trains the segmentation model by minimizing the difference between predicted and actual noise. The recursive process of denoising can be calculated as:
Where (xt-1) is the denoised patch at the previous time step. The terms (αt-1) and (σt) are parameters controlling the denoising process. This equation (11) iteratively refines the patches, reducing noise and improving details at each step. In this process, the final segmentation is accurate and clear, as shown in the equation (12).
This equation represents the final segmented output (P’), as depicted in Figure 4. Where (f) is a function applied to the denoised patches (xt ) with parameter (σ). By accurately segmenting the images, the features can be extracted effectively and improve the performance of classification tasks.
Figure 4.
Final segmented images using the semantic segmentation method.
Algorithm 2. Pseudocode for semantic segmentation
| Objective: To set a category for each pixel in an image |
| Input: Pre-processed Image (P) |
| Output: Segmented image (P’) |
| 1: Initialize αt , σ // Parameters |
| 2: for each pixel xt in P do |
| 3: Calculate q(xt |P) using Eq. (9) |
| 4: Train segmentation model with the loss function L defined in Eq. (10) |
| 5: Update xt-1 using Eq. (11) to refine patches iteratively |
| 6: if the end of the iteration, then |
| 7: Apply function f to xt with parameter σ // Final segmentation |
| 8: Set the corresponding pixel in P’ to the segmented result |
| 9: end if |
| 10: Output the final segmented image P’ using Eq. (12) |
| 11: end for |
| 12: Return P’ // Return segmented image |
Feature extraction
Feature extraction is the most important process in computer vision. This is a process of identifying and extracting the features from an image to facilitate classification. In this framework, an Improved Gray Level Co-occurrence Matrix (I-GLCM), is used to extract the shape and color features from the segmented images. GLCM captures second-order statistical information by evaluating the spatial relationships between the image pixels. However, GLCM cannot capture more intricate patterns and structures present in histopathology images. To fix this, the I-GLCM model is used. It includes higher-order statistics to create matrices that consider relationships among triplets or quadruplets of pixels instead of pairs. I-GLCM is modeled on the following mathematical equations:
Equation (13) represents the mean (μ) of the segmented output (P’). Where (Sab ) is the value of the co-occurrence matrix at position (a) and (b) are pixel intensities. To understand the overall intensity distribution, the mean measures the average pixel value in the segmented region.
Equation (14) represents the standard deviation (τ) of the segmented image to provide a measure of the variation of the pixel intensities. Here, the term (a-μ)2 measures the deviation of each pixel value from the mean. The standard deviation provides information about the spread of pixel values, indicating the contrast and texture of the image.
Equation (15) represents the contrast of the image, which measures the intensity contrast between a pixel and its neighbor over the entire image. This contrast indicates significant variations in the image for identifying different tissue types.
Equation (16) represents the dissimilarity of the image to measure the absolute differences between the pixel pairs. It highlights subtle variations in texture in the differentiation of tissue structures. By using I-GLCM, these equations allow for capturing more intricate patterns and detailed structures in the image to the feature extraction process. The extracted features can be expressed as:
In equation (17), (R) is the extracted features from the segmented images. The set {r1, r2, … r∞} includes features, such as mean, standard deviation, contrast, and dissimilarity, providing information for accurate classification.
Classification
In this process, the extracted features are input into the U-Net model to distinguish malignant tumors and benign tumors, as shown in Figure 5. The U-Net model works by introducing an encoder-decoder structure. The path to the encoder is made up of the convolutional layer, which repeats the function over and over each time, followed by the activation function as a rectified linear unit (ReLU) and the pooling layer to down-sample the input. This path captures the features of the input image. The process followed in the decoder path is the reverse one: at each stage, the feature map will be upsampled in parallel, and then the convolutional layer is used to reconstruct the spatial dimensions.
Figure 5.
U-Net architecture for tumor classification.
To distinguish between malignant and benign tumors using a GRU U-Net structure. Nonetheless, the U-Net architecture works with fixed-size inputs, thus, medical imaging data presents the main possible issue for different whole-slide sizes. With the addition of the network with GRU in the U-Net structure, GRU U-Net could deal with the sequence lengths for images that are not required to be resized or cropped, thus representing the accurate spatial features and time details in the images for a precise classification of different tumors. The mathematical equations for the GRU integrated into the U-Net architecture are:
Equation (18) represents the update gate (Zt) in the GRU. Here, (ρ) denotes the ReLU activation function, (WZ) is the weight matrix, (kt-1) is the hidden state from the previous time step, (R(xt) represents the feature map at the current input step, and (bZ) is the bias term. The update gate controls the past information that needs to be passed to the future. The following equation (19) denotes the reset gate (rt) in the GRU.
Similar to the update gate, the reset gate (r) determines the past information to forget. Following that, Equation (20) represents the candidate’s hidden state () in the GRU.
Here, (tanh) denotes the hyperbolic tangent function, and (ut*ht-1) represents the element-wise multiplication of the reset gate with the previous hidden state with the current feature map R(xt). This candidate’s hidden state captures the new data to be added to the hidden state. The final hidden state (kt) in the GRU is:
This equation (21) combines the previous hidden state (kt-1) and the candidate’s hidden state (), weighted by the update gate (zt). This state captures the relevant data needed for the current time step. The output (yi) of the GRU U-Net is represented as:
The (yi ) represents the classification predictions produced by the GRU U-Net model (malignant or benign tumors) on the input image, as shown in Figure 6. The ReLU activation function holds a non-linear model for learning the complexity of the data. In this way, the tumor interface features are explicitly detected in the GRU U-Net, giving higher accuracy in malignant and benign tumors. Finally, XAI visual explanations of this model help clinicians understand the model’s decision-making process.
Figure 6.
Cancerous tumors (malignant) and non-cancerous tumors (benign) for 8 different classes from the BreakHis dataset.
Algorithm 3. Pseudocode for tumor classification
| Objective: To predict the classification type as benign or malignant |
| Input: Segmented image features (|R|) |
| Output: Classification predictions (yi ) |
| 1: Feature extraction using Improved GLCM (I-GLCM) |
| 2: Compute mean (μ) using Eq. (13) |
| 3: Compute standard deviation (τ) using Eq. (14) |
| 4: Compute contrast using Eq. (15) |
| 5: Compute dissimilarity using Eq. (16) |
| 6: Initialize U-Net architecture |
| 7: for each time step t do |
| 8: Calculate update gate zt using Eq. (18) |
| 9: Calculate reset gate rt using Eq. (19) |
| 10: Calculate the candidate’s hidden state using Eq. (20) |
| 11: Update hidden state kt using Eq. (21) |
| 12: Generate classification predictions: |
| 13: for each output unit i do |
| 14: Compute predictions yi using ReLU activation on the final hidden state kt |
| 15: end for |
| 16: return Classification predictions (yi ) |
Results and discussions
This section presents the experimental results of the study. Implementation is done on a Python platform with CPU-Intel(R) Xeon(R) CPU E5-1650 v3 @ 3.50GHz GPU-NVIDIA Quadro M2000, Python 3.10, Windows 10 Pro for Workstations, 64-bit operating system, x64-based processor. It shows the classification performance according to the relevant training, testing, and validation trends by the use of the BreakHis dataset.
Performance analysis
The performance of the proposed GRU U-Net model in classifying breast cancer is validated by making the BreakHis dataset that consists of tumors of various types. This dataset was divided into training and testing sets. The details division of the dataset into different classes is shown in Table 1.
Table 1.
Dataset distribution for training and testing
| Class | Total | Train | Test |
|---|---|---|---|
| Adenosis | 113 | 79 | 34 |
| Ductal Carcinoma | 890 | 623 | 267 |
| Fibroadenoma | 260 | 182 | 78 |
| Lobular Carcinoma | 170 | 119 | 51 |
| Mucinous Carcinoma | 222 | 155 | 67 |
| Papillary Carcinoma | 142 | 99 | 43 |
| Phyllodes Tumor | 121 | 84 | 37 |
| Tubular Adenoma | 150 | 105 | 45 |
The performance metrics for the proposed Patho-Net model of classifying breast cancer, as shown in Figure 7. Accuracy measures the overall correctness of the model. Precision, sensitivity, and F-measure detecting of cancerous cases by reducing false positives and negatives. Specificity and negative predictive value (NPV) indicate correctly recognizing non-cancer cases. The Matthews correlation coefficient (MCC) further highlights the model performance to class balance. Lastly, the false positive rate (FPR) and false negative rate (FNR) represent the model’s efficiency in reducing classification errors. Table 2 shows the proposed Patho-Net model’s performance metrics for breast cancer classification.
Figure 7.
Performance analysis of the proposed Patho-Net model.
Table 2.
Performance metrics of the proposed Patho-Net model
| Metric | Value |
|---|---|
| Accuracy | 98.90244 |
| Precision | 98.60976 |
| Sensitivity | 98.60976 |
| Specificity | 99.37282 |
| F measure | 98.60976 |
| MCC | 96.98258 |
| NPV | 99.37282 |
| FPR | 0.627178 |
| FNR | 4.390244 |
The training and testing loss curves for the evaluation of how well the model works and improves with time are shown in Figure 8. Loss reduction was a result of the value of being able to learn from the data and thus minimize error classification in breast cancer by the model. Both plots show a decrease in loss. Hence, the model works better and improves in generalization to new data.
Figure 8.

Training and validation of accuracy and loss.
To evaluate the performance of a classification model, a confusion matrix is used, as shown in Figure 9. It shows the estimated classes for a total of eight diseases. The cells diagonal reflects that the prediction is correct ((TP)-True Positives and (TN)-True Negatives, while the cells off-diagonal represent the misclassification ((FP)-False Positives and (FN)-False Negatives. The matrix supports identifying the aspects where the model is capable of differentiating these classes with the highest accuracy. The results of the confusion matrix are shown in Table 3.
Figure 9.
A Confusion matrix to evaluate the performance of a classification model.
Table 3.
Confusion matrix outcomes
| Classes | TP | TN | FP | FN |
|---|---|---|---|---|
| Adenosis | 32 | 748 | 2 | 1 |
| Ductal Carcinoma | 261 | 517 | 5 | 1 |
| Fibroadenoma | 74 | 710 | 1 | 1 |
| Lobular Carcinoma | 40 | 744 | 1 | 1 |
| Mucinous Carcinoma | 67 | 718 | 1 | 1 |
| Papillary Carcinoma | 32 | 748 | 1 | 1 |
| Phyllodes Tumor | 0 | 780 | 0 | 0 |
| Tubular Adenoma | 48 | 732 | 0 | 0 |
Comparative analysis
The primary goal of this study is to measure the effectiveness of breast cancer classification using U-Net-GRU. It is compared to several cutting-edge models like Extreme Gradient Boosting (XGBoost) [27], Support Vector Machine (SVM) [28], A-Network (A-Net) [29], and Residual Network (ResNet50V2) [30].
Figure 10 exhibits the comparative values for the accuracy, precision, recall, and F1-score for each model. Among the other models, the proposed U-Net-GRU model is much better in terms of all indices. This strategy is based on U-Net architecture fused with GRU, which strengthens the model’s capability to capture complex patterns in the histopathology images to improve accuracy and performance. Table 4 provides performance details of the classification models.
Figure 10.
Comparative performance analysis of breast cancer classification models.
Table 4.
Performance comparison of classification models
Table 5 represents the comparison of the proposed model against the literature survey in terms of accuracy. Compared to other models in the literature, the proposed U-Net-GRU model is more accurate in classifying breast cancer diagnosis.
Table 5.
Accuracy comparison with literature models
| References | Model Used | Accuracy (%) |
|---|---|---|
| Reshma et al. [17] (2022) | CAD-CNN | 92.44 |
| Mohanakurup et al. [19] (2022) | CDBN | 92.00 |
| Eltoukhy et al. [20] (2022) | DL-NN | 96.30 |
| Sharma et al. [21] (2022) | SVM | 96.25 |
| Lu et al. [22] (2023) | YOLOv4 | 96.00 |
| Tagnamas et al. [23] (2024) | CNN | 86.00 |
| Hossain et al. [25] (2024) | D-CNN | 87.82 |
| Islam et al. [26] (2024) | BC-CNN | 95.20 |
| Proposed | U-Net-GRU | 98.90 |
Explainable artificial intelligence
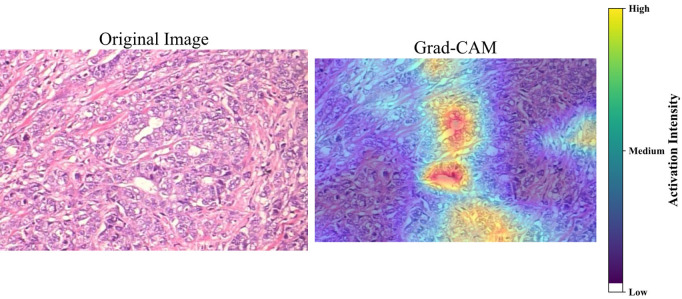
The paper is centered on XAI use to make the U-Net model in breast cancer diagnosis from histopathology images more interpretable. For cancerous tumors, the XAI models used Grad-CAM and XRAI, and they are used for the classes Ductal, Lobular, Mucinous, and Papillary Carcinoma (shown in Figure 11). XAI techniques allow the doctors to visualize the paths of the model’s decision-making processes to medical professionals in bridging the gap between advanced AI technologies and the clinical meaning of it, hence yielding better patient results.
Figure 11.
XAI visualization methods of Grad-CAM and XRAI. It highlights important regions in histopathology images in the diagnosis of cancerous tumors.
Conclusion
In conclusion, this paper proposed a novel Patho-Net method for breast cancer classification. This framework provides a precise classification with clear visual explanations of the predictions. In the Patho-Net model, high-quality input data are ensured through the integration of the GRU network with the U-Net architecture. Also, the Reinhard method and EANLM filtering are used for noise removal in the color normalization process. Additionally, the I-GLCM image texture features-based feature extraction techniques were used to analyze the specific patterns in the tumor regions. Hence, the model can distinguish the cancerous and non-cancerous tissues. Finally, the use of XAI visualizes the model’s decision-making process by highlighting the tumor areas of histopathological images. The experimental results show that Patho-Net is effective in being able to reach an accuracy of 98.90% using the BreakHis dataset. The use of Patho-Net can become significant in the earlier classification of breast cancer treatment of patients to another level in the healthcare field, thus creating healthcare for better results and improved patient care. However, this model does not analyze the stage of the tumor. Future work will focus on improving the model to classify the tumor stage into intermediate, advanced stages, or deadly cancers.
Disclosure of conflict of interest
None.
References
- 1.Batool A, Byun YC. Toward improving breast cancer classification using an adaptive voting ensemble learning algorithm. IEEE Access. 2024;12:12869–12882. [Google Scholar]
- 2.Taheri S, Golrizkhatami Z, Basabrain AA, Hazzazi MS. A Comprehensive study on classification of breast cancer histopathological images: binary versus multi-category and magnification-specific versus magnification-independent. IEEE Access. 2024;12:50431–50443. [Google Scholar]
- 3.Koshy SS, Anbarasi LJ. LMHistNet: levenberg-marquardt based deep neural network for classification of breast cancer histopathological images. IEEE Access. 2024;12:52051–52066. [Google Scholar]
- 4.Wakili MA, Shehu HA, Sharif MH, Sharif MHU, Umar A, Kusetogullari H, Ince IF, Uyaver S. Classification of breast cancer histopathological images using DenseNet and transfer learning. Comput Intell Neurosci. 2022;2022:8904768. doi: 10.1155/2022/8904768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phasukkit P. Non-ionic deep learning-driven IR-UWB multiantenna scheme for breast tumor localization. IEEE Access. 2022;10:4536–4549. [Google Scholar]
- 6.AmeliMojarad M, AmeliMojarad M, Pourmahdian A. The inhibitory role of stigmasterol on tumor growth by inducing apoptosis in Balb/c mouse with spontaneous breast tumor (SMMT) BMC Pharmacol Toxicol. 2022;23:42. doi: 10.1186/s40360-022-00578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael E, Ma H, Li H, Qi S. An optimized framework for breast cancer classification using machine learning. Biomed Res Int. 2022;2022 doi: 10.1155/2022/8482022. 8482022, 18 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashmi R, Prasad K, Udupa CBK. Breast histopathological image analysis using image processing techniques for diagnostic purposes: a methodological review. J Med Syst. 2021;46:7. doi: 10.1007/s10916-021-01786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta SR. Prediction time of breast cancer tumor recurrence using machine learning. Cancer Treat Res Commun. 2022;32:100602. doi: 10.1016/j.ctarc.2022.100602. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Wang N, Du X, Mei K, Zhou Y, Cai G. Classification prediction of breast cancer based on machine learning. Comput Intell Neurosci. 2023;2023:6530719. doi: 10.1155/2023/6530719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niyas S, Bygari R, Naik R, Viswanath B, Ugwekar D, Mathew T, Kavya J, Kini JR, Rajan J. Automated molecular subtyping of breast carcinoma using deep learning techniques. IEEE J Transl Eng Health Med. 2023;11:161–169. doi: 10.1109/JTEHM.2023.3241613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank SJ. A deep learning architecture with an object-detection algorithm and a convolutional neural network for breast mass detection and visualization. Healthcare Analytics. 2023;3:100186. [Google Scholar]
- 13.Anas M, Ul Haq I, Husnain G, Jaffery SAF. Advancing breast cancer detection: enhancing YOLOv5 network for accurate classification in mammogram images. IEEE Access. 2024;12:16474–16488. [Google Scholar]
- 14.Ortega-Ruíz MA, Karabağ C, Roman-Rangel E, Reyes-Aldasoro CC. DRD-UNet, a UNet-like architecture for multi-class breast cancer semantic segmentation. IEEE Access. 2024;12:40412–40424. [Google Scholar]
- 15.You G, Yang X, Lee X, Zhu K. EfficientUNet: an efficient solution for breast tumour segmentation in ultrasound images. IET Image Process. 2024;18:523–534. [Google Scholar]
- 16.Xu M, Huang K, Qi XA. Regional-attentive multi-task learning framework for breast ultrasound image segmentation and classification. IEEE Access. 2023;11:5377–5392. [Google Scholar]
- 17.Reshma VK, Arya N, Ahmad SS, Wattar I, Mekala S, Joshi S, Krah D. Detection of breast cancer using histopathological image classification dataset with deep learning techniques. Biomed Res Int. 2022;2022:8363850. doi: 10.1155/2022/8363850. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Shah D, Khan MAU, Abrar M. Reliable breast cancer diagnosis with deep learning: DCGAN-driven mammogram synthesis and validity assessment. Applied Computational Intelligence and Soft Computing. 2024;2024:1122109. [Google Scholar]
- 19.Mohanakurup V, Parambil Gangadharan SM, Goel P, Verma D, Alshehri S, Kashyap R, Malakhil B. Breast cancer detection on histopathological images using a composite dilated backbone network. Comput Intell Neurosci. 2022;2022:8517706. doi: 10.1155/2022/8517706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eltoukhy MM, Hosny KM, Kassem MA. Classification of multiclass histopathological breast images using residual deep learning. Comput Intell Neurosci. 2022;2022:9086060. doi: 10.1155/2022/9086060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S, Kumar S. The Xception model: a potential feature extractor in breast cancer histology images classification. ICT Express. 2022;8:101–108. [Google Scholar]
- 22.Lu Y, Zhang J, Liu X, Zhang Z, Li W, Zhou X, Li R. Prediction of breast cancer metastasis by deep learning pathology. IET Image Process. 2023;17:533–543. [Google Scholar]
- 23.Tagnamas J, Ramadan H, Yahyaouy A, Tairi H. Multi-task approach based on combined CNN-transformer for efficient segmentation and classification of breast tumors in ultrasound images. Vis Comput Ind Biomed Art. 2024;7:2. doi: 10.1186/s42492-024-00155-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hossain MS, Rahman MS, Ahmed M, Alfaz N, Shifat SM, Syeed MM, Hussen MA, Uddin MF. Residual tumor cellularity assessment of breast cancer after neoadjuvant therapy using image transformer. IEEE Access. 2024;12:86083–86095. [Google Scholar]
- 25.Islam MR, Rahman MM, Ali MS, Nafi AAN, Alam MS, Godder TK, Miah MS, Islam MK. Enhancing breast cancer segmentation and classification: an ensemble deep convolutional neural network and U-net approach on ultrasound images. Mach Learn Appl. 2024;16:16100555. [Google Scholar]
- 26.Thakur A, Gupta M, Sinha DK, Mishra KK, Venkatesan VK, Guluwadi S. Transformative breast cancer diagnosis using CNNs with optimized ReduceLROnPlateau and early stopping enhancements. International Journal of Computational Intelligence Systems. 2024;17:14. [Google Scholar]
- 27.Laghmati S, Hamida S, Hicham K, Cherradi B, Tmiri A. An improved breast cancer disease prediction system using ML and PCA. Multimed Tools Appl. 2024;83:33785–33821. [Google Scholar]
- 28.Hemalatha B, Karthik B, Reddy CK, Gokulakrishnan D. Cervical cancer classification: optimizing accuracy, precision, and recall using SMOTE preprocessing and t-SNE feature extraction. 2024 International Conference on Intelligent and Innovative Technologies in Computing, Electrical and Electronics (IITCEE). IEEE; 2024. pp. 1–7. [Google Scholar]
- 29.Thwin SM, Malebary SJ, Abulfaraj AW, Park HS. Attention-based ensemble network for effective breast cancer classification over benchmarks. Technologies. 2024;12:16. [Google Scholar]
- 30.Sharmin S, Ahammad T, Talukder MA, Ghose P. A hybrid dependable deep feature extraction and ensemble-based machine learning approach for breast cancer detection. IEEE Access. 2023:87694–87708. [Google Scholar]