Abstract
Introduction: Ischemic Stroke (IS) is characterized by complex molecular alterations involving disruptions in lipid metabolism and immune interactions. However, the roles of lipid metabolism-associated genes in the pathogenesis of IS through immune regulation interaction are rarely explored. In this study, we aimed to explore the intricate correlation between lipid metabolism-associated immune changes and IS through a machine-learning algorithm. Materials and methods: We downloaded the GSE16561, GSE22255, and GSE37587 datasets from NCBI. Using the GSE16561 dataset, we analyzed differential gene expression profiles related to lipid metabolism with the “Limma” R package. We constructed a diagnostic model employing techniques such as Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression and Random Forest (RF), which was further validated using the independent GSE22255 and GSE37587 datasets. Correlations between model genes and immune cell percentages were examined by Spearman analysis. We further validated the diagnostic value of these model genes in 28 clinical samples using RT-qPCR. Results: We identified 26 lipid metabolism genes with significant expression disparities between normal and diseased groups, closely linked to immune cell populations. Seven signature genes (ACSS1, ADSL, CYP27A1, MTF1, SOAT1, STAT3, and SUMF2) were identified using LASSO and RF algorithms for a potential diagnostic model, effectively distinguishing healthy and IS samples in both training and validation (AUC = 0.725) datasets. The mRNA expression levels of these model genes were further validated as a blood biomarker for IS patients in our clinical samples. Single-cell analysis further revealed high expression of Cyp27a1 in dendritic cells and macrophages, and decreasing expression of Soat in progenitor cells as the disease progressed. The expression of Stat3 in most immune cells was upregulated in progenitor cells as the disease progressed. Additionally, a regulatory network identified transcription factors regulating genes such as STAT3. Conclusion: This study identified novel lipid metabolism biomarkers for IS, enhancing our understanding of IS by shedding light on lipid metabolism and immune interactions. This may facilitate innovative diagnostic approaches to IS.
Keywords: Ischemic stroke, machine learning, gene signature, lipid metabolism, immune cells
Introduction
Ischemic Stroke (IS), a neurological ailment, is a significant global health issue associated with high morbidity and mortality rates [1,2]. IS, characterized by restricted blood supply to brain tissue, triggers a cascade of events leading to cellular damage and neurological dysfunction [3]. Inflammation plays a significant role in elevating the risk of stroke through a multitude of interconnected mechanisms [4]. Recent research suggests that immunomodulation is affected by cellular metabolism [5], including lipid metabolism, also termed as immunometabolism [6,7]. Rapid and accurate biomarkers regarding lipid metabolism-modulated immune cells are imperative to develop personalized treatment regimens for IS patients.
The intersection of lipid metabolism and inflammation in the pathogenesis of IS [8-10], uncovered a robust association between immune-related dyslipidemia and the risk of IS development. A study examining the TLR8 rs3764880 polymorphism found an association with IS susceptibility linked to inflammatory response-related lipid metabolism [11]. IL-1β was positively correlated with high-density lipoprotein (HDL) levels in IS patients [12,13]. Additionally, all vegetable oils and trans fats, except sesame oil, were found to increase body weight and body fat, possibly elevating the risk of IS in rats [14]. Serum HDL cholesterol concentrations were a protective factor in IS patients [15]. Plasma homocysteine and lipoprotein cholesterol levels affect early recovery from neurological disorders, which was also associated with immune responses [16]. Recent studies explored lipid profiles and neuroprotective biomarkers in patients with hemorrhagic stroke, as well as neuroprotective approaches in preclinical studies and the interaction of lipid mediators in IS [17]. These studies reveal an association between IS pathogenesis and different characteristics of immune regulation and lipid metabolism, which will help design individualized treatment for IS patients.
Peripheral whole blood is widely recognized as a biomarker for systemic inflammation and may play a role in IS processes [5]. Due to its non-invasive nature, low risk, and ease of repetitive sampling, peripheral blood has become increasingly attractive to researchers for the development of blood-based biomarkers [18], especially for patient selection and treatment monitoring [19]. Machine learning algorithms have significantly advanced biomedical research by facilitating the analysis of complex datasets and extracting patterns from extensive omics data [20]. The Least Absolute Shrinkage and Selection Operator (LASSO) imposes penalties on regression coefficients, promoting the selection of informative genes as potential biomarkers [21]. The Random Forest (RF) algorithm excels in handling complex and noisy data. Integration of these algorithms empowers researchers to navigate the intricacies of inflammation and lipid metabolism interactions in IS.
In this study, we aimed to explore the intricate correlation between lipid metabolism-associated immune changes and IS through a machine-learning algorithm. We acquired two datasets from the Gene Expression Omnibus (GEO) to compare the gene expression profiles from peripheral whole blood cells between individuals with IS and healthy controls. By intersecting these differentially expressed genes (DEGs) with genes related to lipid metabolism, we identified vital lipid metabolism genes and their associations with immune cell subtypes. Employing the RF and Lasso approaches, we developed an IS classification model and evaluated its performance on independent validation sets and our clinical samples. This study fills the gap between lipid metabolism, immunomodulation, and diagnostic precision in IS.
Materials and methods
Data and preprocessing
Transcriptomic data expression data and clinical parameters were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo). In selecting a dataset from the GEO database for this study, the following criteria were applied to ensure its appropriateness: the dataset must include both control (healthy) and disease (ischemic stroke, IS) samples for comparative analysis; each group must contain more than 20 samples to ensure sufficient statistical power and reliability; the gene expression profiling platform using microarray must be suitable for our study; and the dataset must primarily consist of blood samples, which are the focus of our research. Transcriptomic data, expression data, and clinical data were downloaded from the GEO database. Microarray datasets GSE16561, GSE22255 and GSE37587, which contained the transcriptomic data from whole blood cell samples, were included in this study. The dataset GSE16561 comprises 39 IS samples and 24 healthy samples, serving as the training set. The validation dataset (GSE22255 and GSE37587) includes 34 IS samples and 39 healthy samples, intended for validation purposes.
Differentially expressed genes analysis
The “Limma” R package was employed to identify differentially expressed genes (DEGs) between the control and disease groups within the training dataset of IS patients (GSE16561), with an adjusted p-value threshold of < 0.05. Furthermore, the DEGs were subjected to Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis using R’s clusterProfiler package.
Functional enrichment analysis
To explore the biological functions linked with the identified differential genes, we performed functional enrichment analysis employing the “clusterProfile” R package. Initially, we pinpointed genes enriched in both Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, assessing their significance by adjusted p-values, with a threshold set at < 0.05. Subsequently, we visualized the enriched GO terms and KEGG pathways utilizing the R package “ggplot2”.
Screening of lipid metabolism-related genes
The lipid metabolism-related genes were obtained from the Molecular Signature Database (MsigDB, v7.5.1, https://www.gsea-msigdb.org/), including the following pathways: glycerophospholipid metabolism, adipocytokine signing pathway, PPAR signaling pathway, glycerolipid metabolism, regulation of lipolysis in adipocytes, fatty acid metabolism, arachidonic acid metabolism, sphingolipid metabolism, cholesterol metabolism, fatty acid degradation, ether lipid metabolism, steroid hormone biosynthesis, fatty acid elongation, fat digestion and absorption, biosynthesis of unsaturated fatty acids, steroid biosynthesis, linoleic acid metabolism, alpha-linolenic acid metabolism, primary bile acid biosynthesis. The lipid metabolism-related genes were further filtered by intersecting with DEGs between healthy and IS samples in the GEO dataset.
Establishment and evaluation of the model
The LASSO (Least Absolute Shrinkage and Selection Operator) Cox regression analysis was conducted using the “Glmnet” R package. This technique served a dual purpose of alleviating overfitting concerns and enabling the creation of a predictive gene signature for prognostic purposes. The “randomForest” R package was utilized to analyze RF. In summary, the IS gene expression profile data from the training set were sorted based on the mean decrease in accuracy, leading to the selection of feature genes. The decision tree count was established at 500. Through five-fold cross-validation, the optimal sample size for RF was determined, effectively minimizing model error. The optimal gene combination was screened by integrating LASSO and RF analysis results.
To further assess the reliability of this signature’s prognostic potential, both a training set and an externally distinct validation cohort were employed. The formula used for calculating the prognostic score was:
The receiver operating characteristic (ROC) curve of the model and genes were plotted, and the area under the curve (AUC) of the ROC curve evaluated their performance. Subsequently, the Wilcoxon-test was employed to assess the expression differences of the genes encompassed within the model. This evaluation included comparisons between IS and normal conditions, as well as comparisons between different feature groups.
Evaluation of immune microenvironment in patients with ischemic stroke
The immune cell proportions within each sample were calculated using TIMER (http://timer.cistrome.org/) and CIBERSOR (https://cibersortx.stanford.edu/) [22]. The correlation between the model genes and immune cell proportions was analyzed using Spearman Correlation analysis, demonstrated with Holm’s adjustment p value.
Patient population and sample collection
The study was conducted in Tongji Hospital, affiliated with Tongji University, and approved by the Institutional Review Board (IRB) of the Tongji Hospital authority (IRB number: 2020-KYSB-190-XZ-210526). Newly diagnosed IS patients or healthy controls with signed informed consent were eligible for this study. Clinical factors were collected, including age, disease history, and gender. 5 mL of peripheral venous blood was collected in EDTA-anticoagulant blood tubes from the newly diagnosed IS patients and immediately stored at -80°C. The Monarch Total RNA Miniprep Kit (NEB #T2010) was used to extract the RNA from the frozen whole blood samples.
Reverse transcription-quantitative PCR (RT-qPCR) assay
RNA was used for reverse transcription (RT) using Servicebio® RT First Strand cDNA Synthesis Kit (Cat: G3330). qPCR was conducted using a Stepone plus qPCR system (ABI) and a 2×SYBR Green qPCR Master Mix (High ROX, Cat: G3321) to determine the mRNA expression levels of the ACSS1 (Acyl-CoA synthetase short-chain family member 1), ADSL (Adenylosuccinate lyase), CYP27A1 (Cytochrome P450 family 27 subfamily A member 1), MTF1 (Metal regulatory transcription factor 1), SOAT1 (Sterol O-acyltransferase 1), STAT3 (Signal transducer and activator of transcription 3), and SUMF2 (Sulfatase modifying factor 2). The thermocycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 58°C for 15 s, and 72°C for 10 s on a 384 plate with LCM 480 Detector (Roche). Each reaction was performed in duplicates, and expression levels were normalized to those of GAPDH. Quantitation was performed as 2-ΔΔCt method. The primers that were used for qPCR are listed in Table 1.
Table 1.
Primer sequences
| Target Gene | Forward Primer (5’-3’) | Reverse Primer (5’-3’) |
|---|---|---|
| ACSS1 | CACAGGACAGACAACAAGGTC | CCTGGGTATGGACGATGCC |
| ADSL | GCTGGAGGCGATCATGGTTC | TGATAGGCAAACCCAATGTCTG |
| CYP27A1 | GGTGCTTTACAAGGCCAAGTA | TCCCGGTGCTCCTTCCATAG |
| MTF1 | CACAGTCCAGACAACAACATCA | GCACCAGTCCGTTTTTATCCAC |
| SOAT1 | CAAGGCGCTCTCTCTTAGATG | GGTCCAAACAACGGTAGGAAA |
| STAT3 | CAGCAGCTTGACACACGGTA | AAACACCAAAGTGGCATGTGA |
| SUMF2 | CAGAACAACTACGGGCTCTATG | CAGTGACCCTAGAAGGCTTTTC |
Single cell analysis of gene expression pattern
The data resource website (https://anratherlab.shinyapps.io/strokevis/) explored the expression pattern of these genes in immune cells to further demonstrate this.
Regulatory network analysis
Aiming to uncover the transcription factors (TFs) regulating the model genes, interaction pairs between transcription factors and their target genes were explored from the TRRUST v2 database (https://www.grnpedia.org/trrust/).
Statistical analysis
All statistical analyses were performed using the R software (version 4.3.0). Heatmaps were generated and visualized using the “pheatmap” package, while the “ggvenn” package was utilized for creating Venn diagrams. “pROC” package was employed to visualize the receiver operating characteristic (ROC) curves. The difference in mRNA expression from the GEO database was compared using the Wilcoxon rank-sum test. The unpaired Student’s T-test was used for mRNA expression level by qPCR comparison analysis. The correlation among immune cells and genes was evaluated by Spearman correlation analysis. Data were visualized using the R package “ggplot2” or “plot” unless otherwise specified. P < 0.05 was considered significant. The significance level was denoted as follows: NS, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Differences in molecular characteristics among ischemic stroke patients
In our analysis, we initially employed the limma package to the genes that exhibited significant differential expression (adjusted p-value < 0.05) within the GSE16561 dataset, differentiating between the healthy and disease groups based on criteria of |log2FC| > 1 and p-value < 0.05. Ultimately, this process yielded a set of 441 differentially expressed genes (Figure 1A; Supplementary Data). To further elucidate the characteristics of the differential genes, we utilized the clusterProfiler package in R to perform comprehensive functional enrichment analyses involving Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations. Our results indicated that the differential genes were significantly enriched in various biological processes, notably the apoptotic process, positive transcription regulation, and positive cell proliferation regulation (Figure 1B; Supplementary Table 1). The genes were enriched in molecular functions such as protein binding, RNA binding and ATP binding (Figure 1B; Supplementary Table 1). Among biological functions, these genes were associated with “positive regulation of transcription from a DNA template”, “positive regulation of cell proliferation”, “protein phosphorylation”, “intracellular signal transduction”, “immune response”, “inflammatory response”, and “regulation of the cell cycle” (Figure 1B). KEGG pathway analysis revealed that these genes were enriched in pathways related to cancer, Epstein-Barr virus infection, Kaposi’s sarcoma-associated herpesvirus infection, and human cytomegalovirus infection (Figure 1C; Supplementary Table 1). In terms of both biological functions and KEGG pathway analysis, the differentially expressed genes were predominantly enriched in immune system-related functions and pathways, particularly those involving cancer and viral infections.
Figure 1.
Differential gene analysis of GSE16561. A. Volcano plot of differentially expressed genes in GSE16561. B. Gene Ontology (GO) enrichment Analysis of Differential Genes in GSE16561. C. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differential genes in GSE16561. GO and KEGG enrichment analyses were performed on the differentially expressed genes within the GSE16561 dataset. D. Venn plot of lipid metabolism associated differential expressed genes in GSE16561. LMDEG: lipid metabolism-associated differential expressed genes.
Furthermore, we performed an intersection analysis between lipid metabolism-associated genes and the DEGs, resulting in 26 Lipid Metabolism Differentially Expressed Genes (LMDEGs) (Figure 1D). The enrichment of these LMDEGs in lipid metabolism pathways was associated with the GO pathway. The positive regulation of transcription, protein binding, RNA binding, and ATP binding suggests that they play crucial roles in lipid synthesis, degradation, and transport, revealing these genes’ potential importance in lipid-related biological processes.
Immune cell and LMDEGs associations
Initially, immune cell proportions for each sample were calculated using TIMER and CIBERSORTx methodologies (Supplementary Table 1). Subsequently, the Spearman correlation coefficients were used to explore the correlation between LMDEGs and immune cell infiltration (Supplementary Table 1). Remarkably, a majority of the LMDEGs displayed significant and strong correlations with multiple immune cell types (Figure 2A). ACSS1 was significantly associated with resting memory CD4+ T cell, gamma delta T cell, M2 macrophage, eosinophil and activated myeloid dendritic cell. CYP27A1 was significantly associated with type I macrophage. DEGS1 was significantly associated with memory B cells, activated NK cells, neutrophils, monocytes, eosinophils, naïve CD4+ T cells, and resting myeloid dendritic cells. GLB1 was significantly associated with neutrophils, memory B cells and activated NK cells. MTF1 was significantly associated with memory B cells, activated NK cells, monocytes, naïve CD4+ T cells, type I macrophage and resting myeloid dendritic cells (Figure 2B).
Figure 2.
Immune cell and LMDEGs associations in ischemic stroke patients. A. Heatmap depicting the correlation between immune cells and lipid metabolism differentially expressed genes by TIMER. B. Heatmap depicting the correlation between immune cells and lipid metabolism differentially expressed genes by TIMER by CIBERSORTx. *, **, and *** denote significance levels: P < .05, P < .01, P < .001, respectively. The color gradient shifts from purple to yellow, indicating correlation coefficients from 1 to -1.
Model establishment and evaluation
We initiated LASSO regression within the training dataset, aiming to construct a robust classification model using the LMDEGs previously identified (Figure 3A). Subsequently, we selected the vital genes using the RF method, in which these first 12 variables can explain the total variation in samples (Figure 3B). The RF importance plot identified 12 vital genes using mean decrease accuracy and GINI analysis (Figure 3C). We further constructed the model by the integration of 7 specific genes (Supplementary Table 1). The calculation formula for this model is as follows: Score = ACSS1 × 3.14 + ADSL × 0.41 + CYP27A1 × 0.71 + MTF1 × 2.17 + SOAT1 × 0.51 + STAT3 × 1.94 + SUMF2 × 0.64 (Supplementary Table 1).
Figure 3.
Model development and evaluation. A. LASSO coefficient analysis. Vertical dashed lines are plotted at the best lambda. B. Cumulative variance is explained by a number of variables from the random forest model. C. Random forest variable importance plot. D. Mean decrease accuracy plot: the measure of the performance of the model without each gene. A higher value indicates the importance of that gene in predicting the group (Ischemic Stroke vs. healthy). E. ROC curves show the diagnostic value of the model genes in the training dataset. F. ROC curves show the diagnostic value of the model genes in the validation dataset. ROC: Receiver operating characteristic; AUC: Area Under the ROC Curve.
The model’s performance on both the training and validation sets was characterized by ROC curves. Notably, the AUC for the training set reached approximately 1.0. Subsequently, we assessed the predictive accuracy of individual gene expressions for the disease. The genes exhibited high predictive accuracy, specifically: ACSS1 (AUC: 0.66), ADSL (AUC: 0.68), CYP27A1 (AUC: 0.70), MTF1 (AUC: 0.76), SOAT (AUC: 0.70), SUMF2 (AUC: 0.69), and STAT3 (AUC: 0.80) (Figure 3D). Furthermore, we validated the gene-signature model in the external GEO dataset (GSE 22255). ROC curves indicated that the model could distinguish IS samples from healthy samples (AUC: 0.725) (Figure 3E). The genes exhibited high predictive accuracy, specifically: ACSS1 (AUC: 0.70), ADSL (AUC: 0.65), CYP27A1 (AUC: 0.68), MTF1 (AUC: 0.69), SOAT (AUC: 0.69), SUMF2 (AUC: 0.72), and STAT3 (AUC: 0.68) (Figure 3F).
Differential analysis of model gene expression pattern
Initially, we analyzed individual gene expression profiles of the model genes in both public database and our clinical samples. This analysis revealed elevated expression levels of CYP27A1, MTF1, SOAT1, and STAT3 in IS patients, whereas SUMF2 and ADSL exhibited lower expression levels in IS patients. ACSS1 expression remained unchanged (Figure 4A).
Figure 4.
Differential analysis of model gene mRNA expression level. A. Box plot showing the different levels of ACSS1, ADSL, CYP27A1, MTF1, SOAT1, STAT3, and SUMF2 mRNA levels in datasets. B. Box plot showing quantification of ACSS1, ADSL, CYP27A1, MTF1, SOAT1, STAT3, and SUMF2 mRNA level, with values normalized to GAPDH by qRT PCR. Student T-test was used for statistical analysis. Ns: no significance; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 compared to the healthy group.
To corroborate further the possible association between the mRNA expression of model genes in whole blood cells and IS, we performed quantitative polymerase chain reaction (Q-PCR) assays on blood samples obtained from 16 IS patients and 12 healthy controls (Supplementary Table 2). The results indicated that the mRNA expression levels of CYP27A1, MTF1, SOAT1, SUMF2, and STAT3 were significantly higher in IS patient samples compared to those of healthy controls (P < 0.05; Figure 4B), consistent with our bioinformatic analysis findings. Conversely, significantly lower levels of ADSL were observed, while ACSS1 expression remained unchanged between the IS and healthy control groups.
Single-cell analysis of model gene expression pattern
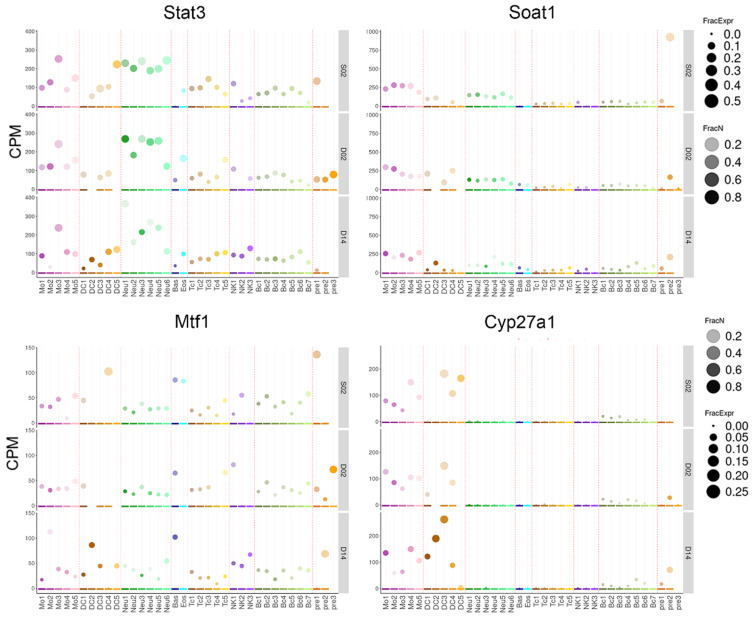
Subsequently, the single-cell dataset related to the progression of IS was employed to analyze the expression patterns of model genes across different immune cell types from mice to help further elucidate the potential interaction between these model genes and immune cell functions. Cyp27a1 was highly expressed in dendritic cells and macrophages. Mtf1 was overexpressed in dendritic cells (group 2) and eosinophils/basophils. During disease progression, the expression of Mtf1 also increased in progenitor cells. Soat was expressed in progenitor cells (group 1), and its level decreased as the disease progressed. Finally, Stat3 was abundantly expressed in most immune cells and upregulated in progenitor cells as the disease progressed (Figure 5).
Figure 5.
Single-cell RNA analysis of model gene expression during IS progression. Dot plot illustrating contrasting distribution patterns of Immune Response Gene Sets between ischemic stroke and healthy control subjects. Y-axis (CPM): Counts per million transcripts within the cluster; only expression levels of genes detected in 0.1% of cells within the cluster and clusters with at least 10 cells are plotted. Size of circles (FracExpr): fraction of cells in the cluster expressing the gene (1 = all cells express it). Color saturation (FracN): cell number distribution at different time points (1 = all cells of the cluster). S02 = Two Day Sham; D02 = Two Day Stroke; D14 = Fourteen Day Stroke. Bc: B cells; DC: dendritic cells; Eos.Bas: Eosinophils/Basophils; Mo: monocytes; Neu: neutrophils; NK: NK cells; pre: various progenitors; Tc: T cells.
Regulatory network analysis
To elucidate the underlying regulatory mechanisms by which these model genes modulate the IS progression, we examined whether these genes could regulate or be regulated by transcription factors (TFs) from the TRUST database. Among the genes analyzed, CYP27A1, MTF1, and STAT3 were identified in the TRUST database. CYP27A1 is regulated by TFs including NR0B2, SP1, and SP3 (Figure 6A). MTF1 can target TFs such as ACO1, GCLC, MCAT, and PGF (Figure 6B). STAT3 can target genes like A2M, ABCA1, AKAP12, AKT1, and BCL2 (Figure 6C), while also being regulated by TFs such as BCL6, BRCA1, CEBPA, and HDAC1 (Figure 6D).
Figure 6.
A multifactor regulatory network of model genes. A-D. Regulatory network features of model genes CYP27A1, MTF1, and STAT3, respectively.
Discussion
Our investigation into the molecular characteristics of IS patients has shed light on the intricate landscape of lipid metabolism and immunomodulation. First, we identified a set of 26 lipid metabolic genes that exhibited significant differential expression between normal and disease groups. Subsequently, we pinpointed a substantial pool of potential marker genes that was selected to establish a refined potential predictive diagnositc model integrating 7 specific genes, which demonstrated remarkable performance in distinguishing between normal and IS samples in both training dataset and our clinical samples. Additionally, our exploration of gene expression differences within the model highlighted genes with significant expression disparities between healthy and IS samples. Beyond the gene-centric analyses, we observed a significant association between the immune cell populations and the model genes. Lastly, the expression pattern and the dynamic change of these model genes as disease progressed were demonstrated. These findings may deepen our scientific understanding of the diagnostic roles of lipid metabolism genes in the pathogenesis of IS.
IS triggers a cascade of events, including lipid metabolism. This initial step allowed us to focus on specific genes that may be pivotal in IS pathogenesis. Enrichment analysis found that cell apoptosis and proliferation indicate the central role of these pathways in responding to ischemic stress and potential implications for cellular survival. Furthermore, the functional enrichment analysis highlights the protein/RNA/ATP binding functions of differentially expressed genes [23]. Overall, functional enrichment analysis provides a systematic and structured approach to interpreting the biological implications of differentially expressed genes, helping us piece together the puzzle of how lipid metabolism genes are involved in IS pathogenesis.
A particularly noteworthy discovery in our study was the identification of the specific gene signature as significantly correlated with lipid metabolism. The inclusion of a carefully curated set of genes that are intricately connected to lipid metabolism and its associated pathways enhances the model’s sensitivity and specificity [24]. These genes likely reflect the intricate molecular changes occurring in response to IS, capturing the underlying molecular signatures associated with this condition. Furthermore, the adoption of LASSO regression, a well-established technique for feature selection, avoided overfitting and generalizes well to new samples, resulting in its robust performance on both the training and validation dataset [25]. The exceptional performance of the model by leveraging the expression profiles of the selected genes associated with lipid metabolism is of significant clinical importance.
CYP27A1, MTF1, SOAT and STAT3 are model genes that showed significant expression differences between healthy and IS samples. Their differential expression underscores possible roles as crucial players in the molecular landscape of IS pathogenesis. CYP27A1 is a cytochrome P450 enzyme that plays a key role in cholesterol metabolism. It catalyzes the conversion of cholesterol to 27-hydroxycholesterol, which is then further metabolized to bile acids in the liver [26]. Recent studies have suggested a potential link between CYP27A1 and stroke risk [27,28]. MTF1 is a transcription factor that plays a key role in regulating the expression of genes involved in metal ion homeostasis and antioxidant responses [29]. While there is no direct evidence that MTF1 is directly involved in stroke pathogenesis, MTF1 may play a role in stroke risk or outcome through its regulation of metal ion homeostasis and antioxidant responses, which have been implicated in the pathogenesis of stroke [30]. STAT3 is a transcription factor that plays a crucial role in various biological processes, including cell growth, survival, differentiation, and inflammation [31]. In the context of stroke, STAT3 activation has been shown to promote neuroprotective signaling pathways that limit neuronal damage and enhance repair mechanisms [32]. This is achieved by inducing the expression of neurotrophic factors, such as brain-derived neurotrophic factor (BDNF), which support neuronal survival and regeneration [33,34]. Overall, the identification of CYP27A1, MTF1, and STAT3 as model genes with regulatory roles in IS highlights their significance as critical mediators of the molecular responses to cerebral ischemia.
As lipid metabolism contributes to modulating inflammation, its dysregulation might influence the extent of damage and recovery post-stroke [35]. The correlations observed between molecular signatures and immune cells in the context of IS hold substantial implications for our understanding of the intricate interplay between genetics and immune pathways. By exploring the relationships between specific genes and immune cell types, we gain insight into the mechanisms through which genetic variations can influence immune cell behavior in the ischemic brain [36,37]. Consistently, It was found that STAT3 may be vital in generating pathogenic Th1 and Th17 cells mediated by macrophage in inflammatory bowel diseases [38]. STAT3 restrained NK cell-mediated cytotoxic immune activity through the AMPK signaling [39]. Besides, STAT3 can also contribute to neuroinflammatory responses that exacerbate stroke damage. Following stroke, activated microglia and astrocytes release inflammatory cytokines and chemokines that activate STAT3 in neurons and other cells [40]. MTF1 activates the inflammatory response by functioning as a competing endogenous RNA to specifically promote IL-6 expression by sponging let-7a [41]. CYP27A1 promote neutrophil cell survival [42], which may be the biological target in the ischemic brain. These correlations suggest that specific genes may serve as regulators or modulators of immune cell populations, influencing the balance between pro-inflammatory and anti-inflammatory responses.
Although our studies reveal a new diagnostic model containing lipid metabolism-associated genes for IS diagnosis, there are several limitations. Further in vitro and in vivo studies into their specific roles and interactions within the context of IS could offer valuable insight into the underlying mechanisms of this complex neurological condition. Additionally, in all the datasets analyzed, the patients had already been diagnosed with stroke prior to the investigation. Prospective validation in a cohort of patients will be necessary to enhance the broad applicability of this model.
In conclusion, our studies constructed a new disease classification model for IS diagnosis using the lipid metabolism expression profile in whole blood cells. These findings not only contribute to the scientific knowledge base but also lay the groundwork for potential personalized treatment advancements in the realm of IS.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen B, Wang G, Li W, Liu W, Lin R, Tao J, Jiang M, Chen L, Wang Y. Memantine attenuates cell apoptosis by suppressing the calpain-caspase-3 pathway in an experimental model of ischemic stroke. Exp Cell Res. 2017;351:163–172. doi: 10.1016/j.yexcr.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 2.Cash D, Easton AC, Mesquita M, Beech J, Williams S, Lloyd A, Irving E, Cramer SC. GSK249320, a monoclonal antibody against the axon outgrowth inhibition molecule myelin-associated glycoprotein, improves outcome of rodents with experimental stroke. J Neurol Exp Neurosci. 2016;2:28–33. [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, Wang Z, Huang ZX, Liu Z. Biomarkers and the outcomes of ischemic stroke. Front Mol Neurosci. 2023;16:1171101. doi: 10.3389/fnmol.2023.1171101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W, Hu M, Li C, Wu R, Lu D, Xie C, Zhang W, Li T, Shen S, Huang H, Qiu W, Liu Q, Lu Y, Lu Z. FOXP3+ macrophage represses acute ischemic stroke-induced neural inflammation. Autophagy. 2023;19:1144–1163. doi: 10.1080/15548627.2022.2116833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H, Hu S, Li Y, Sun Y, Xiong X, Hu X, Chen J, Qiu S. Interleukins and ischemic stroke. Front Immunol. 2022;13:828447. doi: 10.3389/fimmu.2022.828447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan J, Horng T. Lipid metabolism in regulation of macrophage functions. Trends Cell Biol. 2020;30:979–989. doi: 10.1016/j.tcb.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Chi H. Immunometabolism at the intersection of metabolic signaling, cell fate, and systems immunology. Cell Mol Immunol. 2022;19:299–302. doi: 10.1038/s41423-022-00840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haley MJ, White CS, Roberts D, O’Toole K, Cunningham CJ, Rivers-Auty J, O’Boyle C, Lane C, Heaney O, Allan SM, Lawrence CB. Stroke induces prolonged changes in lipid metabolism, the liver and body composition in mice. Transl Stroke Res. 2020;11:837–850. doi: 10.1007/s12975-019-00763-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura A, Sakai S, Taketomi Y, Tsuyama J, Miki Y, Hara Y, Arai N, Sugiura Y, Kawaji H, Murakami M, Shichita T. PLA2G2E-mediated lipid metabolism triggers brain-autonomous neural repair after ischemic stroke. Neuron. 2023;111:2995–3010. e2999. doi: 10.1016/j.neuron.2023.06.024. [DOI] [PubMed] [Google Scholar]
- 10.Sidorov EV, Xu C, Garcia-Ramiu J, Blair A, Ortiz-Garcia J, Gordon D, Chainakul J, Sanghera DK. Global metabolomic profiling reveals disrupted lipid and amino acid metabolism between the acute and chronic stages of ischemic stroke. J Stroke Cerebrovasc Dis. 2022;31:106320. doi: 10.1016/j.jstrokecerebrovasdis.2022.106320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Cui Y, Feng J, Guo Y. Identifying the pattern of immune related cells and genes in the peripheral blood of ischemic stroke. J Transl Med. 2020;18:296. doi: 10.1186/s12967-020-02463-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Zhan F, Wang Y. Evaluation of monocyte-to-high-density lipoprotein cholesterol ratio and monocyte-to-lymphocyte ratio in ischemic stroke. J Int Med Res. 2020;48:300060520933806. doi: 10.1177/0300060520933806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joung KH, Kim JM, Choung S, Lee JH, Kim HJ, Ku BJ. Association between IL-1beta and cardiovascular disease risk in patients with newly diagnosed, drug-naïve type 2 diabetes mellitus: a cross-sectional study. Ann Transl Med. 2020;8:225. doi: 10.21037/atm.2020.01.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H, Ban YH, Cha Y, Kim TS, Lee SP, Suk An E, Choi J, Woom Seo D, Yon JM, Choi EK, Kim YB. Comparative effects of plant oils and trans-fat on blood lipid profiles and ischemic stroke in rats. J Biomed Res. 2017;31:122–129. doi: 10.7555/JBR.31.20160095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackam DG, Hegele RA. Cholesterol lowering and prevention of stroke. Stroke. 2019;50:537–541. doi: 10.1161/STROKEAHA.118.023167. [DOI] [PubMed] [Google Scholar]
- 16.Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. Nat Med. 2011;17:796–808. doi: 10.1038/nm.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andone S, Farczádi L, Imre S, Bălașa R. Fatty acids and lipid paradox-neuroprotective biomarkers in ischemic stroke. Int J Mol Sci. 2022;23:10810. doi: 10.3390/ijms231810810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullimore MA, Ritchey ER, Shah S, Leveziel N, Bourne RRA, Flitcroft DI. The risks and benefits of myopia control. Ophthalmology. 2021;128:1561–1579. doi: 10.1016/j.ophtha.2021.04.032. [DOI] [PubMed] [Google Scholar]
- 19.Tsai YT, Schlom J, Donahue RN. Blood-based biomarkers in patients with non-small cell lung cancer treated with immune checkpoint blockade. J Exp Clin Cancer Res. 2024;43:82. doi: 10.1186/s13046-024-02969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun W, Liu H, Dong L, Sun R, Guo L, Zhang H. Cognition of postoperative lymphedema among breast cancer patients in Lianyungang area. Chin J Clin Res. 2020;33:856–859. [Google Scholar]
- 22.Zhang X, Zhang S, Yan X, Shan Y, Liu L, Zhou J, Kuang Q, Li M, Long H, Lai W. m6A regulator-mediated RNA methylation modification patterns are involved in immune microenvironment regulation of periodontitis. J Cell Mol Med. 2021;25:3634–3645. doi: 10.1111/jcmm.16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trivedi P, Pandey M, Kumar Rai P, Singh P, Srivastava P. A meta-analysis of differentially expressed and regulatory genes with their functional enrichment analysis for brain transcriptome data in autism spectrum disorder. J Biomol Struct Dyn. 2023;41:9382–9388. doi: 10.1080/07391102.2022.2143900. [DOI] [PubMed] [Google Scholar]
- 24.York AG, Skadow MH, Oh J, Qu R, Zhou QD, Hsieh WY, Mowel WK, Brewer JR, Kaffe E, Williams KJ, Kluger Y, Smale ST, Crawford JM, Bensinger SJ, Flavell RA. IL-10 constrains sphingolipid metabolism to limit inflammation. Nature. 2024;627:628–635. doi: 10.1038/s41586-024-07098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muthukrishnan R, Rohini R. LASSO: A feature selection technique in predictive modeling for machine learning. In 2016 IEEE International Conference on Advances in Computer Applications (ICACA).2016. [Google Scholar]
- 26.Gupta RP, Patrick K, Bell NH. Mutational analysis of CYP27A1: assessment of 27-hydroxylation of cholesterol and 25-hydroxylation of vitamin D. Metabolism. 2007;56:1248–1255. doi: 10.1016/j.metabol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, Li W, Hibshoosh H, Loda M, Rabadan R, Shen MM. A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. Elife. 2020;9:e59465. doi: 10.7554/eLife.59465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, Cai L, Chen P, Kuang W. A study of the correlation between stroke and gut microbiota over the last 20years: a bibliometric analysis. Front Microbiol. 2023;14:1191758. doi: 10.3389/fmicb.2023.1191758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han H, Nakaoka HJ, Hofmann L, Zhou JJ, Yu C, Zeng L, Nan J, Seo G, Vargas RE, Yang B, Qi R, Bardwell L, Fishman DA, Cho KWY, Huang L, Luo R, Warrior R, Wang W. The Hippo pathway kinases LATS1 and LATS2 attenuate cellular responses to heavy metals through phosphorylating MTF1. Nat Cell Biol. 2022;24:74–87. doi: 10.1038/s41556-021-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludhiadch A, Sharma R, Muriki A, Munshi A. Role of calcium homeostasis in ischemic stroke: a review. CNS Neurol Disord Drug Targets. 2022;21:52–61. doi: 10.2174/1871527320666210212141232. [DOI] [PubMed] [Google Scholar]
- 31.Tolomeo M, Cascio A. The multifaced role of STAT3 in cancer and its implication for anticancer therapy. Int J Mol Sci. 2021;22:603. doi: 10.3390/ijms22020603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai AS, Berry K, Beneyto MM, Gaudilliere D, Ganio EA, Culos A, Ghaemi MS, Choisy B, Djebali K, Einhaus JF, Bertrand B, Tanada A, Stanley N, Fallahzadeh R, Baca Q, Quach LN, Osborn E, Drag L, Lansberg MG, Angst MS, Gaudilliere B, Buckwalter MS, Aghaeepour N. A year-long immune profile of the systemic response in acute stroke survivors. Brain. 2019;142:978–991. doi: 10.1093/brain/awz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Liang Y, He Z, An Y, Zhao W, Wu J. Autocrine activity of BDNF induced by the STAT3 signaling pathway causes prolonged TrkB activation and promotes human non-small-cell lung cancer proliferation. Sci Rep. 2016;6:30404. doi: 10.1038/srep30404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis CM, Lyon-Scott K, Varlamov EV, Zhang WH, Alkayed NJ. Role of endothelial STAT3 in cerebrovascular function and protection from ischemic brain injury. Int J Mol Sci. 2022;23:12167. doi: 10.3390/ijms232012167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie N, Zhang L, Gao W, Huang C, Huber PE, Zhou X, Li C, Shen G, Zou B. NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct Target Ther. 2020;5:227. doi: 10.1038/s41392-020-00311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu X, Yu Z, Qian T, Jin Y, Xu G, Li J, Gu J, Li M, Tao K. Transcriptomic analysis identifies the shared diagnostic biomarkers and immune relationship between atherosclerosis and abdominal aortic aneurysm based on fatty acid metabolism gene set. Front Mol Biosci. 2024;11:1365447. doi: 10.3389/fmolb.2024.1365447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Tan S, Chen F, An Y. Identification of immune-related biomarkers co-occurring in acute ischemic stroke and acute myocardial infarction. Front Neurol. 2023;14:1207795. doi: 10.3389/fneur.2023.1207795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, Sun L, Wang M, Liu J, Zhong S, Li R, Li P, Guo L, Fang A, Chen R, Ge WP, Wu Q, Wang X. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020;18:e3000705. doi: 10.1371/journal.pbio.3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Liu W, Zhuang D, Hong S, Chen J. Sestrin2 and sestrin3 suppress NK-92 cell-mediated cytotoxic activity on ovarian cancer cells through AMPK and mTORC1 signaling. Oncotarget. 2017;8:90132–90143. doi: 10.18632/oncotarget.21487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wicks EE, Ran KR, Kim JE, Xu R, Lee RP, Jackson CM. The translational potential of microglia and monocyte-derived macrophages in ischemic stroke. Front Immunol. 2022;13:897022. doi: 10.3389/fimmu.2022.897022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C, Li X, Xue B, Yu C, Wang L, Deng R, Liu H, Chen Z, Zhang Y, Fan S, Zuo C, Sun H, Zhu H, Wang J, Tang S. RasGRP1 promotes the acute inflammatory response and restricts inflammation-associated cancer cell growth. Nat Commun. 2022;13:7001. doi: 10.1038/s41467-022-34659-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoyratty TE, Ai Z, Ballesteros I, Eames HL, Mathie S, Martín-Salamanca S, Wang L, Hemmings A, Willemsen N, von Werz V, Zehrer A, Walzog B, van Grinsven E, Hidalgo A, Udalova IA. Distinct transcription factor networks control neutrophil-driven inflammation. Nat Immunol. 2021;22:1093–1106. doi: 10.1038/s41590-021-00968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.