Abstract
The RNA-editing enzyme ADAR1 modifies adenosines by deamination and produces A-to-I mutations in mRNA. ADAR1 was recently demonstrated to function in host defense and in embryonic erythropoiesis during fetal liver development. The mechanisms for these phenotypic effects are not yet known. Here we report a novel function of ADAR1 in the regulation of gene expression by interacting with the nuclear factor 90 (NF90) proteins, known regulators that bind the antigen response recognition element (ARRE-2) and have been demonstrated to stimulate transcription and translation. ADAR1 upregulates NF90-mediated gene expression by interacting with the NF90 proteins, including NF110, NF90, and NF45. A knockdown of NF90 with small interfering RNA suppresses this function of ADAR1. Coimmunoprecipitation and double-stranded RNA (dsRNA) digestion demonstrate that ADAR1 is associated with NF110, NF90, and NF45 through the bridge of cellular dsRNA. Studies with ADAR1 deletions demonstrate that the dsRNA binding domain and a region covering the Z-DNA binding domain and the nuclear export signal comprise the complete function of ADAR1 in upregulating NF90-mediated gene expression. These data suggest that ADAR1 has the potential both to change information content through editing of mRNA and to regulate gene expression through interacting with the NF90 family proteins.
TheRNA-editing enzyme ADAR (adenosine deaminase acting on RNA) is a typical double-stranded RNA (dsRNA) binding protein (DRBP) that modifies dsRNA and pre-mRNA by adenosine deamination (20, 36, 51). Homologous genes occur in organisms from unicellular protozoa to humans, indicating a widespread conservation in evolution. In mammals, two homologous genes encoding ADAR1 and ADAR2 have been identified, each of which is endowed with domains for dsRNA binding and adenosine deaminase. Both render site-specific editing on pre-mRNA and preferably convert adenosines in dsRNA. A third homologue, ADAR3 or RED2, is a candidate as an A-to-I editing enzyme that has both the dsRNA binding domain and the adenosine deaminase domain, but it does not demonstrate editing activity on dsRNA (19).
ADAR1 exhibits several features that are different from those of ADAR2 and ADAR3; it comprises two putative Z-DNA binding domains (7), three dsRNA binding repeats, and an interferon-inducible promoter (29, 45). Unlike ADAR2 and ADAR3, which are predominately found in the brain, ADAR1 is ubiquitously expressed in immune organs. Thus, the function of ADAR1 has been implicated in host defense mechanisms (37, 44). ADAR1 is upregulated in response to viral and microbial infections (31, 37, 50). An upregulation of ADAR1 was observed in vitro in T lymphocytes and macrophages stimulated with gamma interferon (IFN-γ). Recent studies demonstrated that ADAR1 is critical for embryonic erythropoiesis and subserves critical steps in developing nonnervous tissue (9, 43). ADAR1-deficient embryonic stem cells failed to contribute to the development of the liver, bone marrow, spleen, thymus, and blood in adult chimeric mice. In mice homozygous for an ADAR1 null mutation, widespread apoptosis was detected in many tissues (44). Fibroblasts derived from ADAR1−/− embryos were prone to apoptosis induced by serum deprivation.
ADAR1 modifies highly structured pre-mRNAs such as those that encode the glutamate and serotonin receptors, resulting in mutations of the encoded proteins and alterations in their biological functions (18). Widespread inosines occur in mouse brain mRNAs (30). We have observed a significant induction of A-to-I mutations in mRNAs during infection (49). Extensive A-to-I modifications have been observed in viral RNA during the late stages of polyomavirus infection (15), and biased hypermutations were found in single-stranded RNA viral genomes during lytic and persistent infections (1). Thus, the modification of highly structured mRNA or viral RNA is one way for ADAR1 to participate in the host response during infections. However, too few ADAR1-produced protein variations have been identified to account entirely for the biological phenotypes of ADAR1.
Because ADAR1 preferably edits dsRNA, ADAR1 may interfere with other DRBPs by sequestering the cellular dsRNA components. The aims of this study were to analyze the proteins that are associated with ADAR1 in vivo and to elucidate the role of ADAR1 in interacting with these proteins. We demonstrate that ADAR1 interacts with the nuclear factor 90 (NF90) family proteins and upregulates NF90-mediated gene expression.
MATERIALS AND METHODS
Cell culture and transfection.
The cell lines used for this study include HEK 293T and N18. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin-streptomycin-amphotericin B. When cells were grown to ∼50% confluence, transfection was performed in 6-cm tissue culture dishes or 24-well plates (Falcon) using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen, CA). The transfected cells were washed with warm DMEM containing 10% FBS 6 h after transfection and were cultured for 48 to 60 h in the same medium.
Immunoprecipitation.
293T or mouse N18 cells (3 × 106) were transfected with double-tagged ADAR1 or ADAR1 fragments for 48 h. Cells were lysed in a buffer containing 150 mM NaCl, 25 mM Tris-HCl, pH 7.45, 0.5% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (pH 7.5), and a protease inhibitor cocktail (Roche). Cell lysates were cleared with rabbit or mouse immunoglobulin G (IgG) immobilized to protein A-Sepharose for 1 h at 4°C and mixed with anti-His-, anti-NF90-, or anti-p68-coated protein A-Sepharose (Amersham) at 4°C overnight. The agarose was extensively washed with a high-stringency wash buffer containing 0.1% SDS, and the associated proteins were stored at −80°C for further analysis.
Protein identification by mass spectrometry.
The coprecipitated proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with SYPRO Ruby (Bio-Rad). The visible bands were excised, proteins were digested with modified trypsin, and peptides were subjected to liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis (Yale Keck Facility). The most abundant ions were obtained, and the MS/MS spectra were directly searched against the National Center for Biotechnology Information's nonredundant protein database with the SEQUEST database search algorithm.
Immunoblotting.
Cell lysates (approximately 80 μg of protein) or immunoprecipitated proteins on agarose beads were heated to 95°C for 5 min in 100 mM dithiothreitol and 2% SDS. The proteins were resolved by 5 to 12% SDS-PAGE and transferred onto a nitrocellulose membrane. The membranes were blocked with 5% nonfat dry milk and stained with a primary antibody against NF90, NF45, ADAR1, the His6 tag, the Myc epitope (Santa Cruz Biotechnology), or eukaryotic initiation factor 2a (eIF2a; Cell Signaling Technology). The secondary antibodies included donkey anti-rabbit and donkey anti-mouse horseradish peroxidase-linked IgG (Jackson Immunoresearch Laboratories Inc.).
Electrophoretic mobility shift assay.
A modified native gel was applied to detect the formation and subgrouping of ADAR1-DRBP complexes. Since 0.05% SDS did not affect the interaction of ADAR1 with NF110, NF90, and NF45, it was included in all electrophoretic mobility shift assays. Protein samples were resuspended in the loading buffer with or without DTT and directly loaded onto SDS-5% PAGE gels containing 0.05% SDS. The resolved proteins were transferred and analyzed by immunoblotting using proper antibodies as described above.
RNase V1 digestion.
The immunoprecipitated proteins on agarose beads were washed twice with RNase V1 digestion buffer (Ambion) and resuspended in the same buffer containing 0 to 0.4U/μl of RNase V1 (Ambion). The samples were incubated at 37°C for 25 min and quickly pulled down by centrifugation. The beads containing the still-bound proteins and the supernatant containing the released proteins were mixed with the loading buffer, heated to 95°C for 5 min, and resolved by SDS-PAGE. The blots were analyzed by immunoblotting as described above.
Luciferase reporter assay.
293T or N18 cells were seeded in 24-well plates in triplets (n = 3) and cotransfected with a plasmid containing ADAR1 cDNA and the luciferase reporter. The reporters included c-fos-luciferase (a gift from J. Ye, Yale University), IFN-β-luciferase (a gift from John Hiscott, McGill University), simian virus 40 (SV40)-luciferase (pGL3-control; Promega), Rous sarcoma virus (RSV)-luciferase, and cytomegalovirus (CMV)-luciferase (pGL3-CMV; a gift from Stratford May, University of Florida). Typically, 0.6 μg of the reporter DNA and 0.8 μg of pcDNA3-ADAR1 were mixed with 2.7 μg of Lipofectamine 2000 in 80 μl of serum-free medium, incubated at room temperature for 30 min, and diluted with 340 μl of medium. The cell medium was replaced with the transfection cocktail. The transfected cells were incubated for 6 h and then continuously cultured for 48 h in fresh DMEM containing 10% FBS. Cells were washed with phosphate-buffered saline and lysed in 150 μl of reporter lysis buffer (Promega) for 30 min. The luciferase activity in 20 μl of lysate was determined and normalized to the protein level (Promega).
siRNAs.
Two pairs of complementary oligoribonucleotides, specific for the sequences GGCAGAGUCCGAUAACAUGTT (positions 443 to 461; GenBank accession no. NM004516) and GGUUUCCAGUACUACUUGCTT (positions 426 to 406; GenBank accession no. NM001111), were chemically synthesized and annealed to form small interfering RNAs (siRNAs) against the 5′ end of human NF110/NF90 and the 5′ end of ADAR1 p150, respectively. A siRNA targeting a bacterial RNA (Ambion) was used as a negative control. 293T cells (2 × 105) were seeded in 24-well plates and allowed to adhere overnight. A 120 nM concentration of NF90, ADAR1, or control siRNA was cotransfected alone with 0.6 μg of the SV40-luciferase reporter. Cells were harvested 60 h after transfection. Western blotting was performed to examine ADAR1 and NF90 expression, and a luciferase assay was performed to examine SV40-luciferase expression.
ADAR1 constructs.
The constructs of ADAR1 variants used for transfection were made by PCR. To facilitate cloning, an XhoI or BamHI cleavage site was added to the 5′ primer or 3′ primer, respectively. The pCRII-ADAR1 plasmid (GenBank accession no. AF291050) was used to amplify the fragments from positions 1 to 3459 (LF or p150), 748 to 2459 (p110), 1561 to 3466 (SF or p80), 1 to 2263 (Dcat), 1 to 748 (Z-DBD/NES), and 748 to 2263 (dsRBD). Each fragment was cleaved with BamHI and XhoI and subcloned into the pcDNA3.1-myc-his vector (Invitrogen) for transient expression. The expression of each protein fragment was confirmed by the transfection of 293T cells and detected by immunoblotting using an antibody against the Myc epitope.
RESULTS
NF90 proteins are associated with ADAR1.
To understand the molecular basis of ADAR1 function, we analyzed the proteins that physically interact with ADAR1 in vivo. Because the dsRNA binding domain (dsRBD) stabilizes protein-protein interactions between DRBPs (35), we speculated that ADAR1 might interact with other dsRNA-dependent proteins through cellular dsRNA substrates. The long and short forms of ADAR1, p150 and p80 (49), were tagged with Myc-His epitopes at their C termini and transiently expressed in human or mouse N18 cells. Cell lysates were subjected to immunoprecipitation using an anti-His antibody immobilized on agarose. The proteins associated with ADAR1 were extensively washed with a high-stringency buffer containing 0.1% SDS to remove nonspecific proteins thoroughly. The remaining proteins were resolved by SDS-PAGE. Other than the transiently expressed ADAR1 protein, we found three protein bands, of approximately 45, 90, and 110 kDa, that were specifically associated with ADAR1 p150 to the exclusion of the short variant p80 (unpublished data). These proteins were excised, digested with trypsin directly in the gel slices, and subjected to LC-ESI-MS/MS protein identification. The MS/MS spectra were searched against the National Center for Biotechnology Information nonredundant protein database with the SEQUEST database search algorithm. As shown in Table 1, the mass spectra for the 45-kDa band matched five peptides of NF45 (GI:1082855). The other two bands matched eight peptides of NF90 (GI:9714266) and four peptides of NF110 (GI:9081982), respectively. These data suggest that NF45, NF90, and NF110 are specifically associated with ADAR1 p150.
TABLE 1.
ADAR1 regulates NF90-mediated gene expression
| GenBank no. | Characteristic of peptide matching MS/MS spectraa
|
||
|---|---|---|---|
| Mass ± errorb | Sequencec | Positions | |
| GI:1082855 (NF45) | 1,040.63 + 0.05 | VLQSALAAIR | 187-196 |
| 1,235.76 + 0.07 | ILITTVPPNLR | 175-185 | |
| 1,408.83 + 0.08 | LLPTLEAVAALGNK | 128-141 | |
| 1,370.91 + 0.09 | VKPAPDETSFSEALLK | 44-59 | |
| 2,581.38 + 0.14 | INNVIDNLIVAPGTFEVQIEEVR | 81-103 | |
| GI:9714266 (NF90) | 1,019.57 + 0.18 | AYAALAALEK | 448-457 |
| 1,071.56 + 0.19 | VLGMPLPSK + oxic | 199-208 | |
| 1,294.94 + 0.23 | FVMEVEVDGQK + oxic | 425-435 | |
| 1,359.63 + 0.25 | AEPPQAMNALMR | 263-274 | |
| 1,414.02 + 0.26 | LFPDTPLALDANK | 458-470 | |
| 1,458.99 + 0.27 | VLQDMGLPTGAEGR + oxic | 327-340 | |
| 1,615.81 + 0.28 | SIGTANRPMGAGEALR | 124-139 | |
| 1,908.98 + 0.34 | VLAGETLSVNDPPDVLDR | 49-66 | |
| GI:9081982 (NF110)d | 1,019.57 + 0.19 | AYAALAALEK | 389-400 |
| 1,295.61 − 0.67 | FVMEVEVDGQK + oxic | 368-378 | |
| 1,413.75 + 0.27 | LFPDTPLALDANK | 401-413 | |
| 1,458.71 + 0.27 | VLQDMGLPTGAEGR + oxic | 274-287 | |
Three proteins corresponding to molecular masses of 45, 90, and 110 kDa that were specifically associated with ADAR1 p150 were digested with trypsin and subjected to LC-ESI-MS/MS for identification. The peptides that match the MS/MS spectra are listed.
The error is difference between the calculated and detected molecular masses.
Matches the molecular mass with oxidized methionine.
The molecular mass of this protein band is approximately 110 kDa.
Interactions between ADAR1 and the NF90 proteins.
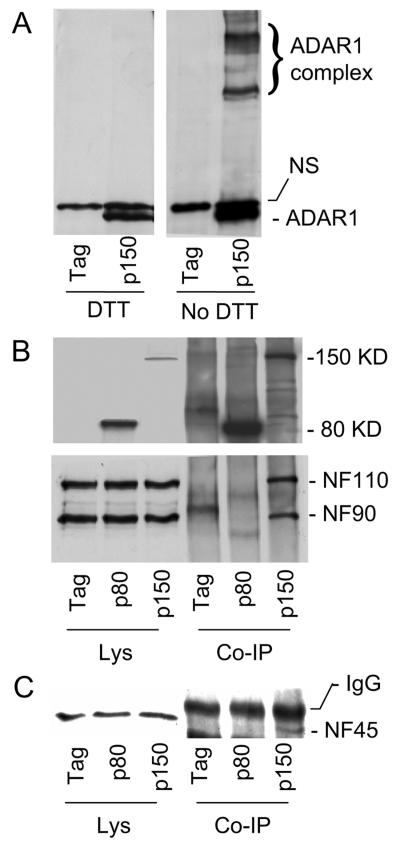
We next examined the formation of ADAR1 complexes by using an electrophoretic mobility shift assay. Because ADAR1 and NF90 proteins were tightly associated even in the presence of 0.1% SDS, we reasoned that the ADAR1 complex might remain intact during polyacrylamide gel electrophoresis. The cell lysates were then precipitated with Myc-agarose beads; associated proteins were mixed with loading buffer without a reducing reagent and separated by SDS-5% PAGE (0.1% SDS). Mobility shifts of the ADAR1 complexes were detected with an anti-Myc antibody. A few slow-migrating ADAR1 complexes were detected (Fig. 1A). Under the same conditions, however, these complexes were completely disrupted by DTT, indicating that the complexes were supported with disulfide bonds. To confirm the association of NF110, NF90, and NF45 in the ADAR1 complexes, we next performed coimmunoprecipitation using an immobilized antibody against the His-tagged ADAR1 protein. The associated proteins were analyzed by immunoblotting using antibodies against NF90 or ADAR1 (Myc or His tagged). In agreement with the mass spectrometry study, NF110 and NF90 were equally detected in association with ADAR1 p150 to the exclusion of the short form, ADAR1 p80, or the Myc-His tag (Fig. 1B). The NF45 protein was also confirmed to be present in the complex (Fig. 1C). Because NF45 is usually associated with NF90 (12), this result suggests that ADAR1 p150 interacts with the NF110/NF90/NF45 complex. In contrast, neither NF110/NF90 nor NF45 was detected in association with ADAR1 p80 or the Myc-His tag, indicating the specificity of the interaction.
FIG. 1.
Interactions between ADAR1 and NF90 proteins. (A) Electrophoretic mobility shift assay. Human 293T cells were transfected with Myc-His-tagged long-form ADAR1 (p150) or the Myc-His tag (tag). Cell lysates were coimmunoprecipitated with an anti-His antibody. Associated proteins were resuspended in the loading buffer, with or without DTT, and resolved by SDS-PAGE (0.1% SDS). Protein complexes containing ADAR1 were identified by immunoblotting with antibodies against the Myc epitope. NS, nonspecific. (B) Coimmunoprecipitation of NF110 and NF90 with ADAR1. Cell lysates (Lys) were coimmunoprecipitated using an immobilized anti-His antibody; the associated proteins (Co-IP) were denatured in loading buffer containing 100 mM DTT and analyzed by immunoblotting using antibodies against NF90 (lower panels) or Myc (upper panels). (C) Coimmunoprecipitation of NF45 with ADAR1. The same membrane was detected with an antibody against NF45.
Interactions occur under endogenous conditions.
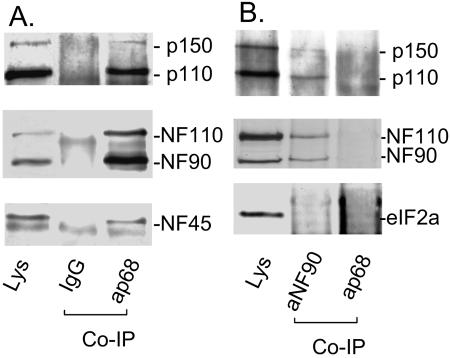
To address whether these interactions occur between endogenous ADAR1 and NF90, we analyzed NF90-associated proteins in human SGC7901 cells (5) without the transient expression of ADAR1. SGC7901 cells were used because of their high levels of ADAR1 p150. Cell lysates were analyzed with affinity columns containing immobilized antibodies against NF90, rabbit IgG, or an unrelated protein, p68 (human RNA helicase p68). The associated proteins were eluted and detected by immunoblotting using antibodies against ADAR1, NF110/90, NF45, or another unrelated protein, eIF2a. In anti-NF90 antibody-associated proteins, we detected endogenous NF110, NF90, and NF45 as well as endogenous ADAR1 p150 (Fig. 2). ADAR1 p110, an N-terminally truncated form of ADAR1 p150 that still contains three dsRBDs and the entire C-terminal end (10, 36, 50), was also immunoprecipitated. In contrast, none of these proteins were detected when affinity purification was performed with an antibody against p68 or rabbit IgG; eIF2a was also not detected in the NF90-associated proteins. These data support the observation that the interactions between ADAR1 p150 and the NF110/90/45 complex are specific.
FIG. 2.
Interaction between endogenous ADAR1 and NF90. (A) Cell lysates (Lys) of SGC7901 cells (human gastric cancer cells) were separated with immobilized rabbit IgG or antigen-purified anti-p68 antibody (ap68). Associated proteins were eluted and analyzed by immunoblotting using antibodies against ADAR1 (upper panel), NF90 (middle panel), or NF45 (the lower panel). (B) For negative controls, the cell lysates were immunoprecipitated with an antibody against an unrelated protein, RNA helicase p68 (ap68), followed by immunoblotting to detect ADAR1 (upper panel), NF110/NF90 (middle panel), or another unrelated protein, eIF2a (lower panel).
ADAR1-NF90 complexes are bridged by cellular dsRNA components.
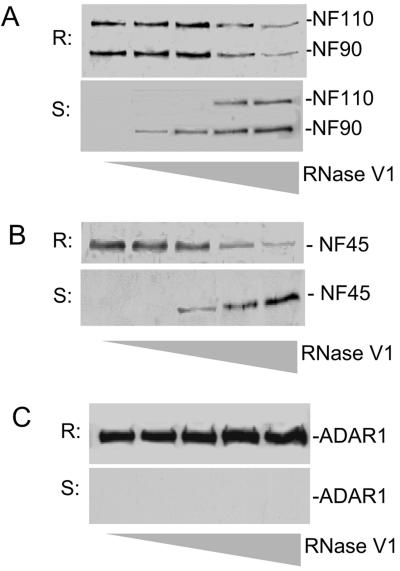
Because ADAR1 and NF90 proteins are all DRBPs, it was essential to establish whether the observed ADAR1 complexes are formed by protein-protein interactions or bridged through cellular dsRNA. This question was addressed by immunoprecipitation and dsRNA digestion. For this purpose, ADAR1-NF90 complexes were coimmunoprecipitated and digested with RNase V1. Complexes that were sensitive or resistant to RNase V1 digestion were analyzed by SDS-PAGE and detected with antibodies against ADAR1, NF90/NF110, or NF45. We confirmed that NF110, NF90, and NF45 were detected in association with ADAR1 before RNase V1 digestion. When the complexes were digested with RNase V1, however, the NF90, NF110, and NF45 proteins were quantitatively released from the complex in a concentration-dependent manner (Fig. 3A and B). Similarly, the dsRNA bridge is required for the interactions between ADAR1 and NF90 proteins under endogenous conditions, because RNase V1 digestion disrupted the complex in SGC7901 cells without transfection (data not shown). As a negative control, RNase V1 digestion did not affect immobilized ADAR1 (Fig. 3C). Treatments with RNase S1 or DNase I did not affect the ADAR1 complexes. Thus, cellular dsRNA is an essential component of the ADAR1-NF90 complexes. Because dsRNA is known to be the substrate of ADAR1 and the effector of NF90, this result suggests a dynamic interplay among ADAR1, NF90 proteins, and dsRNA and reveals a unique regulatory mechanism of ADAR1 in regulating NF90-mediated gene expression.
FIG. 3.
dsRNA components in ADAR1-NF90 complex. ADAR1-NF90 complexes immunoprecipitated with an anti-His-tag antibody were digested with 0, 0.006, 0.025, 0.1, or 0.4 units/μl of RNase V1 (lanes 1 to 5, respectively). Proteins resistant (R) or sensitive (S) to RNase V1 digestion were analyzed by immunoblotting using antibodies against NF90 (A), NF45 (B), or ADAR1 (C).
ADAR1 regulates NF90-mediated gene expression.
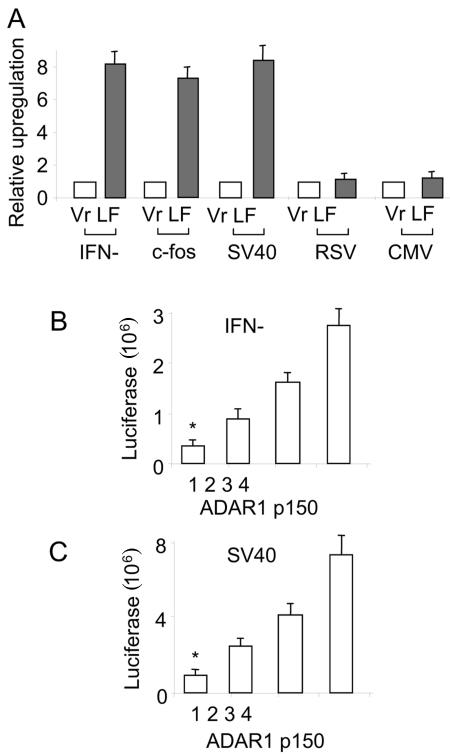
To elucidate possible functions of the interaction, we examined whether ADAR1 affects NF90-mediated gene expression. The NF90 proteins have been demonstrated to stimulate gene expression at multiple levels, including transcription (32, 34), translation (47, 48), posttranscriptional stabilization (41, 42), and nuclear exportation (42). In particular, the NF90 proteins stimulate the activation of the cellular antiviral expression cascade and are important mediators for gene expression during viral infection (12, 14, 34). We tested whether ADAR1 affects the expression of the promoters that are regulated by NF90 proteins. It is known that NF90 upregulates the SV40 promoter (33, 34, 39); however, it has less effect on CMV and no effect on the RSV promoter. IFN-β was included because NF90 activates both IFN-β and dsRNA-inducible promoters (14, 26) and because IFN-β is a typical dsRNA-inducible gene (8). The c-fos gene was tested because it is regulated through protein kinase R (PKR) (4, 21) and because NF90 interacts with PKR (14, 16, 26). Thus, a luciferase reporter driven by the IFN-β, c-fos, SV40, RSV, or CMV promoter was cotransfected into 293T cells along with pcDNA3-ADAR1. The effects of ADAR1 on the expression of these promoters were examined. While ADAR1 p150 markedly enhanced luciferase expression driven by the IFN-β, c-fos, or SV40 promoter, it had only a minor effect on luciferase reporter expression driven by the RSV or CMV promoter (Fig. 4A). This result was confirmed by a dose-dependent assay, in which IFN-β- or SV40-luciferase responded to ADAR1 in a concentration-dependent manner (Fig. 4B and C). Since the expression of SV40, IFN-β, and c-fos is regulated by the NF90 proteins (14, 34), these data suggest that ADAR1 upregulates NF90-mediated gene expression.
FIG. 4.
ADAR1 upregulates NF90-mediated gene expression. (A) Effects of ADAR1 on different promoters. A plasmid containing the IFN-β-, c-fos-, SV40-, RSV-, or CMV-luciferase reporter was cotransfected into 293T cells along with pcDNA3-ADAR1 p150 (LF) or pcDNA3 vector DNA (Vr) for 48 h. Luciferase activities were determined and normalized to the amount of protein in cell lysates. Relative upregulation was calculated in comparison with the vector. Data are expressed as means ± standard deviations (n = 3). (B) ADAR1 upregulates IFN-β-luciferase. 293T cells were cotransfected with the IFN-β-luciferase reporter along with a mixture of pcDNA3-ADAR1 and pcDNA3 at a ratio of 0:0.6, 0.2:0.4, 0.4:0.2, or 0.6:0 (μg:μg). Luciferase activities were determined and normalized as described above. *, P < 0.05 for lane 1 versus lane 2, 3, or 4. (C) ADAR1 upregulates SV40-luciferase. 293T cells were cotransfected with the SV40-luciferase reporter along with a mixture of pcDNA3-ADAR1 and pcDNA3 at a ratio of 0:0.6, 0.2:0.4, 0.4:0.2, or 0.6:0 (μg:μg). Luciferase activities were determined and normalized as described above. *, P < 0.05 for lane 1 versus lane 2, 3, or 4.
ADAR1-mediated gene expression requires NF90.
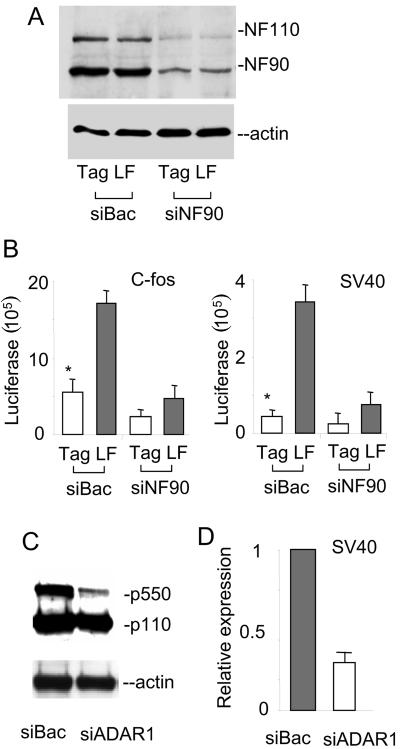
To confirm the role of NF90 proteins in the ADAR1-induced upregulation of gene expression, we examined the c-fos- and SV40-luciferase reporters by silencing the expression of NF90 using siRNA. Because NF110 and NF90 are translated from a differentially spliced mRNA (38), a single siRNA was chemically synthesized to target both NF110 and NF90. To eliminate nonspecific effects, a 21-mer siRNA against a bacterial RNA was used as a control. In the cells treated with siRNA, a reduced expression of NF110 and NF90 expression was confirmed by immunoblotting using an anti-NF90 antibody (Fig. 5A). The total expression levels of the c-fos- and SV40-luciferase reporters were suppressed when the endogenous NF90 was silenced; this confirmed that the expression of these genes is regulated by NF90. While ADAR1 stimulated the expression of the c-fos-luciferase reporter in cells with normal NF90 expression, this function of ADAR1 was suppressed when the endogenous NF90 was silenced by siRNA (Fig. 5B). Specifically, ADAR1 stimulated increases in SV40 expression of approximately eight- and threefold in the absence and presence of siRNA, respectively. In support of these data, a reciprocal experiment with a knockdown of the endogenous ADAR1 showed a decrease in SV40-luciferase activity (Fig. 5C and D). Thus, the NF90 proteins are important for ADAR1-mediated gene expression. This supports the observation that the interaction between ADAR1 and the NF90 proteins plays a role in regulating NF90-mediated gene expression.
FIG. 5.
NF90 is required for ADAR1-mediated gene expression. (A) A siRNA specific for a bacterial RNA (siBac) or the NF110/NF90 mRNA (siNF90) was cotransfected into 293T cells along with pcDNA3-ADAR1 p150 (LF) or Myc-His-tagged vector (tag). The protein levels of ADAR1, NF110, NF90, and β-actin were determined by immunoblotting. (B) c-fos- and SV40-luciferase expression was determined and normalized to the amounts of protein in cell lysates. All data are expressed as means ± standard deviations (n = 3). *, P < 0.05 for LF versus tag. (C) A siRNA specific for the 5′ end of ADAR1 p150 (siADAR1) or the control siRNA (siBac) was cotransfected into 293T cells along with the SV40-luciferase reporter plasmid. The protein levels of ADAR1 and β-actin were determined by immunoblotting. (D) SV40-luciferase expression in transfected cells was determined and normalized to the amounts of protein in cell lysates. The relative expression affected by siADAR1 was normalized to that of the siBac control.
The function of ADAR1 is independent of RNA editing.
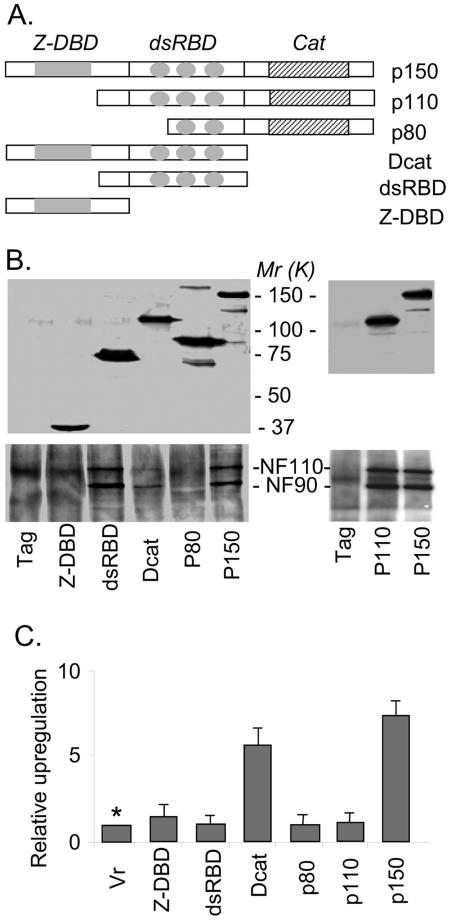
Finally, we characterized the selectivity and hence the biological significance of the interaction and function of ADAR1. A few tagged ADAR1 fragments were constructed (Fig. 6A), and their expression in 293T cells was confirmed by transfection (Fig. 6B, top panels). Interactions of these fragments with NF110 and NF90 were determined by coimmunoprecipitation and immunoblotting analysis (Fig. 6B, bottom panels), and their function of upregulating the SV40-luciferase reporter was examined (Fig. 6C). Consistently, ADAR1 p150 interacted with the NF90 proteins and promoted SV40-luciferase expression. While ADAR1 p110 that lacked the Z-DNA binding domain (Z-DBD)/NES (10, 50) still interacted with the NF90 complex (Fig. 2 and 6B), it lost its function (Fig. 6C). The short form, ADAR1 p80, which deleted the Z-DBD/NES and the first dsRBD, lost both the interaction and the function. Because the dsRBD fragment alone interacted with the NF90 complex as efficiently as ADAR1 p150 (Fig. 6B), we reasoned that the dsRBD fragment is essential for interaction but not sufficient for function. A regulatory domain, likely the Z-DBD/NES region, is suggested to comprise the complete function of ADAR1 in regulating NF90-mediated gene expression. In support of this, neither the dsRBD fragment nor the Z-DBD/NES alone had detectible activity. However, the Dcat fragment containing both the Z-DBD/NES and the dsRBDs was active (Fig. 6C). This demonstrated that the dsRBD fragment and the Z-DBD/NES region together comprise sufficient components to promote NF90-mediated gene expression. Markedly, the deaminase domain or RNA-editing activity of ADAR1 was not required for its interaction or function.
FIG.6.
Selectivity of ADAR1 interaction and function. (A) Double-tagged ADAR1 constructs used for interaction and function assays. p150, p80, Dcat, dsRBD, and Z-DBD/NES are ADAR1 fragments amplified from positions 1 to 3466, 1561 to 3466, 1 to 2263, 748 to 2263, and 1 to 748 of the ADAR1 DNA (GenBank accession no. AF291050), respectively. (B) Expression and interaction of ADAR1 fragments. The double-tagged ADAR1 fragments were transfected into 293T cells, and their expression was detected by immunoblotting using an anti-Myc antibody (upper panels). Interactions of ADAR1 fragments with NF110/NF90 were determined by coimmunoprecipitation using an immobilized anti-His antibody followed by immunoblotting using an anti-NF90 antibody (lower panels). The expression and interaction of the transient ADAR1 p110 were determined and are shown in the right panels. (C) Upregulation of SV40-luciferase by ADAR1 fragments. Luciferase activities in the cell lysates were determined and normalized to the amounts of protein. Relative upregulation was calculated compared to the tag control. Data are expressed as means ± standard deviations (n = 3). *, P < 0.05 for Vr versus Dcat or p150.
DISCUSSION
It is intriguing that ADAR1 has the potential both to change information content through editing mRNA and to regulate gene expression through interacting with the NF90 family proteins. Because the deaminase domain of ADAR1 is not required for the interaction and the observed function, our data reveal a novel mechanism of ADAR1 in regulating NF90-mediated gene expression independently of RNA editing. This function requires the dsRBDs and a regulatory domain of ADAR1.
Notably, ADAR1 and the NF90 proteins are typical DRBPs. A common feature of the DRBPs is that they all contain dsRBDs, which are known to stabilize homo- or heterotypic protein-protein interactions between different DRBPs and often subserve a regulatory function between these proteins (3, 27, 35, 50). ADAR1 possesses this feature and has been shown to dimerize in order to target and edit the dsRNA substrate (2). We demonstrated that NF110 and NF90 are associated with ADAR1. The interaction is specific to ADAR1 p150 and occurs between the endogenous proteins, suggesting that the biological functions of NF90 are dynamically affected by ADAR1 or vice versa. Consistent with this, the ADAR1-NF90 complexes are bridged through not-yet-defined cellular dsRNA components. In addition, dsRNA is the preferential substrate of ADAR1 that can be efficiently unwound. The dsRNA binding property is also required for the function of NF90, since mutations that reduced the dsRNA binding ability of NF110 reduced the ability of NF110 to stimulate gene expression (32). Our results show that the enzyme, the substrate/effector, and the transcription regulators are spontaneously associated, supporting a dynamic interplay between these components that plays a regulatory role in NF90-mediated gene expression.
The NF90 family consists of a group of closely related proteins (6, 32, 38, 48), which are also known as interleukin enhancer binding factor 3 (ILF3) (6), translation control protein 80 (TCP80) (47), dsRNA-binding nuclear protein 76 (DRBP76) (28), and nuclear factor associated with dsRNA (NFAR) (38). The NF90 proteins have been demonstrated to stimulate gene expression at the transcription (32) and translation levels (47, 48) in vertebrates. The mechanisms of these functions are still under investigation. It was previously reported that TCP/NF90 interacts with beta-glucosidase mRNA and inhibits its translation in vitro and ex vivo (47, 48). It also interacts with interleukin-2 mRNA and increase its stabilization (42). At the transcription level, NF110 is proposed to interact with a putative regulatory RNA or the nascent transcript to stimulate transcription initiation or elongation (32). Particularly, NF110 upregulates gene expression driven by the IFN-β or SV40 promoter/enhancer (12, 14, 34). We show evidence here that ADAR1 stimulates the expression of IFN-β- or SV40-luciferase and regulates NF90-mediated gene expression. Further evidence of this conclusion comes from knockdown experiments, in which the expression of NF90 and NF110 was reduced by siRNA. The ADAR1-mediated upregulation of SV40-luciferase decreased accordingly when the expression of NF90 and NF110 was reduced. It remains to be addressed whether ADAR1 regulates NF90-mediated gene expression at the transcription or translation level. Because the NF90 proteins have been shown to either promote (41, 42) or inhibit (47, 48) translation by stabilizing mRNA, we propose that ADAR1 is involved in translation control by facilitating interactions between the NF90 proteins and the dsRNA components of highly structured mRNAs. Furthermore, since these highly structured mRNAs themselves can potentially be cellular activators of PKR (23, 25), the ADAR1/NF90/dsRNA complex may also affect protein synthesis through PKR-mediated signaling (46).
We also demonstrated that the dsRBDs of ADAR1 are required for both its interaction and function. ADAR1 variants that lack the first dsRBD will not form complexes with NF90 or stimulate NF90-mediated expression. Instead, the complete function requires the N-terminal region, including the Z-DBD/NES. ADAR1 variants that contain the dsRBDs but lack the Z-DBD/NES region still interact with NF90 but do not stimulate its function. A regulatory domain and the dsRBD fragment comprise the essential components needed to sufficiently upregulate NF90-mediated gene expression. Our data indicate that the interaction domain of ADAR1, the dsRBD region, interacts with NF90 through the dsRNA components; the regulatory domain, the Z-DBD and/or NES domain, facilitates the function of NF90 through a mechanism that has yet to be determined. Notably, ADAR1 p150 is predominantly cytoplasmic, and the Dcat fragment is exclusively cytoplasmic (7, 22). These observations suggest that ADAR1 might act in the cytoplasm by stimulating translation, which is consistent with the function of NF90 in stabilizing mRNA and promoting protein translation (42).
Most DRBPs, including the NF90 proteins, are known to modulate antiviral responses (11, 14, 17, 35, 37, 40). ADAR1 is ubiquitously expressed in immune organs and upregulated in response to viral and microbial infections and is thus suggested to be important for host defense mechanisms (31, 37, 44, 50). The data presented here suggest a possible mechanism for how ADAR1 participates in antiviral responses through the NF90 proteins. The link is further emphasized by the involvement of cellular dsRNA components in the ADAR1-NF90 complexes. This is because the presence of foreign dsRNA in infected cells will signal for the defense mechanisms of the innate immune system against viral pathogens. In the case of human adenovirus, the highly structured viral RNA counteracts a cellular antiviral defense mechanism by blocking the activation of PKR that would otherwise lead to the shutoff of protein synthesis in infected cells (13, 24). The viral RNA also binds NF110b and modulates its activity (17), possibly by competing with the cellular ligand and thereby inhibiting its activity and blocking gene expression. Therefore, it is possible that the role of ADAR1 is to prevent the counteraction of viral RNAs by interacting with other DRBPs in addition to its primary role in RNA editing. By forming complexes with the NF90 proteins, ADAR1 consequently sustains gene expression critical for antiviral responses. Furthermore, NF90 is also associated with and phosphorylated by PKR (14, 26). Whether ADAR1 is involved in the activation of PKR requires further investigation.
Acknowledgments
We thank Yan Li for assistance with experiments, Stratford May, John Hiscott, and Jianjiang Ye for providing constructs for luciferase assays, Ralf Janknecht for providing the anti-p68 antibody, Brenda Bass for providing the anti-ADAR1 antibody, and Alfred Bothwell and members of Yang's laboratory for critically reading the manuscript.
This work was supported by grants GM-60426 and AI060701 from the National Institutes of Health to J.-H. Yang.
REFERENCES
- 1.Cattaneo, R. 1994. Biased (A 3 I) hypermutation of animal RNA virus genomes. Curr. Opin. Genet. Dev. 4:895-900. [DOI] [PubMed] [Google Scholar]
- 2.Cho, D. S., W. Yang, J. T. Lee, R. Shiekhattar, J. M. Murray, and K. Nishikura. 2003. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278:17093-17102. [DOI] [PubMed] [Google Scholar]
- 3.Cosentino, G. P., S. Venkatesan, F. C. Serluca, S. R. Green, M. B. Mathews, and N. Sonenberg. 1995. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl. Acad. Sci. USA 92:9445-9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deb, A., M. Zamanian-Daryoush, Z. Xu, S. Kadereit, and B. R. Williams. 2001. Protein kinase PKR is required for platelet-derived growth factor signaling of c-fos gene expression via Erks and Stat3. EMBO J. 20:2487-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du, J., Y. Pan, Y. Shi, C. Guo, X. Jin, L. Sun, N. Liu, T. Qiao, and D. Fan. 2005. Overexpression and significance of prion protein in gastric cancer and multidrug-resistant gastric carcinoma cell line SGC7901/ADR. Int. J. Cancer 113:213-220. [DOI] [PubMed] [Google Scholar]
- 6.Duchange, N., J. Pidoux, E. Camus, and D. Sauvaget. 2000. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene 261:345-353. [DOI] [PubMed] [Google Scholar]
- 7.Eckmann, C. R., and M. F. Jantsch. 1999. The RNA-editing enzyme ADAR1 is localized to the nascent ribonucleoprotein matrix on Xenopus lampbrush chromosomes but specifically associates with an atypical loop. J. Cell Biol. 144:603-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodbourn, S. 1990. The regulation of beta-interferon gene expression. Semin. Cancer Biol. 1:89-95. [PubMed] [Google Scholar]
- 9.Hartner, J. C., C. Schmittwolf, A. Kispert, A. M. Muller, M. Higuchi, and P. H. Seeburg. 2004. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 279:4894-4902. [DOI] [PubMed] [Google Scholar]
- 10.Herbert, A., S. Wagner, and J. A. Nickerson. 2002. Induction of protein translation by ADAR1 within living cell nuclei is not dependent on RNA editing. Mol. Cell 10:1235-1246. [DOI] [PubMed] [Google Scholar]
- 11.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao, P. N., L. Chen, G. Brock, J. Ng, J. Kenny, A. J. Smith, and B. Corthesy. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 269:20691-20699. [PubMed] [Google Scholar]
- 13.Kitajewski, J., R. J. Schneider, B. Safer, S. M. Munemitsu, C. E. Samuel, B. Thimmappaya, and T. Shenk. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45:195-200. [DOI] [PubMed] [Google Scholar]
- 14.Krasnoselskaya-Riz, I., A. Spruill, Y. W. Chen, D. Schuster, T. Teslovich, C. Baker, A. Kumar, and D. A. Stephan. 2002. Nuclear factor 90 mediates activation of the cellular antiviral expression cascade. AIDS Res. Hum. Retrovir. 18:591-604. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, M., and G. G. Carmichael. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. USA 94:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langland, J. O., P. N. Kao, and B. L. Jacobs. 1999. Nuclear factor-90 of activated T-cells: a double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38:6361-6368. [DOI] [PubMed] [Google Scholar]
- 17.Liao, H. J., R. Kobayashi, and M. B. Mathews. 1998. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. USA 95:8514-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomeli, H., J. Mosbacher, T. Melcher, T. Hoger, J. R. Geiger, T. Kuner, H. Monyer, M. Higuchi, A. Bach, and P. H. Seeburg. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266:1709-1713. [DOI] [PubMed] [Google Scholar]
- 19.Melcher, T., S. Maas, A. Herb, R. Sprengel, M. Higuchi, and P. H. Seeburg. 1996. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 271:31795-31798. [DOI] [PubMed] [Google Scholar]
- 20.Melcher, T., S. Maas, M. Higuchi, W. Keller, and P. H. Seeburg. 1995. Editing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J. Biol. Chem. 270:8566-8570. [DOI] [PubMed] [Google Scholar]
- 21.Mundschau, L. J., and D. V. Faller. 1995. Platelet-derived growth factor signal transduction through the interferon-inducible kinase PKR. Immediate early gene induction. J. Biol. Chem. 270:3100-3106. [DOI] [PubMed] [Google Scholar]
- 22.Nie, Y., Q. Zhao, Y. Su, and J. H. Yang. 2004. Subcellular distribution of ADAR1 isoforms is synergistically determined by three nuclear discrimination signals and a regulatory motif. J. Biol. Chem. 279:13249-13255. [DOI] [PubMed] [Google Scholar]
- 23.Nussbaum, J. M., S. Gunnery, and M. B. Mathews. 2002. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 30:1205-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Malley, R. P., T. M. Mariano, J. Siekierka, and M. B. Mathews. 1986. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 44:391-400. [DOI] [PubMed] [Google Scholar]
- 25.Osman, F., N. Jarrous, Y. Ben-Asouli, and R. Kaempfer. 1999. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 13:3280-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker, L. M., I. Fierro-Monti, and M. B. Mathews. 2001. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J. Biol. Chem. 276:32522-32530. [DOI] [PubMed] [Google Scholar]
- 27.Patel, R. C., and G. C. Sen. 1998. Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol. Cell. Biol. 18:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, R. C., D. J. Vestal, Z. Xu, S. Bandyopadhyay, W. Guo, S. M. Erme, B. R. Williams, and G. C. Sen. 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 274:20432-20437. [DOI] [PubMed] [Google Scholar]
- 29.Patterson, J. B., D. C. Thomis, S. L. Hans, and C. E. Samuel. 1995. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210:508-511. [DOI] [PubMed] [Google Scholar]
- 30.Paul, M. S., and B. L. Bass. 1998. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 17:1120-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabinovici, R., K. Kabir, M. Chen, Y. Su, D. Zhang, X. Luo, and J. H. Yang. 2001. ADAR1 is involved in the development of microvascular lung injury. Circ. Res. 88:1066-1071. [DOI] [PubMed] [Google Scholar]
- 32.Reichman, T. W., and M. B. Mathews. 2003. RNA binding and intramolecular interactions modulate the regulation of gene expression by nuclear factor 110. RNA 9:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichman, T. W., L. C. Muniz, and M. B. Mathews. 2002. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol. Cell. Biol. 22:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichman, T. W., A. M. Parrott, I. Fierro-Monti, D. J. Caron, P. N. Kao, C. G. Lee, H. Li, and M. B. Mathews. 2003. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 332:85-98. [DOI] [PubMed] [Google Scholar]
- 35.Romano, P. R., F. Zhang, S. L. Tan, M. T. Garcia-Barrio, M. G. Katze, T. E. Dever, and A. G. Hinnebusch. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rueter, S. M., C. M. Burns, S. A. Coode, P. Mookherjee, and R. B. Emeson. 1995. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science 267:1491-1494. [DOI] [PubMed] [Google Scholar]
- 37.Samuel, C. E. 1998. Reoviruses and the interferon system. Curr. Top. Microbiol. Immunol. 233:125-145. [DOI] [PubMed] [Google Scholar]
- 38.Saunders, L. R., V. Jurecic, and G. N. Barber. 2001. The 90- and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics 71:256-259. [DOI] [PubMed] [Google Scholar]
- 39.Saunders, L. R., D. J. Perkins, S. Balachandran, R. Michaels, R. Ford, A. Mayeda, and G. N. Barber. 2001. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 276:32300-32312. [DOI] [PubMed] [Google Scholar]
- 40.Sharp, T. V., F. Moonan, A. Romashko, B. Joshi, G. N. Barber, and R. Jagus. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302-315. [DOI] [PubMed] [Google Scholar]
- 41.Shi, L., G. Zhao, D. Qiu, W. R. Godfrey, H. Vogel, T. A. Rando, H. Hu, and P. N. Kao. 2005. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 280:18981-18989. [DOI] [PubMed] [Google Scholar]
- 42.Shim, J., H. Lim, J. R. Yates, and M. Karin. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10:1331-1344. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Q., J. Khillan, P. Gadue, and K. Nishikura. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290:1765-1768. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Q., M. Miyakoda, W. Yang, J. Khillan, D. L. Stachura, M. J. Weiss, and K. Nishikura. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279:4952-4961. [DOI] [PubMed] [Google Scholar]
- 45.Weier, H. U., C. X. George, K. M. Greulich, and C. E. Samuel. 1995. The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1-21.2. Genomics 30:372-375. [DOI] [PubMed] [Google Scholar]
- 46.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed]
- 47.Xu, Y. H., C. Busald, and G. A. Grabowski. 2000. Reconstitution of TCP80/NF90 translation inhibition activity in insect cells. Mol. Genet. Metab. 70:106-115. [DOI] [PubMed] [Google Scholar]
- 48.Xu, Y. H., and G. A. Grabowski. 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 68:441-454. [DOI] [PubMed] [Google Scholar]
- 49.Yang, J. H., X. Luo, Y. Nie, Y. Su, Q. Zhao, K. Kabir, D. Zhang, and R. Rabinovici. 2003. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology 109:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, J. H., Y. Nie, Q. Zhao, Y. Su, M. Pypaert, H. Su, and R. Rabinovici. 2003. Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J. Biol. Chem. 278:45833-45842. [DOI] [PubMed] [Google Scholar]
- 51.Yang, J. H., P. Sklar, R. Axel, and T. Maniatis. 1995. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature 374:77-81. [DOI] [PubMed] [Google Scholar]