Abstract
The target of rapamycin (TOR) protein kinases, Tor1 and Tor2, form two distinct complexes (TOR complex 1 and 2) in the yeast Saccharomyces cerevisiae. TOR complex 2 (TORC2) contains Tor2 but not Tor1 and controls polarity of the actin cytoskeleton via the Rho1/Pkc1/MAPK cell integrity cascade. Substrates of TORC2 and how TORC2 regulates the cell integrity pathway are not well understood. Screening for multicopy suppressors of tor2, we obtained a plasmid expressing an N-terminally truncated Ypk2 protein kinase. This truncation appears to partially disrupt an autoinhibitory domain in Ypk2, and a point mutation in this region (Ypk2D239A) conferred upon full-length Ypk2 the ability to rescue growth of cells compromised in TORC2, but not TORC1, function. YPK2D239A also suppressed the lethality of tor2Δ cells, suggesting that Ypks play an essential role in TORC2 signaling. Ypk2 is phosphorylated directly by Tor2 in vitro, and Ypk2 activity is largely reduced in tor2Δ cells. In contrast, Ypk2D239A has increased and TOR2-independent activity in vivo. Thus, we propose that Ypk protein kinases are direct and essential targets of TORC2, coupling TORC2 to the cell integrity cascade.
Cell growth and proliferation are tightly linked with the cell's perception of its nutritional environment. Tor (target of rapamycin) proteins belong to a family of phosphatidylinositol kinase-like protein kinases and play a central role in controlling cell growth (12, 17). Specifically, TOR signaling couples nutrient signals to various growth-related processes, including protein synthesis, gene expression, uptake of amino acids, actin organization, endocytosis, and autophagy.
The budding yeast Saccharomyces cerevisiae has two homologous TOR genes, TOR1 and TOR2, the protein products of which regulate at least two essential signaling branches (17). In one branch, Tor1 and Tor2 act redundantly to control temporal aspects of cell growth; i.e., when adequate nutrients are available Tor promotes the accumulation of mass by increasing translation (2, 5, 25) and ribosome biogenesis (30) and by repressing autophagy (20). TOR signaling in this branch is sensitive to rapamycin. Tor2 but not Tor1 also functions in a second essential signaling branch. In this branch Tor2 controls spatial aspects of cell growth by regulating the cell cycle-dependent polarization of the actin cytoskeleton. Polarization of the actin cytoskeleton is thought to orient the secretory apparatus such that newly synthesized macromolecules are efficiently delivered to the major site of growth-the bud. A distinct TOR complex operates in each of these signaling branches (26). TOR complex 1 (TORC1) contains Kog1, Tco89, Lst8, and either Tor1 or Tor2. TORC1 regulates the temporal aspects of cell growth, and Tor in TORC1 is bound and inhibited by rapamycin. TORC2 contains Avo1, Avo2, Avo3, Bit61, Lst8, and Tor2 but not Tor1. TORC2 regulates the spatial aspects of cell growth, and Tor2 in TORC2 is not bound by rapamycin; therefore, TORC2 function is insensitive to this drug.
In wild-type yeast, actin patches are concentrated in small- and medium-sized buds and are simultaneously absent from the mother cell. Disruption of TOR2 or TORC2 results in a loss of this actin patch polarization (34, 35). Recently, receptor internalization was also observed to be impaired in tor2 mutants (7). TORC2 acts upstream of the Rho1 GTPase, presumably by regulating the activity of the Rho1 GTP/GDP exchange factor Rom2 (26, 34). Rho1 has several known effectors. One Rho1 effector is the yeast protein kinase C homologue Pkc1. Pkc1 acts upstream of the “cell integrity” mitogen-activated protein kinase (MAPK) pathway (21), and this pathway is required for TORC2/Rho1 to regulate actin polarization (13). A second Rho1 effector is the glucan synthase Fks1. Fks1 appears to act downstream of TORC2/Rho1 to regulate receptor internalization (7). However, the mechanisms by which TORC2 regulates Rom2 are unknown.
Substrates of mammalian TOR (mTOR) have been reported, the best characterized being the translation regulators 4E-BP1 and S6K1 (12). S6K1 belongs to a family of related kinases known as AGC kinases. Recently, Matsuo et al. reported that overexpression of the AGC kinase gad8 rescues the mating deficiency elicited by a tor1 mutation in fission yeast (27). It is important to note that fission yeast strains harboring mutations in TOR1, or in genes encoding proteins homologous to budding yeast TORC2 components, all share very similar phenotypes. For example, ste20 (= Sc AVO3), sin1 (= Sc AVO1), and tor1 strains all have very similar mating defects and all are hypersensitive to various stresses (14, 22, 37, 38). These observations suggest that fission yeast tor1 more closely resembles budding yeast Tor2 and may therefore function primarily in a TORC2-like complex.
Budding yeast encodes a pair of GAD8 orthologues, YPK1 and YPK2, that perform a redundant, essential function (4). The protein kinase activities of the Ypks are dependent upon the PKH1 and PKH2 genes (31), and Ypk1 is directly phosphorylated and activated by Pkh1 (3). PKH genes encode PDK1-like protein kinases (15). While mammal PDK1 requires 3-phosphoinositide as an activating cofactor, Pkhs are activated by the presence of sphingolipids (8). Overexpression of YPK1 confers resistance to ISP-1, an analog of sphingosine that inhibits the serine-palmityl transferase LCB1 and thus blocks sphingolipid production, confirming that Ypks function downstream of Pkhs (36). Recently, several reports have demonstrated that Ypks and TORC2 share similar downstream readouts. For example, receptor internalization and fluid-phase endocytosis are impaired in ypk mutants (6) and loss of Ypk function results in loss of actin polarization (31, 33). Furthermore, overexpression of Rho1 exchange factor (TUS1) suppresses the lethality of both ypk mutants and TORC2 mutants, suggesting that Ypks act in parallel or downstream of TORC2 signaling (33).
In a screen for multicopy suppressors of the lethality of a tor2 mutant, we obtained a 5′-truncated allele of YPK2. A point mutation near the truncated region (e.g., D239A mutation) conferred upon full-length YPK2 the ability to act as a tor2 suppressor. We further demonstrated that immunopurified Tor2 directly phosphorylates Ypk2 in vitro. In vivo, phosphorylation of Ypk2 by Tor2 is required for both Ypk2 kinase activity and function. In contrast, Ypk2D239A has increased and TOR2-independent activity, and YPK2D239A can suppress the lethality resulting from loss of TORC2 function but not loss of TORC1 function. Together, these results indicate that Ypk2 is a substrate of TORC2 and is required for TORC2 to regulate spatial aspects of cell growth.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
Standard techniques were used for yeast manipulation (19). Yeast strains used in this study are listed in Table 1. YYK241 was created by chemical mutagenesis of YMW1. Mutation in the TOR2 locus was confirmed by integration mapping (data not shown). Deletion constructs to disrupt YPK2 and TOR1 with KanMX were amplified by PCR from BY4741-based deletion mutants (10). YYK346 was constructed by crossing DL201 and W303-1A three times into the W303 genetic background. Deletion TOR2 with HIS3 was constructed as follows. A 2.6-kb fragment containing 2.4 kb of the 3′ region of TOR2 open reading frame was amplified by PCR and cloned into BamHI and XhoI sites of pBluescriptII KS+. The HIS3 gene (1.3 kb) was then used to replace a 0.9-kb SalI-BglII fragment, yielding a tor2Δ::HIS3 cassette. The resultant deletion cassette was transformed into a W303 diploid strain. Similarly, a TOR1 deletion cassette (tor1Δ::KanMX) was transformed into W303 to make YYK339. YYK339 was transformed with pRS316[TOR2], and its segregants generated YYK354 and YYK357. Site-directed mutagenesis was done using a QuikChange site-directed mutagenesis kit (Stratagene).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YMW1 | MATα ade2-1 ade3-Δ22 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Laboratory stock |
| YYK241 | YMW1 tor2 p414GAL1[Tor2 ADE3] | This study |
| W303 | MATa/α ade2 his3 leu2 trp1 ura3 can1 | Y. Mukai |
| W303-1A | MATα ade2 his3 leu2 trp1 ura3 can1 | Y. Mukai |
| W303-1B | MATaade2 his3 leu2 trp1 ura3 can1 | Y. Mukai |
| DL201 | MATahis4 leu2 ura3 ypk1Δ::TRP1 | D.E. Levin |
| YYK346 | W303-1B ypk1Δ::TRP1 | This study |
| YYK373 | W303-1B ypk1Δ::TRP1 ypk2Δ::KanMX p415GAL1[Ypk2-244] | This study |
| YYK339 | W303 tor1Δ::/KanMX/Tor1 tor2Δ::HIS3/Tor2 | This study |
| YYK354 | W303-1B tor2Δ::HIS3 pRS316[Tor2] | This study |
| YYK357 | W303-1B tor1Δ::KanMX tor2Δ::HIS3 pRS316[Tor2] | This study |
| JK9-3da | MATahis4 leu2-3, 112 ura3-52 trp1 rme1 GAL+ | 13 |
| SH100 | JK9-3da tor2Δ::ADE2-3 YCplac111[Tor2] | 13 |
| SH121 | JK9-3da tor2Δ::ADE2-3 YCplac111[tor2-21ts] | 13 |
| SH221 | JK9-3da tor1Δ::HIS3-3 tor2 Δ::ADE2-3 YCplac111[tor2-21ts] | 13 |
| INA28-1B | MATaade1 his2 leu2 trp1 ura3 pkh2Δ::LEU2 | 15 |
| INA106-3B | INA21-1B pkh1-D398Gts | 15 |
| TB50a | MATahis3 leu2-3,112 trp1 ura3 rme1 | Laboratory stock |
| RL23-1c | TB50a [KanMX4]-GAL1p-AVO1 | 26 |
| RL93a | TB50a [KanMX4]-GAL1p-KOG1 | 26 |
Multicopy suppressor screening.
The YYK241 strain was transformed with a yeast genomic DNA library constructed in the YEp24 multicopy plasmid, and the transformants were grown on selective medium (SC)-galactose-Ura plates. We screened approximately 24,000 colonies, and we picked 27 white sectoring colonies as the first candidates. We further examined their growth at 37°C (nonpermissive temperature of YYK241 cells) on YEPD as a second screening. We were left with six candidates as multicopy suppressors of YYK241 cell. Genomic library-derived plasmids were rescued, and the genes contained in these plasmids were identified by sequencing both ends of the inserts. Deletion and subcloning analyses revealed that the 5′-truncated YPK2 gene was a suppressor.
Protein kinase assay of Tor2 protein.
SH121cell harboring pRS314[HATOR2] plasmid (18) grown in yeast extract-peptone-dextrose (YEPD) medium were collected, washed once with distilled water, and suspended in ice-cold TOR lysis buffer (16 μl/optical density at 600 nm cell) (1× phosphate-buffered saline [PBS] [pH 7.4], 10% glycerol, 4 mM Na3VO4, 50 mM KF, 15 mM Na-PPi [pH 7.5], 15 mM p-nitrophenylphosphate [pNPP], 20 μg/ml leupeptin, 20 μg/ml benzamidine, 10 μg/ml pepstatin A, 40 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF]). An equal volume of glass beads (425 to 600 μm) was added to this suspension, and cells were broken by vigorous vortexing for 10 min at 4°C. A half volume of lysis buffer containing 2% Tween 20 was added, and the mixture was further incubated for 10 min with mild rotation. The beads and cell debris were removed by centrifugation for 10 min at 10,000 × g at 4°C, and the supernatant was further clarified by an additional 10-min centrifugation. Glycerol was added to the resulting lysate to a final concentration of 30% before storage at −20°C. Protein concentration was estimated using a BCA kit (Pierce). For immunoprecipitation of HATor2 protein, 1 mg of cell lysate was brought up to 1 ml with immunoprecipitation buffer (TOR lysis buffer containing 0.5% Tween 20 and 0.25% gelatin) and incubated with 1 μl of antihemagglutinin (anti-HA) ascites (16B12; BabCO) for 2 h at 4°C with gentle rotation. To the extract/antibody mixture was added 20 μl of 50% suspension of protein G-Sepharose 4FF (Amersham), followed by a further rotation for 1 h at 4°C. Immunocomplexes were transferred into fresh microcentrifuge tubes and washed four times with 1 ml of immunoprecipitation buffer (without gelatin) with gentle rotation and twice with 1 ml of 25 mM HEPES-KOH (pH 7.5). The resultant immunocomplexes were resuspended in 36 μl of TOR kinase assay buffer (25 mM HEPES-KOH [pH 7.5], 50 mM NaCl, 10 mM MnCl2, 15 mM pNPP) containing 4 μg of substrates (4E-BP1 [Santa Cruz] or recombinant Ypk2). This mixture was preincubated for 3 min at 30°C before the reaction was initiated by adding 4 μl of 0.5 mM [γ-32P]ATP (Amersham) (20 μCi/sample). After incubation for 60 min at 30°C, the reaction was terminated by addition of 40 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and incubation for 5 min at 65°C. Samples were subjected to 7.5 to 12.5% SDS-PAGE gels. After electrophoresis, gels were immersed in 12.5% trichloroacetic acid for 30 min, followed by several washes in distilled water. Proteins were detected by Coomassie staining (GelCode Blue; Pierce). Phosphorylated proteins were detected by autoradiography.
Protein kinase assay of Ypk2 protein.
C-terminal hemagglutinin (HA) tagging of Ypk2 (Ypk2HA) was conducted as follows. The EcoRI-EcoRI fragment (2.4 kb) of YEp352[YPK2] plasmid (4) was cloned into pUC18. A NotI site was created immediately 5′ to the translational terminator by use of a QuikChange kit (Stratagene). Next, a NotI cassette of 3× HA was subcloned into the newly created NotI site of YPK2. For immunoprecipitation experiments, cells harboring YEp352[YPK2HA] plasmid grown in YEPD were collected, washed once with distilled water, and suspended in ice-cold YPK lysis buffer (1× PBS [pH 7.4], 1 mM EDTA, 1 mM EGTA, 4 mM Na3VO4, 50 mM KF, 15 mM Na-PPi [pH 7.5], 15 mM pNPP, 20 μg/ml leupeptin, 20 μg/ml benzamidine, 10 μg/ml pepstatin A, 40 μg/ml aprotinin, 1 mM PMSF). Cells were broken by vortexing with glass beads as described above. The beads and cell debris were removed by centrifugation for 10 min at 10,000 × g at 4°C, and the supernatant was further clarified by an additional 10-min centrifugation. Glycerol was added to the resulting lysate to a final concentration of 30% before storage at −20°C. Ypk2HA protein was immunoprecipitated as described above with several modifications. The immunoprecipitation buffer used was YPK lysis buffer containing 0.5% Tween 20 (and it did not contain gelatin), immunoprecipitation was conducted with 100 to 1,000 μg protein in a 500-μl volume, and incubation time with HA antibody was 5 h. Immunoprecipitated Ypk2HA protein was suspended with 18 μl of YPK kinase assay buffer (50 mM MOPS-KOH [pH 7.5], 10 mM MgCl2, 1 mM Na3VO4) containing 4 μg 2× crosstide (GRPRTSSFAEGGRPRTSSFAEG). This mixture was preincubated for 3 min at 30°C, and the reaction was initiated by adding 2 μl of 1 mM [γ-32P]ATP (Amersham) (10 μCi/sample). After incubation for 30 min at 30°C, the reaction was terminated by addition of 30 μl of 2× SDS-PAGE sample buffer and incubation for 5 min at 65°C. Samples were subjected to 15% SDS-PAGE gel. After electrophoresis, gels were immersed in 12.5% trichloroacetic acid for 30 min, followed by several washes in distilled water. Proteins were detected by Coomassie staining (GelCode Blue; Pierce). Phosphorylated proteins were detected by autoradiography or BAS2000 (Fuji Film), the amount of immunoprecipitated Ypk2HA was monitored by immunoblotting (LAS1000; Fuji Film), and the specific activity of Ypk2 was estimated from these values.
Actin staining.
YEPD-grown cells were shifted from 30°C to 37°C for 2 h. Cells were then fixed for 30 min by the direct addition of 37% formaldehyde stock to a final concentration of 5%. Fixed cells were collected, washed with 1× PBS three times, and then 0.1 U/μl of rhodamine-phalloidin (Sigma) was added for 2 h at room temperature to stain actin as described previously (19). Cells were observed, and the images were acquired as described previously (28) using a rhodamine filter.
Preparation of recombinant Ypk2 protein.
We used modified pGEX-6p-1 (Amersham) for expression of recombinant protein. An annealed oligonucleotide (5′-TCGACTCCACCATCATCATCATCATTAATGC-3′) was inserted into the SalI-NotI sites of pGEX-6p-1, and we designated the created plasmid pGEX-His. A YPK2 open reading frame was amplified with primers 5′-GGGGATCCATGCATTCCTGGCGAATATC-3′ and 5′-ACGCGTCGACCACTAATGCTTCTCCCCTGC-3′, with YEp352[YPK2] as a template. The PCR product was cloned into BamHI-SalI sites of pGEX-His (designated pGEX[YPK2-His]). To create pGEX[YPK2S641A T659A-His]), a QuikChange kit (Stratagene) was used. The plasmids of YPK2 with an amino-terminal glutathione S-transferase-tagged and a carboxyl-terminal hexahistidine-tagged construct were transformed into Escherichia coli BL21(DE3). The cells were disrupted by sonication at 4°C in PBS (pH 7.4)-1 mM EDTA-5 mM dithiothreitol-0.1 mM PMSF. The protein was applied on a glutathione-Sepharose 4B column (Amersham) equilibrated with PBS (pH 7.4). The bound protein was eluted with 50 mM Tris-HCl (pH 8.0) and 5 mM reduced glutathione. The amino-terminal glutathione S-transferase tags in proteins were then removed by incubation with PreScission protease (Amersham) for 6 h at 4°C, and the digested proteins with a carboxyl-terminal His tag were applied on an Ni-nitrilotriacetic acid (NTA) column (QIAGEN) equilibrated with 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 10 mM imidazole. The bound protein was eluted with 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 200 mM imidazole. Fractions containing proteins were purified on a Superdex 200 gel filtration column (Amersham) and eluted with 20 mM Tris-HCl (pH 8.0), 2 mM dithiothreitol, and 150 mM NaCl.
RESULTS
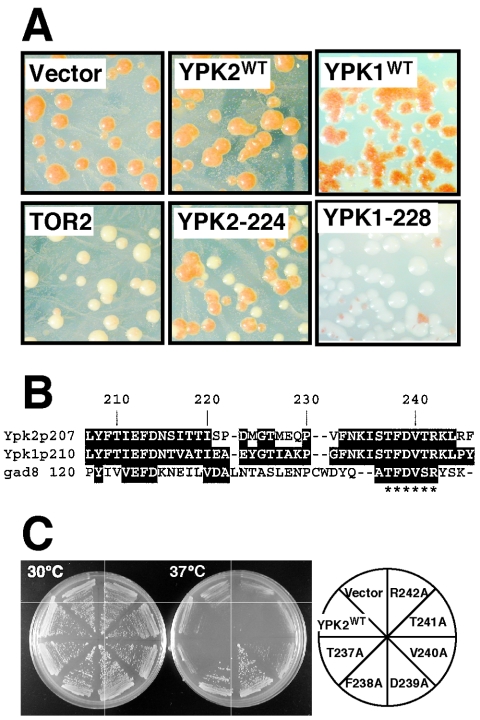
To identify a novel factor(s) involved in TOR signaling, we screened for multicopy suppressors of the lethality of a tor2 mutant. The yeast strain YYK241 (see Table 1) has mutations in the ADE2, ADE3, and TOR2 loci and harbors plasmid p414GAL1[TOR2 ADE3]. This strain requires maintenance of the plasmid for growth and thus forms nonsectoring red colonies (Fig. 1A; ade2 cells accumulate a red pigment, ade2 ade3 cells do not). In galactose-containing medium TOR2 is overexpressed, and YYK241 cells grow like wild-type cells. In glucose-containing medium YYK241 cells have a temperature-sensitive cell lysis phenotype (at 37°C; data not shown). We transformed YYK241 with a yeast genomic library, and from 24,000 transformants we obtained seven colonies that could form white sectors. By DNA sequencing and subcloning, we determined that a 5′ truncated YPK2 gene suppressed the tor2 mutant phenotype (Fig. 1A). The 5′-most methionine codon in the open reading frame of the truncated YPK2 is M224, so we assume that the polypeptide produced starts at position 224. Hereafter we refer to this suppressor as YPK2-224. The remaining candidates contained the TOR2 gene (data not shown).
FIG. 1.
N-truncated Ypk2 suppresses tor2 mutants. A. YYK241 (ade2 ade3 tor2 p414GAL1[TOR2 ADE3]) was transformed with YEp352 (vector), YEp352[full-length YPK2] (YPK2FL), YEp352[N-truncated YPK2] (YPK2-224), and YEp352[N-truncated ypk2-K373A (kinase dead)] (YPK2-224KD), as indicated. The transformants were grown on YEPD plates for 4 days at 30°C. Sectoring colonies showed independence of TOR2 plasmid, i.e., suppression of tor2 mutation. B. Alignment of the amino acid sequences around the M224 of Ypk2. Ypk1, Ypk2 (S. cerevisiae), and Gad8 (S. pombe) are shown. The arrowhead highlights M224 of Ypk2, and asterisks denote the conserved region shared among these protein kinases. C. YPK2 genes (cloned into YEp352) harboring a point mutation in the conserved region as indicated (right panel) were transformed into YYK241, and the resultant transformants were grown on YEPD plates for 2 days at 30°C (left) or 37°C (right). Note that these YPK2 genes expressed full-length of Ypk2 protein under the control of their own promoters.
We expected that overexpression of full-length YPK2 would also suppress the tor2 phenotypes. However, to our surprise, full-length YPK2 did not act as a multicopy suppressor (YPK2FL; Fig. 1A). Similarly, a kinase-dead (ATP-binding site) mutant, YPK2-224-K373A (YPK2-224KD), was also unable to suppress tor2 phenotypes, indicating that kinase activity of Ypk2-224 is required for the suppression. We further tried to examine kinase activity of Ypk2-224, but the expression level of the protein was too low for biochemical analyses (data not shown). Ypk2 functions redundantly with Ypk1. We therefore examined whether a 5′-truncated YPK1 could also act as a tor2 multicopy suppressor. Full-length and 5′-truncated YPKs were expressed using the GAL1 promoter in YYK241. Expression of 5′-truncated YPK1 permitted sectoring, but overexpression of full-length YPK1 did not (data not shown). As the truncated versions of YPK1 and YPK2 behave similarly, we focused on only the YPK2 gene in subsequent experiments.
We hypothesized that a cis-inhibitory motif is compromised in the YPK2-224 allele, which results in a polypeptide with increased kinase activity. We aligned the Ypk1, Ypk2, and Gad8 sequences to see whether there are any regions around Ypk2 aa224 that may be highly conserved. Indeed, there is a six-amino-acid residue sequence (237TFDVT243R in Ypk2) nearly completely conserved between these three sequences (Fig. 1B). To examine the function of this sequence we individually mutated each amino acid in this sequence to an alanine in the context of otherwise full-length, wild-type Ypk2. We then expressed these constructs in a multicopy vector under the control of the YPK2 promoter and asked whether or not these alleles could suppress tor2 phenotypes (Fig. 1C). The ability to suppress a tor2 mutation (YYK241) was conferred upon full-length Ypk2 by substitution of F238 or D239 (or 240V, albeit weakly) to alanine. We provide further evidence (below) that this region does indeed inhibit the kinase activity of Ypk2. There is a similar sequence around F213 (F213DNSI217) in Ypk2 (Fig. 1B); however, when we replaced F213 with an alanine, we did not observe suppression (data not shown).
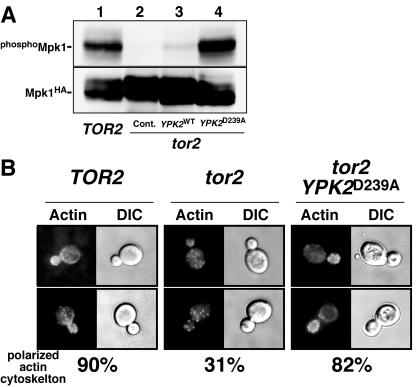
The TORC2 signaling pathway plays an important role in regulating polarization of the actin cytoskeleton via the Rho1/Pkc1/MAPK (cell integrity) cascade. We therefore wished to examine the effect of expression of the YPK2D239A allele has on the cell integrity cascade. Compared to TOR2 cell results, phosphorylation of Mpk1, the MAPK in this cascade, was diminished in tor2 mutants (YYK241), confirming that the Rho1/Pkc1/Mpk1 cell integrity pathway is compromised in tor2 cells (Fig. 2A). Overexpression of YPK2D239A fully rescued Mpk1 phosphorylation, while the wild-type allele rescued poorly. YYK241 cells also showed a cell lysis phenotype, presumably because of inactivation of the Pkc1/Mpk1 cascade resulting in fragility of the cell wall. Expression of YPK2D239A suppressed this phenotype (data not shown). We further asked whether our multicopy YPK2 alleles were able to suppress the actin polarization defect of tor2 cells. At 37°C 90% of TOR2 cells displayed normal actin localization. In contrast, in tor2-21 cells, actin patches were most often not restricted to the small bud but were observed in both the mother cell and the bud (only 31% of cells displayed normal action organization; Fig. 2B). Overexpression of YPK2D239A and YPK2 in tor2-21 cells restored proper actin organization in 82% and 56% of cells, respectively (Fig. 2B and data not shown).
FIG. 2.
YPK2D239A restores Mpk1 activation and suppresses actin cytoskeleton organization defects in tor2 mutant cells. A. Wild-type cells (YMW1) and tor2 mutant cells (YYK241) harboring indicated YPK2 plasmids were transformed with pRS424[HA-tagged MPK1 (MPK1HA)] to express Mpk1HA protein. Cells were grown at 37°C, and phosphorylation of Mpk1HA was determined by immunoblotting with anti-diphospho-p44/p42 MAPK (Cell Signaling Technology) (top panel). Total Mpk1 was determined by immunoblotting with anti-HA antibody (bottom panel). B. Wild-type (SH100) and tor2-21 mutant (SH121) cells harboring YEp352[YPK2D239A] grown in YEPD at 30°C were shifted to 37°C for 6 h. The actin cytoskeleton was stained with rhodamine-phalloidin as described in Materials and Methods. The percentage of cells exhibiting a polarized actin patch is also shown (bottom).
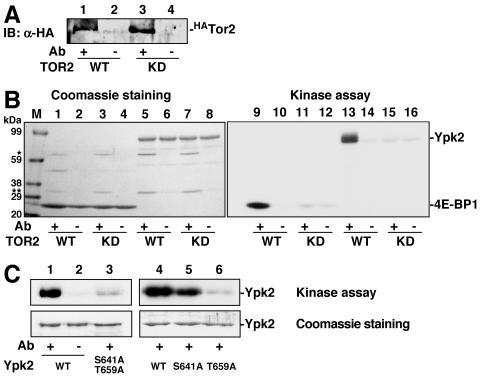
Given that overexpression of alleles of YPK2 can suppress tor2 phenotypes and that the phosphorylation of two other AGC kinases is regulated by TOR (Gad8 by Tor1 in Schizosaccharomyces pombe and S6K1 by mTOR in mammals), we hypothesized that Ypk2 may be directly phosphorylated by Tor2. Gad8 and S6K1 are phosphorylated in a TOR-dependent manner on serine and threonine residues residing in a so-called hydrophobic motif and the turn motif (T659 and S641 in Ypk2, respectively). TOR-dependent phosphorylation of these residues is indispensable for the kinase function of Gad8 and S6K1 (16, 27). Therefore, we examined whether Tor2 directly phosphorylates these C-terminal residues in Ypk2. Specifically, we assayed whether immunoprecipitated N-terminally hemagglutinin (HA)-tagged Tor2 protein (HATor2) (Fig. 3A) could phosphorylate recombinant Ypk2 in vitro (Fig. 3B). HATor2 indeed phosphorylated Ypk2 (lane 13) as well as 4E-BP (a substrate of mTOR and positive control in this experiment) (lane 9), suggesting that Tor2 directly phosphorylates Ypk2. Immunoprecipitated HATor2D2298E (kinase dead mutant) displayed no detectable kinase activity toward 4E-BP or Ypk2 (lanes 11 and 15), excluding the possibility that a co-immunoprecipitated protein kinase phosphorylates Ypk2. As shown in Fig. 3C, the turn- and hydrophobic-motif mutant (Ypk2S641A T659A) was only marginally phosphorylated compared to wild-type Ypk2. We also tested Ypk2S641A and Ypk2T659A mutants as substrates. Ypk2S641A was phosphorylated (but significantly more weakly than Ypk2WT) by Tor2, while phosphorylation of Ypk2T659A was largely diminished (Fig. 3C). These results indicate that Tor2 directly phosphorylates Ypk2 at S641 in the turn motif and T659 in the hydrophobic motif.
FIG. 3.
Tor2 directly phosphorylates Ypk2 in vitro. A. Detection of immunoprecipitated HA-tagged Tor2 (HATOR2). HATor2wild type (WT; lanes 1 and 2) or HATor2D2298E (kinase dead mutant [KD]; lanes 3 and 4) were immunoprecipitated in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of anti-HA antibody. Immunocomplexes were subjected to 7% SDS-PAGE gels, and HATor2 protein was detected by immunoblotting with anti-HA antibody. B. In vitro Tor2 kinase assay. Immunoprecipitated HATor2 (wild type [WT] or kinase dead [KD]) was incubated with [γ-32P]ATP and 4E-BP (lanes 1 to 4 and lanes 9 to 12) or recombinant Ypk2 (lanes 5 to 8 and lanes 13 to 16) at 30°C for 60 min and stopped with sample buffer, and proteins were separated by SDS-PAGE. The results of Coomassie staining (left) and autoradiography (kinase assay) (right) are shown. * and **, migration of heavy and light chains of the immunoprecipitation antibody. C. Tor2 phosphorylates Ypk2 at the turn motif and the hydrophobic motifs. In vitro kinase assays were carried out using the wild type (WT; lanes 1, 2, and 4) and the turn- and hydrophobic-motif mutants (S641A T659A, S641A, and T659A; lanes 3, 5, and 6, respectively) of Ypk2.
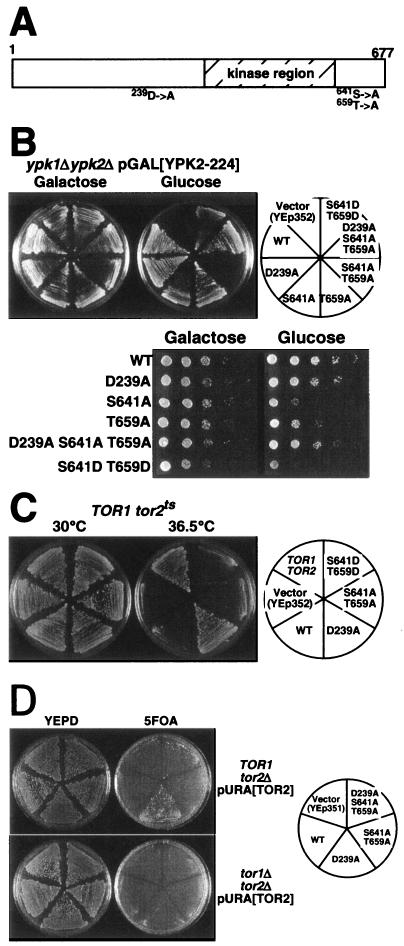
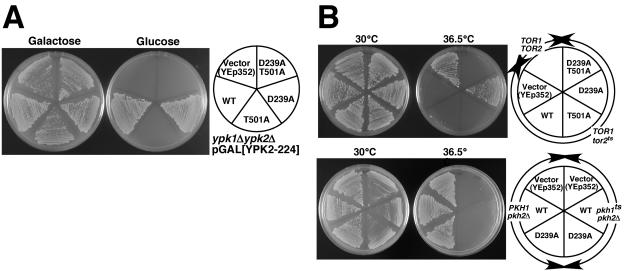
In contrast to wild-type YPK2, expression of YPK2S641A T659A failed to restore growth to a strain lacking YPK1 and YPK2 (ypkΔ), suggesting that phosphorylation of the turn and hydrophobic motifs is essential for Ypk function (Fig. 4B, top panel). The YPK2S641A and YPK2T659A alleles restored slow growth to ypkΔ cells (Fig. 4B, bottom panel). In contrast, the YPK2D239A mutant allele could fully complement this ypkΔ strain, and moreover the YPK2D239A S641A T659A (combination of suppressor allele with phosphorylation mutations) allele could also complement (Fig. 4B). Overexpression of TOR2 could not suppress the growth defect of ypkΔ cells, confirming that Ypks act downstream of Tor2 (data not shown).
FIG. 4.
YPK2D239A overcomes mutation in cis, Tor2-dependent phosphorylation sites. A. Mutation sites of Ypk2. D239 is in a “TOS-like” conserved region, and S641 and T659 are in the turn motif and the hydrophobic motif (sites phosphorylated by Tor2), respectively. B. (Top) YPK2 genes (cloned into YEp352) harboring mutation at D239 or/and Tor2-phosphorylation sites as indicated (right) were transformed into YYK373 (ypksΔ p415GAL1[YPK2-224]). The transformants were grown on YEP galactose (Ypk2-224 expressed; left) or YEPD (Ypk2-224 not expressed; center) plates for 2 days at 30°C.(Bottom) Serial dilutions of the indicated transformants were spotted on SCgalactose-URA (left) and SCglucose-URA (right) and grown for 2 days at 30°C. C. A temperature-sensitive tor2-21 (SH121 [TOR1 tor2-21]) mutant was transformed with YEp352-based YPK2 plasmids as indicated (right) and grown on YEPD for 2 days at 30°C (left) or 36.5°C (center). Wild-type control cells (SH100 [TOR1 TOR2]) are also shown. D. YPK2 mutant genes were transformed into YYK353 (TOR1 tor2Δ pRS316[TOR2]) (top panel) and YYK357 (tor1Δ tor2Δ pRS316[TOR2]) (bottom panel) as indicated in the right panel, and the resultant transformants were grown on YEPD (left) and 5-fluoro-orotic acid (5FOA) plate (center) for 2 days at 30°C. On 5FOA plates, cells must lose the pRS316[TOR2] (harboring URA3 marker) plasmid to grow (19).
Next, we tested the ability of a YPK2S641D T659D mutant to restore growth to ypkΔ cells. Mutation of these residues to aspartate was expected to mimic phosphorylation. YPK2S641D T659D restored very slow growth to ypkΔ cells (Fig. 4B, bottom panel). However, it did not rescue the temperature-sensitive growth of tor2-21 cells (Fig. 4C). This suggests that these mutations cannot fully substitute for phosphoserine/threonine in the activation of Ypk2. From studies whose results were consistent with these, Matsuo et al. (27) reported that gad8-S527D/S546D (a mutation equivalent to YPK2S641D T659D) only partially rescued the mating defect of a gad8Δ mutant, and Casamayor et al. (3) reported that Ypk1T504D T662D displayed no detectable activity.
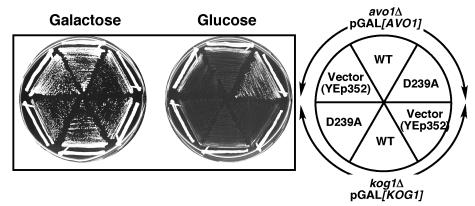
Tor2 functions in two distinct complexes; we therefore wanted to determine which Tor2 complex regulates Ypk. Overexpression of YPK2D239A or YPK2D239A S641A T659A suppressed the lethality of a tor2 null mutant but not a tor1Δ tor2Δ double mutant, suggesting that Ypk2D239A could compensate for the essential function of Tor2 but not of both Tor1 and Tor2 (Fig. 4C). Furthermore, overexpression of YPK2D239A suppressed the growth defects of an avo1 (TORC2) mutant but not a kog1 (TORC1) mutant (Fig. 5). Together these results suggest that Ypks function downstream of TORC2 and not downstream of TORC1.
FIG. 5.
YPK2D239A suppresses growth defects associated with TORC2 disruption but not TORC1 disruption. GAL1 promoter-KOG1 (a component of TORC1; RL93a)- and GAL1 promoter-AVO1 (a component of TORC2; RL23-1c)-containing cells harboring YPK2 plasmids as indicated (right) were grown in SCgalactose glycerol-Ura (left) or SCglucose-Ura (center).
It is known that Ypks are downstream of Pkh1 and Pkh2, the yeast orthologues of the PDK1 protein kinase (15). Pkhs act as effectors of sphingoid-base signaling, playing important roles in cell integrity, endocytosis, actin organization, and activation of Pkc1 and Ypk2 (15). Phosphorylation of T501 in Ypk2 by Pkhs is indispensable for Ypk2 function, because YPK2T501A or YPK2D239A T501A did not complement the lethality of ypkΔ cells (Fig. 6A, top panel). YPK2D239A T501A did not suppress tor2ts cells, either (Fig. 6A, bottom panel). We also tested whether the tor2-suppressing YPK2D239A allele could suppress pkh phenotypes. Neither YPK2WT nor YPK2D239A suppressed the lethality of pkh1ts cells at the nonpermissive temperature (Fig. 6B). At a semipermissive temperature (35°C) both YPK2 alleles weakly suppressed temperature sensitivity (data not shown). One interpretation of these results is that the Pkhs have other essential targets in addition to the Ypks (Pkc1, for example).
FIG. 6.
YPK2D239A acts as a tor2-specific suppressor. A. (Top) YPK2 alleles (cloned into YEp352) harboring mutations at D239 or/and T501 (Pkh phosphorylation site) were transformed into YYK373 (ypksΔ p415GAL1[YPK2-224]). The transformants were grown on YEPgalactose (Ypk2-224 expressed; left) or YEPD (Ypk2-224 not expressed; center) plates for 2 days at 30°C. (Bottom) A temperature-sensitive tor2-21 mutant (SH121 [TOR1 tor2-21]) was transformed with YEp352-based YPK2 plasmids as indicated (right) and grown on YEPD for 2 days at 30°C (left) or 36.5°C (center). Wild-type control cells (SH100 [TOR1 TOR2]) are also shown. B. Temperature-sensitive pkh mutant cells (INA106-3B [pkh1D398G pkh2Δ]) and wild-type control cells (INA28-1B [PKH1 pkh2Δ]) were transformed with YEp352-based YPK2 plasmids as indicated (right) and grown on YEPD for 2 days at 30°C (left) or 36.5°C (center).
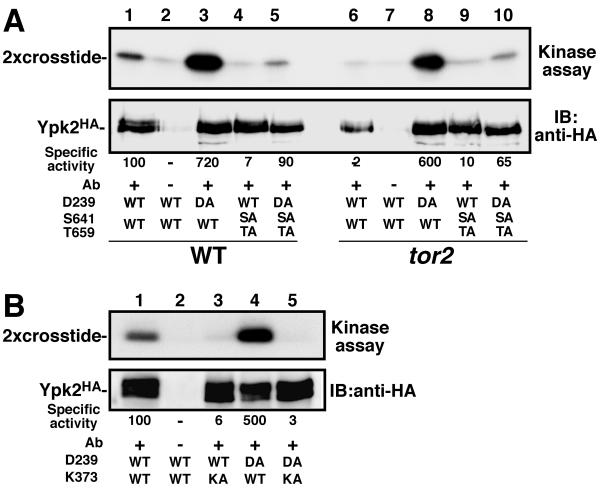
Lastly, we assayed the kinase activity of C-terminally HA-tagged Ypk2 immunopurified from TOR2 or tor2-21 cells by using a peptide (2× crosstide) as a substrate (3) (Fig. 7A). The activity of Ypk2WT was significantly reduced when isolated from tor2-21 cells compared to wild-type cell results (decreased to 2% of wild type; Fig. 7A, lanes 1 and 6), confirming that Ypk kinase activity is regulated by TOR2. We also found that Ypk2D239A was hyperactive (sevenfold activation compared to Ypk2WT; lane 3), and this activity was independent of TOR2 (lane 8). In contrast Ypk2S641A T659A possessed a greatly reduced kinase activity (about 10% of Ypk2WT; lanes 4 and 9), which was partially restored (up to 65% of Ypk2WT) by introduction of the D239A mutation (lanes 5 and 10). As a control, we also performed in vitro protein kinase assays with a kinase dead Ypk2 mutant (Ypk2K373A). The protein kinase activity of Ypk2K373A and Ypk2D239A K373A was essentially lost, confirming that the protein kinase activity detected was an intrinsic property of Ypk2 (Fig. 7B). These results correspond well with the above-presented conclusions that Ypk activity and function are regulated by Tor2 via direct phosphorylation of S641 and T659 and that the D239A substitution potentially disrupts an inhibitory region in Ypk and thereby increases basal activity to a level which is sufficient to compensate for the loss of activating signals from Tor2.
FIG. 7.
Protein kinase activity of Ypk2 is regulated by TOR2. A. Wild-type (SH100) and tor2-21 mutant (SH121) cells harboring YEp352-based HA-tagged YPK2 plasmids as indicated were grown in YEPD at 30°C. Ypk2HA was immunoprecipitated from cell lysates and subjected to in vitro kinase assays (Kinase assay; top panel). Immunoprecipitated Ypk2HA was monitored by immunoblot (IB; bottom). Radioactivity and intensity of immunoblot signal were measured by BAS2000 and LAS1000 (Fuji Film), respectively, and specific activity of each sample was calculated (shown in the figure). B. Ypk2 kinase activity was assessed as described above. A kinase dead Ypk2 mutant (K373A) displayed no detectable protein kinase activity (lanes 3 and 5).
DISCUSSION
We have shown that Ypk2 is a direct substrate of Tor2 and likely TORC2. This observation is consistent with the recent report of Matsuo et al. (27) that the fission yeast orthologue of Ypk1/2, Gad8, is also downstream of Tor in this organism. However, whether Gad8 is directly phosphorylated by Tor has not been elucidated. Both Ypk and TORC2 have been shown to function upstream of Rho1 to mediate actin polarization and endocytosis (6, 7, 34). Thus, Ypk appears to be a missing link between TORC2 and Rho1 activation. Furthermore, given that YPK2D239A could suppress the temperature sensitivity and actin defects of tor2-21 mutants, even when it is integrated in the genome (data not shown), and that it suppressed the growth defect of tor2Δ cells, suggests that the Ypks are the essential target of TORC2. How Ypk signaling regulates Rho1 activation remains to be determined. Recently, Audhya et al. reported that the essential homologous protein pair, Slm1 and Slm2, are also directly phosphorylated by TORC2 (1). Like Ypks, the Slms are required for polarization of the actin cytoskeleton and seem to operate upstream of the cell integrity pathway. The functions of the Slms are not known, and the precise biochemical connection between the Slms and Ypks remains to be determined. Our results suggest that Ypks that act downstream of TORC2 but not TORC1-YPK2D239A could not suppress the growth defects resulting from the loss of TORC1 function. However, Gelperin et al. reported that ypk mutant cells are rapamycin hypersensitive (9). Also, rapamycin treatment appears to enhance Mpk1 activity (24). These findings are curious, because TORC2, unlike TORC1, is rapamycin insensitive. Cross talk between TORC1 and TORC2 could provide one explanation for these observations.
We also found that YPK2D239A has TOR2-independent kinase activity and function but is still dependent upon PKH genes, encoding counterparts of mammalian PDK. PDK directly phosphorylates the activation loop of AGC kinases, such as S6K (mammal) and Gad8 (fission yeast). Thus, Ypks could integrate TORC2 and Pkh-mediated sphingolipid signals to regulate and coordinate actin polarization and endocytosis.
How does D239A relieve TORC2 dependency? We propose that Ypk has two regions that mediate TOR dependency, the N-terminal inhibitory region (containing D239) and the C-terminal phosphorylation domain (the turn and hydrophobic motifs). Interestingly, mutations in the former region confer TOR-independent activity, while mutations in the latter result in a loss of TOR-dependent activation. It is possible that these regions interact with each other, although this remains to be determined. Curiously, D239 of Ypk2 resides in a sequence, 237TFDVT243R, which resembles the TOS motif in mammalian TOR (mTOR) target proteins (32). In mammals, mTOR forms a complex with Raptor (11) (23). Raptor binds to TOS motifs and thereby recruits substrates, i.e., S6K and 4E-BP, to mTOR (29). Thus, the TOS motif plays a positive role in mTOR signaling. In yeast, the Raptor orthologue, Kog1, is a component of TORC1 (26), and it is therefore unlikely that Kog1 recognizes the “TOS-like” regions of Ypks. Moreover, the “TOS-like” region in Ypks antagonizes TORC2 signaling. Therefore, although the TOS motifs are apparently conserved among mammalian and yeast targets of TOR, the function of this sequence does not seem to be conserved. Rather, it is reasonable that the “TOS-like” region in Ypks plays an inhibitory role in regulating Ypk activity, such as the pseudosubstrate domain in protein kinase C. Our current model of Ypk activation is as follows. When inactive, the catalytic domain of Ypk is sheltered by the “TOS-like” regulatory region. Next, TORC2 directly phosphorylates inactive Ypk at the turn and hydrophobic motifs. The possibility is not fully excluded that another protein kinase(s) may phosphorylate Ypk at these sites, but we demonstrated here that its phosphorylation by Tor2 but not by any other kinase is indeed required for activation of Ypk. Phosphorylation at the turn and hydrophobic motifs by TORC2 releases the “TOS-like” regulatory region, exposing the Ypk kinase domain to phosphorylation by Pkhs. Phosphorylation in the activation loop by Pkhs results in fully active Ypk (3).
Acknowledgments
We thank Kazuma Tanaka, Konomi Fujimura-Kamada, David E. Levin, Yukio Mukai, Kenji Irie for strains and plasmids; Akira Matsuura, Kazuyoshi Yonezawa, Maho Hamasaki, Tomoko Kawamata, Kuninori G. Suzuki, and members of our laboratory for helpful discussion; and Yuko Imamura, Chika Kondo, National Institute for Basis Biology Center for Analytical Instruments for technical assistance.
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
This paper is dedicated to the memory of Shoshi Muto, whose mentorship meant a great deal to Y.K.
REFERENCES
- 1.Audhya, A., R. Loewith, A. B. Parsons, L. Gao, M. Tabuchi, H. Zhou, C. Boone, M. N. Hall, and S. D. Emr. 2004. Genome-wide lethality screen identifies new PI4,5P(2) effectors that regulate the actin cytoskeleton. EMBO J. 23:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casamayor, A., P. D. Torrance, T. Kobayashi, J. Thorner, and D. R. Alessi. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9:186-197. [DOI] [PubMed] [Google Scholar]
- 4.Chen, P., K. S. Lee, and D. E. Levin. 1993. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol. Gen. Genet. 236:443-447. [DOI] [PubMed] [Google Scholar]
- 5.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 17:859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.deHart, A. K., J. D. Schnell, D. A. Allen, and L. Hicke. 2002. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.deHart, A. K., J. D. Schnell, D. A. Allen, J. Y. Tsai, and L. Hicke. 2003. Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol. Biol. Cell 14:4676-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friant, S., R. Lombardi, T. Schmelzle, M. N. Hall, and H. Riezman. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20:6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelperin, D., L. Horton, A. DeChant, J. Hensold, and S. K. Lemmon. 2002. Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics 161:1453-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 11.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 12.Harris, T. E., and J. C. Lawrence, Jr. 2003. TOR signaling. Sci. STKE 2003:re15. [DOI] [PubMed] [Google Scholar]
- 13.Helliwell, S. B., I. Howald, N. Barbet, and M. N. Hall. 1998. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilti, N., D. Baumann, A. M. Schweingruber, P. Bigler, and M. E. Schweingruber. 1999. Gene ste20 controls amiloride sensitivity and fertility in Schizosaccharomyces pombe. Curr. Genet. 35:585-592. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki, M., T. Schmelzle, K. Yamaguchi, K. Irie, M. N. Hall, and K. Matsumoto. 1999. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol. Cell. Biol. 19:8344-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274:34493-34498. [DOI] [PubMed] [Google Scholar]
- 17.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser, C., S. Michaelis, A. Mitchell, and Cold Spring Harbor Laboratory. 1994. Methods in yeast genetics: a Cold Spring Harbor laboratory course manual, 1994 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, M., A. Nakashima, M. Ueno, T. Ushimaru, K. Aiba, H. Doi, and M. Uritani. 2001. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39:166-174. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 24.Krause, S. A., and J. V. Gray. 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12:588-593. [DOI] [PubMed] [Google Scholar]
- 25.Kubota, H., T. Obata, K. Ota, T. Sasaki, and T. Ito. 2003. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2 alpha kinase GCN2. J. Biol. Chem. 278:20457-20460. [DOI] [PubMed] [Google Scholar]
- 26.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo, T., Y. Kubo, Y. Watanabe, and M. Yamamoto. 2003. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22:3073-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima, N., A. Yamamoto, M. Matsui, T. Yoshimori, and Y. Ohsumi. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 30.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roelants, F. M., P. D. Torrance, N. Bezman, and J. Thorner. 2002. Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13:3005-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 33.Schmelzle, T., S. B. Helliwell, and M. N. Hall. 2002. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22:1329-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt, A., M. Bickle, T. Beck, and M. N. Hall. 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88:531-542. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt, A., J. Kunz, and M. N. Hall. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, Y., R. Taniguchi, D. Tanoue, T. Yamaji, H. Takematsu, K. Mori, T. Fujita, T. Kawasaki, and Y. Kozutsumi. 2000. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol. 20:4411-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisman, R., and M. Choder. 2001. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276:7027-7032. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson, M. G., T. S. Pino, S. Tournier, V. Buck, H. Martin, J. Christiansen, D. G. Wilkinson, and J. B. Millar. 1999. Sin1: an evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 18:4210-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]