Abstract
Activation of tumor suppressor p53 in response to genotoxic stress imposes cellular growth arrest or apoptosis. We identified Cdc6, a licensing factor of the prereplication complex, as a novel target of the p53 pathway. We show that activation of p53 by DNA damage results in enhanced Cdc6 destruction by the anaphase-promoting complex. This destruction is triggered by inhibition of CDK2-mediated CDC6 phosphorylation at serine 54. Conversely, suppression of p53 expression results in stabilization of Cdc6. We demonstrate that loss of p53 results in more replicating cells, an effect that can be reversed by reducing Cdc6 protein levels. Collectively, our data suggest that initiation of DNA replication is regulated by p53 through Cdc6 protein stability.
Initiation of DNA replication is a tightly regulated process, which is highly conserved in eukaryotes. Key to this process is the licensing of replication origins for DNA replication in which Cdc6 plays a major role (36). Cdc6 is recruited to the origins by the origin recognition complex during G1, where it serves, together with Cdt1, as a loading factor for the minichromosome maintenance (MCM) complex, a putative replicative helicase. This prereplication complex (pre-RC) renders the genome competent for replication, but initiation of replication only occurs after activation of the complex by cyclin-dependent kinases (CDKs) at the onset of S phase. This promotes the loading of Cdc45, replication protein A, and DNA polymerase α onto chromatin. After initiation of DNA replication, the pre-RCs are converted to post-RCs, which are present in the S, G2, and M phases and are not competent for initiation of replication (6, 16, 45).
Cdc6 has been shown to be essential for loading of the MCM complex onto chromatin and therefore for the initiation of DNA replication in several eukaryotic organisms. First, mutant studies with yeast have demonstrated that association of MCMs with origins is dependent on Cdc6 (4, 46). Second, it was shown in Xenopus laevis that immunodepletion of Cdc6 led to loss of MCM origin association (11). Last, ectopic expression of Cdc6 was sufficient to induce stable association of endogenous MCM protein with chromatin in mammalian cells with low Cdc6 levels as a result of serum deprivation (12). In addition, immunodepletion of Cdc6 in human cells by microinjection of anti-Cdc6 antibody could block initiation of DNA replication, which suggests that Cdc6 is also limiting for DNA synthesis in human cells (21, 50). Conversely, recombinant Cdc6 protein induced premature entry into S phase in a mammalian cell-free system (44) and ectopic expression of Cdc6 together with Cdt1 induces rereplication and polyploidy (48). Thus, Cdc6 is crucial for MCM loading and therefore involved in regulation of S-phase entry.
In support of an essential function for Cdc6 in initiation of DNA replication, Cdc6 protein abundance is tightly regulated during the cell cycle. In human cells, Cdc6 transcription is regulated by the E2F transcription factors (21, 50). In addition, Cdc6 is targeted for ubiquitin-mediated proteolysis in early G1 by the E3 ligase anaphase-promoting complex (APC) (38). This is mediated by Cdh1, a protein required for both substrate recognition and activation of the APC in late mitosis-early G1 (40, 41). Typically, Cdh1 associates with its substrates via their N termini and requires a well-defined RXXL destruction box (D box) or KEN box (39, 40). APCcdh1-dependent proteolysis of Cdc6 requires both a D box and a KEN box (38).
Several studies have shown that mammalian Cdc6 can be phosphorylated in vitro by both cyclin E/CDK2 and cyclin A/CDK2, and an interaction between the cyclins and Cdc6 could be detected (22, 26, 37). Moreover, Cdc6 can be phosphorylated in vivo at three N-terminal sites (ser54, ser74, and ser106) that are phosphorylated by CDK2 in vitro (26). Together, this strongly suggests that mammalian Cdc6 is an in vivo CDK2 substrate. However, the role of this phosphorylation in Cdc6 regulation remains controversial. Based on studies with ectopically expressed and tagged wild-type (wt) Cdc6 or Cdc6 that was mutated in several potential CDK2 phosphorylation sites, it has been proposed that phosphorylation of Cdc6 by cyclin A/CDK2 in S phase results in translocation from the nucleus to the cytosol and subsequent degradation (15, 22, 26, 37, 42). Recently it has become evident that only ectopically expressed Cdc6 or the soluble endogenous form is translocated to the cytosol, whereas the chromatin-bound form persist through the S and G2 phases (14, 30). Furthermore, by using a phosphospecific antibody it has been shown that ser54-phosphorylated Cdc6 maintains a high affinity for chromatin during S phase (3).
The DNA damage response is essential for maintenance of genome stability. Genotoxic stress activates checkpoints that account for a rapid and prolonged cell cycle arrest, DNA repair, or apoptosis (25). Primary among the checkpoint genes is tumor suppressor p53 (19). In response to DNA damage, such as that induced by ionizing radiation (IR), transcription factor p53 is activated by the ATM-CHK2 pathway. Rapid phosphorylation at serines 15 and 20 leads to release of Mdm2 and stabilization and activation of p53 (43). p53 is required for maintenance of the G1 arrest through activation of its transcriptional target gene, p21cip1, and induction of apoptosis and for the sustained arrest of cells prior to M phase (10, 19, 32). In addition, DNA damage initiates a rapid cell cycle arrest by inactivation of proteins that promote cell cycle progression, such as cyclin D1 and CDC25A (2, 18).
In this study, we determined the effect of DNA damage on Cdc6 protein levels. We observed a rapid degradation of Cdc6 in response to IR. Interestingly, this irradiation-induced Cdc6 destruction was p53 dependent. Activation of p53 caused inhibition of cyclin E/CDK2 through induction of the cell cycle inhibitor p21cip1. This resulted in loss of Cdc6 serine 54 phosphorylation, which led to accelerated proteolysis by APCCdh1.
MATERIALS AND METHODS
Materials and antibodies.
Irradiation was performed with a 2 × 415-Ci 137Cs source. The proteasome inhibitor MG-132 was purchased from Sigma and used at a final concentration of 10 μM. Cycloheximide (CHX; Sigma) was used at a final concentration of 25 μg/ml and roscovitin (Sigma) at 25 μM. The small interfering RNA (siRNA) oligonucleotides against luciferase and CDK2 were described previously (17, 47). The antibodies used in this study were directed against human Cdc6 (180.2), p-Cdc6 (Ser 54), p53 (DO-1), p21 (C-19), cyclin D1 (M-20), CDK4 (C-22), Cdc20 (H-175), cyclin E (HE12), and CDK2 (M-20) from Santa Cruz. Antibodies Kip1/p27 (Transduction Laboratories), Cdh1 (Abcam), ORC1 (Abcam), and MCM2 (Abcam) were also used.
Constructs.
Cdc6 expression constructs were cloned by PCR in pcDNA3.1 vector (Invitrogen), pGL3 (Promega), or pEGFP-N3 (Clontech). Cdc6 mutants were generated by site-directed mutagenesis using PCR, and constructs were verified by DNA sequence analysis. We used pSuper (pS) constructs targeting p53, Cdh1, and Cdc20 that were described previously (9). The other 19-nucleotide target knockdown sequences used were p21cip1 (GACCATGTGGACCTGTCAC), p27kip1 (GGGCAGCTTGCCCGAGTTC), Cdc6#1 (ATGTCCAAACCGTAACCTG), and Cdc6#2 (CCTATGCAACACTCCCCAT). For stable expression, siRNAs constructs were cloned in the retroviral 480-puro vector (49).
Cell culture and transfection.
MCF-7 and BJ cells were grown in Dulbecco modified Eagle medium and 10% fetal bovine serum. MCF-7 cells carrying the murine ecotropic receptor were infected with ecotropic retroviral supernatants as previously described (8) to generate a polyclonal pool of cells. MCF-7 cells were transfected by electroporation as previously described (2), and a transfection efficiency of about 90% was obtained. Cells were electroporated and seeded, washed 16 h later, and harvested 48 h after washing.
Western blotting, cell cycle profile analysis, and luciferase and immunoprecipitation kinase assays.
For Western blot assays, whole-cell extracts were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene difluoride membranes (Millipore). Western blot assays were developed using enhanced chemiluminescence (Amersham Bioscience, Inc.). Densitometric quantification of Western blot assays was done by using Tina 2.09 software. For bromodeoxyuridine (BrdU) labeling, cells were incubated with 2.5 μM BrdU 40 min prior to harvesting. Cells were fixed at 4°C in ethanol. Fixed cells were treated 30 min at 37°C with RNase A (0.5 mg/ml), washed with phosphate-buffered saline, incubated 20 min in 5 M HCl-0.5% Triton solution, and neutralized with 0.1 M Na2B4O7. Cells were sequentially stained with anti-BrdU antibodies (DAKO) and fluorescein isothiocyanate-conjugated goat anti-mouse antibodies (Molecular Probes). Prior to flow cytometric analysis, cells were resuspended in 50 μg/ml propidium iodide in phosphate-buffered saline. In each assay, 10,000 single cells were analyzed using the Cell Quest program (Becton Dickinson). Firefly luciferase and Renilla luciferase (internal control) activities were measured by employing the Dual Luciferase Reagent Assay Kit (Promega). Immunoprecipitation kinase assays were performed as described in references 2 and 18.
Northern blot analysis.
RNA was isolated from cells using Trizol (Invitrogen), Northern blot analysis was performed exactly as previously described (1), and membranes were hybridized with a probe made from the Cdc6 cDNA.
RESULTS
p53-dependent down-regulation of Cdc6 following DNA damage.
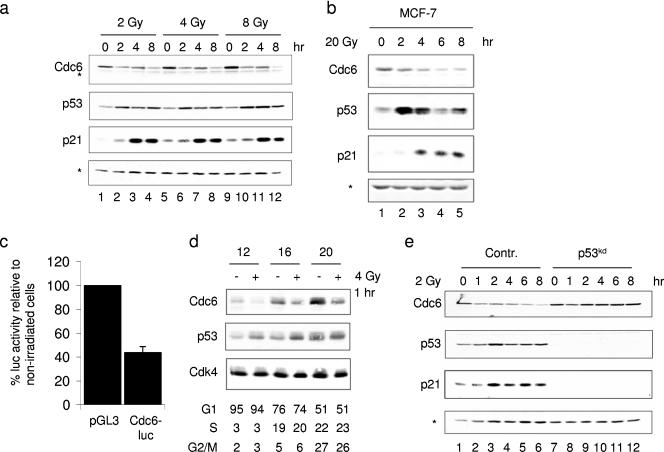
Since the licensing factor Cdc6 is critical for formation of the pre-RC and initiation of DNA replication, we examined the effect of IR on Cdc6 protein levels. Primary human BJ fibroblasts were treated with increasing amounts of IR, and the cells were harvested at various time points. We found reduced Cdc6 protein levels within 2 h after exposure to IR, and 2 Gy was sufficient to elicit this response (Fig. 1a). Control immunostainings showed an increase in the p53 transcription factor and its target, p21cip1, at these doses, demonstrating that p53 was both stabilized and activated after DNA damage in these cells. Similar down-regulation of Cdc6 was observed in other cell types, such as MCF-7 cells (Fig. 1b). However, this cancer cell line required a higher dose of 5 to 20 Gy to activate a DNA damage response that resulted in both Cdc6 down-regulation and stabilization of p53 (data not shown). Furthermore, Cdc6 down-regulation following irradiation in MCF-7 cells was observed 2 to 4 h following IR and at these time points no significant change in the cell cycle profile was observed (see Fig. S1a in the supplemental material). To estimate the quantity of Cdc6 protein reduction following IR, we constructed a firefly Cdc6-luciferase fusion construct driven by the simian virus 40 promoter and transfected it into MCF-7 cells together with a Renilla luciferase control. Treatment with 20 Gy irradiation resulted in a 60% reduction in Cdc6-luciferase activity after 4 h, whereas no change in control luciferase activity was observed (Fig. 1c). Collectively, these results indicate that the Cdc6 protein level is reduced in response to DNA damage.
FIG. 1.
p53-dependent regulation of Cdc6. (a) Primary human BJ fibroblasts were subjected to 2, 4, and 8 Gy of IR and harvested 0, 2, 4, and 8 h later. Whole-cell extracts were made, separated by 10% SDS-PAGE, and immunoblotted to detect Cdc6, p53, and p21 proteins. Asterisks mark background bands. (b) MCF-7 cells irradiated with 20 Gy were treated as described for panel a. (c) MCF-7 cells were transfected with Cdc6-luciferase and control pGL3-promoter vector constructs (n = 3). At 72 h after transfection, cells were irradiated (20 Gy) and harvested 2 h later. Luciferase activities were measured, normalized to cotransfected Renilla luciferase, and compared to nonirradiated cells. (d) Primary human BJ fibroblasts were serum starved for 72 h, released with addition of 10% fetal calf serum, and irradiated (4Gy) 1 h prior to harvest at the indicated time points. In parallel, cells were collected for flow cytometric analysis (supplemental Fig. S1). (e) Polyclonal pools of primary human BJ fibroblasts stably carrying the retroviral vector pRetroSuper (pRS) or pRS-p53kd, which mediates the inhibition of p53 expression through RNAi, were subjected to 2 Gy of irradiation. Whole-cell extracts were prepared at the indicated time points, and levels of the Cdc6, p53, and p21 proteins were examined by immunoblotting. Contr., control.
Next, we assessed whether Cdc6 protein levels are reduced in the G1 phase of the cell cycle when pre-RCs are formed. We synchronized primary human BJ fibroblasts in G0 by serum starvation and then released them to enter synchronously G1 phase around 12 h later and S phase around 16 and 20 h later (see Fig. S1c in the supplmental material). Cells were irradiated (4 Gy) 12, 16, and 20 h after the release and harvested 1 h later, and Cdc6 protein levels were compared to untreated cells by immunoblot analysis. Clearly, potent down-regulation of Cdc6 following IR occurred when cells were almost exclusively in the G1 phase of the cell cycle (12 h), indicating that Cdc6 protein levels are reduced when it is functional (Fig. 1d). Also here, Cdc6 down-regulation preceded any detectable cell cycle changes induced by IR, suggesting that Cdc6 down-regulation in G1 contributes to the IR-induced cellular responses (Fig. 1d; see also Fig. S1c in the supplemental material).
The p53 protein is a central player in the DNA damage response. We therefore investigated whether the decrease in the Cdc6 protein level following IR is p53 dependent. We transduced primary human BJ cells with pRetroSuper (pRS)-p53kd, which stably suppresses p53 expression through RNA interference (RNAi) (49), and monitored the Cdc6 protein level following IR. Interestingly, immunoblot analysis showed that IR-induced Cdc6 down-regulation was completely abrogated in p53kd cells (Fig. 1e). Control immunolabeling showed the absence of p53 activation and induction of its transcriptional target, p21cip1, following IR in the p53kd cells. Quantification of the Cdc6 reduction in control BJ cells revealed that the Cdc6 protein level was reduced to 10 to 30% following 2 Gy of irradiation, whereas no reduction in the Cdc6 level was observed in p53kd cells (data not shown). These results demonstrate that the p53 pathway controls the Cdc6 protein level in human cells in response to DNA damage.
Enhanced Cdc6 proteolysis following DNA damage.
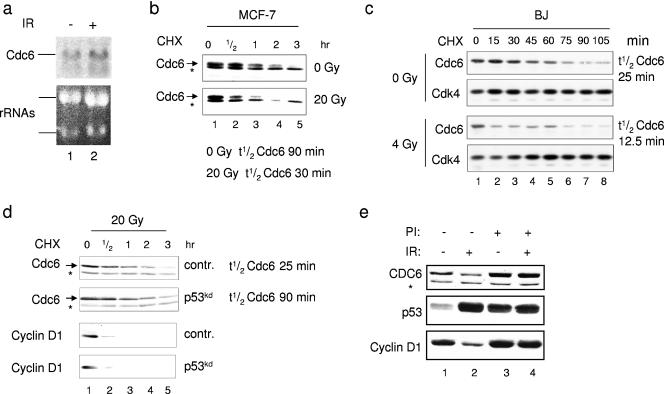
As a transcription factor, p53 can directly repress gene expression (33) and may therefore switch off Cdc6 transcription when activated. However, no changes in the Cdc6 mRNA level after IR were found by Northern blot analysis (Fig. 2a). Furthermore, Cdc6 protein expressed from a heterologous cytomegalovirus promoter was reduced after DNA damage to a similar extent as the endogenous protein (data not shown). These results indicate that the IR-induced Cdc6 down-regulation occurs at the posttranscriptional level.
FIG. 2.
Increased Cdc6 proteolysis following DNA damage. (a) MCF-7 cells were subjected to IR treatment (20 Gy) and collected for Northern blot analysis 4 h later. Blots were hybridized with a Cdc6 cDNA probe to detect Cdc6 mRNA. (b) Irradiated (20 Gy) or nonirradiated MCF-7 cells were treated with CHX 2 h after IR. Cells were harvested at the indicated time points after CHX addition and immunoblotted against Cdc6. Asterisks indicate background bands. Half-life values were determined by quantification analysis. (c) Serum-starved BJ cells were released for 13 h, treated as described above, and immunoblotted to detect Cdc6 and CDK4. Quantification of the Cdc6 protein level was used to determine the half-life. (d) pRS and pRS-p53kd cells were irradiated and treated with CHX 2 h later. Cells were harvested at the indicated time points, and quantifications of half-life were calculated. (e) MCF-7 cells were treated with or without 20 Gy of IR in the presence or absence of proteasome inhibitors (PI). Protease inhibitors were added 30 min prior to the IR treatment. Whole-cell extracts were immunoblotted against Cdc6, p53, and cyclin D1. A cross-reacting band (asterisk) was used as a loading control.
Next, we tested whether the rapid decrease in the Cdc6 protein level after IR is a result of increased proteolysis. We used the protein synthesis inhibitor CHX to monitor Cdc6 half-life. To allow a proper cell cycle response to DNA damage, CHX was added to cells 2 h after the IR treatment. If CHX was added right after the radiation treatment, Cdc6 stability remained unchanged following IR (see Fig. S2 in the supplemental material). In MCF-7 cells, the Cdc6 protein level decreased more rapidly in irradiated cells compared to nonirradiated cells (Fig. 2b) and quantification analysis showed that the half-life of Cdc6 was reduced from 90 min to 30 min upon IR treatment (see Fig. S3a in the supplemental material). Also in synchronized G1/S primary BJ fibroblasts we found that the half-life of Cdc6 protein is reduced from 25 min to 12.5 min following DNA damage (Fig. 2c; see also Fig. S3b in the supplemental material). Since Cdc6 down-regulation following IR appeared to be p53 dependent, we monitored Cdc6 protein stability in control and p53kd MCF-7 cells 2 h after IR (Fig. 2d; see also Fig. S3c in the supplemental material). Indeed, Cdc6 maintained its stability following IR in p53kd MCF-7 cells (half-life, 90 min) compared to control irradiated cells (half-life, 25 min). The stability of cyclin D1 protein following IR was not increased by the reduced level of p53, thereby showing the specificity of Cdc6 stabilization (Fig. 2d). Last, this p53-dependent Cdc6 degradation was mediated by the proteasome since it was abrogated when MCF-7 cells were irradiated in the presence of the proteasome inhibitor MG-132 (Fig. 2e). Taken together, these results demonstrate that the proteasome machinery mediates the accelerated proteolysis of Cdc6 following genotoxic stress and that Cdc6 protein abundance following IR is mainly controlled by p53.
Enhanced destruction of Cdc6 following DNA damage is mediated by the E3 ligase APCcdh1.
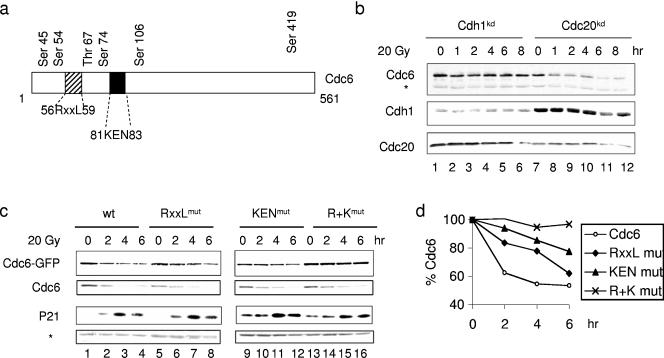
In early G1, the N-terminal KEN and RXXL (D box) destruction motifs of Cdc6 (Fig. 3a) mediate its degradation by the E3 ligase APC (38). To determine whether Cdc6 destruction following IR involves the APC, we suppressed the expression of Cdh1 and Cdc20 by RNAi. Both proteins act as substrate recognition and activating modules for the APC (41). Inhibition of Cdh1 expression completely abrogated IR-induced Cdc6 destruction, whereas the Cdc20 knockdown had no effect (Fig. 3b). The reduced expression of both Cdh1 and Cdc20 was not accompanied by any significant change in the cell cycle profile or p53 activation following IR (see Fig. S4a in the supplemental material; also data not shown).
FIG. 3.
Enhanced destruction of Cdc6 following DNA damage is mediated by the E3 ligase APCcdh1. (a) Schematic representation of the Cdc6 protein with its five putative CDK2 N-terminal phosphorylation sites and the APC destruction boxes, RXXL and KEN motifs. (b) MCF-7 cells were transiently electroporated with pS and RNAi constructs that target Cdh1 and Cdc20. Cells were subjected to IR treatment 60 h after transfection and subsequently harvested for immunoblotting analysis to detect Cdc6, Cdh1, and Cdc20 at the indicated time points. The asterisk marks a background band. (c) The same experimental settings as in panel b, only that cells were electroporated with Cdc6-GFP chimera (wt) or mutant forms of the RXXL and KEN box destruction motifs. R+K contains mutations in both the RXXL and KEN box motifs. Whole-cell extracts were immunoblotted against Cdc6 and p21. The asterisk indicates a background band and shows equal loading. (d) Quantification analysis of the different Cdc6 proteins shown in panel c. mut, mutant.
The involvement of APCcdh1 in the p53-dependent regulation of Cdc6 suggests that also the KEN or RXXL destruction motif plays a role in IR-induced Cdc6 destruction. Therefore, we constructed Cdc6-green fluorescent protein (GFP) chimeras with mutations in these destruction motifs. Similar to endogenous Cdc6, the stability of the transfected Cdc6-GFP fusion protein is reduced upon IR treatment (Fig. 3c, lanes 1 to 4; quantified in Fig. 3d). Mutating the KEN motif to AAA (KENmut) or the RXXL motif to AXXA (RXXLmut) partly prevented IR-induced Cdc6 destruction (lanes 5 to 12 and Fig. 3d). However, mutating both destruction motifs (R+K) completely abrogated Cdc6 destruction in response to IR (lanes 13 to 16). This demonstrates that both the KEN and RXXL destruction motifs are involved in the p53-dependent Cdc6 destruction following IR through APCCdh1.
Inhibition of CDK2 activity results in enhanced Cdc6 proteolysis.
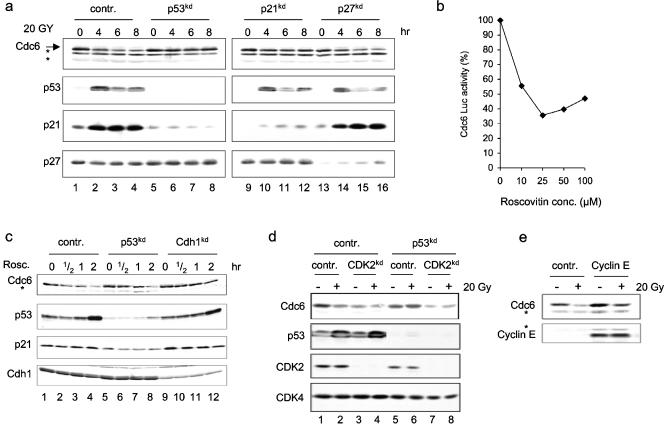
Activation of the p53 transcription factor following genotoxic stress results in up-regulation of the cell cycle inhibitor p21Cip1. We therefore asked whether p21Cip1 is required for the p53-dependent destruction of Cdc6 and transfected MCF-7 cells with knockdown constructs targeting p21cip1, p27kip1 (a homologue of p21cip1 that is not regulated by p53), and p53. Suppression of p21cip1 expression, but not p27kip1, completely abrogated Cdc6 destruction after IR (Fig. 4a, lanes 9 to 16). This effect correlated with CDK2 activity, as p21cip1, but not p27kip1, knockdown cells maintained significant CDK2 activity following IR treatment (see Fig. S4b in the supplmental material). The protection from IR-induced destruction of Cdc6 in cells with suppressed expression of p21cip1 was comparable to that observed in p53kd cells (Fig. 4a, lanes 4 to 12). This effect was specific to Cdc6 as the enhanced degradation of cyclin D1, which is p53 independent, was still sustained in the p21cip1 knockdown cells (data not shown).
FIG. 4.
Inhibition of CDK2 activity results in enhanced Cdc6 proteolysis. (a) MCF-7 cells were electroporated with pS, pS-p53kd, pS-p21kd, and pS-p27kd and subjected to IR treatment 60 h after transfection. Whole-cell extracts were prepared at the indicated time points, and immunoblot analysis was performed to detect the levels of the Cdc6, p53, p21, and p27 proteins. Asterisks indicate cross-reacting bands. (b) MCF-7 cells expressing the Cdc6-luciferase chimera protein were treated with the indicated amounts of roscovitin (Rosc.) and harvested for luciferase analysis 1 h later. conc., concentration. (c) MCF-7 cells were electroporated with a control (contr.) construct and RNAi constructs targeting p53 and Cdh1 and treated with roscovitin 60 h later. Whole-cell extracts were prepared at several time points after treatment, and immunoblot analysis was performed to detect the indicated proteins. (d) MCF-7 and MCF-7-p53kd cells were electroporated with CDK2-siRNA oligonucleotides, irradiated 60 h later (20 Gy), and harvested 4 h later. Whole-cell extracts were made and immunoblotted with the indicated antibodies. (e) MCF-7 cells were electroporated with cyclin E and control expression vectors. Whole-cell extracts were made and immunoblotted to detect Cdc6 and cyclin E proteins. Asterisks indicate cross-reacting bands.
In response to IR, accumulation of p21cip1 inhibits CDK2 activity. It was reported that mammalian Cdc6 is phosphorylated in vivo at three N-terminal sites (S54, S74, and S106) (26), which can be phosphorylated by the cyclin E/CDK2 complex in vitro (22, 26, 37). To address the role of CDK2 in regulating Cdc6 stability, we treated cells expressing the Cdc6-luciferase chimera protein with increasing amounts of the CDK2 inhibitor roscovitin and monitored luciferase activity 1 h later. Roscovitin treatment resulted in a rapid reduction of luciferase activity, suggesting a direct connection between CDK2 activity and Cdc6 protein stability (Fig. 4b). Furthermore, the reduction in Cdc6 levels following IR in MCF-7 cells correlated with the inhibition of CDK2 activity (Fig. 1b; see also Fig. S1b in the supplemental material). Interestingly, Cdc6 reduction preceded the accumulation of p21cip1, which is likely to be the result of p53-independent DNA damage responses, such as cyclin D1 and Cdc25A destruction, operating early after DNA damage to inhibit CDK2 activity (2, 18). In a second experiment, we transfected MCF-7 cells with control, pS-p53kd, or pS-Cdh1kd RNAi vectors, treated them with roscovitin, and harvested the cells for immunoblot analyses at various time points. Figure 4c shows that also endogenous Cdc6 was rapidly reduced upon roscovitin treatment in control-transfected cells (lanes 1 to 4). In addition, Cdc6 was down-regulated upon roscovitin treatment in p53kd cells (lanes 5 to 8), although these cells are impaired in the ability to enhance degradation of Cdc6 following IR (Fig. 1e and 4a). This indicates that CDK2 is downstream of p53 in IR-induced Cdc6 degradation. In contrast, Cdc6 remained stable upon roscovitin treatment in cells with suppressed Cdh1 levels (lanes 9 to 12), suggesting that APCCdh1 is downstream of CDK2 in this pathway. Notably, this experiment also demonstrates that the resistance of Cdc6 to IR in p53kd cells is not a result of increased mRNA levels.
To further substantiate the role of CDK2 in Cdc6 destruction following DNA damage, we assessed the effect of IR on Cdc6 protein levels in MCF-7 and MCF-7-p53kd cells transfected with control or CDK2 siRNAs. In agreement with the observed Cdc6 protein reduction upon roscovitin treatment, the Cdc6 protein level was reduced in response to low CDK2 levels in both control and p53kd cells (Fig. 4d, lanes 1, 3, 5, and 7), indicating that CDK2 plays an essential role in maintaining the Cdc6 protein level. Furthermore, Cdc6 protein was down-regulated in control cells within 2 h after irradiation, whereas it remained unaffected in irradiated CDK2kd cells (Fig. 4d, lanes 1 to 4). As expected, no change in Cdc6 levels was observed in p53kd cells after irradiation (lanes 5 to 8). Last, our model predicts that ectopic activation of CDK2 should confer significant resistance to Cdc6 from the enhanced destruction following DNA damage. To test this hypothesis, we overexpressed cyclin E, an activator of CDK2, and treated the cells with IR. Figure 4e shows that cyclin E overexpression results in accumulation of Cdc6 in untreated cells and significant resistance to DNA damage effects. Altogether, these results demonstrate the direct requirement of CDK2 inhibition in the p53-dependent destruction of Cdc6 protein after IR.
Phosphorylation of serine 54 by CDK2 inhibits p53-dependent Cdc6 proteolysis.
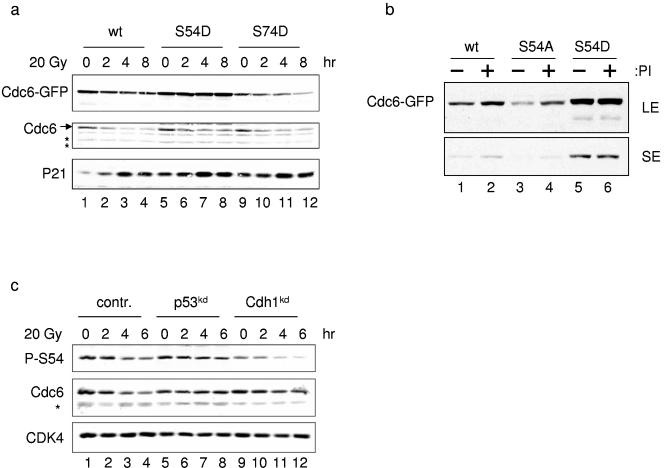
It has been proposed that phosphorylation of Cdc6 results in translocation from the nucleus to the cytosol (15, 22, 26, 37, 42). In stark contrast, our results indicate that phosphorylation of Cdc6 by CDK2 leads to its stabilization. Therefore, we aimed to map this stabilizing phosphorylation site by mutating putative CDK2 phosphorylation sites to aspartic acid, thereby mimicking their phosphorylated state. The N terminus of Cdc6 harbors three sites that can be phosphorylated in vivo, serines 54, 74, and 106 (26). We chose to mutate S54 and S74, as they are the sites nearest to the destruction motifs (Fig. 3a). We made Cdc6-GFP fusion constructs that contained S54D and S74D mutations and examined them in a transient transfection assay in MCF-7 cells. Mutation of S54 to aspartic acid (S54D) completely abrogated the p53-induced Cdc6 destruction, whereas the S74D mutant was degraded to an extent that was comparable to wt Cdc6-GFP and to the endogenous protein (Fig. 5a; see also Fig. S4c in the supplemental material). Importantly, the expression of the S54D mutant did not affect the upstream p53 response, as p21cip1 levels accumulated in these cells after DNA damage (Fig. 5a). This result suggests that the phosphorylation of S54 is also critical for Cdc6 stability in nonstressed cells. To examine this, we mutated serine 54 to alanine (S54A), a change that mimics an unphosphorylated state of Cdc6 at this position. Introduction of S54A into cells as a GFP chimera protein resulted in very little expression compared with wt Cdc6 and its stable S54D mutant (Fig. 5b, lanes 1, 3, and 5). Treatment with the proteasome inhibitor MG-132 resulted in the accumulation of both wt and S54A mutant Cdc6 protein, whereas S54D remained unchanged (lanes 2, 4, and 6). This result indicates that in nonstressed cells, phosphorylation at serine 54 protects Cdc6 from rapid destruction.
FIG. 5.
Phosphorylation of serine 54 by CDK2 inhibits p53-dependent Cdc6 proteolysis. (a) MCF-7 cells were electroporated with wt Cdc6-GFP or S54D and S74D mutant Cdc6 constructs, and the experiment was performed as described in the legend to Fig. 3c. (f) MCF-7 cells were electroporated with pS, pS-p53kd, and pS-Cdh1kd; irradiated (20 Gy); and harvested at the indicated times. Whole-cell extracts were immunoblotted with anti-phospho-S54 Cdc6, Cdc6, or control (contr.) Cdk4 antibodies. The asterisk marks a nonspecific band. (b) MCF-7 cells were electroporated with wt and S54A and S54D mutant Cdc6-GFP constructs. Sixty hours after transfection, the proteasome inhibitor (PI) MG-132 was added, the mixture was incubated for 4 h, and whole-cell extracts were separated by 10% SDS-PAGE and immunoblotted to detect Cdc6. For comparison, long and short exposures (LE and SE, respectively) are shown. (c) MCF-7 cells were electroporated as described in the legend to Fig. 4d, and whole-cell extracts were immunoblotted to detect Cdc6 and serine 54-phosphorylated Cdc6.
To further substantiate the connection between S54 phosphorylation and the p53-dependent destruction of Cdc6, we used an anti-phospho-S54-specific Cdc6 antibody and monitored the phosphorylation state of endogenous Cdc6. We transfected MCF-7 cells with pS, pS-p53kd, or pS-Cdh1kd constructs and subjected them to IR treatment. Importantly, we detected S54-phosphorylated Cdc6 in whole-cell extracts from cycling control-transfected cells and these levels were reduced following IR (Fig. 5c, lanes 1 to 4). In the cells transfected with either pS-p53kd or pS-Cdh1kd, the Cdc6 protein level remained high following DNA damage; however, the level of phosphorylated S54 was only stable after IR in the p53kd cells (lanes 4 to 12). In the cdh1kd cells, the level of S54-phosphorylated Cdc6 was still reduced. This result is in full agreement with a pathway consisting of upstream p53-dependent regulation of Cdc6 stability by phosphorylation and downstream APCcdh1-mediated degradation. Last, we examined the effect of cyclin E overexpression or Cdh1kd on the stability of S54D mutant Cdc6 in the absence of DNA damage. Figure S4d in the supplemental material shows that the S54D mutant protein was unaffected by both treatments, suggesting that S54 phosphorylation is a major determinant of Cdc6 stability. Collectively, these results reveal that inhibition of S54 phosphorylation is the mechanism by which p53 regulates Cdc6 stability following DNA damage. The importance of this pathway is further extended by the fact that both Cdc6 S54 and the RXXL and KEN destruction boxes are evolutionarily conserved in mice.
p53-dependent regulation of Cdc6 protein stability controls cellular proliferation.
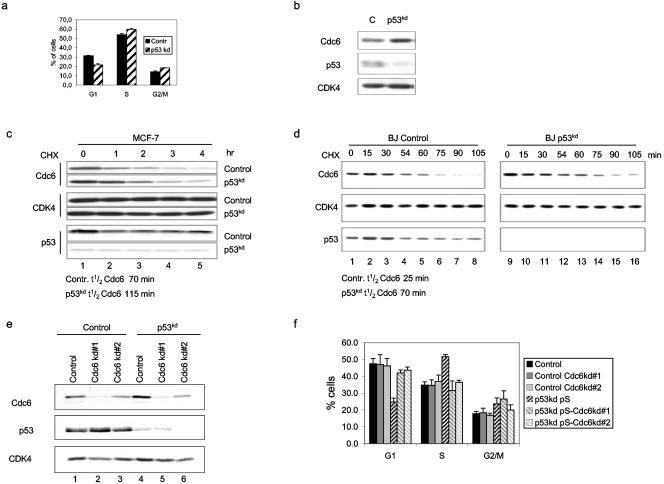
Apart from its role in DNA damage responses, p53 regulates cellular proliferation in nonstressed cells as well, since its suppression by RNAi accelerates proliferation of primary human cells (49) and MCF-7 cells (data not shown). Therefore, we examined the effect of reduced p53 expression on the cell cycle profile of MCF-7 cells. Also here we observed that suppression of endogenous p53 expression resulted in more proliferating cells (6% increase in S phase and 10% decrease in G1) (Fig. 6a). This larger fraction of the S-phase population could be due to earlier assembly of the pre-RC in G1. Since Cdc6 is regulated by p53 in the DNA damage response, we examined the level of Cdc6 in proliferating cells with suppressed p53 expression. Interestingly, we found a higher level of Cdc6 protein in pRS-p53kd cell extracts compared to control cells (Fig. 6b). This suggests that Cdc6 is more stable in p53 knockdown cells. To test this directly, we treated MCF-7 cells with CHX and observed that the protein turnover of Cdc6 is slower in p53kd cells (115 min) compared to control cells (70 min) (Fig. 6c). Control immunoblot analyses of CDK4 and p53 showed equal loading and the extent of the p53 knockdown. The same qualitative result was also seen in synchronized G1-phase primary BJ cells, where Cdc6 stability was raised from 25 min in normal cells to 70 min in BJ-p53kd cells (Fig. 6d). To investigate whether Cdc6 stability is important for the increased proliferation of p53kd cells, we transfected control and stable polyclonal p53 knockdown cells with two effective knockdown constructs targeting Cdc6. Immunoblot analysis showed a reduction of Cdc6 expression in pS-Cdc6kd-transfected cells of both types (Fig. 6e). Notably, Cdc6kd had no significant effect on p53 levels. Flow cytometric analysis of the same cell populations repeatedly revealed that inhibition of Cdc6 expression altered the cell cycle profile of only p53kd cells by increasing the fraction of cells in G1 and decreasing the fraction of cells in S phase (Fig. 6f). In contrast, no significant change was detected in the cell cycle profile of the control MCF-7 cells. This indicates that the increased Cdc6 levels observed in the absence of p53 are required for the enhanced proliferation of these cells. Collectively, our data imply that Cdc6 is part of a p53 protein network that regulates cellular proliferation.
FIG. 6.
p53-dependent regulation of Cdc6 protein stability controls cellular proliferation. (a) Cell cycle analysis of polyclonal control (Contr) or p53 knockdown (kd) MCF-7 cells. BrdU-labeled cells were analyzed by fluorescence-activated cell sorting. The result is the mean of three independent experiments. (b) Whole-cell lysates of control and p53kd MCF-7 cells were immunoblotted to detect Cdc6, p53, and CDK4 proteins. (c) pRS and pRS-p53kd MCF-7 cells were treated with CHX as described in the legend to Fig. 2b. Whole-cell extracts were labeled with Cdc6, p53, and control CDK4 antibodies. (d) Serum-starved BJ cells that were released for 13 h were treated as described in the legend to Fig. 2c. (e) The polyclonal pools of pRS and pRS-p53kd cells were electroporated with Cdc6 knockdown constructs (#1 and #2), harvested after 72 h, and immunoblotted to detect Cdc6, p53, and as a loading control CDK4 proteins. (f) The same experimental setting as in Fig. 5e, but cells were subjected to flow cytometric analysis. The result is the mean of three independent experiments.
DISCUSSION
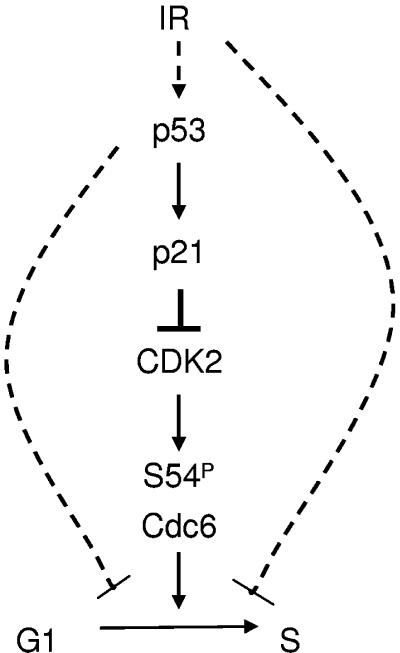
In response to genotoxic stress, cells activate checkpoints that prevent DNA replication and cell cycle progression (5). Our results establish a novel connection between DNA damage and the formation of pre-RCs in G1 phase (Fig. 7). Cdc6, an essential protein of the pre-RC, is rapidly degraded upon genotoxic stress. The enhanced degradation of Cdc6 following IR is not complete, which can be the result of a small population of Cdc6 that is protected from degradation or due to restriction of the accelerated proteolysis to certain phases of the cell cycle. A central role in this destruction pathway is played by the p53 tumor suppressor gene, which controls the rate by which Cdc6 is degraded by APCcdh1. Cdc6 is a licensing factor of DNA replication, and once bound to the origin recognition complex at origins of replication it recruits, together with Cdt1, the MCM complex. Our data imply that IR-induced down-regulation of Cdc6 contributes to a checkpoint that prevents DNA replication in cells with damaged DNA. Interestingly, DNA damage also induces the destruction of licensing factor Cdt1, but in a p53-independent manner (23, 24, 27). Therefore, we propose that the destruction of both Cdc6 and Cdt1 acts in concert to execute tight regulation of pre-RC assembly following genotoxic stress.
FIG. 7.
Schematic model depicting the p53 pathway that regulates Cdc6, and thereby S-phase entry, both under normal tissue culture conditions (solid lines) and following IR (dashed lines).
We also found that p53-dependent regulation of Cdc6 protein stability plays a role in nongenotoxic stressed cells. Under normal tissue culture conditions, Cdc6 protein stability is increased in p53kd cells. We show that reducing the Cdc6 protein level by siRNAs in p53kd cells results in a reduction of replicating cells and a cell cycle distribution that is comparable to cells with functional p53. However, reducing Cdc6 protein levels in control MCF-7 cells did not affect their proliferation, which suggests that cells without functional p53 are more susceptible to a reduction in Cdc6 levels.
It is generally thought that initiation of DNA replication is regulated in a two-step process to ensure that the DNA is replicated only once in each cell cycle (6). First, pre-RCs are assembled at origins of replication, and second, the origins are fired. Notably, the regulation of these events varies in different organisms. In mammals, CDKs appear to have an inhibiting role in pre-RC assembly. For both Cdt1 and Orc1, it has been proposed that they are targeted for proteolysis in a CDK-dependent manner (28, 31, 34). In addition, it has been proposed that cyclin A/CDK2 activity results in Cdc6 translocation from the nucleus to the cytosol and subsequent proteolysis (15, 22, 26, 37, 42). In contrast to these observations, we found that phosphorylation of Cdc6 by cyclin E/CDK2 results in stabilization of the protein. We established that one amino acid (serine 54) mediates p53-induced Cdc6 destruction. Once serine 54 is phosphorylated by CDK2, Cdc6 becomes more resistant to APCCdh1 degradation. Our results are in agreement with a report showing that only ectopically expressed Cdc6 is translocated to the cytosol and that serine 54-phosphorylated Cdc6 remains chromatin bound in S phase (3). Therefore, we propose that phosphorylation of S54 by cyclin E/CDK2 is a novel stabilizing modification of the Cdc6 protein which can be modulated by the p53 pathway.
Our data imply that cyclin E/CDK2 activity might have a positive role in DNA replication licensing by stabilizing Cdc6. This is consistent with in vitro studies which show that cyclin E cooperates with Cdc6 to make mammalian G1 nuclei competent for replication (13). Furthermore, Cdc6 and cyclin E appear to have a synergistic effect on inducing S-phase entry upon cotransfection into human cells (12, 21). In addition, it was shown that overexpression of cyclin E could stabilize Cdc6 protein during megakaryocytic endoreplication (7). In these cells, cyclin E could not stabilize an unphosphorylatable Cdc6 mutant in which all five N-terminal putative CDK2 phosphorylation sites were mutated to alanines. Significantly, it was also shown that cyclin E ablation in mice results in DNA replication defects (20, 35). Cells that re-enter the cell cycle from quiescence fail to load MCM2-7, and endoreplication in trophoblast giant cells and megakaryocytes is impaired. In view of our results, it will be interesting to examine whether these defects are a consequence of reduced Cdc6 stabilization in these cells.
Both CDKs and the APC are key regulators of initiation of DNA replication (16). Intriguingly, proteolysis of both licensing factor Cdc6 and geminin, the inhibitor of licensing factor Cdt1, is regulated by APC (29). It is unclear how the APC-dependent degradation of an activator and that of an inhibitor of DNA replication are coordinated to ensure that Cdc6 and Cdt1 are present in the same time frame to allow efficient pre-RC assembly. Our results shed new light on this. Similar to geminin, Cdc6 is degraded in the end of G2/M and in the early G1 phase of the cell cycle (38). We propose that the increased activity of cyclin E/CDK2 during G1 protects Cdc6 from APCCdh1-dependent destruction, whereas geminin is still degraded. This provides the cells with a time frame in which both Cdt1 and Cdc6 can bind to the origins and recruit the MCM complex to assemble the pre-RC.
In conclusion, our findings give novel insights into the regulation of Cdc6 in the assembly of pre-RCs. We established that phosphorylation of Cdc6 at S54 by cyclin E/CDK2 stabilizes the protein by protecting it from APCCdh1-mediated destruction. The rapid and p53-dependent destruction of Cdc6 in response to DNA damage suggests that it guards the genome for mutations by blocking new origin firing.
Supplementary Material
Acknowledgments
We thank Mathijs Voorhoeve for the p21 and p27 knockdown constructs and Judith Kaufmann and Eva Bogaards for assistance during their student project. We also thank Ingrid Kolfschoten, Eithan Zlotorynski, Mathijs Voorhoeve, and Jacques Neefjes for critical reading of the manuscript and all members of the Agami lab for stimulating discussions.
This work was supported by the Dutch Cancer Society (KWF), the Centre for Biomedical Genetics, and the European Young Investigator Award (EURYI) to R.A.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agami, R., and R. Bernards. 2002. Convergence of mitogenic and DNA damage signaling in the G1 phase of the cell cycle. Cancer Lett. 177:111-118. [DOI] [PubMed] [Google Scholar]
- 2.Agami, R., and R. Bernards. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55-66. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrow, M. G., and J. L. Hamlin. 2004. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol. 24:1614-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 5.Bartek, J., and J. Lukas. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13:738-747. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 7.Bermejo, R., N. Vilaboa, and C. Cales. 2002. Regulation of CDC6, geminin, and CDT1 in human cells that undergo polyploidization. Mol. Biol. Cell 13:3989-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [Online.] [DOI] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 10.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, T. R., P. B. Carpenter, and W. G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 87:53-63. [DOI] [PubMed] [Google Scholar]
- 12.Cook, J. G., C. H. Park, T. W. Burke, G. Leone, J. DeGregori, A. Engel, and J. R. Nevins. 2002. Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc. Natl. Acad. Sci. USA 99:1347-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coverley, D., H. Laman, and R. A. Laskey. 2002. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 4:523-528. [DOI] [PubMed] [Google Scholar]
- 14.Coverley, D., C. Pelizon, S. Trewick, and R. A. Laskey. 2000. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113(Pt. 11):1929-1938. [DOI] [PubMed] [Google Scholar]
- 15.Delmolino, L. M., P. Saha, and A. Dutta. 2001. Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 276:26947-26954. [DOI] [PubMed] [Google Scholar]
- 16.Diffley, J. F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778-R786. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 18.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 19.Fei, P., and W. S. El-Deiry. 2003. P53 and radiation responses. Oncogene 22:5774-5783. [DOI] [PubMed] [Google Scholar]
- 20.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114:431-443. [DOI] [PubMed] [Google Scholar]
- 21.Hateboer, G., A. Wobst, B. O. Petersen, L. Le Cam, E. Vigo, C. Sardet, and K. Helin. 1998. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 18:6679-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbig, U., J. W. Griffith, and E. Fanning. 2000. Mutation of cyclin/cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells. Mol. Biol. Cell 11:4117-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higa, L. A., I. S. Mihaylov, D. P. Banks, J. Zheng, and H. Zhang. 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5:1008-1015. [DOI] [PubMed] [Google Scholar]
- 24.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003-1009. [DOI] [PubMed] [Google Scholar]
- 25.Iliakis, G., Y. Wang, J. Guan, and H. Wang. 2003. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene 22:5834-5847. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, W., N. J. Wells, and T. Hunter. 1999. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 96:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo, T., M. Kobayashi, J. Tanaka, A. Yokoyama, S. Suzuki, N. Kato, M. Onozawa, K. Chiba, S. Hashino, M. Imamura, Y. Minami, N. Minamino, and M. Asaka. 2004. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J. Biol. Chem. 279:27315-27319. [DOI] [PubMed] [Google Scholar]
- 28.Liu, E., X. Li, F. Yan, Q. Zhao, and X. Wu. 2004. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 279:17283-17288. [DOI] [PubMed] [Google Scholar]
- 29.McGarry, T. J., and M. W. Kirschner. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93:1043-1053. [DOI] [PubMed] [Google Scholar]
- 30.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez, J., X. H. Zou-Yang, S. Y. Kim, M. Hidaka, W. P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9:481-491. [DOI] [PubMed] [Google Scholar]
- 32.Motoyama, N., and K. Naka. 2004. DNA damage tumor suppressor genes and genomic instability. Curr. Opin. Genet. Dev. 14:11-16. [DOI] [PubMed] [Google Scholar]
- 33.Nakade, K., H. Zheng, G. Ganguli, G. Buchwalter, C. Gross, and B. Wasylyk. 2004. The tumor suppressor p53 inhibits Net, an effector of Ras/extracellular signal-regulated kinase signaling. Mol. Cell. Biol. 24:1132-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishitani, H., Z. Lygerou, and T. Nishimoto. 2004. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 279:30807-30816. [DOI] [PubMed] [Google Scholar]
- 35.Parisi, T., A. R. Beck, N. Rougier, T. McNeil, L. Lucian, Z. Werb, and B. Amati. 2003. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22:4794-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelizon, C. 2003. Down to the origin: Cdc6 protein and the competence to replicate. Trends Cell Biol. 13:110-113. [DOI] [PubMed] [Google Scholar]
- 37.Petersen, B. O., J. Lukas, C. S. Sorensen, J. Bartek, and K. Helin. 1999. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18:396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. L. Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfleger, C. M., and M. W. Kirschner. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655-665. [PMC free article] [PubMed] [Google Scholar]
- 40.Pfleger, C. M., E. Lee, and M. W. Kirschner. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 15:2396-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed, S. I. 2003. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell. Biol. 4:855-864. [DOI] [PubMed] [Google Scholar]
- 42.Saha, P., J. Chen, K. C. Thome, S. J. Lawlis, Z. H. Hou, M. Hendricks, J. D. Parvin, and A. Dutta. 1998. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 18:2758-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 44.Stoeber, K., A. D. Mills, Y. Kubota, T. Krude, P. Romanowski, K. Marheineke, R. A. Laskey, and G. H. Williams. 1998. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 17:7219-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takisawa, H., S. Mimura, and Y. Kubota. 2000. Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr. Opin. Cell Biol. 12:690-696. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 47.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 48.Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D. S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11:997-1008. [DOI] [PubMed] [Google Scholar]
- 49.Voorhoeve, P. M., and R. Agami. 2003. The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4:311-319. [DOI] [PubMed] [Google Scholar]
- 50.Yan, Z., J. DeGregori, R. Shohet, G. Leone, B. Stillman, J. R. Nevins, and R. S. Williams. 1998. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 95:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.