Abstract
Hedgehog (Hh) signaling plays a major role in multiple aspects of embryonic development. To understand how a single Hh signal is capable of generating distinct readouts in Hh-responsive cells requires elucidation of the signal transduction cascade at the molecular level. Key components that mediate Hh signal transduction downstream of the receptor include Fused (Fu), Suppressor of fused (Sufu), and Costal-2 (Cos2) or the vertebrate homologs Kif27/Kif7. Studies with both invertebrates and vertebrates have led to a model in which a protein complex composed of Fu, Sufu, and Cos2 controls the processing, activity, and subcellular distribution of the Ci/Gli transcription factors responsible for Hh target gene activation. These converging results obtained with different species reaffirm the prevailing view of pathway conservation during evolution. Genetic studies of Fu, Sufu, and Kif27/Kif7 in mice are required to provide further verification of Hh pathway conservation. To this end, we generated a gene-targeted allele of Fu in mice. Surprisingly, our analysis indicates that Fu-deficient mice do not exhibit any embryonic phenotypes indicative of perturbed Hh signaling. This could be due to either functional redundancy or Hh pathway divergence and clearly indicates greater complexity of Hh signaling in vertebrates.
Hedgehog (Hh) signaling plays a key role in inductive interactions in various tissues during animal development, and deregulation of the pathway in humans is associated with various congenital anomalies and tumors (33, 53, 87). The main players in the Hh signaling pathway appear to be conserved between invertebrates and vertebrates (29, 33, 48, 53). Moreover, many aspects of Hh signaling characterized so far are also conserved (29, 33, 48, 53). This leads to the view that little change in Hh pathway design has occurred over millions of years of evolution, and it is widely believed that principles derived from studies with simpler organisms will be applicable to more complex animal models.
In comparison to other major signaling pathways, the Hh pathway possesses several unique and unconventional features, specifically, lipid modification and transport of the ligand and signal transduction at the cell surface (33, 48, 50, 53). Hh signaling is initiated through Hh binding to Patched (Ptch) (51, 85), a 12-pass membrane protein of the sterol-sensing domain family (42). This interaction relieves Ptch repression of Smoothened (Smo), a seven-pass transmembrane protein (1, 85, 94), and allows Smo to activate the Hh signal transduction cascade. Despite intense studies, surprisingly little is known about the biochemical mechanism by which Smo activity is regulated by Ptch (14, 88).
Elucidating the signaling cascade downstream of Smo is crucial to our understanding of how distinct Hh readouts are generated in diverse developmental contexts, and a major player in this process is the putative serine/threonine kinase Fused (Fu) (73). Initially identified in genetic screens by its segment polarity phenotype (62), the essential role of Fu in invertebrate Hh signaling was firmly established through genetic studies with Drosophila and the nature of several classes of fu mutants was clarified through genetic interactions with Suppressor of fused (Sufu) (2, 25, 74, 91, 93). Interestingly, Sufu, which encodes a novel PEST domain protein (70), is dispensable for viability in flies and was identified as an extragenic suppressor of fu mutations rather than through mutant phenotypes (72, 74). Fu was shown to function in concert with the atypical kinesin gene Costal-2 (Cos2) (75, 82), the transcription factor gene Cubitus interruptus (Ci) (65), and Sufu in transducing the Hh signal. The amino-terminal kinase domain of Fu (mutated in class I fu mutant flies) is proposed to primarily counteract Sufu (2, 25, 74, 91, 93), while the carboxyl-terminal domain (truncated in class II fu mutant flies) directly associates with Cos2 to oppose its activity (3, 4, 57). A cytoplasmic complex composed of Fu, Ci, Cos2, and a small amount of Sufu was also shown to be associated with Smo via Cos2 in an Hh-dependent manner through both biochemical and genetic studies (35, 49, 63, 76, 83, 84). In this model, Hh signal transduction leads to recruitment of the cytoplasmic protein complex to Smo and subsequent inhibition of Ci proteolysis, which would otherwise produce a transcriptional repressor of Hh target gene expression (48). Instead, Ci is converted into an activator by unknown mechanisms to activate Hh target genes (64). Fu and Sufu were proposed to exert opposite effects in controlling the processing, activity, and shuttling of Ci between the nucleus and the cytoplasm (43, 55, 97, 98). Sufu is believed to tether Ci in the cytoplasm and repress Hh signaling, which could be antagonized by Fu. While kinase activity of Fu has not been documented, Fu, as well as Sufu and Cos2, is phosphorylated in response to Hh signaling (49, 61, 92), raising the possibility that posttranslational modifications of Hh pathway components play a key role in regulating multiple steps of Hh signal transduction. Despite these insights, the central issues of how Ci proteolysis and activation are executed and controlled, the composition and transport of the cytoplasmic complex in response to Hh signaling, and the biochemical nature and consequences of protein-protein interaction in the protein complex remain largely unexplained.
In parallel with Drosophila studies, vertebrate homologs of Fu (58, 100), Sufu (19, 20, 41, 69, 86, 100), Cos2 (37, 38, 89), and Ci (32, 39, 40, 78, 95) were identified through either sequence analysis or functional studies. While mammals appear to contain a single Fu gene and a single Sufu gene, multiple vertebrate Ci and Cos2 homologs have been reported, adding greater complexity to vertebrate Hh signaling. The activator and repressor activities of Ci are differentially distributed among three Gli proteins, the Ci homologs in mammals. Gli1 is a transcriptional activator, while Gli2 and Gli3 function as both activators and repressors of transcription (5, 17, 77, 80, 81). The relative contributions and combinatorial effects of Gli activator and repressor activities in distinct developmental contexts remain to be fully characterized (6-8, 13, 21, 31, 44, 52, 56, 68), but one may speculate that the complex genetic interactions and regulation of the three Gli proteins may be linked to modifications of Hh pathway design. Multiple members of the kinesin family (Kif), including Kif27 and Kif7, are thought to be the vertebrate homologs of Cos2 (37, 38; data not shown). This notion was further supported by biochemical and morpholino-mediated knockdown studies of Kif7 in zebra fish, in which Kif7 was shown to function as an intracellular repressor of Hh signaling in conjunction with Sufu (89).
In vitro studies have independently revealed a role of vertebrate Fu in Hh signaling. Vertebrate Fu was shown to weakly synergize with Gli1/2 in activation of an Hh-responsive reporter (18, 58, 66), to affect Gli protein localization (58), and to antagonize the activity of Sufu (58). Moreover, a protein complex composed of Fu, Sufu, and Gli was identified, as well as the demonstration of opposite effects on the activity and subcellular distribution of Gli proteins exerted by Fu and Sufu (20, 22, 41, 54, 58, 69, 86). These independent studies with invertebrates and vertebrates strengthened the notion of conservation of the Hh pathway, and it is conceivable that genetic studies of Fu, Sufu, and Kif27/Kif7 in mice will simply provide a final proof. It is expected that loss of mammalian Fu will recapitulate many aspects of reduced Hh signaling while mutations in Sufu will exhibit little or no effect, given the converging evidence from flies, zebra fish, and mice. To definitively test the conservation of the Hh signaling cascade downstream of Smo during evolution and to understand the role Fu plays in Hh signaling, we took a genetic approach in this study to inactivate vertebrate Fu in mice.
MATERIALS AND METHODS
Molecular biology.
Standard molecular biology techniques, including molecular cloning, genomic DNA preparation, RNA isolation, reverse transcription (RT)-PCR, Southern analysis, and Northern analysis, were performed as previously described (59, 79). The sequences of the oligonucleotides (derived from mouse Fu genomic sequences) used in RT-PCRs are 5′ GTTTCTAGGCAACTGAAGACTCTAGATCCTTT 3′ (Fu exon 1, sense strand), 5′ CTGGTCTTCAGGAAGTTTTCCATCATCTTCCAGAA 3′ (Fu exon 5, antisense strand), 5′ AGCTCTGTACTACCTGCATTCCCACCGCATCCTA 3′ (Fu exon 6, sense strand), 5′ CTTCTGCTTGGCTTCTTCAGCCATGAGTTTAC 3′ (Fu exon 9, antisense strand), 5′ CCACATCAACCTGGAGTGTGAACAAGGCTTCCC 3′ (Fu exon 11, sense strand), 5′ CTGCTCGCCAGTCCCGCGCAGCTGACTCTGGATG 3′ (Fu exon 12, antisense strand), 5′ CCGTCCACAGCTTCACACAAGACAGCAAAGCA 3′ (Fu exon 16, antisense strand), 5′ TGCTTTGCTGTCTTGTGTGAAGCTGTGGACGGA 3′ (Fu exon 16, sense strand), 5′ GTGAAAGTAGCAGATTGGGAAGAGTCCACTGA 3′ (Fu exon 20, sense strand), 5′ ACCAACCACATCCCTGATCAGGCCAGGCTTCCC 3′ (Fu exon 24, antisense strand), 5′ GGGATGTGGTTGGTTCAGAGGTGTGGACCATTCT 3′ (Fu exon 24, sense strand), and 5′ CTGATTCCCCAAAGAGAGCAAGGCTAACTTCTCA 3′ (Fu exon 28, antisense strand).
Generation of targeted Fu mutant mice.
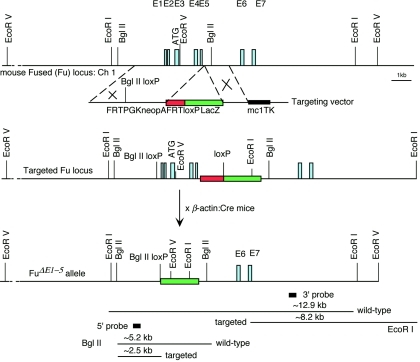
To construct a positive-negative targeting vector for removing exons 1 to 5 of the Fu gene (the resulting allele is designated FuΔE1-5), a 4-kb fragment containing genomic sequences of the first 142 bp of intron 5 and upstream sequences was used as the 5′ region of homology (see Fig. 3). This region encompasses the first five exons, as well as sequences approximately 1.8 kb upstream of exon 1. Oligonucleotides containing one loxP site (5′ ATAACTTCGTATA GCATACAT TATACGAAGTTAT 3′) were inserted into the SalI site, located ∼110 bp upstream of the 5′ untranslated region (exon 1) in the 5′ region of homology (see Fig. 3), resulting in destruction of the SalI site and creation of a BglII site to facilitate the identification of targeted embryonic stem (ES) cell clones. A 1.4-kb fragment containing genomic sequences (intron 5) immediately downstream of the 5′ region of homology was used as the 3′ region of homology and was inserted upstream of the MC1-tk-pA cassette (see Fig. 3). A cassette comprising FRT/PGK-neo-pA/FRT/loxP and β-galactosidase was inserted between the 5′ and 3′ homology regions (see Fig. 3). One loxP site was included at the 3′ end of FRT/PGK-neo-pA/FRT. E14Tg2A.4 (E14) feeder-independent ES cells (60) were electroporated with a SalI-linearized targeting vector and selected in G418 and fialuridine as previously described (36). Targeted ES clones were identified by Southern blotting using the 5′ and 3′ probes. The targeting frequency was approximately 2%. Heterozygous E14 ES cells were injected into blastocysts of C57BL/6 strain mice to generate germ line chimeras. Chimeric males were mated with β-actin::Cre mice (45) to remove sequences between the two loxP sites (including FRT/PGK-neo-pA/FRT) to generate FuΔE1-5 heterozygous animals. As a result, β-galactosidase was brought under the control of putative upstream Fu enhancers. Heterozygotes were identified by Southern blotting of tail tip DNA and were mated with C57BL/6, 129/Sv, 129/Ola, or Swiss-Webster females to maintain the Fu mutant allele in different genetic backgrounds. The majority of the analyses were performed in the Swiss-Webster mixed background. Homozygous mice were produced by crossing heterozygotes and identified by Southern blotting.
FIG. 3.
Targeted disruption of the mouse Fu gene. The schematic diagram shows the Fu genomic locus, the targeting vector, and the mutant allele. The top line shows a partial restriction map of the mouse Fu genomic locus on chromosome 1. The Fu genomic locus consists of 28 exons (E1 to E28), and only the first 7 exons are shown for simplicity. The translation start ATG is predicted to reside in the third exon (E3). The regions between the dotted lines represent the 5′ and 3′ regions of homology used in gene targeting, respectively, and the symbol × indicates events of homologous recombination. Germ line-transmitting chimeric males carrying the targeted Fu locus were mated with β-actin::Cre mice to remove sequences between the two loxP sites (including the PGK-neo-pA selection cassette) and generate the FuΔE1-5 allele in which the first five exons of Fu are removed. As a result, β-galactosidase was brought under the control of putative upstream Fu regulatory elements. The locations of the fragments used as the 5′ or 3′ external probes in Southern blotting are shown, as well as the sizes of the restriction fragments detected for wild-type and targeted FuΔE1-5 alleles. The Rnf25 (ring finger protein 25) gene is divergently transcribed immediately upstream of Fu, suggesting that targeted disruption of Fu could potentially remove regulatory elements that control Rnf25 expression.
Histology and in situ hybridization.
Embryo collection, histological analysis, whole-mount in situ hybridization using digoxigenin-labeled probes, and section in situ hybridization using 33P-labeled riboprobes were performed as previously described (59, 99).
RESULTS
Mouse Fu is broadly expressed during embryogenesis.
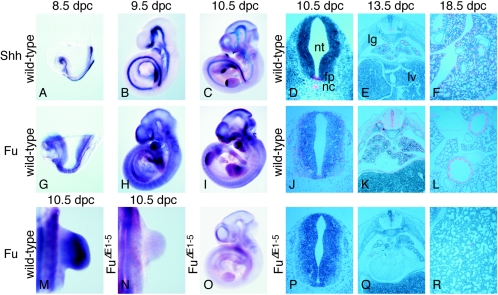
To better understand the potential role that Fu plays in vertebrate Hh signaling, we examined the temporal and spatial expression patterns of Fu in mouse embryos collected from 8.5 days postcoitus (dpc) to 18.5 dpc and compared the expression to that of other Hh pathway members such as Shh (23). By 8.5 dpc, when Shh is expressed at the axial midline including the notochord (Fig. 1A) (23), Fu is widely expressed at low levels (Fig. 1G and data not shown). At ∼9.5 dpc, Shh is activated in the zone of polarizing activity of the forelimb (Fig. 1B) and its expression levels increase from 9.5 to 10.5 dpc, with expression extending to both limbs (Fig. 1C) (23). Fu continues to be broadly expressed (Fig. 1H), and at 10.5 dpc Fu expression can be observed in tissues including the limb (Fig. 1M), the neural tube (Fig. 1I and J), the somite (Fig. 1J), and the branchial arches (Fig. 1I). Fu expression persists in many tissues through later stages of embryogenesis (Fig. 1K and L and data not shown). Taken together, these findings suggest a potential role for Fu in Hh signaling since its expression domain overlaps those of Hh-responsive cells.
FIG. 1.
Expression of Fu during mouse embryonic development. (A to C, G to I, and M to O) Whole-mount in situ hybridization using digoxigenin-labeled riboprobes on wild-type embryos at 8.5 (A and G), 9.5 (B and H), and 10.5 (C, I, and M) dpc and FuΔE1-5 embryos at 10.5 dpc (N and O). All views are lateral, with the exception of M and N, which are dorsal views of the forelimb. Shh is expressed in several signaling centers, while Fu is broadly expressed in wild-type embryos, including in domains of Hh-responsive cells. Only background Fu signal could be detected in FuΔE1-5 embryos. Embryos in panels A and G; B and H; C, I, and O; and M and N were photographed at the same magnification, respectively. (D to F, J to L, and P to R) Isotopic in situ hybridization using [33P]UTP-labeled riboprobes (pink) on paraffin sections of 10.5-dpc wild-type (D and J) and FuΔE1-5 (P) embryos at the forelimb-heart level, 13.5-dpc wild-type (E and K) and FuΔE1-5 (Q) embryos, and 18.5-dpc wild-type (F and L) and FuΔE1-5 (R) lungs. Panels D, J, and P; E, K, and Q; and F and R were photographed at the same magnification, respectively. Panel L was photographed at a higher magnification than panels F and R. Fu signal is absent in FuΔE1-5 embryos. The Fu probe used for in situ hybridization is derived from Fu cDNA sequences that correspond approximately to exons 2 to 9. Probes derived from several other regions of Fu cDNA, including exons 2 to 5 and exons 27 and 28, gave identical expression patterns (data not shown). The Fu sense probes did not yield signals above the background (data not shown). nt, neural tube; fp, floor plate; nc, notochord; lg, lung; lv, liver.
The Fu transcripts are alternatively spliced during mouse development.
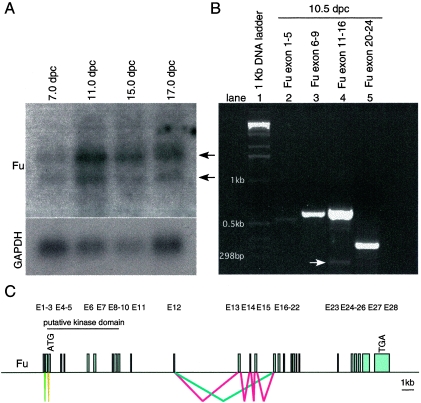
The mouse Fu genomic locus, located on chromosome 1, consists of 28 predicted exons, and the translational start ATG codon of the Fu transcripts is predicted to reside in the third exon (Fig. 2C). The putative serine/threonine kinase domain located at the N terminus of Fu is encoded approximately by exons 3 to 9 (Fig. 2C). Analysis of mouse Fu cDNA sequences revealed multiple Fu splice variants. For instance, exons 13 to 15 are alternatively spliced and are not included in the shorter Fu transcripts (Fig. 2B and C). Interestingly, the first and second exons, which contain the 5′ untranslated region, are utilized alternatively in different Fu cDNA clones (Fig. 2B and C). We performed RT-PCR on RNA obtained from 10.5 dpc wild-type mouse embryos to verify the splice variants of the Fu transcripts and to determine their relative abundance during embryonic development. We found that longer Fu transcripts, containing exons 13 to 15, constitute the major species while transcripts without exons 13 to 15 (arrow in Fig. 2B, lane 4) are coexpressed at a much lower level. Alternative splicing of exon 8 or exon 23, previously reported for human Fu cDNA (66), was not detected using mouse 10.5 dpc RNA (Fig. 2B, lane 3 and 5). These results suggest that the Fu transcript, which includes all coding exons and encodes a protein of 1,262 amino acids, is the major product during mouse embryonic development.
FIG. 2.
Alternative splicing of Fu transcripts during mouse embryogenesis. (A) Northern blot analysis of poly(A)+ RNA isolated from 7-, 11-, 15-, and 17-dpc wild-type mouse embryos (Clontech). Two major species of Fu transcripts, approximately 4.7 and 4.1 kb, respectively (arrows), were detected. In adult mice, Fu is mainly expressed in testis (data not shown). The Fu probe used for hybridization is derived from Fu cDNA sequences that correspond approximately to exons 2 to 9. The same membrane was rehybridized with a GAPDH probe, which serves as the loading control. (B) RT-PCR using RNA derived from wild-type 10.5-dpc mouse embryos in combination with primers spanning the indicated exons. Alternative splicing of the region containing exons 11 to 16 was detected by PCR. This was subsequently verified to be alternative splicing of exons 13 to 15 by sequencing. The white arrow points to the PCR product without exons 13 to 15, which is much less abundant than the upper band containing exons 13 to 15. No alternative splicing of exon 8 or exon 23, as reported for human Fu transcripts, was detected in this assay. A PCR product was amplified with primers derived from exons 1 and 5, respectively. Sequence analysis of the PCR product revealed that it contains exons 1 and 3 to 5, and whether it represents the dominant form during mouse embryogenesis remains to be further investigated. RT-PCR using primers derived from exons 2 and 5 failed to produce a product (data not shown). (C) Schematic diagram of the Fu genomic locus and possible alternative splicing. The Fu genomic locus consists of 28 exons (E1 to E28). The ATG translation start codon is predicted to reside in the third exon (E3), and the TGA stop codon is in the 28th exon (E28). Alternative splicing of exons 13 to 15 is labeled in red and blue, respectively, and potential alternative splicing of exons 1 and 2 is labeled with lime and olive, respectively. Splicing of common exons is not labeled. The longer Fu transcript, containing exons 13 to 15, appears to constitute the major species during mouse embryonic development based on the relative abundance of the transcripts.
Targeted disruption of mouse Fu results in postnatal lethality.
To define the role Fu plays in vertebrate Hh signaling, we generated a gene-targeted allele of Fu in mice. We decided to delete the first five exons, since exons 3 to 5 are shared by different Fu splice variants and encode both the translational start and a portion of the highly conserved N-terminal kinase domain (Fig. 2C and 3) (58). In addition, ∼110 bp upstream of exon 1 were also deleted in this targeting strategy, which could potentially contain the upstream promoter and regulatory elements (Fig. 3). We anticipated that no functional Fu transcripts or protein would be produced from the resulting targeted allele (designated FuΔE1-5).
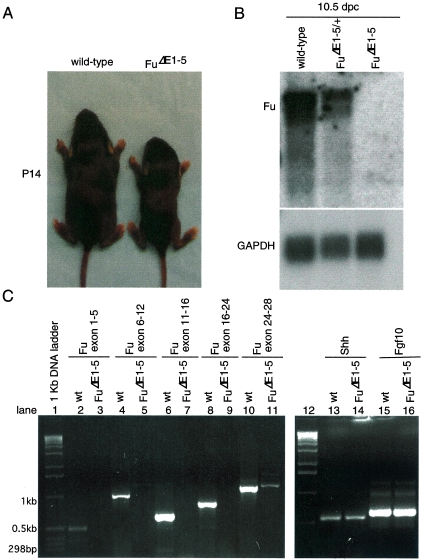
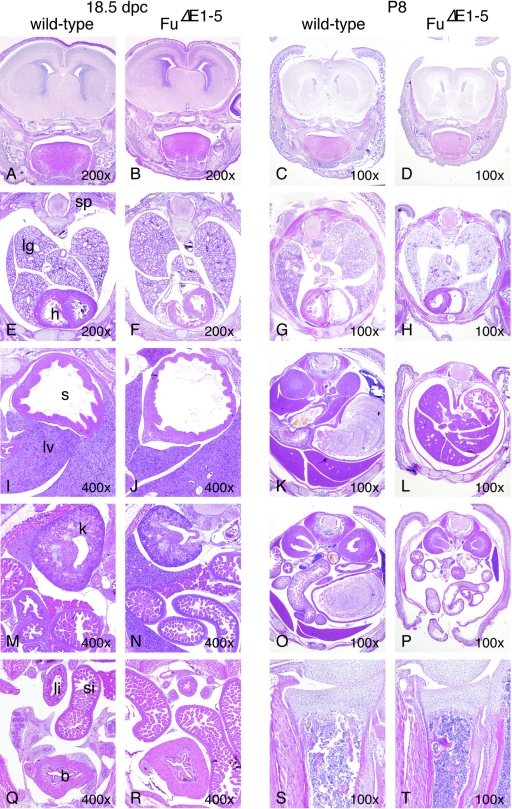
To our surprise, animals homozygous for FuΔE1-5 were born alive and their appearance could not be distinguished from that of their wild-type littermates. The ratio of wild-type to FuΔE1-5/+ to FuΔE1-5 newborn pups approximates a 1:2:1 Mendelian distribution (data not shown). Homozygous FuΔE1-5 animals continued to thrive after birth, but the majority of them became visibly smaller than their wild-type littermates around postnatal day 7 (data not shown and Fig. 4A). These runts failed to gain weight and appeared starved and emaciated, and the majority of them died before they reached 3 weeks of age. The lethality of FuΔE1-5 animals appears to be completely penetrant (with more than 50 animals examined so far). Examination of major organs did not reveal any obvious pathology in FuΔE1-5 mutants (Fig. 5, right panel, and data not shown). The findings of standard biochemical analyses of blood samples, including glucose levels, lipid profiles, electrolytes, and liver and renal function tests, are consistent with changes associated with starvation in FuΔE1-5 pups (data not shown). The cause of death of FuΔE1-5 animals remains to be further investigated.
FIG. 4.
FuΔE1-5 animals exhibit growth retardation and postnatal lethality, and Fu transcript levels are barely detectable in FuΔE1-5 embryos. (A) Wild-type and FuΔE1-5 animals photographed at postnatal day 14. The mutant is significantly smaller than its wild-type littermate. Some toes in both animals were clipped for numbering and genotyping. (B) Northern blot analysis of poly(A)+ RNA isolated from 10.5-dpc wild-type, FuΔE1-5/+, and FuΔE1-5 embryos. The two Fu probes used for hybridization gave identical results, and they correspond approximately to the last 775 bp of Fu transcripts and the 624-bp genomic sequences (exons 2 to 5) deleted in the gene-targeted FuΔE1-5 allele (data not shown). A ∼4.7-kb upper band with stronger intensity and a ∼4.1-kb lower band were detected in both wild-type and FuΔE1-5/+ embryos but were completely absent in FuΔE1-5 embryos. The expression level of the housekeeping gene GAPDH serves as the loading control with the same membrane reprobed. (C) RT-PCR using RNA derived from 10.5-dpc wild-type (wt) and FuΔE1-5 mouse embryos in combination with primers spanning the exons indicated. While PCR products of the predicted sizes were detected from RNA derived from wild-type embryos, no PCR products were generated using RNA obtained from FuΔE1-5 mutants. The same number of PCR cycles (37 cycles) was employed for all the PCRs shown in this panel. The faint band, corresponding to exons 24 to 28, seen in FuΔE1-5 was not detected when a lower number of PCR cycles (less than 33) was applied (data not shown). PCR products corresponding to other genes such as Shh, Fgf10, and GAPDH (not shown) were detected at similar levels using RNA produced from wild-type or FuΔE1-5 mutants, suggesting that the absence of a Fu signal in FuΔE1-5 was not due to technical difficulties in RNA preparation or RT-PCR.
FIG. 5.
Development of major tissues and organs does not appear to be affected in FuΔE1-5 mutants. (A to T) Hematoxylin-and-eosin-stained sections of major tissues and organs of wild-type and FuΔE1-5 animals at embryonic dpc 18.5 and postnatal day P8. A to D, coronal sections through the forebrain; E to H, cross sections through the thoracic cavity; I to R, cross sections through the abdominal cavity; S and T, longitudinal sections through the proximal ulna. Although FuΔE1-5 mutants were significantly smaller than their wild-type littermates at day P8, no obvious developmental defects or pathological changes in major tissues and organs were detected in FuΔE1-5 mutants. Multiple wild-type and FuΔE1-5 embryos were examined. In certain regions of the sections, the minor differences in morphology between wild-type and FuΔE1-5 animals were due to planes of sections. sp, spinal cord; lg, lung; h, heart; s, stomach; si, small intestine; li, large intestine; lv, liver; k, kidney; b, bladder. The magnification of each section is indicated.
Fu transcripts are barely detectable in gene-targeted FuΔ1-5 mice.
The unexpected phenotypes of Fu-deficient mice prompted us to investigate whether the targeted FuΔE1-5 allele represents a null allele. Since the 5′ untranslated region of Fu and 110 bp of upstream sequence were deleted, we expected that Fu transcript levels would be greatly reduced or even completely absent in FuΔE1-5 embryos. Consistent with this, in situ hybridization of FuΔE1-5 embryos using a probe derived from genomic sequences deleted in FuΔE1-5 detected only background signals of Fu mRNA (Fig. 1N to R) compared to those of their wild-type littermates (Fig. 1M and I to L). Furthermore, Northern analysis of mRNA isolated from 10.5 dpc embryos revealed that Fu transcripts were not detectable in FuΔE1-5 embryos, contrasted with wild-type littermates (Fig. 4B). We also failed to detect any truncated Fu transcripts that could have resulted from aberrant splicing of the targeted FuΔE1-5 allele (Fig. 4B). To rule out the possibility that a trace amount of Fu transcript is produced but is beyond the detection limit of Northern analysis, we employed a sensitive PCR-based assay designed to detect any residual Fu transcripts. RT-PCR was performed using RNA isolated from 10.5 dpc embryos deficient in Fu or from wild-type controls, in combination with multiple primer pairs spanning different regions of the Fu transcripts (Fig. 4C). While PCR products of the predicted sizes were detected from RNA derived from wild-type embryos, no PCR products were generated using RNA obtained from FuΔE1-5 embryos (Fig. 4C). In contrast, PCR products corresponding to other genes such as Shh and Fgf10 were detected at similar levels using RNA produced from either wild-type or FuΔE1-5 embryos (Fig. 4C). Nevertheless, it should be noted that a minute amount of PCR product, corresponding to a Fu transcript starting approximately at exon 19, could be detected when a higher number of PCR cycles was applied (Fig. 4C, lane 11, and data not shown). This could potentially represent a trace amount of truncated transcripts generated from the FuΔE1-5 allele, but whether any residual truncated Fu protein was generated is not known. Taken together, these results indicate that Fu transcript levels are greatly reduced (or even absent) in FuΔE1-5 mutants, suggesting that little or no functional Fu protein was produced in FuΔE1-5 embryos.
Mice deficient in Fu do not exhibit phenotypes indicative of perturbed Hh signaling during embryogenesis.
Mice homozygous for FuΔE1-5 appear to develop normally through embryogenesis, and their appearance cannot be distinguished from that of wild-type littermates. We performed a detailed histological analysis of mouse embryos collected between 9.5 and 18.5 dpc (Fig. 5, left panel, and data not shown). In contrast to Shh mutants, which exhibit defects in multiple tissues, including the developing neural tube, limb, hair follicle, gut, lung, pancreas, and kidney, due to defective Hh signaling (53), no significant morphological changes can be discerned in FuΔE1-5 animals compared to wild-type littermates (Fig. 5, left panel). There are also no skeletal defects due to defective Ihh signaling (53) in Fu-deficient embryos (Fig. 5, left panel, and data not shown). These results suggest that Hh signaling is not disrupted in FuΔE1-5 embryos.
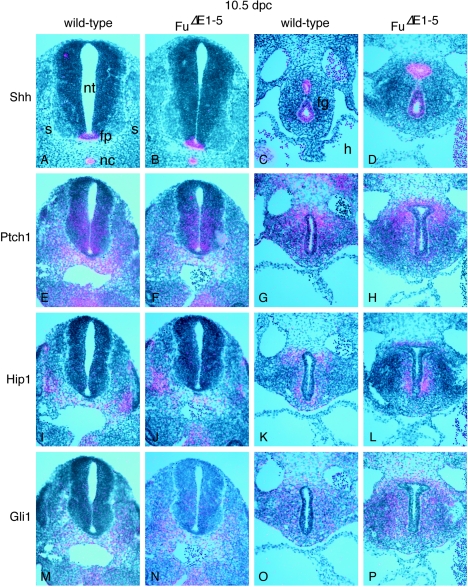
It is possible that the Hh pathway is perturbed at molecular levels but the effects are subtle and cannot be discerned by our phenotypic analysis. To test this hypothesis, we performed in situ hybridization on wild-type and FuΔE1-5 mouse embryos collected between 9.5 and 18.5 dpc, focusing on Hh target genes including Ptch1, Hip1, and Gli1 (16, 28, 32, 71). These genes are expressed in Hh-responsive tissues and are up-regulated in response to Hh signaling. For instance, while Shh is expressed in midline structures (23) (Fig. 6A), including the notochord and floor plate, Ptch1, Hip1, and Gli1 are all expressed in the ventral neural tube (Fig. 6E, I, and M). Similarly, Shh is expressed in the epithelium of many developing organs, such as the gut (Fig. 6C) (9), while Ptch1, Hip1, and Gli1 are expressed in the surrounding mesenchyme (Fig. 6G, K, and O). Expression patterns of Hh target genes in Fu-deficient embryos (Fig. 6F, J, N, H, L, and P) cannot be distinguished from those of their wild-type littermates, indicating that the Hh pathway is not perturbed due to loss of Fu during embryonic development.
FIG. 6.
Expression of Hh targets is not perturbed in FuΔE1-5 mutants. (A to P) Isotopic in situ hybridization using [33P]UTP-labeled riboprobes (pink) on paraffin sections of wild-type (A, C, E, G, I, K, M, and O) and FuΔE1-5 (B, D, F, H, J, L, N, and P) 10.5-dpc embryos at the forelimb-heart level (A, B, C, D, G, H, K, L, O, and P) or the hind limb level (E, F, I, J, M, and N). The axis for orientation of the specimens is dorsal upward and ventral downward. Shh is expressed in several signaling centers such as the notochord and floor plate, while Hh targets, including Ptch1, Hip1, and Gli1, are expressed in the ventral neural tube and the somite in response to Hh signaling. Shh is also expressed in the epithelium of many developing organs, including the foregut endoderm, while Ptch1, Hip1, and Gli1 are expressed in the surrounding mesenchyme. Multiple sections were examined in multiple rounds of in situ hybridization, and expression of Hh targets does not appear to be affected in FuΔE1-5 mutants. nt, neural tube; fp, floor plate; nc, notochord; s, somite; fg, foregut; h, heart.
DISCUSSION
The lack of embryonic phenotypes in FuΔE1-5-targeted mice came as a surprise, given the critical role that Hh signaling plays in embryonic development and the observation that a similar set of molecules appears to be employed for Hh signal transduction in both invertebrates and vertebrates (29, 33). It is clear that vertebrates contain more Hh pathway members due to gene duplication and in some cases even vertebrate-specific players that modulate Hh signaling (11, 16, 24, 30, 34). Nevertheless, many aspects of Hh signaling characterized so far are conserved (33). One can argue that the key issue of how lipid-modified Hh ligand is transported in the morphogenetic field requires further exploration to clarify the similarities and differences between vertebrates and invertebrates (12, 15, 27, 46, 67). It is, however, generally accepted that Hh signal transduction through Ptch and Smo on the cell surface is conserved as well as Hh target gene activation by the Ci/Gli family of transcriptional factors (33). The signal transduction cascade between Smo and Gli in vertebrates has been investigated to some extent in vitro (20, 22, 41, 58, 69, 86). An essential role of vertebrate Fu in Hh signaling was predicted from characterization of Drosophila Fu (2, 25, 26, 74, 75, 91-93) and further strengthened by studies of cultured cells in which vertebrate Fu was shown to weakly synergize with Gli1/2 in Hh activation (18, 58, 66), to affect Gli protein localization (58), and to antagonize the activity of Sufu (58). Moreover, morpholino-mediated knockdown of zebra fish Fu activities produces muscle phenotypes consistent with reduced Hh signaling (100). All of the available studies provided no evidence to suggest that vertebrates utilize a different strategy for Hh signal transduction downstream of Smo. In this regard, the lack of embryonic phenotypes in FuΔE1-5 mice is particularly puzzling.
It is possible that the lack of phenotypes in FuΔE1-5 embryos was simply due to the failure to generate a null allele of Fu since only the first 5 exons (out of 28) were deleted. However, several lines of evidence suggest that FuΔE1-5 is likely a null allele. No wild-type Fu transcripts were detected in FuΔE1-5 embryos by in situ hybridization, Northern analysis, or RT-PCR. Consistent with this observation, we failed to detect any β-galactosidase staining in FuΔE1-5 embryos (data not shown), suggesting that no functional Fu transcript was made. Nonetheless, it should be noted that a trace amount of truncated Fu transcript, possibly generated through alternative downstream transcriptional start signal or aberrant splicing, was detected from the FuΔE1-5 allele. It follows that a small amount of truncated Fu protein containing the distant region of Fu could potentially be produced, albeit in this case the Fu activity is unlikely to be preserved. The truncated Fu protein expressed in cultured cells displayed no effect in activating multimerized Gli reporters or opposing the activities of Sufu (N. Gao and P.-T. Chuang, unpublished data). The truncated Fu protein is also unlikely to exert dominant negative effects since overexpression of a truncated Fu protein lacking the N-terminal kinase domain failed to generate noticeable phenotypes in mice (R. Sutherland and P.-T. Chuang, unpublished data), which is different from the dominant negative effects exerted by a similar mutation in Drosophila Fu (3). Despite a lack of embryonic phenotypes in FuΔE1-5 mutants, these animals became emaciated after birth and eventually died of unknown causes. Whether the lethality reflects an essential requirement of Hh signaling during postnatal life or perturbation of other signaling pathways remains to be investigated.
The lack of embryonic phenotypes in FuΔE1-5 could also be attributed to functional redundancy between Fu and other potential Fu homologs. However, sequence analysis of vertebrate Fu has failed to identify additional Fu homologs. The similarity between fly and vertebrate Fu sequences predominantly resides in the putative serine/threonine kinase domain located at the N terminus (58). It is possible that the regions of Fu outside the kinase domain could have diverged extensively during evolution and sequence analysis alone may not be sufficient to identify additional Fu homologs in vertebrates. Instead, functional studies will be required to test the ability of any potential Fu homologs, which exhibit exceedingly limited sequence similarity to Fu, to compensate for the loss of Fu function in vitro and in vivo. Alternatively, loss of Fu could be compensated for by changes in expression levels or activities of other Hh pathway members, providing that the more complex vertebrate Hh pathway contains additional self-correcting mechanisms. For instance, compensatory down-regulation of Sufu or up-regulation of Gli in Fu-deficient mice may restore normal embryonic development. Further genetic and molecular analyses are required to test these hypotheses.
The design of the Hh pathway may have diverged during evolution. In the extreme case, Fu may have no role in vertebrate Hh signaling and Sufu has assumed a more prominent role in Hh signaling. Consistent with this notion, mice deficient in Sufu display dramatic embryonic phenotypes indicative of elevated Hh signaling (10; C. C. Hui, personal communication). Likewise, the function of vertebrate Cos2 homologs may also have diverged during evolution, especially given the limited sequence similarity between Kif27/Kif7 proteins and Drosophila Cos2 (10, 37, 38, 89). In this scenario, the mechanism by which the Hh signal is transduced downstream of Smo would be significantly different between invertebrates and vertebrates (10). One can also surmise that the evolution of three Gli proteins with different activator and repressor functions and modified circuitry of Gli regulation has led to a corresponding change in the mechanics of Hh signaling downstream of Smo.
Another possibility of Hh pathway divergence would be that Fu is involved in Hh signaling but its relative contribution to Hh signaling differs between invertebrates and vertebrates, perhaps even between different vertebrate species. The ratio of combined Gli repressor and activator forms and their regulation may differ among species and one form may even predominate in certain tissues (47, 90, 96). This could contribute to an altered balance of Fu and Sufu function in Hh signaling in vertebrates to ensure that proper Hh responses are generated in diverse developmental contexts. Further experiments are required to distinguish between these models and to elucidate the mechanisms of Hh signal transduction in vertebrates.
Acknowledgments
We thank Julie Qiao for assistance with histology, Rachel Sutherland for molecular cloning, and Rhodora Gacayan for mouse husbandry and genotyping. We thank members of the Chuang laboratory for helpful discussion; Chris Wilson, Tony Gerber, and C. C. Hui for critical reading of the manuscript; and Frederic de Sauvage for communicating results prior to publication.
Work in the Chuang laboratory was supported by an NIH grant (HL67822).
REFERENCES
- 1.Alcedo, J., M. Ayzenzon, T. Von Ohlen, M. Noll, and J. E. Hooper. 1996. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86:221-232. [DOI] [PubMed] [Google Scholar]
- 2.Alves, G., B. Limbourg-Bouchon, H. Tricoire, J. Brissard-Zahraoui, C. Lamour-Isnard, and D. Busson. 1998. Modulation of Hedgehog target gene expression by the Fused serine-threonine kinase in wing imaginal discs. Mech. Dev. 78:17-31. [DOI] [PubMed] [Google Scholar]
- 3.Ascano, M., Jr., K. E. Nybakken, J. Sosinski, M. A. Stegman, and D. J. Robbins. 2002. The carboxyl-terminal domain of the protein kinase fused can function as a dominant inhibitor of hedgehog signaling. Mol. Cell. Biol. 22:1555-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascano, M., Jr., and D. J. Robbins. 2004. An intramolecular association between two domains of the protein kinase Fused is necessary for Hedgehog signaling. Mol. Cell. Biol. 24:10397-10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aza-Blanc, P., H. Y. Lin, A. Ruiz i Altaba, and T. B. Kornberg. 2000. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development 127:4293-4301. [DOI] [PubMed] [Google Scholar]
- 6.Bai, C. B., W. Auerbach, J. S. Lee, D. Stephen, and A. L. Joyner. 2002. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129:4753-4761. [DOI] [PubMed] [Google Scholar]
- 7.Bai, C. B., and A. L. Joyner. 2001. Gli1 can rescue the in vivo function of Gli2. Development 128:5161-5172. [DOI] [PubMed] [Google Scholar]
- 8.Bai, C. B., D. Stephen, and A. L. Joyner. 2004. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6:103-115. [DOI] [PubMed] [Google Scholar]
- 9.Bitgood, M. J., and A. P. McMahon. 1995. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 172:126-138. [DOI] [PubMed] [Google Scholar]
- 10.Briscoe, J., and P. Therond. 2005. Hedgehog signaling: from the Drosophila cuticle to anti-cancer drugs. Dev. Cell 8:143-151. [DOI] [PubMed] [Google Scholar]
- 11.Bulgakov, O. V., J. T. Eggenschwiler, D. H. Hong, K. V. Anderson, and T. Li. 2004. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development 131:2149-2159. [DOI] [PubMed] [Google Scholar]
- 12.Burke, R., D. Nellen, M. Bellotto, E. Hafen, K. A. Senti, B. J. Dickson, and K. Basler. 1999. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99:803-815. [DOI] [PubMed] [Google Scholar]
- 13.Buttitta, L., R. Mo, C. C. Hui, and C. M. Fan. 2003. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development 130:6233-6243. [DOI] [PubMed] [Google Scholar]
- 14.Casali, A., and G. Struhl. 2004. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature 431:76-80. [DOI] [PubMed] [Google Scholar]
- 15.Chen, M. H., Y. J. Li, T. Kawakami, S. M. Xu, and P.-T. Chuang. 2004. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 18:641-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang, P.-T., and A. P. McMahon. 1999. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397:617-621. [DOI] [PubMed] [Google Scholar]
- 17.Dai, P., H. Akimaru, Y. Tanaka, T. Maekawa, M. Nakafuku, and S. Ishii. 1999. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274:8143-8152. [DOI] [PubMed] [Google Scholar]
- 18.Daoud, F., and M. F. Blanchet-Tournier. 2005. Expression of the human FUSED protein in Drosophila. Dev. Genes Evol. 215:230-237. [DOI] [PubMed] [Google Scholar]
- 19.Delattre, M., S. Briand, M. Paces-Fessy, and M. F. Blanchet-Tournier. 1999. The Suppressor of fused gene, involved in Hedgehog signal transduction in Drosophila, is conserved in mammals. Dev. Genes Evol. 209:294-300. [DOI] [PubMed] [Google Scholar]
- 20.Ding, Q., S. Fukami, X. Meng, Y. Nishizaki, X. Zhang, H. Sasaki, A. Dlugosz, M. Nakafuku, and C. Hui. 1999. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 9:1119-1122. [DOI] [PubMed] [Google Scholar]
- 21.Ding, Q., J. Motoyama, S. Gasca, R. Mo, H. Sasaki, J. Rossant, and C. C. Hui. 1998. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125:2533-2543. [DOI] [PubMed] [Google Scholar]
- 22.Dunaeva, M., P. Michelson, P. Kogerman, and R. Toftgard. 2003. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 278:5116-5122. [DOI] [PubMed] [Google Scholar]
- 23.Echelard, Y., D. J. Epstein, B. St-Jacques, L. Shen, J. Mohler, J. A. McMahon, and A. P. McMahon. 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75:1417-1430. [DOI] [PubMed] [Google Scholar]
- 24.Eggenschwiler, J. T., E. Espinoza, and K. V. Anderson. 2001. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature 412:194-198. [DOI] [PubMed] [Google Scholar]
- 25.Forbes, A. J., Y. Nakano, A. M. Taylor, and P. W. Ingham. 1993. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev. Suppl. 1993:115-124. [PubMed] [Google Scholar]
- 26.Fukumoto, T., R. Watanabe-Fukunaga, K. Fujisawa, S. Nagata, and R. Fukunaga. 2001. The fused protein kinase regulates Hedgehog-stimulated transcriptional activation in Drosophila Schneider 2 cells. J. Biol. Chem. 276:38441-38448. [DOI] [PubMed] [Google Scholar]
- 27.Gallet, A., R. Rodriguez, L. Ruel, and P. P. Therond. 2003. Cholesterol modification of hedgehog is required for trafficking and movement, revealing an asymmetric cellular response to hedgehog. Dev. Cell 4:191-204. [DOI] [PubMed] [Google Scholar]
- 28.Goodrich, L. V., R. L. Johnson, L. Milenkovic, J. A. McMahon, and M. P. Scott. 1996. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 10:301-312. [DOI] [PubMed] [Google Scholar]
- 29.Hooper, J. E., and M. P. Scott. 2005. Communicating with Hedgehogs. Nat. Rev. Mol. Cell. Biol. 6:306-317. [DOI] [PubMed] [Google Scholar]
- 30.Huangfu, D., A. Liu, A. S. Rakeman, N. S. Murcia, L. Niswander, and K. V. Anderson. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426:83-87. [DOI] [PubMed] [Google Scholar]
- 31.Hui, C. C., and A. L. Joyner. 1993. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 3:241-246. [DOI] [PubMed] [Google Scholar]
- 32.Hui, C. C., D. Slusarski, K. A. Platt, R. Holmgren, and A. L. Joyner. 1994. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 162:402-413. [DOI] [PubMed] [Google Scholar]
- 33.Ingham, P. W., and A. P. McMahon. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15:3059-3087. [DOI] [PubMed] [Google Scholar]
- 34.Izraeli, S., L. A. Lowe, V. L. Bertness, S. Campaner, H. Hahn, I. R. Kirsch, and M. R. Kuehn. 2001. Genetic evidence that Sil is required for the Sonic Hedgehog response pathway. Genesis 31:72-77. [DOI] [PubMed] [Google Scholar]
- 35.Jia, J., C. Tong, and J. Jiang. 2003. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 17:2709-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyner, A. L. 2000. Gene targeting: a practical approach, second edition. University Press, Oxford, United Kingdom.
- 37.Katoh, Y., and M. Katoh. 2004. Characterization of KIF7 gene in silico. Int. J. Oncol. 25:1881-1886. [PubMed] [Google Scholar]
- 38.Katoh, Y., and M. Katoh. 2004. KIF27 is one of orthologs for Drosophila Costal-2. Int. J. Oncol. 25:1875-1880. [PubMed] [Google Scholar]
- 39.Kinzler, K. W., S. H. Bigner, D. D. Bigner, J. M. Trent, M. L. Law, S. J. O'Brien, A. J. Wong, and B. Vogelstein. 1987. Identification of an amplified, highly expressed gene in a human glioma. Science 236:70-73. [DOI] [PubMed] [Google Scholar]
- 40.Kinzler, K. W., J. M. Ruppert, S. H. Bigner, and B. Vogelstein. 1988. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature 332:371-374. [DOI] [PubMed] [Google Scholar]
- 41.Kogerman, P., T. Grimm, L. Kogerman, D. Krause, A. B. Unden, B. Sandstedt, R. Toftgard, and P. G. Zaphiropoulos. 1999. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1:312-319. [DOI] [PubMed] [Google Scholar]
- 42.Kuwabara, P. E., and M. Labouesse. 2002. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 18:193-201. [DOI] [PubMed] [Google Scholar]
- 43.Lefers, M. A., Q. T. Wang, and R. A. Holmgren. 2001. Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev. Biol. 236:411-420. [DOI] [PubMed] [Google Scholar]
- 44.Lei, Q., A. K. Zelman, E. Kuang, S. Li, and M. P. Matise. 2004. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development 131:3593-3604. [DOI] [PubMed] [Google Scholar]
- 45.Lewandoski, M., E. N. Meyers, and G. R. Martin. 1997. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harbor Symp. Quant. Biol. 62:159-168. [PubMed] [Google Scholar]
- 46.Lewis, P. M., M. P. Dunn, J. A. McMahon, M. Logan, J. F. Martin, B. St-Jacques, and A. P. McMahon. 2001. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105:599-612. [DOI] [PubMed] [Google Scholar]
- 47.Litingtung, Y., R. D. Dahn, Y. Li, J. F. Fallon, and C. Chiang. 2002. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418:979-983. [DOI] [PubMed] [Google Scholar]
- 48.Lum, L., and P. A. Beachy. 2004. The Hedgehog response network: sensors, switches, and routers. Science 304:1755-1759. [DOI] [PubMed] [Google Scholar]
- 49.Lum, L., C. Zhang, S. Oh, R. K. Mann, D. P. von Kessler, J. Taipale, F. Weis-Garcia, R. Gong, B. Wang, and P. A. Beachy. 2003. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol. Cell 12:1261-1274. [DOI] [PubMed] [Google Scholar]
- 50.Mann, R. K., and P. A. Beachy. 2004. Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 73:891-923. [DOI] [PubMed] [Google Scholar]
- 51.Marigo, V., R. A. Davey, Y. Zuo, J. M. Cunningham, and C. J. Tabin. 1996. Biochemical evidence that patched is the Hedgehog receptor. Nature 384:176-179. [DOI] [PubMed] [Google Scholar]
- 52.Matise, M. P., D. J. Epstein, H. L. Park, K. A. Platt, and A. L. Joyner. 1998. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 125:2759-2770. [DOI] [PubMed] [Google Scholar]
- 53.McMahon, A. P., P. W. Ingham, and C. J. Tabin. 2003. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53:1-114. [DOI] [PubMed] [Google Scholar]
- 54.Merchant, M., F. F. Vajdos, M. Ultsch, H. R. Maun, U. Wendt, J. Cannon, W. Desmarais, R. A. Lazarus, A. M. de Vos, and F. J. de Sauvage. 2004. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol. Cell. Biol. 24:8627-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Methot, N., and K. Basler. 2000. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127:4001-4010. [DOI] [PubMed] [Google Scholar]
- 56.Mo, R., A. M. Freer, D. L. Zinyk, M. A. Crackower, J. Michaud, H. H. Heng, K. W. Chik, X. M. Shi, L. C. Tsui, S. H. Cheng, A. L. Joyner, and C. Hui. 1997. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124:113-123. [DOI] [PubMed] [Google Scholar]
- 57.Monnier, V., K. S. Ho, M. Sanial, M. P. Scott, and A. Plessis. 2002. Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the kinesin-related protein Costal2. BMC Dev. Biol 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murone, M., S. M. Luoh, D. Stone, W. Li, A. Gurney, M. Armanini, C. Grey, A. Rosenthal, and F. J. de Sauvage. 2000. Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell Biol. 2:310-312. [DOI] [PubMed] [Google Scholar]
- 59.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual, third edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Nichols, J., E. P. Evans, and A. G. Smith. 1990. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development 110:1341-1348. [DOI] [PubMed] [Google Scholar]
- 61.Nybakken, K. E., C. W. Turck, D. J. Robbins, and J. M. Bishop. 2002. Hedgehog-stimulated phosphorylation of the kinesin-related protein Costal2 is mediated by the serine/threonine kinase fused. J. Biol. Chem. 277:24638-24647. [DOI] [PubMed] [Google Scholar]
- 62.Nüsslein-Volhard, C., and E. Wieschaus. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795-801. [DOI] [PubMed] [Google Scholar]
- 63.Ogden, S. K., M. Ascano, Jr., M. A. Stegman, L. M. Suber, J. E. Hooper, and D. J. Robbins. 2003. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr. Biol. 13:1998-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohlmeyer, J. T., and D. Kalderon. 1998. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396:749-753. [DOI] [PubMed] [Google Scholar]
- 65.Orenic, T. V., D. C. Slusarski, K. L. Kroll, and R. A. Holmgren. 1990. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 4:1053-1067. [DOI] [PubMed] [Google Scholar]
- 66.Osterlund, T., D. B. Everman, R. C. Betz, M. Mosca, M. M. Nothen, C. E. Schwartz, P. G. Zaphiropoulos, and R. Toftgard. 2004. The FU gene and its possible protein isoforms. BMC Genomics 5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panakova, D., H. Sprong, E. Marois, C. Thiele, and S. Eaton. 2005. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 435:58-65. [DOI] [PubMed] [Google Scholar]
- 68.Park, H. L., C. Bai, K. A. Platt, M. P. Matise, A. Beeghly, C. C. Hui, M. Nakashima, and A. L. Joyner. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127:1593-1605. [DOI] [PubMed] [Google Scholar]
- 69.Pearse, R. V., II, L. S. Collier, M. P. Scott, and C. J. Tabin. 1999. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev. Biol. 212:323-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pham, A., P. Therond, G. Alves, F. B. Tournier, D. Busson, C. Lamour-Isnard, B. L. Bouchon, T. Preat, and H. Tricoire. 1995. The Suppressor of fused gene encodes a novel PEST protein involved in Drosophila segment polarity establishment. Genetics 140:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Platt, K. A., J. Michaud, and A. L. Joyner. 1997. Expression of the mouse Gli and Ptc genes is adjacent to embryonic sources of hedgehog signals suggesting a conservation of pathways between flies and mice. Mech. Dev. 62:121-135. [DOI] [PubMed] [Google Scholar]
- 72.Preat, T. 1992. Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132:725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Preat, T., P. Therond, C. Lamour-Isnard, B. Limbourg-Bouchon, H. Tricoire, I. Erk, M. C. Mariol, and D. Busson. 1990. A putative serine/threonine protein kinase encoded by the segment-polarity fused gene of Drosophila. Nature 347:87-89. [DOI] [PubMed] [Google Scholar]
- 74.Preat, T., P. Therond, B. Limbourg-Bouchon, A. Pham, H. Tricoire, D. Busson, and C. Lamour-Isnard. 1993. Segmental polarity in Drosophila melanogaster: genetic dissection of fused in a Suppressor of fused background reveals interaction with costal-2. Genetics 135:1047-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robbins, D. J., K. E. Nybakken, R. Kobayashi, J. C. Sisson, J. M. Bishop, and P. P. Therond. 1997. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90:225-234. [DOI] [PubMed] [Google Scholar]
- 76.Ruel, L., R. Rodriguez, A. Gallet, L. Lavenant-Staccini, and P. P. Therond. 2003. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 5:907-913. [DOI] [PubMed] [Google Scholar]
- 77.Ruiz i Altaba, A. 1999. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development 126:3205-3216. [DOI] [PubMed] [Google Scholar]
- 78.Ruppert, J. M., K. W. Kinzler, A. J. Wong, S. H. Bigner, F. T. Kao, M. L. Law, H. N. Seuanez, S. J. O'Brien, and B. Vogelstein. 1988. The GLI-Kruppel family of human genes. Mol. Cell. Biol. 8:3104-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 80.Sasaki, H., C. Hui, M. Nakafuku, and H. Kondoh. 1997. A binding site for Gli proteins is essential for HNF-3β floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124:1313-1322. [DOI] [PubMed] [Google Scholar]
- 81.Sasaki, H., Y. Nishizaki, C. Hui, M. Nakafuku, and H. Kondoh. 1999. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126:3915-3924. [DOI] [PubMed] [Google Scholar]
- 82.Sisson, J. C., K. S. Ho, K. Suyama, and M. P. Scott. 1997. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell 90:235-245. [DOI] [PubMed] [Google Scholar]
- 83.Stegman, M. A., J. A. Goetz, M. Ascano, Jr., S. K. Ogden, K. E. Nybakken, and D. J. Robbins. 2004. The Kinesin-related protein Costal2 associates with membranes in a Hedgehog-sensitive, Smoothened-independent manner. J. Biol. Chem. 279:7064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stegman, M. A., J. E. Vallance, G. Elangovan, J. Sosinski, Y. Cheng, and D. J. Robbins. 2000. Identification of a tetrameric hedgehog signaling complex. J. Biol. Chem. 275:21809-21812. [DOI] [PubMed] [Google Scholar]
- 85.Stone, D. M., M. Hynes, M. Armanini, T. A. Swanson, Q. Gu, R. L. Johnson, M. P. Scott, D. Pennica, A. Goddard, H. Phillips, M. Noll, J. E. Hooper, F. de Sauvage, and A. Rosenthal. 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384:129-134. [DOI] [PubMed] [Google Scholar]
- 86.Stone, D. M., M. Murone, S. Luoh, W. Ye, M. P. Armanini, A. Gurney, H. Phillips, J. Brush, A. Goddard, F. J. de Sauvage, and A. Rosenthal. 1999. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J. Cell Sci. 112(Pt. 23):4437-4448. [DOI] [PubMed] [Google Scholar]
- 87.Taipale, J., and P. A. Beachy. 2001. The Hedgehog and Wnt signalling pathways in cancer. Nature 411:349-354. [DOI] [PubMed] [Google Scholar]
- 88.Taipale, J., M. K. Cooper, T. Maiti, and P. A. Beachy. 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418:892-897. [DOI] [PubMed] [Google Scholar]
- 89.Tay, S. Y., P. W. Ingham, and S. Roy. 2005. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development 132:625-634. [DOI] [PubMed] [Google Scholar]
- 90.te Welscher, P., A. Zuniga, S. Kuijper, T. Drenth, H. J. Goedemans, F. Meijlink, and R. Zeller. 2002. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science 298:827-830. [DOI] [PubMed] [Google Scholar]
- 91.Therond, P., G. Alves, B. Limbourg-Bouchon, H. Tricoire, E. Guillemet, J. Brissard-Zahraoui, C. Lamour-Isnard, and D. Busson. 1996. Functional domains of fused, a serine-threonine kinase required for signaling in Drosophila. Genetics 142:1181-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Therond, P. P., J. D. Knight, T. B. Kornberg, and J. M. Bishop. 1996. Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc. Natl. Acad. Sci. USA 93:4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Therond, P. P., B. Limbourg Bouchon, A. Gallet, F. Dussilol, T. Pietri, M. van den Heuvel, and H. Tricoire. 1999. Differential requirements of the fused kinase for hedgehog signalling in the Drosophila embryo. Development 126:4039-4051. [DOI] [PubMed] [Google Scholar]
- 94.van den Heuvel, M., and P. W. Ingham. 1996. Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature 382:547-551. [DOI] [PubMed] [Google Scholar]
- 95.Walterhouse, D., M. Ahmed, D. Slusarski, J. Kalamaras, D. Boucher, R. Holmgren, and P. Iannaccone. 1993. gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev. Dyn. 196:91-102. [DOI] [PubMed] [Google Scholar]
- 96.Wang, B., J. F. Fallon, and P. A. Beachy. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100:423-434. [DOI] [PubMed] [Google Scholar]
- 97.Wang, G., K. Amanai, B. Wang, and J. Jiang. 2000. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 14:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang, Q. T., and R. A. Holmgren. 2000. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development 127:3131-3139. [DOI] [PubMed] [Google Scholar]
- 99.Wilkinson, D. G., and M. A. Nieto. 1993. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225:361-373. [DOI] [PubMed] [Google Scholar]
- 100.Wolff, C., S. Roy, and P. W. Ingham. 2003. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebra fish embryo. Curr. Biol. 13:1169-1181. [DOI] [PubMed] [Google Scholar]