Abstract
The Drosophila Fused (Fu) kinase is an integral component of the Hedgehog (Hh) pathway that helps promote Hh-dependent gene transcription. Vertebrate homologues of Fu function in the Hh pathway in vitro, suggesting that Fu is evolutionarily conserved. We have generated fused (stk36) knockout mice to address the in vivo function of the mouse Fu (mFu) homologue. fused knockouts develop normally, being born in Mendelian ratios, but fail to thrive within 2 weeks, displaying profound growth retardation with communicating hydrocephalus and early mortality. The fused gene is expressed highly in ependymal cells and the choroid plexus, tissues involved in the production and circulation of cerebral spinal fluid (CSF), suggesting that loss of mFu disrupts CSF homeostasis. Similarly, fused is highly expressed in the nasal epithelium, where fused knockouts display bilateral suppurative rhinitis. No obvious defects were observed in the development of organs where Hh signaling is required (limbs, face, bones, etc.). Specification of neuronal cell fates by Hh in the neural tube was normal in fused knockouts, and induction of Hh target genes in numerous tissues is not affected by the loss of mFu. Furthermore, stimulation of fused knockout cerebellar granule cells to proliferate with Sonic Hh revealed no defect in Hh signal transmission. These results show that the mFu homologue is not required for Hh signaling during embryonic development but is required for proper postnatal development, possibly by regulating the CSF homeostasis or ciliary function.
The Hedgehog (Hh) signaling pathway is highly conserved in most animals, helping to shape cell fate decisions and proper embryonic development (for a review, see reference 33). Improper activation or disruption of the Hh pathway is associated with a variety of development abnormalities, as well as several types of cancer (for reviews, see references 51, 65, and 85). Genetic and biochemical studies using Drosophila melanogaster have helped to identify the molecular components of the Hh pathway and potential mechanisms by which they interact to transmit the Hh signal. Many of these components are conserved in vertebrates (33), suggesting that the mechanism by which the Hh signal is generated and transmitted is fundamentally similar in invertebrates and vertebrates.
At the cell surface, the Hh receptor, Patched (Ptch), a 12-pass transmembrane protein with homology to bacterial proton-driven molecular transporter proteins (86), acts to repress the pathway in the absence of ligand through inhibition of Smoothened (Smo), a seven-pass transmembrane protein (8, 33). The mechanism by which Ptch represses Smo is poorly understood but appears to be catalytically mediated (86). Upon Hh binding, Ptch-mediated repression of the pathway is relieved, leading to a posttranscriptional increase in Smo protein levels, Smo phosphorylation by PKA and CKI, and stabilization of Smo on the cell surface (1, 14, 34, 38, 95, 98). The carboxy-terminal tail of Smo is thought to direct downstream signaling through Hh signaling complexes (HSC), composed of the atypical microtubule-binding kinesin Costal 2 (Cos2), the zinc finger transcription factor Cubitus Interruptus (Ci), and the Drosophila serine-threonine kinase Fused (dFu) (37, 48, 57, 72, 74, 80, 81).
Following its discovery as a segment polarity gene (56, 67), dFu was found to encode a serine-threonine kinase involved in transmitting the Hh signal (69). While dFu plays a critical role in inducing the expression of several Hh target genes (2, 58, 76), some Hh-dependent signals appear to be transmitted independently of dFu (83, 90). The dFu protein consists of an amino-terminal kinase domain followed by a long carboxy-terminal regulatory domain. Homozygous dFu mutants are not viable, but dFu mutants carried in a hemizygous state display a “fused wing vein” phenotype, reflecting disruptions in Hh-dependent patterning of the wing (69). The severity of the dFu phenotypes correlates with the type of mutation, with mutations in the kinase domain causing less severe wing vein fusion phenotypes than mutations in the regulatory domain (70, 89). Interestingly, suppressor of fused [su(fu)], was identified in a modifier screen as a gene which, when mutated, is capable of rescuing the phenotypes of some of the dFu mutants (68, 70).
Despite somewhat limited sequence similarity, potential vertebrate counterparts have been identified for all of the Drosophila Hh signaling components. Evidence for functional conservation has been firmly established in vitro and in vivo through the use of knockout mice for many of the mammalian genes, including smo, ptch, the Hh-releasing protein gene dispatched, and the ci homologues gli1, gli2, and gli3 (3, 5, 16, 17, 21, 42, 49, 50, 53, 64, 97). However, the role of several intracellular components of the pathway still needs to be clarified in mammals. This is especially the case for components involved in the regulation of Gli activity, downstream of Smo, such as Fu, Su(fu), and Cos2. At least two kinesin-like candidates have been identified as potential orthologs of Cos2 and remain to be fully characterized in vitro and in vivo (40, 41, 87). In contrast, single Su(fu) (15, 82) and Fu genes (54) are readily identifiable in the mouse and human genomes. Su(fu) is highly conserved among all species, and its capacity to repress Gli activity has been well characterized in vitro (13, 15, 66, 82, 93).
While the mammalian Fu sequences are the most closely related genes to dFu identified in the genomes, the homology at the protein level is restricted to the kinase domain, where human Fused (hFu) and dFu share 52% identity and 20% similarity. The carboxy-terminal regions in mammalian Fu homologues are roughly 500 additional amino acids longer and show limited homology outside of short stretches of peptides (12). Similar to the mammalian Fu homologues, the zebra fish Fu (zfFu) homologue is only conserved at the sequence level with dFu within the kinase domain. The regulatory domain of zfFu is more closely related to the mammalian Fu homologues; however, the homology remains weak (21% identity, 12% homology). In vitro, hFu was shown to interact with Su(fu) and the Gli proteins and was shown to activate the Hh pathway, at least in part through its ability to counteract the cytoplasmic tethering function of Su(fu) (54, 60). To date, only the zfFu homologue has been characterized in vivo and found to play a role in regulating Hh signaling in the myotome, where it is required for the Hh-dependent specification of muscle pioneer (MP) cells (93). As in Drosophila, loss of both Su(fu) and zfFu abrogates the effect loss of zfFu alone has on MP cells (93), suggesting that the Fu homologues may be functionally conserved in vertebrates.
To validate the role mFu plays in the Hh pathway, we generated fused knockout mice. fused knockout mice are born in Mendelian ratios and appear normal at birth. However, within the first week, fused knockout mice fail to thrive and display a severe growth-retarded phenotype characterized by a communicating (nonobstructive) form of hydrocephalus to which they eventually succumb by 2 weeks of age. fused knockout mice also display bilateral suppurative rhinitis, a massive inflammation in the nasal cavity, a phenotype observed in other mice with secretory defects and hydrocephalus. High expression of fused in the brain, spinal cord, and nasal epithelium correlates with the observed pathologies, implicating mFu in the homeostasis of secretory and/or ciliated tissues. Interestingly, loss of mFu does not appear to impact the transmission of the Hh signal in vitro or in vivo. Implications for the nature of Hh signaling and related diseases are discussed.
MATERIALS AND METHODS
Abbreviations.
The abbreviations used in this report are as follows: β-Arr2, β-arrestin 2; Ci, cubitus interruptus; CK-Iα, casein kinase Iα; Cos2, Costal-2; CNS, central nervous system; CSF, cerebral spinal fluid; dFu, Drosophila Fused; EGL, external germinal layer; GNP, granule neuronal precursors; GRK2, G protein-coupled receptor kinase 2; GSK3, glycogen synthase kinase 3; HFH4, hepatocyte nuclear factor/forkhead transcription factor 4; Hh, Hedgehog protein; hh, Hedgehog gene; HSC, Hedgehog signaling complex; hFu, human Fused; IFT, intraflagellar transport protein; Kif, kinesin superfamily protein; Mdnah5, mouse axonemal dynein heavy chain 5; Mf1, mesoderm/mesenchyme forkhead 1; MRI, magnetic resonance imaging; Msi1, Musashi 1; PKA, protein kinase A; Ptch, Patched; Shh, Sonic Hedgehog; mFu, mouse Fused; Smo, Smoothened; SPAG6, sperm-associated antigen 6; Su(fu), suppressor of Fused; TM, transmembrane; 5-HT, 5-hydroxytryptophan; zfFu, zebra fish Fused.
Gene targeting.
To generate the fused knockout-targeting construct, a 129/Sv mouse genomic library (Stratagene, Inc.) was screened for sequences homologous to hFu. Genomic constructs were then used to clone a 5′ short arm and a 3′ long arm on either side of a PGK-neomycin cassette and a herpes simplex virus thymidine kinase cassette flanking the 3′ end of the long arm (Fig. 1A). The short arm consists of a 1,159-bp BamHI-NcoI fragment spanning 5′ noncoding exons 1 and 2 and part of exon 3 up to the initiation codon (Fig. 1A). The long arm consisted of a 6,812-bp PacI-NotI fragment spanning exons 13 to 15 (Fig. 1A). Properly integrated, this construct removes most of exon 3 through exon 12 of the mFu gene (Fig. 1A).
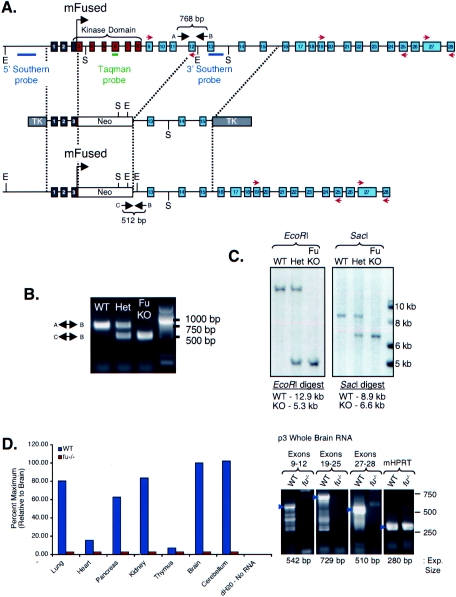
FIG. 1.
Generation of fused knockout mice. (A) Schematic of the fused gene targeting construct. The wild-type mouse fused locus is depicted (top), with dashed lines indicating the regions of homology contained in the targeting construct (middle) and the resulting knockout locus following proper recombination (bottom). Exons 1 and 2 are 5′ noncoding (gray), exons 3 to 9 encode the kinase domain of mFu (red), and exons 10 to 28 encode the long carboxy-terminal regulatory region (blue). Insertion of the neo cassette into the fused locus results in insertion after the initiation codon in exon 3, resulting in a nonsense transcript. Restriction enzyme sites EcoRI (E) and SacI (S) are denoted, the locations of 5′ and 3′ Southern blotting probes are depicted in blue, the location of the TaqMan primer-probe set is indicated in green, the positions of RT-PCRoligonucleotides are indicated as red arrows above and below the exons to which they anneal, and lettered arrows indicate the locations of oligonucleotides used to genotype fused knockout mice. (B) Genotyping of fused knockout (KO) mice. The upper band corresponds to a 768-bp product formed with oligonucleotides A and B from the intact fused locus, while the lower band corresponds to a 512-bp product formed with oligonucleotides C an B from the recombined fused locus. WT, wild type; Het, heterozygous. (C) Southern blot assay confirmation of homologous recombination. EcoRI digestion and Southern blotting using the 5′ probe confirmed proper recombination of the short arm, while SacI digestion and Southern blotting using the 3′ probe confirmed the proper recombination of the long arm. (D) Expression analysis of fused in wild-type (WT, blue) and fused mutant (fu−/−, red) p7 tissues by quantitative RT-PCR. Data are graphed as a percentage, relative to the expression levels to the ubiquitous transcript RPL19, with brain RNA samples set to 100%. Standard RT-PCR results are shown for exons 9 to 12, 19 to 25, and 27 to 28 and mouse hypoxanthine phosphoribosyltransferase amplified from p3 brain RNAs. The expected RT-PCR products are indicated by blue arrowheads. dH2O, distilled water; Exp., expected.
The BamHI-NotI linearized construct was electroporated into 129/Sv CJ7 embryonic stem (ES) cells (84) that were seeded on feeder layers of gamma-irradiated (3,000 cGy) mouse embryo fibroblasts and treated with 300 μg/ml of G418 and 2 μM ganciclovir. Roughly 1 in 100 ES cell clones contained a disrupted fused gene, 5 of which were correctly integrated. Two correctly recombined knockout ES cell lines were injected into C57BL/6 C2 blastocysts to generate chimeric animals. Two independent lines (M16 and 20.7.8) passed the mutation of fused to their offspring.
Mice and animal husbandry.
Founder lines were backcrossed onto the C57BL/6 background. Intercrosses between heterozygous offspring from each generation were used to generate fused knockout mice and analyzed for phenotype. Ptch1D11 is a weak ptch1 allele that was made during the generation of Ptch1KO1 (21, 59). Ptch1D11 mice are viable, and LacZ staining from the associated β-galactosidase cassette is a faithful measure of endogenous ptch1 expression. Pthh1D11 mice were bred to fused heterozygous females, and Ptch1D11 fused heterozygotes (Ptch1D11 fu+/−) were generated. These animals were bred to fused heterozygous animals in timed pregnancies to generate Ptch1D11 fu+/+ and Ptch1D11 fu−/− embryos for analyzing the expression of ptch1 by monitoring β-galactosidase activity during embryonic development.
The Genentech Institutional Animal Care and Use Committee approved all animal protocols. Mice were maintained in a barrier facility at Genentech, Inc., conforming to California State legal and ethical standards of animal care. All animals were properly anesthetized during any treatment and, when required, euthanized by ethically acceptable means.
Genotyping.
Genotyping was initially verified in each line by Southern blotting on the short arm with an EcoRI digestion that reveals a 12.9-kb product in wild-type mice and a 5.3-kb product in fused knockout mice and on the long arm with a SacI digestion that reveals an 8.9-kb product in wild-type mice and a 6.6-kb product in fused knockout mice (Fig. 1C). DNA was extracted and prepared by proteinase K digestion, followed by phenol-chloroform extraction and ethanol (EtOH) precipitation. Restriction enzyme digestions were performed according to the manufacturer's recommendations (NEB BioLabs, Inc.), and samples were run on 0.8% agarose gels, followed by standard Southern blotting using [32P]dCTP random primed DNA labeled probes for either the short or the long arm of the fused knockout construct (9).
Genotyping was more routinely performed by a PCR wherein both DNA preparation and PCR amplification were done using the Extract-N-Amp kit (Sigma, Inc.). Genotyping of fused knockout mice made use of three primers, two forward and one reverse, resulting in two differential products. Primers Fu-1 and Fu-2 (Fig. 1A) produce a 768-bp product in wild-type and fused heterozygous DNA samples. Primer Fu-3 maps to the neo cassette and pairs with primer Fu-2 (Fig. 1A) to produce a 512-bp product in fused knockout and heterozygous DNA samples. PtchD11 mice were genotyped with primers for the lacZ cassette, producing a product of 501 bp. Primer sequences were as follows: Fu-1, 5′-GGA GGT TTT ACT GAA TCG AGG G-3′; Fu-2, 5′-CCA GCA CTC AGG AGG CAA GTA C-3′; Fu-3, 5′-CCA CTT GTG TAG CGC CAA GTG C-3′; LacZ-forward, 5′-CGG TGA TGG TGC TGC GTT GG-3′; LacZ-reverse, 5′-GAA TCA GCA ACG GCT TGC CG-3′.
Gene expression analysis. (i) Reverse transcription (RT)-PCR.
Tissues were dissected from postnatal day 1 (p1), p3, p5, and p7 neonates and snap-frozen in liquid nitrogen. RNA was prepared using 1.2 ml of RLT buffer (QIAGEN) to lyse samples and homogenized using a Polytron PT1200 tissue homogenizer (Kinematica). One milliliter of homogenized sample was then used for subsequent RNA preparation using the RNeasy kit (QIAGEN). Total RNA was used to generate first-strand cDNAs using PowerScript reverse transcriptase (Clontech) according to the manufacturer's recommendations. Standard PCRs were then set up using the cDNA from the RT reaction and oligonucleotides spanning exons in the mouse fused gene. The sequences of oligonucleotides used for RT-PCR analysis are as follows: exon 8 forward, GCA CCC CAT TTA CTA GTC GCC T-3′; exon 11 reverse, 5′-GCT GAC TCT GGA TGC GCT GGC A-3′; exon 18 forward, 5′-GAG CGC CTG TGC CAT ATT CTG-3′; exon 24 reverse, 5′-GCT CAG GTG GTG CAA GAA GCT-3′; exon 26 forward, 5′-GCA CTA CAG AGT CAG TCA GGA-3′; exon 27 reverse, 5′-GCT GTG GGT TGG CCT CAG GAG TT-3′; mHPRT forward, 5′-GCT GGT GAA AAG GAC CTC T-3′; mHPRT reverse, 5′-CAC AGG ACT AGA ACA CCT GC-3′.
(ii) Real-time PCR for quantitative gene analysis.
Fifty micrograms was analyzed using the real-time RT-PCR ABI Prism 7700 sequence detection system (Applied Biosystems, Inc.) according to the manufacturer's recommendations. Levels of assayed genes were normalized to mRPL19 RNA abundance. The sequences of the amplification primers and TaqMan probes (labeled with carboxyfluoroscein and N,N′-tetramethylrhodamine) are as follows: mFu forward, 5′-GTA TGG GGC GCT CTT ACG-3′, mFu reverse, 5′-ATT GAA GGT GCA CGA AAA GTA G-3′; probe, 5′-CCT GAA CCA CAG CCA CCA GGT C-3′; mPtch1 forward, 5′-GGA GTG GGT CCA TGA CAA A-3′, mPtch1 reverse, 5′-CTC TGC TGC TGG GAT TCT C-3′; probe, 5′-CGA CTA CAT GCC AGA GAC CAG GCT-3′; mGli1 forward, 5′-GCA GTG GGT AAC ATG AGT GTC T-3′, mGli1 reverse, 5′-AGG CAC TAG AGT TGA GGA ATT GT-3′, probe, 5′-CTC TCC AGG CAG AGA CCC CAG C-3′; mSmo forward, 5′-GGC TGG GAT CCA TTC ATT-3′; mSmo reverse, 5′-GTC CGA GTC TGC ATC CAA-3′; probe, 5′-CCG CAC TAA CCT AAT GGA GGC TGA GAT-3′.
Histology.
Tissues were routinely fixed with 10% neutral buffered formalin overnight. Tissues were then dehydrated through a series of EtOH solutions, delipidated in mixed xylenes, and embedded in paraffin (55). Tissues were then sectioned at 5 to 20 μm and mounted on Superfrost-Plus slides (Fisher Scientific). For histological staining, sections were deparaffinized and stained with hematoxylin and eosin (H&E) using standard histological protocols. Slides were photographed using a Zeiss Axioplan microscope and a SPOT camera (Diagnostic Instruments).
β-Galactosidase staining.
β-Galactosidase activity was assayed as previously described (55). Briefly, samples were fixed with a glutaraldehyde fixative (0.1 M phosphate buffer [pH 7.3], 0.2% glutaraldehyde [Sigma G6257], 5 mM EGTA [from a 0.1 M stock at pH 8.0], 2 mM MgCl2) for 20 min at 4°C for embryos under E11.5 or 60 to 90 min for larger samples. Samples were washed three times for 10 min each at room temperature in X-Gal wash buffer (0.1 M phosphate buffer [pH 7.3], 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 [Sigma N 6507]). Samples were then incubated into X-Gal staining solution (0.1 M phosphate buffer [pH 7.3], 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-Gal) at 37°C until the desired color change was apparent (2 h to overnight). Samples were washed in X-Gal wash buffer three times for 10 min each at room temperature and then postfixed in 4% paraformaldehyde in 1× phosphate-buffered saline (PBS) for 2 h at room temperature. Samples were transferred to 80% glycerol in 1× PBS for storage and imaging. Embryos were imaged on a Leica MZFLIII microscope using a SPOT camera (Diagnostic Instruments). Alternatively, samples were sectioned at 5 μm after paraffin embedding, as outlined above.
Radioisotopic in situ hybridization.
Tissues were fixed in 10% neutral buffered formalin and paraffin embedded. Five-micrometer sections were deparaffinized, deproteinized in 4 μg/ml of proteinase K for 30 min at 37°C, and then further processed for in situ hybridization as previously described (27, 47). [33P]dUTP-labeled sense and antisense probes were hybridized to the sections at 55°C overnight. Nonhybridized probe was removed by incubation in 20 μg/ml RNase A for 30 min at 37°C, followed by a high-stringency wash at 55°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 h and dehydration through graded EtOH solutions. The slides were dipped in NBT2 nuclear track emulsion (Eastman Kodak), exposed in sealed plastic slide boxes containing desiccant for 4 to 6 weeks at 4°C, developed, and counterstained with H&E.
Probe templates for mouse ptch1, gli1, and fused were PCR amplified using the following primer sets: mPtch1 upper primer, 5′-CCA ATG GCC TAA ACC GAC TGC-3′; mPtch1 lower primer, 5′-CCC ACG GCC TCT CCT CAC A-3′ (generating a 771-bp probe corresponding to nucleotides 3518 to 4289 of the sequence with accession number U46155); mGli1 upper primer, 5′-GCT GAA GTC AGA GCT GGA TAT G-3′; mGli1 lower primer, 5′-GAC AGC CTT CAA ACG TGC AC-3′ (generating a 393-bp probe corresponding to nucleotides 703 to 1096 of the sequence with accession number AF026305); mFused upper primer, 5′-GCA TCA GCT CTA GGC AAT CTG-3′; mFused lower primer, 5′-GG AAA TCT TGG CAA CAG TAG G-3′ (generating a 369-bp probe corresponding to nucleotides 3697 to 4065 of the sequence with accession number BC061470). In addition, the upper and lower primer pairs had 27-nucleotide extensions appended to the 5′ ends encoding T7 RNA polymerase and T3 RNA polymerase promoters, respectively, for generation of sense and antisense transcripts. Tissue sections were processed and hybridized as previously described (27).
Immunofluorescence assay.
For analysis of the neural tube, embryos were collected at embryonic day 10.5 (E10.5), fixed in 4% paraformaldehyde in 1× PBS for 20 min, and then allowed to sink in 30% sucrose in 1× PBS overnight. Subsequently, embryos were embedded in OCT solution (Tissue Tek) and snap-frozen. Frozen sections were taken using a Leica cryotome at 5 μm per section. Immunofluorescence assay of tissue sections was done as previously described (28-30), using mouse anti-MNR2, mouse anti-Lim1/2, mouse anti-Lim3, mouse anti-Islet1, mouse anti-Pax6, mouse anti-Pax7, mouse anti-Nkx2.2, and rabbit anti-Nkx6.1 (Developmental Studies Hybridoma Bank, University of Iowa) to stain for specification of neural populations in the developing neural tube. Hybridomas and monoclonal antibodies developed by T. Jessell, J. Jensen, and A. Joyner were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa Department of Biological Sciences (Iowa City). Sections were subsequently stained with Cy3-conjugated donkey anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.), followed by mounting with Vectashield mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc.), and photos were taken using an Olympus BX61 microscope using MetaMorph analysis software (Universal Imaging).
Whole-mount immunofluorescence was done on E13.5 embryos that were collected, fixed in 4% paraformaldehyde in 1× PBS for 20 min, and subsequently stained with rabbit anti-5-HT (Serotec) to stain serotonergic neurons. Embryos were subsequently stained with Cy3-conjugated donkey anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, Inc.), and photos were taken using a Leica MZFLIII microscope using a SPOT camera (Diagnostic Instruments).
Skeletal analysis.
Mouse embryos and neonates were euthanized by CO2 asphyxiation and the skin and organs removed. Skeletal samples were then fixed in 99% EtOH for 24 h then transferred to acetone for an additional 24 h. Skeletons were then stained with alcian blue and alizarin red (1 volume 0.3% alcian blue [in 70% EtOH], 1 volume 0.1% alizarin red [in 95% EtOH], 1 volume glacial acetic acid, 17 volumes of 70% EtOH) for 3 days at 37°C. Skeletons were rinsed thoroughly with water and then transferred to 1% KOH at room temperature. Samples were monitored for clearance of tissue, changing the KOH as needed. Following clearance of the tissue, the skeletons were transferred through a series of glycerol (20%, 50%, and then 80%)-1% KOH washes. Measurements of bone lengths were taken, and skeletons were photographed on a Leica MZFLIII microscope using a SPOT camera (Diagnostic Instruments).
μCT.
The mouse embryos were imaged with a μCT40 (SCANCO Medical, Basserdorf, Switzerland) X-ray micro-computed tomography (μCT) system. A sagittal scout image, comparable with a conventional planar X-ray, was obtained to define the start and end points for the axial acquisition of a series of μCT image slices. The locations and number of axial images were chosen to provide complete coverage of the embryo. The embryos were imaged with air as the background medium. The μCT images were generated by operating the X-ray tube at an energy level of 50 kV, a current of 160 μA, and an integration time of 300 ms. Axial images were obtained at an isotropic resolution of 16 μm.
The relationship between the image intensity values and bone mineral density was assumed to be linear and was obtained by scanning a 97% pure hydroxyapatite sample (2.91 g/cm3). Three-dimensional surface renderings were created from the μCT data using the Analyze software package (AnalyzeDirect Inc., Lenexa, KS). Applying a bone mineral density threshold of 0.45 g of hydroxyapatite/cm3 generated bone surface renderings.
MRI.
MRI experiments were performed with a Varian 7T Unity Inova MR imaging system (Varian Inc., Palo Alto, CA) equipped with ±100 G/cm self-shielding gradients and a quadrature birdcage coil. Mice were anesthetized with isoflurane, and their body temperature was maintained at 37°C with an automated warm airflow system. MRI data consisted of 30 contiguous, sagittal, 0.33-mm-thick slices (field of view, 20 by 35 mm; pixel resolution, 256 by 256). A T2-weighted spin echo imaging sequence was employed to image the mice (parameters were repetition time = 3 s, echo time = 40 ms, and number of excitations = 4). Brain regions that are hyperintense in these images are consistent with the presence of CSF and indicative of ventricle size.
Granule cell proliferation assays.
Cerebellar granule cells were isolated as described previously (73). Briefly, cerebella from p5 neonates were dissected in HHGN buffer (1× Hanks' balanced salt solution, 0.25% glucose, 3 mg/ml bovine serum albumin [fraction V], 15 mM HEPES, 4.1 mM sodium bicarbonate, 1.5 mM MgSO4, pH 7.4) and meninges removed and then washed three times with HHGN buffer. Cerebella were then transferred to 10 ml dissociation buffer (HHGN buffer with 10 mg/ml trypsin [Sigma T5266] sterilized with a 0.2-μm-pore-size filter) for 15 min at room temperature, followed by three washes with HHGN buffer. Samples were then thoroughly triturated to produce a single-cell suspension in DNase buffer (HHGN buffer, DNase I type IV [Sigma], 3 mM MgSO4) and then centrifuged at 1,000 rpm for 5 min and resuspended in plating medium (Neurobasal medium [GIBCO], 1× B27 supplement [GIBCO], 25 mM KCl, 2 mM glutamine, 1× penicillin-streptomycin). Cells were counted and then seeded at 3 × 105 cells/cm2 on poly-d-lysine-coated plates in the presence or absence of Shh-N modified by the addition of an eight-carbon octyl chain (N-octylmaleimide) to the N-terminal cysteine (octyl-Shh) (88).
Cells were incubated for 24 and 48 h and pulsed with [3H]thymidine 5 h prior to harvesting onto UniFilter-96,GF/C filters (Perkin-Elmer) using the Filtermate 196 (Packard). Filters were dried overnight and read on a Packard TopCount microplate scintillation counter following addition of 40 μl of MicroScint 0 (Packard). Data were averaged and normalized to untreated controls.
Gli-luciferase reporter assays and siRNA knockdown.
The 9x-Gli-binding site (BS)-Luciferase reporter assay in mouse C3H/10T1/2 cell lines has been previously described (52). C3H/10T1/2-derived S12 cells stably expressing the 9x-Gli-BS-Luciferase reporter (18) growing in fibroblast medium (high-glucose Dulbecco modified Eagle medium with 10% fetal bovine serum, 10 mM HEPES, and 2 mM glutamine) were transfected with 60 nM small interfering RNA (siRNA) using Lipofectamine 2000 (Invitrogen) in a 96-well format. After 64 h, the medium was replaced with low-serum fibroblast medium (0.5% fetal bovine serum albumin) with or without 200 ng/ml of octyl-Shh (88). Luciferase assays were conducted 24 h later using Steadylite (Perkin-Elmer) according to the manufacturer's instructions. The siRNA sequences used to knock down gene expression were mSmo (5′ GAA GAG CAA GAT GAT CGC CAA 3′) and mFu (5′ CAG GAA GAC GAC CTG CTA CTA 3′). Knockdown of gene expression was confirmed by quantitative PCR as indicated above.
RESULTS
Generation and gross analysis of fused knockout mice.
A thorough analysis of the human fused gene found that it maps to chromosome 2, where the protein is encoded by two 5′ noncoding exons and 27 coding exons, several of which appear to be differentially utilized (60). The mouse fused gene maps to chromosome 1 and is likewise predicted to have two 5′ noncoding exons, followed by 26 coding exons (Fig. 1). We used homologous recombination target mutagenesis in ES cells to generate mice lacking the 3′ coding portion of exon 3 through exon 12 of the fused gene, a deletion that removes the entire kinase domain and part of the putative regulatory domain (see Materials and Methods and Fig. 1). Two knockout lines (M16 and 20.7.8) were produced with identical phenotypes (Fig. 1; all data presented are from line M16). Quantitative RT-PCR from p7 neonatal tissue indicated that fused is expressed weakly in the heart and thymus; at moderate to high levels in a number of tissues, including the lungs, pancreas, and kidneys; more highly expressed in the brain and cerebellum; and very highly expressed in the testes (Fig. 1D). These data were consistent with the radioisotopic in situ hybridization for murine fused transcripts (data not shown) (54). Expression of fused mRNA in fused knockouts was analyzed by standard and quantitative RT-PCRs. Quantitative RT-PCR revealed that expression from exon 6, within the deleted region, was abolished, as expected (Fig. 1D). To determine whether transcripts could be detected from nondeleted exons, standard RT-PCR was performed for several exonic regions throughout the fused mRNA. Standard RT-PCR with p3 brain RNA revealed that no significant expression of 3′ nondeleted fused exons could be detected in fused knockout animals (Fig. 1D).
The fused knockout lines were backcrossed into the C57BL/6 background. Upon each successive generation into the C57BL/6 background, fused heterozygous mice were intercrossed and analyzed for phenotype. The fused knockout mice were produced at Mendelian ratios and appeared healthy for the first 2 to 3 days following birth; however, the mice displayed a failure-to-thrive phenotype characterized by severe growth retardation with prominent doming of the head (Fig. 2A and B). Upon ambulation, the knockout mice were ataxic, stabilizing themselves with the tail and splayed legs, and had a slight head tremor. Detailed histological analysis of fused knockout neonates ranging from p1 to p9 revealed an early onset of hydrocephalus at p3 (data not shown).
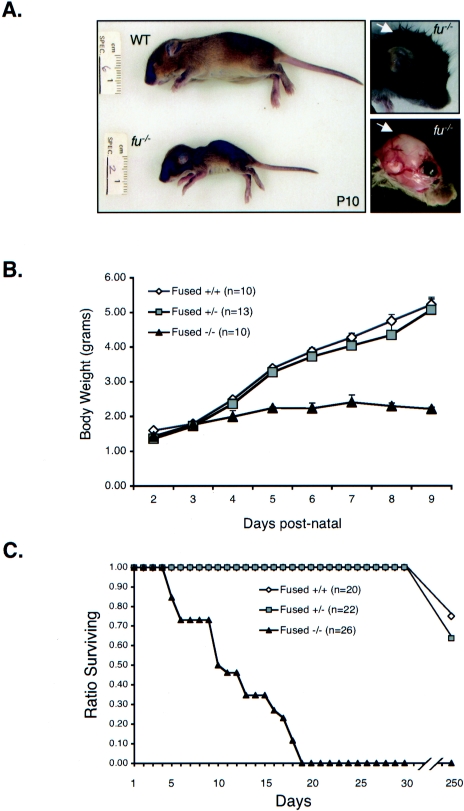
FIG. 2.
Gross observations of fused knockout mice. (A) Physical appearance of the fused knockout mouse at p10. The fused knockouts (fu−/−) are dramatically runty compared to wild-type (WT) littermate controls and display apparent hydrocephalus, as indicated by domed crania (right side). The skin from the neonate in the right top image was removed and photographed (lower right image) to reveal cranial swelling. (B) Body weights of wild-type (WT, open diamonds), fused heterozygous (fu+/−, gray squares), and fused knockout (fu−/−, black triangles) animals following birth. (C) Survival plot of wild-type (WT, open diamonds), fused heterozygous (fu+/−, gray squares), and fused knockout (fu−/−, black triangles) animals following birth.
fused knockout mice often develop overt characteristics of hydrocephalus, as indicated by a domed skull (Fig. 2A and 3A). Figure 3A shows mice generated from the early intercrosses that produced two fused knockout mice that survived over 2 months. Although the hydrocephalus in these mice was not as severe as in later backcrossed generations, these mice remained runted and ataxic. Both surviving knockouts were males and infertile. Once backcrossed into the C57BL/6 background a second time, no escape was observed and the median time of survival of fused knockouts was 10 days (Fig. 2C). Later generations of fused knockout mice (six to eight generations into the C57BL/6 background) had such severe hydrocephalus by p10 that the cranial vault had only a fluid-filled sac, and upon histological evaluation only a thin rim of cerebral tissue was identifiable. Onset of hydrocephalus coincides with the onset of growth arrest and ataxia. As mice were further crossed into the C57BL/6 line, progression to overt hydrocephalus shortened while growth retardation was observed in all genetic backgrounds.
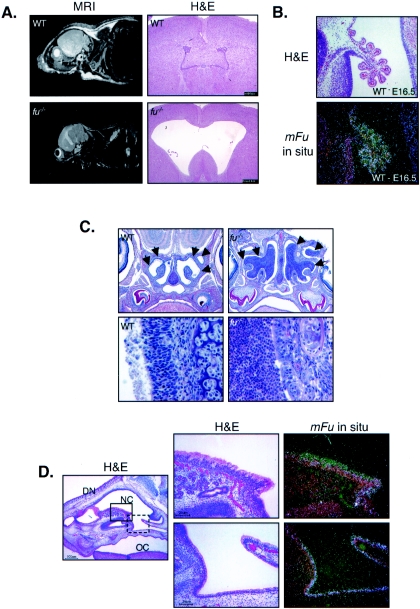
FIG. 3.
Areas of high fused expression correlate with hydrocephalus and suppurative rhinitis. (A) fused knockout mice develop a communicating form of hydrocephalus. Wild-type (WT) and fused knockout (fu−/−) mice are shown by MRI on the left, and brain sections stained with H&E are shown on the right. fu−/− mice develop a progressive, communicating form of hydrocephalus. (B) The fused mRNA is expressed highly in the choroid plexus. The top image shows H&E staining of an E16.5 mouse brain, while the bottom image shows the anti-fused radiolabeled in situexposure (white areas are positively stained with fused antisense probes). (C) fu−/− mice develop bilateral suppurative rhinitis characterized by massive infiltration by neutrophils. The top images show saggital sections of WT and fu−/− nasal cavities at a magnification of ×10, whereas the lower images show the nasal epithelial border in WT and fu−/− mice at a magnification of ×40. WT nasal cavities have a well-defined border at the nasal epithelium with no infiltrating cells (arrows), whereas fu−/− nasal cavities are filled with infiltrating neutrophils (arrows). (D) The fused mRNA is expressed in normal nasal epithelium. The leftmost image is an H&E-stained section at a magnification of ×10. The dorsum of the nose (DN), the nasal cavity (NC), and the oral cavity (OC) are indicated. The solid box corresponds to the H&E and fused radioisotopic in situ hybridization images (magnification, ×40) in the top right images, whereas the dashed box corresponds to the bottom right images.
To determine whether the reduced growth of fused knockout mice was due to behavioral defects or competition with littermates, feeding behavior was monitored. The fused knockout mice fed normally, as confirmed by direct observation and the presence of milk in their stomachs. Furthermore, the reduced growth of fused knockout mice cannot be attributed to competition with healthy littermates, as fused knockouts isolated with their mothers at p1 still suffer a growth crisis and do not survive longer than fused knockouts caged with littermates (data not shown).
Detailed histological analysis of p7 to p9 neonates indicated that most nonneuronal tissues, though smaller than wild-type controls, were normally formed, with the exception of the thymus and spleen, which displayed atrophy with massive apoptosis of lymphocytes, particularly in the thymus (see supplemental material at http://share-qa.gene.com). While Shh signaling has been implicated in driving the development of immature thymocytes (24, 61, 79), mFu is unlikely to play a role in this process as adoptive transfer of fetal liver cells from fused knockout embryos was sufficient to restore all B- and T-cell populations in irradiated recipients (see supplemental material at http://share-qa.gene.com).
The lungs and kidneys of p7 fused knockout mice were immature, consistent with overall growth retardation, and were similar to p2 to p3 wild-type neonatal lung and kidney samples by histology (data not shown). These data may indicate a role for mFu in the proper postnatal development of these tissues. However, it is possible that these defects are secondary to the hydrocephalus, as observed with the thymic hypoplasia, rather than primary defects.
Analysis of skeletal formation in fused knockout mice.
To determine whether the hydrocephalus observed in the fused knockout mice occurs as a result of altered cranial development that would obstruct CSF flow, E16.5 and E18.5 embryos and neonates from fused heterozygous intercrosses were subjected to skeletal analysis, μCT analysis, and standard serial sectioning and histology.
Skeletal preparations from fused knockout embryos and neonates showed no defect in skeletal formation (see supplemental material at http://share-qa.gene.com). As hydrocephalus in other knockout and mutant mice has been found to be associated with defects in skull formation (22, 43), the skulls and spinal columns of fused knockout mice were analyzed for developmental defects. Of particular interest, the bones of the skull were all present and properly located. Furthermore, there were no defects in the fusion or ossification of bones that could account for a blockage in CSF. There were also no defects observed in the axial/appendicular skeleton or in digit formation (see supplemental material at http://share-qa.gene.com), processes that normally depend on Indian Hh and Shh signaling (33).
To further evaluate the skeletal structure of the skull, μCT was performed on E16.5 and E18.5 embryos. Interestingly, the skulls of fused knockout mice are slightly brachycephalic (see supplemental material at http://share-qa.gene.com); however, this subtle effect is unlikely to disrupt the flow of CSF. Expression of fused is observed in the developing skull by in situ hybridization, suggesting that mFu could play a role in the development of the skull (see supplemental material at http://share-qa.gene.com).
Lastly, no obvious physical blockage was observed in mice subjected to MRI analysis (Fig. 3A) or in histological sectioning through several brain and spinal column samples from fused knockout neonates (p1 to p7), despite the dramatic onset of hydrocephalus within this time frame (data not shown). These data indicate that fused knockout mice develop a communicating (nonobstructive) form of hydrocephalus, most likely due to overproduction of and/or disruption in the reabsorption of CSF (20).
fused expression in the brain and nasal cavity is associated with hydrocephalus and rhinitis.
Hydrocephalus could results from either obstructive skeletal deformations, as observed in the Mf1 knockout mice (44), or from secretory or ciliary defects, as seen in E2F-5, p73, Mdnah5, Msi1, and HFH4 knockouts (4, 32, 46, 75, 94). Both E2F-5 and p73 are highly expressed in ependymal cells and the choroid plexus in the brain, tissues involved in CSF production and circulation (20), and deletion of either gene results in progressive hydrocephalus (46, 94). To determine whether the fused gene showed a similar expression pattern in these tissues, radioisotopic in situ hybridizations were performed on brain samples from wild-type mice. Similar to E2F-5 and p73 knockout mice, the mFu gene is expressed at high levels in both the choroid plexus and ependymal cells (Fig. 3B), linking fused expression to CSF-producing tissues.
In addition to defects in the brain, mice deficient in p73, Mdnah5, or Msi1 display suppurative rhinitis or otitis, massive inflammations of the nasal cavity or ear canal characterized by infiltrating neutrophils (32, 75, 94). Similar to these mutants, fused knockout animals display bilateral suppurative rhinitis with infiltrating neutrophils (Fig. 3C). As in the brain, mFu expression is exquisitely associated with the secretory mucosal epithelium lining the nasal cavity, a tissue that is destroyed by the inflammation observed in fused knockouts (Fig. 3D).
Fused is not required for Hh signal transduction in vivo or in vitro.
The fused knockout mice do not display any obvious Hh-related morphological phenotypes, such as holoprosencephaly, or skin and limb patterning defects. However, it is possible that the loss of mFu imparts a more subtle effect on the transmission of Hh signals. To further characterize the Hh-dependent signaling events in fused knockout mice, we analyzed (i) neural tube patterning at E10.5 and specification of serotonergic neurons in the hindbrain at E13.5, (ii) the expression of Hh target genes (ptch1 and gli1) by quantitative RT-PCR and radioisotopic in situ hybridization, (iii) the proliferation of cerebellar granule cells in response to Shh, and (iv) ptch1 expression as monitored by LacZ from PtchD11 at E11.5.
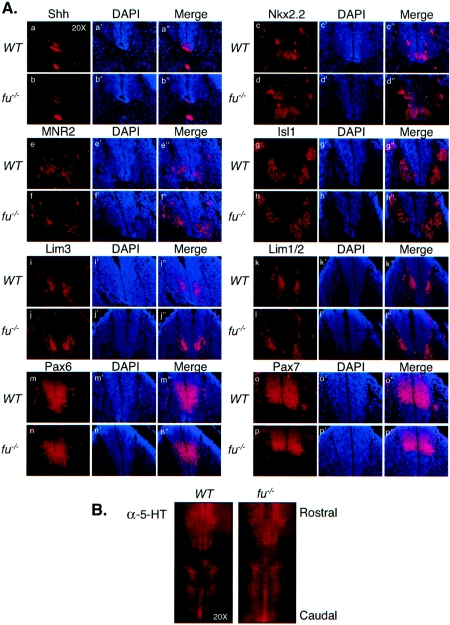
Many lines of evidence indicate that Shh signaling plays a role in the induction of neural progenitor populations in the ventral neural tubes (for a review, see reference 35). Shh signaling is initiated in the notochord (E8.0) and continues later at the ventral midline floor plate cells (E9.5 to E10.5). The expression of specific markers for ventral neural progenitors (Nkx2.2 and Shh) and precursors (MNR2, Isl1, Lim3, Lim1/2, Pax6, and Pax7) was analyzed by immunostaining sections from E10.5 embryos to investigate a potential ventral patterning defect in fused knockout mice. Both wild-type and fused knockout embryos showed comparable expression levels for all markers tested (Fig. 4A). We also examined Hh-dependent neuronal specification at times later than E10.5, near the ventral midline of the hindbrain. Equivalent levels of serotonergic neurons, marked by 5-HT, were detected in E13.5 embryos by whole-mount immunostaining in two clusters flanking the floor plate in both wild-type and fused knockout embryos (Fig. 4B).
FIG.4.
Loss of mFu does not impact Hh-dependent patterning of the ventral neural tube. (A) Specification of neural tube markers is shown for Shh (a and b), Nkx2.2 (c and d), MNR2 (e and f), Isl1 (g and h), Lim3 (i and j), Lim1/2 (k and l), Pax6 (m and n), and Pax7 (o and p). Frozen sections of E10.5 embryos were stained with monoclonal antibodies against the indicated antigens and counterstained with DAPI, and photos were taken at amagnification of ×20. No defects were observed in the specification of any neuronal population in fu−/− mice. (B) Specification of serotonergic neurons is normal in fu−/− embryos. Embryos were collected at E13.5 and stained for serotonergic specified neurons with antibodies against 5-HT. Whole-mount immunostaining of the ventral hindbrain is shown, with the rostral-most region at the top. Specification of serotonergic neurons is seen in two clusters consisting of rhombomeres 2 and 3 and 5 to 7, whereas rhombomere 4 is characteristically negative for 5-HT.
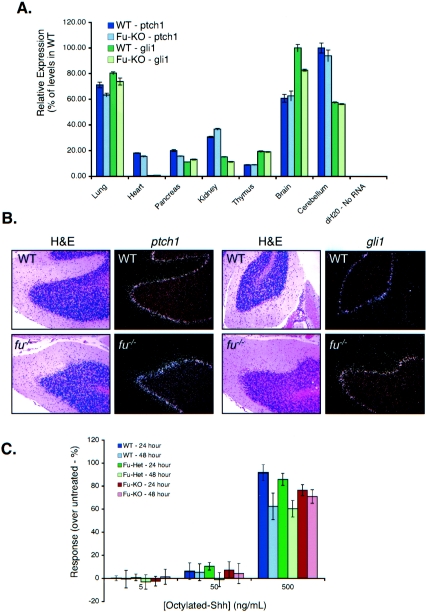
Signaling through the Hh pathway results in the up-regulation of many Hh target genes, including ptch1 and gli1. To address whether loss of mFu alters ptch1 or gli1 expression levels, both quantitative RT-PCR and radioisotopic in situ hybridization were performed. RNAs from various tissues, including the lungs, heart, pancreas, kidneys, thymus, brain, and cerebellum, were collected from wild-type and fused knockout neonates. Quantitative RT-PCR for ptch1 and gli1 showed little difference in expression levels in all tissues analyzed (Fig. 5A). These data indicate that global mRNA levels of ptch1 and gli1 in these whole organs are similar between p7 wild-type and fused knockout mice.
FIG. 5.
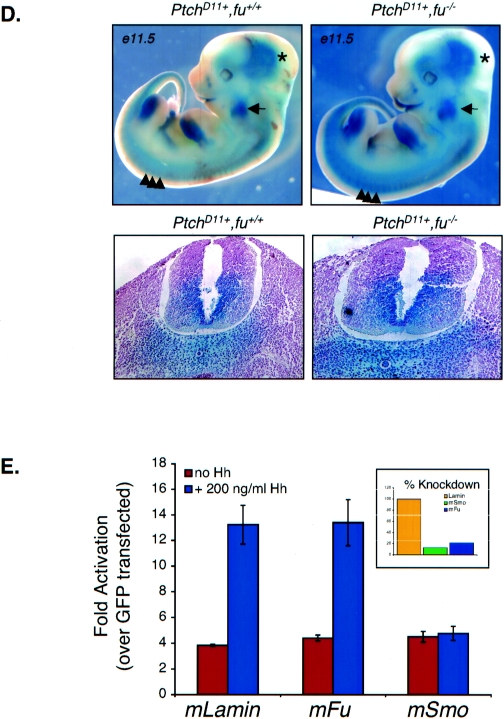
Loss of Fused does not impact Hh signal transduction in vivo or in vitro. (A) Loss of Fused does not impact Hh target gene activation in vivo. Quantitative PCR from various p7 tissues is shown for the ptch1 gene. No effects were observed on ptch1 levels in any tissue. Similar results were observed for the gli1 gene. KO, knockout; dH2O, distilled water. (B) Loss of Fused does not inhibit Hh target gene expression in the cerebellum, as measured by 32P-labeled in situ hybridization to ptch1 and gli1. Cerebellum sections (magnification, ×10) from wild-type (WT) and fu−/− neonates are shown stained with H&E with the comparable radiolabeled signal in the EGL for both ptch1 (left) and gli1 (right). (C) Loss of Fused does not impact the ability of cerebellar granule cells to respond to Shh. Cerebellar granule cells from wild-type, fu+/−, and fu−/− mice were treated in vitro for either 24 h (left image) or 48 h (right image) with octyl-modified Shh at 0, 5, 50, or 500 ng/ml. Cells were pulsed with [3H]thymidine 5 h prior to harvesting, and results are plotted as the percent response over unstimulated cells. (D) Loss of Fused does not alter ptch1 expression in vivo. High expression of ptch1 was observed in the brain (star), branchial arches (arrow), ventral CNS somites (arrowheads), and posterior limb buds of both PtchD11 fu+/+ and PtchD11 fu−/− mice. A cross section through the neural tube of these embryos shows comparable levels of ptch1 expression. (E) siRNA knockdown of mFu does not disrupt Shh-mediated signaling in C3H/10T1/2 S12 cells. Expression of mFu and mSmo was knocked down to approximately 25% and 12% of the normal levels, respectively (inset), as measured by quantitative PCR, and cells were treated with (black bars) and without (white bars) 200 ng/ml of octyl-modified Shh. No effect upon Shh-mediated activation of the 9x-Gli-BS-Luciferase reporter is observed when mFu is knocked down, while similar knockdown of mSmo totally abolishes Shh-mediated signaling. GFP, green fluorescent protein.
To address whether there are subtle effects upon ptch1 and gli1 expression in subsets of cells within fused knockout tissues, radioisotopic in situ hybridization was performed on sections from wild-type and fused knockout embryos and neonates. High ptch1 expression is observed in both wild-type and fused knockout animals in the following embryonic tissues: in hair follicles, in mesenchymal cells adjacent to intestinal mucosa and developing bone, in cartilage, in the brain adjacent to the developing ventricle, and in lung mesenchymal cells adjacent to epithelium lining immature air spaces (data not show). ptch1 expression is also present in both wild-type and fused knockout postnatal tissues as follows: in the lamina propria of the small intestine, in the kidneys in the primary distal portions of the collecting system and in mesenchymal cells underneath the mucosa of the renal pelvis, in hair follicles of the skin, in the thymic medulla, in the liver, in the lungs, in the ovaries, and in the interstitial cells of the testes (data not shown). A good example of ptch1 expression was observed in the developing cerebellum, where very strong expression of ptch1 was observed in the outer aspect of the granular cell layer, including the Purkinje cell layer (Fig. 5B). Likewise, gli1 is expressed highly in both wild-type and fused knockout animals in the following embryonic tissues: bone anlagen, submucosal mesenchymal tissues, lungs, kidneys, and developing hair follicles. In postnatal tissues, gli1 expression is present in the brain (and is particularly strong in the outer granular cell layer of the cerebellum) and kidneys (similar to ptch1), with similar expression in both wild-type and fused knockout animals. Together with the quantitative RT-PCR results, these data indicate that loss of mFu does not dramatically alter the overall expression levels of two established Hh-target genes, ptch1 and gli1.
In the developing cerebellum, Shh provided by Purkinje cells acts as a mitogen to drive the proliferation of GNP in the EGL (91). The mFu gene is expressed in the cerebellum, as measured by quantitative PCR and by radioisotopic in situ hybridization (Fig. 1D) (54), implying that it may function in Shh-mediated postnatal GNP proliferation. GNP from p5 wild-type, fused heterozygous, and fused knockout neonates were isolated and tested for the ability to proliferate in vitro in response to low, moderate, and high levels of octyl-modified Shh. The fused mutant GNP proliferated equivalently to fused heterozygous and wild-type littermate controls at all concentrations of octyl-modified Shh at both 24 and 48 h (Fig. 5C). These data indicate that loss of mFu does not impact Shh-mediated GNP proliferation in vitro.
Expression ptch1 can also be monitored by β-galactosidase expression driven from the ptch1 promoter in the Ptch1D11 mutant (21, 59), providing a sensitive readout of Hh pathway activity in vivo. LacZ staining revealed the expected ptch1 expression pattern in the ventral CNS, branchial arches, oral epithelium, whiskers, and posterior half of the limb buds (Fig. 5D). Cross sections through the neural tube showed high expression of ptch1 in mesenchymal cells surrounding the notochord and the entire ventral half of the neural tube (Fig. 5D). The overall pattern and staining intensity of LacZ were similar between Ptch1D11 fu−/− and Ptch1D11 fu+/+ littermate controls (Fig. 5D), indicating that loss of mFu does not impact the expression of the Hh target gene ptch1 in vivo.
Previous studies have shown that overexpression of the human fused gene is capable of inducing Hh signal transduction in the mouse C3H/10T1/2 cell line (54). Results from fused knockout mice suggest that the function of mFu is not required during embryogenesis for normal Hh signal transduction. To address this apparent discrepancy, we performed siRNA knockdown experiments with C3H/10T1/2 cell lines using siRNAs against mouse fused. The level of fused transcripts was successfully knocked down in C3H/10T1/2 cell lines to less than a quarter of the normal level, as measured by quantitative RT-PCR; however, no effect was observed on Hh signal transduction following treatment with octyl-modified Shh, while similar knockdown of mouse smo abrogates Shh-mediated signaling (Fig. 5E). These data suggest that mFu is not required for Shh signal transmission in vitro.
DISCUSSION
We have shown here that loss of the mouse fused kinase homologue (stk36) results in a postnatal growth defect characterized by a communicating form of hydrocephalus and nasal inflammation, the former of which most likely is responsible for the death of knockout animals within 2 weeks. The nature of the defects in the CNS and nasal cavity remains to be established. While we favor a model wherein loss of mFu results in overproduction of CSF and mucus, it is possible that mFu is playing a critical role in the absorption of these fluids. No defects were observed in the subarachnoid space in the CNS (data not shown), the major site of CSF absorption; however, it is not clear whether absorption of CSF is taking place or not. And while the onset of hydrocephalus is very early and dramatic, the actual cause of death may be related to primary or secondary defects in other organs, such as the lungs or the kidneys, where fused is expressed. However, we believe the growth defects in these organs, usually observed at p7, are secondary to the early onset and progressive nature of the hydrocephalus.
The parallels between the fused knockout phenotype and those of several other mutants with communicating hydrocephalus (e.g., p73 and E2F-5) suggests that these components may impinge upon a common critical postnatal developmental step. The fused gene is expressed at high levels in specialized secretory tissues, including the choroid plexus, ependymal cells, and nasal epithelium. The strong correlation between the expression of fused and p73, E2F-5, HFH4, Msi1, Mdnah5, and SPAG6 in these secretory cell types suggests that these factors are involved in the regulation of CSF production and/or circulation (4, 23, 31, 46, 75, 77, 94).
It is unlikely that the loss of mFu in the nasal lining could result in improper recruitment of neutrophils or modulation of cytokine signaling, as the inflammatory defect is not observed globally despite broad expression of mFu. The inflammation observed probably occurs in response to overproduction of mucus within the nasal cavity, followed by increased trapping of foreign particles. Further studies are required to delineate how mFu is regulating these processes and whether it overlaps any of the other factors implicated in hydrocephalus.
To our surprise, no defect in Hh signal transduction in vivo or in vitro was detected in fused-deficient mice or cells, suggesting either that mFu does not function in the Hh pathway or that its function is redundant or can be compensated for by other kinases. Data from studies largely relying on overexpression of hFu in mammalian cell lines suggested that hFu is capable of inducing Hh target genes (54, 60). Interestingly, kinase-deficient forms of hFu were also capable of inducing Hh target genes; however, this may be an overexpression phenomenon mediated by excess levels of hFu sequestering the mammalian counterparts of Su(fu) or Cos2 and thereby preventing them from repressing Gli (54). Consistent with this idea, ectopic hFu expression in the imaginal wing disk of Drosophila induces novel wing vein phenotypes, indicating that hFu is capable of impinging on the Hh pathway in flies (12). Furthermore, when hFu was expressed in dFu mutant backgrounds it was found to exacerbate rather than rescue the dFu wing vein phenotypes, suggesting that hFu competes with dFu for factors important for Hh signaling but lacks some critical function of dFu (12).
In contrast to our study with mice, knockdown of a fu homologue in zebra fish through antisense morpholino oligonucleotides disrupted the Hh-dependent specification of myotome cell types (93). Similar to dFu, this defect is rescued by knocking down Su(fu) function (93), suggesting that in zebra fish there is a single Fu homologue acting similarly to dFu. The overall homology between the mammalian Fu homologues and zfFu (28% identity, 13% homology) is slightly better than the homology between the mammalian Fu homologues and dFu (15% identity, 11% homology), and the homology of zfFu with dFu is also fairly weak (22% identity, 14% homology).
While database searches do not identify other strong Fu homologue candidates that could play a redundant role with Fu, it is possible that a kinase(s) wholly unrelated to Fu replace the classical function of Fu. Other kinases have been implicated in Hh signaling, including PKA, GSK3β, CKIα, and more recently, GRK2 (6, 7, 25, 36, 38, 71, 92, 95, 96). GRK2 has recently been shown to phosphorylate Smo, allowing β-arrestin 2 to bind Smo, leading to relocalization of the complex to clathrin-coated pits (6, 92). β-Arr2 appears to be required for Hh activity in zebra fish, implying that GRK2 and β-Arr2 function downstream of Smo in vertebrates (92). However, unlike dFu, neither GRK2 nor the other kinases that impinge upon Hh signal transduction are exclusive to the Hh pathway and it remains to be determined whether they or other kinases truly functionally compensate for Fu activity.
If redundancy for the Fu kinase exists, one intriguing possibility is that mFu is acting within the Hh pathway within the choroid plexus and/or ependymal cells, where it regulates CSF production and/or transmission. Indeed, ependymal cells have been identified as potential sources of neural stem cells (10, 39, 45), a population of cells regulated by Hh signaling (19, 26, 62, 63, 78). Furthermore, mutations in Gli2 result in mice born with enlarged ventricles, presumably representing a perinatal hydrocephalus phenotype and suggesting a possible connection between Hh activity and ependymal cell and/or choroid plexus function (63). In support of this idea, expression of shh and gli1 has been reported in the mouse choroid plexus at p3 (11). However, this hypothesis requires further study. Our data show that mFu (stk36) is not required in vivo or in vitro for Hh signaling in mice but functions in specialized secretory tissues that impinge upon proper postnatal development. Future studies are needed to determine if any other kinase plays the role of Fu or can compensate in its absence in the mammalian Hh pathway.
Acknowledgments
We thank Matt Scott for use of the PtchD11 line. We also thank Pao-Tien Chuang and his lab. Thanks also go to the Genentech LAR department and the Sequencing, Flow Cytometry, and Transgenic-Knockout facilities. We also acknowledge Hua Tian, Sarah Craven, Jasvinder Atwal, T'Nay Pham, Weilan Ye, Weidong Wang, Rui Yu, Ji Li, Vivian Barry, Derek Marshall, Tracy Tang, Ajay Malik, Ryan Scott, Deborah Kwok, and Joel Morales for reagents, technical assistance, advice, and help with preparation of the manuscript.
REFERENCES
- 1.Alcedo, J., Y. Zou, and M. Noll. 2000. Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the Hedgehog signaling system. Mol. Cell 6:457-465. [DOI] [PubMed] [Google Scholar]
- 2.Alves, G., B. Limbourg-Bouchon, H. Tricoire, J. Brissard-Zahraoui, C. Lamour-Isnard, and D. Busson. 1998. Modulation of Hedgehog target gene expression by the Fused serine-threonine kinase in wing imaginal discs. Mech. Dev. 78:17-31. [DOI] [PubMed] [Google Scholar]
- 3.Bose, J., L. Grotewold, and U. Ruther. 2002. Pallister-Hall syndrome phenotype in mice mutant for Gli3. Hum. Mol. Genet. 11:1129-1135. [DOI] [PubMed] [Google Scholar]
- 4.Brody, S. L., X. H. Yan, M. K. Wuerffel, S. K. Song, and S. D. Shapiro. 2000. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 23:45-51. [DOI] [PubMed] [Google Scholar]
- 5.Caspary, T., M. J. Garcia-Garcia, D. Huangfu, J. T. Eggenschwiler, M. R. Wyler, A. S. Rakeman, H. L. Alcorn, and K. V. Anderson. 2002. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr. Biol. 12:1628-1632. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W., X. R. Ren, C. D. Nelson, L. S. Barak, J. K. Chen, P. A. Beachy, F. de Sauvage, and R. J. Lefkowitz. 2004. Activity-dependent internalization of smoothened mediated by β-arrestin 2 and GRK2. Science 306:2257-2260. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., N. Gallaher, R. H. Goodman, and S. M. Smolik. 1998. Protein kinase A directly regulates the activity and proteolysis of cubitus interruptus. Proc. Natl. Acad. Sci. USA 95:2349-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., and G. Struhl. 1996. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87:553-563. [DOI] [PubMed] [Google Scholar]
- 9.Chory, J. 1995. Analysis of DNA sequences by blotting and hybridization, p. 2.9.1-2.10.16. In F. M. Ausubel (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., Boston, Mass. [Google Scholar]
- 10.Clarke, D. L. 2003. Neural stem cells. Bone Marrow Transplant. 32(Suppl. 1):S13-S17. [DOI] [PubMed] [Google Scholar]
- 11.Dahmane, N., P. Sanchez, Y. Gitton, V. Palma, T. Sun, M. Beyna, H. Weiner, and A. Ruiz i Altaba. 2001. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 128:5201-5212. [DOI] [PubMed] [Google Scholar]
- 12.Daoud, F., and M. F. Blanchet-Tournier. 2005. Expression of the human FUSED protein in Drosophila. Dev. Genes Evol. 215:230-237. [DOI] [PubMed] [Google Scholar]
- 13.Delattre, M., S. Briand, M. Paces-Fessy, and M. F. Blanchet-Tournier. 1999. The Suppressor of fused gene, involved in Hedgehog signal transduction in Drosophila, is conserved in mammals. Dev. Genes Evol. 209:294-300. [DOI] [PubMed] [Google Scholar]
- 14.Denef, N., D. Neubuser, L. Perez, and S. M. Cohen. 2000. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102:521-531. [DOI] [PubMed] [Google Scholar]
- 15.Ding, Q., S. Fukami, X. Meng, Y. Nishizaki, X. Zhang, H. Sasaki, A. Dlugosz, M. Nakafuku, and C. Hui. 1999. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr. Biol. 9:1119-1122. [DOI] [PubMed] [Google Scholar]
- 16.Ding, Q., J. Motoyama, S. Gasca, R. Mo, H. Sasaki, J. Rossant, and C. C. Hui. 1998. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125:2533-2543. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, N. R., G. E. Winnier, L. K. Hargett, J. J. Schrick, A. B. Fogo, and B. L. Hogan. 1997. Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev. Biol. 188:235-247. [DOI] [PubMed] [Google Scholar]
- 18.Frank-Kamenetsky, M., X. M. Zhang, S. Bottega, O. Guicherit, H. Wichterle, H. Dudek, D. Bumcrot, F. Y. Wang, S. Jones, J. Shulok, L. L. Rubin, and J. A. Porter. 2002. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, H., Y. Qi, M. Tan, J. Cai, X. Hu, Z. Liu, J. Jensen, and M. Qiu. 2003. Molecular mapping of the origin of postnatal spinal cord ependymal cells: evidence that adult ependymal cells are derived from Nkx6.1+ ventral neural progenitor cells. J. Comp. Neurol. 456:237-244. [DOI] [PubMed] [Google Scholar]
- 20.Go, K. G. 1997. The normal and pathological physiology of brain water. Adv. Tech. Stand. Neurosurg. 23:47-142. [DOI] [PubMed] [Google Scholar]
- 21.Goodrich, L. V., L. Milenkovic, K. M. Higgins, and M. P. Scott. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277:1109-1113. [DOI] [PubMed] [Google Scholar]
- 22.Gruneberg, H. 1943. Congenital hydrocephalus in the mouse, a case of spurious pleiotropism. J. Genet. 45:1-21. [Google Scholar]
- 23.Gruneberg, H. 1943. Two new mutant genes in the house mouse. J. Genet. 45:22-28. [Google Scholar]
- 24.Gutierrez-Frias, C., R. Sacedon, C. Hernandez-Lopez, T. Cejalvo, T. Crompton, A. G. Zapata, A. Varas, and A. Vicente. 2004. Sonic hedgehog regulates early human thymocyte differentiation by counteracting the IL-7-induced development of CD34+ precursor cells. J. Immunol. 173:5046-5053. [DOI] [PubMed] [Google Scholar]
- 25.Hammerschmidt, M., M. J. Bitgood, and A. P. McMahon. 1996. Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev. 10:647-658. [DOI] [PubMed] [Google Scholar]
- 26.Ho, K. S., and M. P. Scott. 2002. Sonic hedgehog in the nervous system: functions, modifications and mechanisms. Curr. Opin. Neurobiol. 12:57-63. [DOI] [PubMed] [Google Scholar]
- 27.Holcomb, I. N., R. C. Kabakoff, B. Chan, T. W. Baker, A. Gurney, W. Henzel, C. Nelson, H. B. Lowman, B. D. Wright, N. J. Skelton, G. D. Frantz, D. B. Tumas, F. V. Peale, Jr., D. L. Shelton, and C. C. Hebert. 2000. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 19:4046-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hynes, M., J. A. Porter, C. Chiang, D. Chang, M. Tessier-Lavigne, P. A. Beachy, and A. Rosenthal. 1995. Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron 15:35-44. [DOI] [PubMed] [Google Scholar]
- 29.Hynes, M., K. Poulsen, M. Tessier-Lavigne, and A. Rosenthal. 1995. Control of neuronal diversity by the floor plate: contact-mediated induction of midbrain dopaminergic neurons. Cell 80:95-101. [DOI] [PubMed] [Google Scholar]
- 30.Hynes, M., W. Ye, K. Wang, D. Stone, M. Murone, F. Sauvage, and A. Rosenthal. 2000. The seven-transmembrane receptor smoothened cell-autonomously induces multiple ventral cell types. Nat. Neurosci. 3:41-46. [DOI] [PubMed] [Google Scholar]
- 31.Ibanez-Tallon, I., S. Gorokhova, and N. Heintz. 2002. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum. Mol. Genet. 11:715-721. [DOI] [PubMed] [Google Scholar]
- 32.Ibanez-Tallon, I., A. Pagenstecher, M. Fliegauf, H. Olbrich, A. Kispert, U. P. Ketelsen, A. North, N. Heintz, and H. Omran. 2004. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum. Mol. Genet. 13:2133-2141. [DOI] [PubMed] [Google Scholar]
- 33.Ingham, P. W., and A. P. McMahon. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15:3059-3087. [DOI] [PubMed] [Google Scholar]
- 34.Ingham, P. W., S. Nystedt, Y. Nakano, W. Brown, D. Stark, M. van den Heuvel, and A. M. Taylor. 2000. Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr. Biol. 10:1315-1318. [DOI] [PubMed] [Google Scholar]
- 35.Jessell, T. M. 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1:20-29. [DOI] [PubMed] [Google Scholar]
- 36.Jia, J., K. Amanai, G. Wang, J. Tang, B. Wang, and J. Jiang. 2002. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416:548-552. [DOI] [PubMed] [Google Scholar]
- 37.Jia, J., C. Tong, and J. Jiang. 2003. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 17:2709-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia, J., C. Tong, B. Wang, L. Luo, and J. Jiang. 2004. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 432:1045-1050. [DOI] [PubMed] [Google Scholar]
- 39.Johansson, C. B., S. Momma, D. L. Clarke, M. Risling, U. Lendahl, and J. Frisen. 1999. Identification of a neural stem cell in the adult mammalian central nervous system. Cell 96:25-34. [DOI] [PubMed] [Google Scholar]
- 40.Katoh, Y., and M. Katoh. 2004. Characterization of KIF7 gene in silico. Int. J. Oncol. 25:1881-1886. [PubMed] [Google Scholar]
- 41.Katoh, Y., and M. Katoh. 2004. KIF27 is one of orthologs for Drosophila Costal-2. Int. J. Oncol. 25:1875-1880. [PubMed] [Google Scholar]
- 42.Kawakami, T., T. Kawcak, Y. J. Li, W. Zhang, Y. Hu, and P. T. Chuang. 2002. Mouse dispatched mutants fail to distribute hedgehog proteins and are defective in hedgehog signaling. Development 129:5753-5765. [DOI] [PubMed] [Google Scholar]
- 43.Kume, T., K. Deng, and B. L. Hogan. 2000. Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127:1387-1395. [DOI] [PubMed] [Google Scholar]
- 44.Kume, T., K. Y. Deng, V. Winfrey, D. B. Gould, M. A. Walter, and B. L. Hogan. 1998. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell 93:985-996. [DOI] [PubMed] [Google Scholar]
- 45.Laywell, E. D., P. Rakic, V. G. Kukekov, E. C. Holland, and D. A. Steindler. 2000. Identification of a multipotent astrocytic stem cell in the immature and adult mouse brain. Proc. Natl. Acad. Sci. USA 97:13883-13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindeman, G. J., L. Dagnino, S. Gaubatz, Y. Xu, R. T. Bronson, H. B. Warren, and D. M. Livingston. 1998. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 12:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu, L. H., and N. Gillette. 1994. An optimized protocol for in situ hybridization using PCR-generated 33P-labeled riboprobes. Cell Vision 7:55-64. [Google Scholar]
- 48.Lum, L., C. Zhang, S. Oh, R. K. Mann, D. P. von Kessler, J. Taipale, F. Weis-Garcia, R. Gong, B. Wang, and P. A. Beachy. 2003. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol. Cell 12:1261-1274. [DOI] [PubMed] [Google Scholar]
- 49.Ma, Y., A. Erkner, R. Gong, S. Yao, J. Taipale, K. Basler, and P. A. Beachy. 2002. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111:63-75. [DOI] [PubMed] [Google Scholar]
- 50.Matise, M. P., D. J. Epstein, H. L. Park, K. A. Platt, and A. L. Joyner. 1998. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 125:2759-2770. [DOI] [PubMed] [Google Scholar]
- 51.McMahon, A. P., P. W. Ingham, and C. J. Tabin. 2003. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53:1-114. [DOI] [PubMed] [Google Scholar]
- 52.Merchant, M., F. F. Vajdos, M. Ultsch, H. R. Maun, U. Wendt, J. Cannon, W. Desmarais, R. A. Lazarus, A. M. de Vos, and F. J. de Sauvage. 2004. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol. Cell. Biol. 24:8627-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Motoyama, J., J. Liu, R. Mo, Q. Ding, M. Post, and C. C. Hui. 1998. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat. Genet. 20:54-57. [DOI] [PubMed] [Google Scholar]
- 54.Murone, M., S. M. Luoh, D. Stone, W. Li, A. Gurney, M. Armanini, C. Grey, A. Rosenthal, and F. J. de Sauvage. 2000. Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell Biol. 2:310-312. [DOI] [PubMed] [Google Scholar]
- 55.Nagy, A., M. Gertsenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Nusslein-Volhard, C., and E. Wieschaus. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795-801. [DOI] [PubMed] [Google Scholar]
- 57.Ogden, S. K., M. Ascano, Jr., M. A. Stegman, L. M. Suber, J. E. Hooper, and D. J. Robbins. 2003. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr. Biol. 13:1998-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohlmeyer, J. T., and D. Kalderon. 1998. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396:749-753. [DOI] [PubMed] [Google Scholar]
- 59.Oro, A. E., and K. Higgins. 2003. Hair cycle regulation of Hedgehog signal reception. Dev. Biol. 255:238-248. [DOI] [PubMed] [Google Scholar]
- 60.Osterlund, T., D. B. Everman, R. C. Betz, M. Mosca, M. M. Nothen, C. E. Schwartz, P. G. Zaphiropoulos, and R. Toftgard. 2004. The FU gene and its possible protein isoforms. BMC Genomics 5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Outram, S. V., A. Varas, C. V. Pepicelli, and T. Crompton. 2000. Hedgehog signaling regulates differentiation from double-negative to double-positive thymocyte. Immunity 13:187-197. [DOI] [PubMed] [Google Scholar]
- 62.Palma, V., D. A. Lim, N. Dahmane, P. Sanchez, T. C. Brionne, C. D. Herzberg, Y. Gitton, A. Carleton, A. Alvarez-Buylla, and A. Ruiz i Altaba. 2005. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palma, V., and A. Ruiz i Altaba. 2004. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131:337-345. [DOI] [PubMed] [Google Scholar]
- 64.Park, H. L., C. Bai, K. A. Platt, M. P. Matise, A. Beeghly, C. C. Hui, M. Nakashima, and A. L. Joyner. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127:1593-1605. [DOI] [PubMed] [Google Scholar]
- 65.Pasca di Magliano, M., and M. Hebrok. 2003. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3:903-911. [DOI] [PubMed] [Google Scholar]
- 66.Pearse, R. V., II, L. S. Collier, M. P. Scott, and C. J. Tabin. 1999. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev. Biol. 212:323-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrimon, N., and A. P. Mahowald. 1987. Multiple functions of segment polarity genes in Drosophila. Dev. Biol. 119:587-600. [DOI] [PubMed] [Google Scholar]
- 68.Preat, T. 1992. Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132:725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Preat, T., P. Therond, C. Lamour-Isnard, B. Limbourg-Bouchon, H. Tricoire, I. Erk, M. C. Mariol, and D. Busson. 1990. A putative serine/threonine protein kinase encoded by the segment-polarity fused gene of Drosophila. Nature 347:87-89. [DOI] [PubMed] [Google Scholar]
- 70.Preat, T., P. Therond, B. Limbourg-Bouchon, A. Pham, H. Tricoire, D. Busson, and C. Lamour-Isnard. 1993. Segmental polarity in Drosophila melanogaster: genetic dissection of fused in a Suppressor of fused background reveals interaction with costal-2. Genetics 135:1047-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price, M. A., and D. Kalderon. 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell 108:823-835. [DOI] [PubMed] [Google Scholar]
- 72.Robbins, D. J., K. E. Nybakken, R. Kobayashi, J. C. Sisson, J. M. Bishop, and P. P. Therond. 1997. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90:225-234. [DOI] [PubMed] [Google Scholar]
- 73.Romer, J. T., H. Kimura, S. Magdaleno, K. Sasai, C. Fuller, H. Baines, M. Connelly, C. F. Stewart, S. Gould, L. L. Rubin, and T. Curran. 2004. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/− p53−/− mice. Cancer Cell 6:229-240. [DOI] [PubMed] [Google Scholar]
- 74.Ruel, L., R. Rodriguez, A. Gallet, L. Lavenant-Staccini, and P. P. Therond. 2003. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 5:907-913. [DOI] [PubMed] [Google Scholar]
- 75.Sakakibara, S., Y. Nakamura, T. Yoshida, S. Shibata, M. Koike, H. Takano, S. Ueda, Y. Uchiyama, T. Noda, and H. Okano. 2002. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. USA 99:15194-15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanchez-Herrero, E., J. P. Couso, J. Capdevila, and I. Guerrero. 1996. The fu gene discriminates between pathways to control dpp expression in Drosophila imaginal discs. Mech. Dev. 55:159-170. [DOI] [PubMed] [Google Scholar]
- 77.Sapiro, R., I. Kostetskii, P. Olds-Clarke, G. L. Gerton, G. L. Radice, and I. J. Strauss. 2002. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 22:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seaberg, R. M., and D. van der Kooy. 2002. Adult rodent neurogenic regions: the ventricular subependyma contains neural stem cells, but the dentate gyrus contains restricted progenitors. J. Neurosci. 22:1784-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah, D. K., A. L. Hager-Theodorides, S. V. Outram, S. E. Ross, A. Varas, and T. Crompton. 2004. Reduced thymocyte development in sonic hedgehog knockout embryos. J. Immunol. 172:2296-2306. [DOI] [PubMed] [Google Scholar]
- 80.Sisson, J. C., K. S. Ho, K. Suyama, and M. P. Scott. 1997. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell 90:235-245. [DOI] [PubMed] [Google Scholar]
- 81.Stegman, M. A., J. E. Vallance, G. Elangovan, J. Sosinski, Y. Cheng, and D. J. Robbins. 2000. Identification of a tetrameric hedgehog signaling complex. J. Biol. Chem. 275:21809-21812. [DOI] [PubMed] [Google Scholar]
- 82.Stone, D. M., M. Murone, S. Luoh, W. Ye, M. P. Armanini, A. Gurney, H. Phillips, J. Brush, A. Goddard, F. J. de Sauvage, and A. Rosenthal. 1999. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J. Cell Sci. 112(Pt. 23):4437-4448. [DOI] [PubMed] [Google Scholar]
- 83.Suzuki, T., and K. Saigo. 2000. Transcriptional regulation of atonal required for Drosophila larval eye development by concerted action of eyes absent, sine oculis and hedgehog signaling independent of fused kinase and cubitus interruptus. Development 127:1531-1540. [DOI] [PubMed] [Google Scholar]
- 84.Swiatek, P. J., and T. Gridley. 1993. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 7:2071-2084. [DOI] [PubMed] [Google Scholar]
- 85.Taipale, J., and P. A. Beachy. 2001. The Hedgehog and Wnt signalling pathways in cancer. Nature 411:349-354. [DOI] [PubMed] [Google Scholar]
- 86.Taipale, J., M. K. Cooper, T. Maiti, and P. A. Beachy. 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418:892-897. [DOI] [PubMed] [Google Scholar]
- 87.Tay, S. Y., P. W. Ingham, and S. Roy. 2005. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development 132:625-634. [DOI] [PubMed] [Google Scholar]
- 88.Taylor, F. R., D. Wen, E. A. Garber, A. N. Carmillo, D. P. Baker, R. M. Arduini, K. P. Williams, P. H. Weinreb, P. Rayhorn, X. Hronowski, A. Whitty, E. S. Day, A. Boriack-Sjodin, R. I. Shapiro, A. Galdes, and R. B. Pepinsky. 2001. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40:4359-4371. [DOI] [PubMed] [Google Scholar]
- 89.Therond, P., G. Alves, B. Limbourg-Bouchon, H. Tricoire, E. Guillemet, J. Brissard-Zahraoui, C. Lamour-Isnard, and D. Busson. 1996. Functional domains of fused, a serine-threonine kinase required for signaling in Drosophila. Genetics 142:1181-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Therond, P. P., B. Limbourg Bouchon, A. Gallet, F. Dussilol, T. Pietri, M. van den Heuvel, and H. Tricoire. 1999. Differential requirements of the fused kinase for hedgehog signalling in the Drosophila embryo. Development 126:4039-4051. [DOI] [PubMed] [Google Scholar]
- 91.Wechsler-Reya, R. J., and M. P. Scott. 1999. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22:103-114. [DOI] [PubMed] [Google Scholar]
- 92.Wilbanks, A. M., G. B. Fralish, M. L. Kirby, L. S. Barak, Y. X. Li, and M. G. Caron. 2004. Beta-arrestin 2 regulates zebrafish development through the hedgehog signaling pathway. Science 306:2264-2267. [DOI] [PubMed] [Google Scholar]
- 93.Wolff, C., S. Roy, and P. W. Ingham. 2003. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13:1169-1181. [DOI] [PubMed] [Google Scholar]
- 94.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 95.Zhang, C., E. H. Williams, Y. Guo, L. Lum, and P. A. Beachy. 2004. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc. Natl. Acad. Sci. USA 101:17900-17907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang, W., Y. Zhao, C. Tong, G. Wang, B. Wang, J. Jia, and J. Jiang. 2005. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev. Cell 8:267-278. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, X. M., M. Ramalho-Santos, and A. P. McMahon. 2001. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell 106:781-792. [PubMed] [Google Scholar]
- 98.Zhu, A. J., L. Zheng, K. Suyama, and M. P. Scott. 2003. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 17:1240-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]