Abstract
Keratin genes afford, given their large number (>50) and differential regulation, a unique opportunity to study the mechanisms underlying specification and differentiation in epithelia of higher metazoans. Moreover, the small size and regulation in cis of many keratin genes enable the use of their regulatory sequence to achieve targeted gene expression in mice. Here we show that 2 kilobases of 5′ upstream region from the mouse keratin 17 gene (mK17) confers expression of green fluorescent protein (GFP) in major epithelial appendages of transgenic mice. Like that of mK17, onset of [mK17 5′]-GFP reporter expression coincides with the appearance of ectoderm-derived epithelial appendages during embryonic development. In adult mice, [mK17 5′]-GFP is appropriately regulated within hair, nail, glands, and oral papilla. Tracking of GFP fluorescence allows for the visualization of growth cycle-related changes in hair follicles, and the defects engendered by the hairless mutation, in live skin tissue. Deletion of an internal 48-bp interval, which encompasses a Gli-responsive element, from this promoter results in loss of GFP fluorescence in most appendages in vivo, suggesting that sonic hedgehog participates in K17 regulation. The compact mK17 gene promoter provides a novel tool for appendage-preferred gene expression and manipulation in transgenic mice.
The study of cellular dynamics in vivo exposes the basic processes enabling specific cell populations to be recruited and coordinated to form and maintain complex tissues and how these go awry in the context of disease (14, 32, 33). Keratin genes provide a unique handle to track the fate and differentiation of cells within epithelia (16, 49). Type I and type II keratins, which are encoded by a large family of ∼50 conserved genes, are abundant proteins in epithelial cells, where they heteropolymerize to form cytoplasmic intermediate filaments (11, 12, 25). The pairwise and differentiation-related transcriptional regulation of keratin genes, which are relatively small (<10 kb in size), relies on proximal cis-acting determinants (48, 59, 64, 73). The 5′ upstream sequences from a number of keratin genes have been exploited to direct cell type-specific expression of heterologous open reading frames in transgenic mice. For instance, the K14 promoter- and K5 promoter-based cassettes (46, 59, 62, 73) have been used to create hundreds of transgenic mouse lines which, collectively, have had an extraordinary impact on our understanding of the epidermis and related complex epithelia.
The unusually broad variety of epithelial settings in which type I keratin 17 (K17) occurs likely call for intricate transcriptional regulatory mechanisms. During development, specification of epithelial appendages is marked by the onset of K17 synthesis in the embryonic ectoderm (40). Concomitant with the maturation of these precursor elements into tissues such as hair follicle, nail, gland, and tooth, K17 expression becomes restricted to specific cell types (40, 70). In the hair follicle, which undergoes a developmental cycle throughout life, K17 expression continues to be dynamically modulated in the adult setting (2, 56). K17 also has the rare distinction of being expressed in both soft and hard epithelia, and K17 protein uses various type II keratins (K6, K6hf, and K5) as polymerization partners, depending on the epithelial setting in vivo (39, 76). In addition to this constitutive regulation, K17 is strongly inducible in epidermis following acute challenges such as skin injury or in disease contexts (e.g., viral infection, psoriasis, or basal cell carcinoma [15, 41]). K17 is not transcribed in normal interfollicular epidermis (40, 67).
The green fluorescent protein (GFP) of Aequarea victoria has evolved into an unparalleled tool for visualizing cells in their natural living context and has been successfully exploited to this end in model organisms ranging from unicellular eukaryotes to mice (21, 50, 81). Here, we show that that the 5′ upstream sequence from the mouse K17 gene (mK17) directs the expression of GFP, used as a reporter, in the appropriate cell types within epithelial appendages in transgenic mice. [mK17 5′]-GFP is also faithfully regulated during the embryonic development of skin epithelia and can be exploited to track hair follicle cycling in adult skin. Recently, the mK17 promoter was shown to be highly responsive to the transcription factor Gli2, a terminal effector of sonic hedgehog signaling, in a heterologous system ex vivo (7). Here we show that deletion of a 48-bp fragment within the mK17 promoter reduces Gli2 responsiveness in transfected cells, as well as the expression of [mK17 5′]-GFP transgene in hair follicles and other epithelial appendages in vivo. These findings have conceptual and practical implications for the study of epithelial appendages from a broad variety of angles in vivo.
MATERIALS AND METHODS
DNA constructs.
To generate [mK17 5′]-GFP, the 5′ upstream sequence of mK17 was amplified from phage clone 16-8 (40) by PCR and subcloned, and the ∼1,970-bp amplicon was sequenced. The mK17 promoter was inserted into the multiple cloning site of pEGFP-N1 (Clontech, Palo Alto, CA), using AseI and BamHI restriction digests. This removed the entire simian virus 40 promoter from pEGFP-N1 while preserving the poly(A) signal at the 3′ end. A 48-bp deletion, from −461 to −414 (+1 refers to the ATG start codon), was created as follows. Two promoter fragments, extending from −1974 to −461 (P1/P2 primers) and from −414 to +1 (P4/P6 primers), were generated by PCR using [mK17 5′]-GFP as a template. PCR conditions were 94°C for 1 min (denaturation), 65°C for 1 min (annealing), and 72°C for 1 min (elongation) for 30 cycles. The PCR primers used were as follows and contained suitable restriction enzyme sites as indicated to facilitate subcloning: mK17delA-P1 (sense), 5′-(EcoRV)GCGATATCTTTTTTTGCTTCCCTC-3′; mK17delA-P2 (antisense), 5′-CAAGCTTGATGGGAAATGAGGTAGGG(HindIII)-3′; mK17delA-P4 (sense), 5′-(HindIII)CGCAAGAAGCTTTTCTTGTCCGTATTAGG-3′; and mK17delA-P6 (antisense), 5′-TGGATCCATGGTGGCAGCGGGCAA(BamHI)-3′. PCR products were sequenced and subcloned as EcoRV-BamHI fragments into [mK17 5′]-GFP to generate [mK17 delA 5′]-GFP.
To generate the parent [mK17 −1950/+19]-luciferase construct, the PCR product used to produce [mK17 5′]-GFP was blunt ended with Klenow polymerase and subcloned into SmaI-digested pGL3-Basic vector (Promega, Madison WI). The [mK17 delA 5′]-luciferase construct was obtained by digesting [mK17 delA 5′]-GFP with ApaI and EcoRI and subcloning the excised fragment into the corresponding sites within [mK17 −1950/+19]-luciferase.
Transgenic mouse lines and production of staged mouse embryos.
Studies involving animals were approved by the Johns Hopkins University Animal Care and Use Committee. Transgenic founders were produced by pronuclear injection of GFP reporter DNA constructs in C57BL/6-BALB/c3 embryos (26). Founders were identified using Southern blotting and PCR with strategies aimed at detecting the GFP-coding sequence in genomic DNA. Southern blotting was used to compare transgene copy numbers between lines. Probes specific for the mK16 (2) or mK17 (5) locus were used as internal loading controls. When needed, band densities on computer-scanned Southern blots were quantitated using NIH Image software.
Transgenic lines were established by crossing the [mK17 5′]-GFP and [mK17 delA 5′]-GFP founders with wild-type C57BL/6-BALB/c3 mice. [mK17 5′]-GFP mice were also bred into the hairless background (80) by using the HRS strain available from Jackson Laboratories (Bar Harbor, ME). Once established, [mK17 5′]-GFP mice were screened using live fluorescence imaging (see below). Genomic DNA from [mK17 delA 5′]-GFP mice was screened using PCR.
To obtain embryos at defined developmental stages, timed pregnancies were initiated by mixing males and females and setting 12:00 noon the next day as embryonic day 0.5 (E0.5) (80). Pregnant females were euthanatized by cervical dislocation at noon on the desired day of embryonic development (ranging from E12.5 to E.18.5; term is at ∼E19), and embryos were surgically harvested. Live imaging was conducted or frozen sections were collected for immunolocalization studies.
RNA analyses.
Back skin, paw, whisker pad, tongue, stomach, liver, kidney, lung, spleen, brain, and heart tissues were surgically collected from adult mice, snap frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted by homogenizing these tissues in TRIzol reagent, following the manufacturer's protocol (Life Technologies, Inc., Gaithersburg, MD). RNA was quantitated spectrophotometrically using absorbance at 260 nm. cDNAs were synthesized via reverse transcription from 1 μg of RNA according to the manufacturer's protocol (Advantage for PCR; Clontech, Palo Alto, CA). This cDNA was then used as a template for PCR with either one of the following pairs of primers, specific for the GFP and mK17 transcripts; in both cases the PCR conditions were 94°C for 1 min (denaturation), 66°C for 1 min (annealing), and 72°C for 1 min (elongation) for 25 to 35 cycles: GFP-forward, 5′-GGCGACGTAAACGGCCACAAGTTCAGCG-3′; GFP-reverse, 5′-CGCTTCTCGTTGGGGTCTTTGCTCAGGG-3′; K17-forward, 5′-CCACCTGACTCAGTACAAGCCAAAAG-3′; and K17-reverse, 5′-CTGTGGCCTTTGTTCTGAACACTG-3′.
For Northern blot analyses (79), 10 μg of total RNA was resolved by formaldehyde-agarose gel electrophoresis, transferred to GeneScreen nylon membrane, and probed with a 714-bp-long cDNA probe obtained by digesting the GFP transgene with HphI and XbaI. The blot was then washed and exposed to radioautographic film.
Western blot analyses.
Hair clippings were harvested from the back skin of adult mice, and total proteins were extracted (39). Extracted protein samples (4 μg) were electrophoresed in either 8.5% or 10% SDS-polyacrylamide gels, to look for K17 or GFP immunoreactivity, respectively. Gels were then electroblotted onto Protran nitrocellulose (Schleicher & Schuell, Keene, NH). Bound primary antibodies directed against mouse K17 (40) or GFP (Clontech, Palo Alto, CA) were detected using enhanced chemiluminescence as described by the manufacturer (Amersham Biosciences Piscataway, NJ).
Morphological analysis and live imaging of GFP expression.
A Leica MZ FL III microscope equipped with an excitation filter of 470/40 nm and a barrier filter of 525/50 nm was used to view GFP fluorescence in living skin. Data were collected using a Zeiss AxioCam charge-coupled-device camera connected to a PC running the Zeiss AxioVision software. Animals were anesthetized via intraperitoneal administration of avertin prior to live imaging.
All tissues slated for frozen sectioning were submerged in Optimal Cutting Temperature compound (Tissue-Tek; Sakura Fineteck, Torrance, CA) and frozen at −80°C. Frozen tissue blocks were sectioned (5-μm width) by a Microm International HM 505 E microtome cyrostat operated at −25°C. Cryosections mounted on microscope glass slides were stored in the dark at −20°C until viewed via fluorescence microscopy. To relate the distribution of GFP to that of K17, we used indirect immunofluorescence using a rabbit polyclonal antiserum directed against K17 (40, 67). Consecutive slides were utilized, as the soluble GFP is rapidly washed off from unfixed fresh frozen tissue sections during the antibody staining procedure.
Cell culture and transfection.
308 mouse keratinocyte cells (66) were maintained in mKer medium (61), consisting of 3 parts Dulbecco's minimum essential medium, 1 part Ham's F-12 medium, 10% premium fetal bovine serum, 5 μg/ml insulin, 0.4 μg/ml hydrocortisone, 5 μg/ml transferrin, 2 × 10−9 M 3,3,5′-triiodo-l-thyroxine, 10−10 M cholera toxin,10 ng/ml epidermal growth factor, 60 μg/ml penicillin, and 25 μg/ml gentamicin. Twenty-four-well tissue culture plates (Falcon, Franklin Lakes, NJ) were seeded with 45,000 cells per well and cultured until the cells were ∼80% confluent. pCMVb (Clontech, Palo Alto, CA) (50 ng), which directs β-galactosidase gene expression, was routinely included for standardization. In addition, cells were transfected with 300 ng of an individual luciferase construct along with 150 ng of the mouse Gli2 expression vector (74), diluted in 50 μl of optiMEM medium (Invitrogen, Carlsbad, CA). Two microliters of Lipofectamine 2000 (Invitrogen) was added to 50 μl of optiMEM medium and incubated at room temperature for 5 min, and this mixture was then added to the DNA solution and left for a further 20 min. The mixture was overlaid on plated cells bathing in 400 μl of fresh optiMEM medium. After 12 h, the transfection medium was replaced with fresh mKer medium and cells harvested at 48 h posttransfection. Cell lysates were prepared by washing wells with phosphate-buffered saline, followed by addition of 150 μl of 1× Passive Lysis Buffer (Promega) and a 15-min incubation with agitation.
Analysis of luciferase activity.
Lysates of transfected cells were assayed for luciferase activity using the Luminoskan Ascent luminescent plate reader (Thermo Electron Corporation, Waltham, MA). Twenty microliters of cell lysate was pipetted into the wells of a 96-well microtiter plate (Microlite 1; Thermo Labsystems, Chantilly, VA). One hundred microliters of luciferase assay buffer [20 mM Tricine-NaOH (pH 7.8), 1.07 mM (MgCO3)4Mg(OH)2 · 5H2O, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM dithiothreitol, 270 μM coenzyme A, Li salt, 470 μM d-Luciferin, K salt, 530 μM ATP] was dispensed into each well and the luciferase activity allowed to integrate for 10 s. β-Galactosidase activity was assayed from a separate 20-μl aliquot, to which 100 μl of assay buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgCl2) and 25 μl of 2-mg/ml ortho-nitrophenyl-β-d-galactopyranoside was added. A 5-min incubation at 37°C was followed by addition of 75 μl of 1 M Na2CO3 to quench the reaction. Absorbance at 420 nm was measured on a Bio-Tek ELx800 universal microplate reader (Bio-Tek, Winooski, VT). Each construct was transfected in duplicate, and the data reported (means ± standard errors of the means) are the results of at least three independent experiments. Data were normalized and expressed as fold induction over the value obtained in cotransfection with the pCDNA3 vector.
[mK17 5′]-GFP transgene regulation during wound epithelialization in vivo.
For an analysis of [mK17 5′]-GFP reporter expression in adult transgenic mouse skin subjected to experimental injury or topical application of a chemical inducer and an analysis of the responsiveness of the mK17 5′ upstream sequence to terminal effectors of Wnt signaling, see Fig. S1 in the supplemental material.
RESULTS
The 5′ upstream region from mK17 confers appendage-specific gene expression in mice.
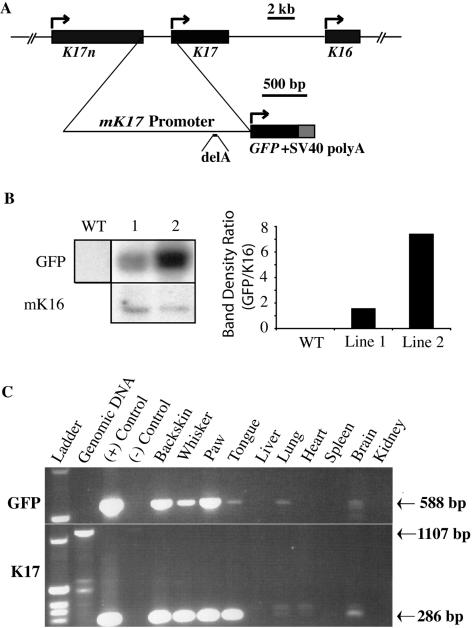
The [mK17 5′]-GFP transgene (Fig. 1A) utilizes, as its central feature, the cloned ∼1,970-bp-long 5′ upstream region from the mK17 gene (40), which corresponds to the entire intergenic region between mK17 and the telomeric mK17n on mouse chromosome 11 (67). Two transgenic mouse lines, designated 1 and 2 (Fig. 1B), were characterized. Line 1 shows stronger expression of GFP than line 2 (even though it has a lower copy number per genome [Fig. 1B]). Otherwise, lines 1 and 2 behave identically in the analyses reported below, which emphasize skin and oral mucosa, two major sites of K17 expression (40, 56, 70). Unless mentioned otherwise, the findings reported are from [mK17 5′]-GFP line 1.
FIG. 1.
Production and characterization of [mK17 5′]-GFP transgenic mouse lines. (A) Top, genomic context in which mK17 is located (data from reference 67). Bottom, structure of the [mK17 5′]-GFP transgene. The entire intergenic sequence between the telomeric mK17n and mK17 was subcloned upstream from the enhanced GFP coding sequence. A simian virus 40 poly(A) signal is used at the 3′end of the construct. (B) Southern blotting-based comparison of transgene copy numbers in [mK17 5′]-GFP lines 1 and 2. The single-copy mK16 is used as a reference. Quantitation was performed by densitometry. WT, wild type. (C) RT-PCR-based survey comparing GFP and K17 mRNA expression in tissues harvested from [mK17 5′]-GFP transgenic mice (line 1). “Ladder” refers to a 1-kb DNA ladder. (+) Control refers to the transgene construct. (−) Control refers to omission of the reverse transcription step. Two micrograms of total RNA was used in the reverse transcription step for each tissue. The expected amplicon products are as follows: GFP mRNA, 588 bp; K17 mRNA, 286 bp; K17 gene (reflecting genomic DNA contamination), 1,107 bp.
In reverse transcription-PCR (RT-PCR) assays applied on total RNA extracted from transgenic F1 progeny samples, [mK17 5′]-GFP mRNA occurs in all tissues also positive for mK17 mRNA, such as back skin, paw, whisker, and tongue (Fig. 1C) and stomach (data not shown). Small amounts of both transcripts occur in the lung (likely reflecting expression in mucous glands [40]) and brain. No expression of [mK17 5′]-GFP was found in tissues negative for mK17 mRNA, such as liver, heart, spleen, and kidney (Fig. 1C). Northern analysis confirmed the occurrence of an appropriately sized GFP transcript in RNA samples prepared from back skin and whisker pad, but not from liver, heart, and kidney, of [mK17 5′]-GFP mice (data not shown).
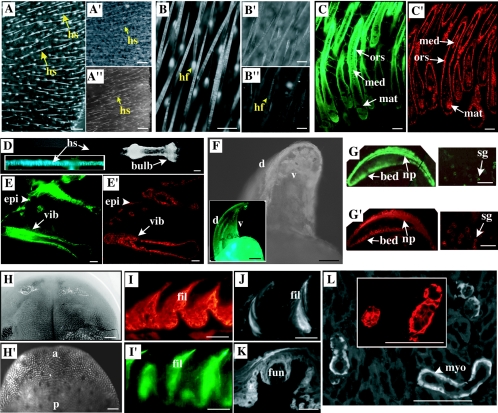
We next analyzed the distribution of GFP fluorescence by using macroscopic live imaging coupled with microscopic analysis on frozen tissue sections (Fig. 2). Intrinsic GFP fluorescence was readily detected through live imaging in tissues known to contain K17-expressing cells, including hair follicles (Fig. 2A [ear] and B [back skin]), vibrissae (Fig. 2D), nail (Fig. 2F), dorsal tongue surface (Fig. 2H), and palate (data not shown). Such fluorescence does not occur in corresponding tissues from wild-type mice (data not shown). In the case of hair follicles, GFP fluorescence occurs in the externally visible hair shaft and beneath its point of emergence at the skin surface (Fig. 2A). Upon tissue sectioning, the distribution of intrinsic GFP fluorescence coincides with that of K17 antigens. Thus, in pelage hair follicles, GFP fluorescence occurs in the outer root sheath, medulla, and matrix compartments (Fig. 2C and C′) (40, 42). Similar results were obtained for vibrissa follicles (Fig. 2 E and E′); for other epithelial appendages known to express K17 (40, 42), including nail (Fig. 2F and G), filiform and fungiform papillae of dorsal tongue epithelium (Fig. 2I to K), sweat glands (Fig. 2G), and the mucous glands embedded in the skeletal muscle of the tongue (Fig. 2L and L′); and for stomach (data not shown). These observations suggest that the [mK17 5′]-GFP transgene faithfully mimics the regulation of the endogenous gene in adult mouse skin, oral epithelia, and stomach.
FIG. 2.
Analysis of GFP expression in live adult skin tissue and in tissue sections. Three-month-old adult mice were used. (A to A") Fluorescence imaging of the ear surface, shown at low magnification, in adult transgenic mice from lines 1 (A) and 2 (A"). Arrows point to protruding hair shafts (hs). A′, bright-field image for frame A. Bars, 500 μm. (B to B") Series similar to panels A to A" (with the exception that panel B" is a nontransgenic control) shown at higher magnification. Arrowheads point to signal located beneath the skin surface, reflecting hair follicles (hf). Bars, 300 μm. (C and C′) Distribution of GFP fluorescence (C) and K17 antigens (C′) in consecutive tissue sections from back skin (line 1). mat, hair matrix; med, medulla of the hair shaft; ors, outer root sheath. Bars, 50 μm. (D) Visualization of GFP fluorescence in a plucked vibrissa (vib) follicle (D) and in a section thereof (inset). Bar, 100 μm. (E and E′) Comparison of the distribution of GFP (E) and K17 (E′) in a newborn whisker pad (line 1). epi, epidermis. Bars, 100 μm. (F) Live fluorescence imaging of nail (low magnification) from an adult transgenic mouse in lines 1 (F) and 2 (inset). d, dorsal side; v, ventral side. Bars, 500 μm. (G and G′) Comparison of GFP (G) and K17 (G′) in an adult digit tip, emphasizing nail and sweat gland histology (line 1). np, nail plate; sg, sweat gland. Bars, 100 μm. (H) Bright-field image of the dorsal surface of the tongue (low magnification) from an adult transgenic mouse (line 1). (H′) Corresponding live fluorescence imaging. a, anterior; p, posterior. Bars, 500 μm. (I to L′) Analysis of tissue sections prepared from a tongue sample from an adult transgenic mouse in line 1. (I and I′) Comparison of K17 (I) and GFP (I′) in sections from dorsal tongue epithelium. (J and K) GFP fluorescence in filiform (fil) and fungiform (fun) papillae. Bars, 50 μm. (L) Comparison of the distributions of GFP (L) and K17 (inset) in glandular tissue embedded within the tongue muscle. Signal is restricted to the myoepithelial layer (myo), as expected. Bars, 50 μm.
The [mK17 5′]-GFP transgene is appropriately regulated during skin development.
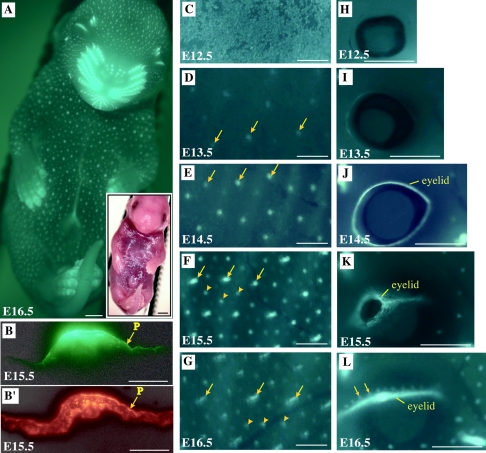
Two interesting aspects of mK17 regulation are the onset of its expression in ectodermal cells instructed to give rise to epithelial appendages during embryogenesis and in the periderm, including the subset of cells participating in the formation of temporary epithelial fusions (38, 40). Live imaging of E16.5 embryos reveals strong GFP fluorescence in all individual vibrissae and nails and in eyelids (Fig. 3). In addition, a geometric pattern of fluorescent dots of various sizes and intensities covers the embryos (Fig. 3A). Tissue sectioning confirmed that these dots are epithelial placodes (Fig. 3B) expressing the K17 antigen (Fig. 3B′), and it also highlighted the presence of GFP in the periderm (Fig. 3B) as expected (Fig. 3B′). The thin nature of periderm cells, along with the strong GFP signal emanating from placodes, likely accounts for the faint fluorescence seen at the surface of embryos subjected to live imaging (Fig. 3A).
FIG. 3.
Analysis of GFP expression in transgenic mouse embryos. All images correspond to GFP-based fluorescence seen in transgenic mouse embryos, unless stated otherwise. (A) Live imaging of an E16.5 embryo shown at low magnification. Inset, bright-field image. Bars, 1 mm. (B and B′) Comparison of the distribution of GFP fluorescence (B) and K17 antigens (B′) in consecutive tissue sections prepared from the back of an E15.5 embryo. A single placode is shown. “p” refers to the thin periderm layer covering the embryo. Bars, 50 μM. (C to L) Temporal series of live GFP fluorescence data collected from the trunk surface (C to G) and eye region (H to L) in embryos ranging from E12.5 to E16.5. Embryo age is indicated at the bottom left. In panels E to G, arrows and arrowheads depict primary and secondary placodes, respectively. In panel L, in which the eyelid is completely fused, arrows indicate sites of future eyelashes. Bars, 500 μm.
Closer examination of E12.5 to E16.5 transgenic embryos shows that GFP-expressing epithelial placodes emerge according to a specific spatiotemporal pattern (Fig. 3C to G). Regularly spaced placodes are first discernible at E13.5 by live imaging, and their fluorescence peaks in intensity by E14.5. At that time, a second set of smaller dots appears within the existing grid pattern. Between E14.5 and E16.5, these dots intensify as well, whereas the initial set of GFP dots become elongated (Fig. 3G), reflecting their maturation into primary hair germs (28, 57, 77). These sequential events correspond to the known formation of an initial set of guard hair follicles, followed by a second set of distinct follicle types (63). Over the same time frame, formation of temporary epithelial fusions at the level of eyelids (Fig. 3H to L), digits, and ear (data not shown) is also reflected by the pattern of GFP fluorescence. For eyelids, a single row of spherical dots can also be distinguished above the terminal fusion, depicting future eyelashes (Fig. 3L). Once again, expression of the [mK17 5′]-GFP transgene reflects known mK17 regulation in developing mouse ectoderm, which was previously characterized in detail (38, 40).
A 48-bp internal deletion in the mK17 promoter abrogates its activity in hair follicles in vivo.
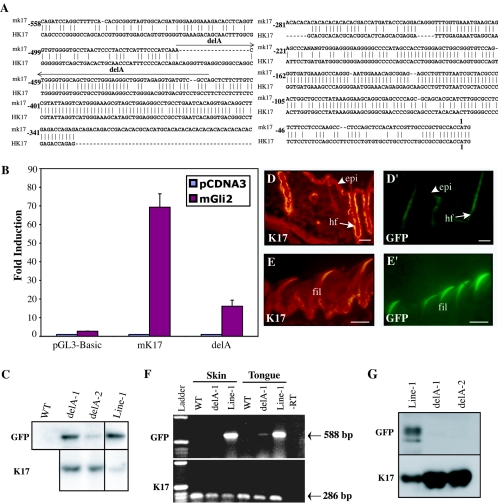
Through luciferase reporter assays performed with human skin keratinocytes, Callahan et al. (7) recently showed that the full-length mK17 promoter can be transactivated by the transcription factor Gli2. This finding provides a likely molecular basis for the consistent induction of mK17 in basal cell carcinoma of the skin, which is often associated with mutations causing deregulated sonic hedgehog signaling (see Discussion). From deletion analyses those authors inferred that a 41-nucleotide-long segment, designated −398/−448 bp (Fig. 1A), was likely to contain a Gli2-responsive element(s) (7). This region of the promoter features potential Gli binding sites that are conserved between mouse and human K17 genes (Fig. 4A).
FIG. 4.
A short internal sequence confers high responsiveness to Gli2 in transfected cells and appendageal expression in vivo. All images shown correspond to GFP-based fluorescence seen in transgenic mouse embryos, unless stated otherwise. (A) Nucleotide sequence alignment (ConSite, http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite/) for the proximal ∼500 bp of 5′upstream sequence from the mouse (mK17) and human K17 (hK17) genes. Vertical lines between the two sequences depict identity. The segment deleted between bp −461 and −414 in mK17 (delA) is identified by a line. (B) Luciferase reporter assays in 308 mouse keratinocytes cotransfected with Gli 2 and either [mK17 5′]-luciferase, [mK17 delA 5′]-luciferase, or pGL3-Basic (empty vector control). Luciferase activity for each construct is expressed as fold induction over the activity registered when cotransfected with pCDNA3. Error bars indicate standard errors of the means. (C) Southern blotting-based comparison of transgene copy number in [mK17 delA 5′]-GFP lines 1 and 2 with that of the parental [mK17 5′]-GFP line 1. The single-copy mK17 is used as a reference. WT, wild-type genomic DNA. (D to E′) Comparison of the distribution of K17 (D and E) and GFP (D′ and E′) fluorescence in consecutive tissue sections prepared from back skin (D and D′) and dorsal tongue epithelium (E and E′). epi, epidermis; hf, hair follicles; fil, filiform papillae. Bars, 50 μm. (F) RT-PCR-based survey comparing GFP mRNA expression in skin and tongue from wild-type (control), [mK17 5′]-GFP (line 1), and [mK17 delA 5′]-GFP (line DelA-1) transgenic mice. “Ladder,” 1-kb DNA ladder. −RT, omission of the reverse transcription step. Expected amplicon products: GFP mRNA, 588 bp; K17 mRNA, 286 bp. (G) Detection, by Western immunoblotting, of GFP (top) and K17 (bottom) antigens in total protein extracts prepared from hair clippings of [mK17 5′]-GFP (“Line-1”), and [mK17 delA 5′]-GFP (“delA-1 and delA-2”) transgenic mice.
To test this hypothesis, we generated an internal 48-bp deletion, Δ−461/−414 bp or delA (Fig. 1A), encompassing these potential Gli binding sites. Under our experimental conditions, the full-length mK17 promoter-driven luciferase construct showed 69-fold activation over baseline when cotransfected with a Gli2 expression vector in a mouse skin keratinocyte cell line (Fig. 4B). In contrast, the delA construct showed a much-reduced, 16-fold activation over baseline when coexpressed with Gli2 (Fig. 4B). When considering intrinsic promoter activity, [mK17 delA 5′]-luciferase is stronger than the parental [mK17 5′]-luciferase construct (data not shown), pointing to the presence of a repressor element within the Δ−461/−414 bp segment. These findings confirm the prediction of Callahan et al. (7) in showing that this short region within the mK17 promoter features an element(s) that contributes to confer strong responsiveness to Gli2.
The region encompassing the delA mutation was subcloned in the [mK17 5′]-GFP DNA construct (Fig. 1A). Microinjection in mouse embryos yielded two transgenic founders and their independent lines (Fig. 4C). In either case, no expression of the [mK17 delA 5′]-GFP transgene could be detected in skin tissue, using either live imaging of tissue sections (Fig. 4D and D′), an RT-PCR-based screen on total RNA samples (Fig. 4F), or Western immunoblotting of hair protein extracts (Fig. 4G). Likewise, no GFP fluorescence could be detected in various types of K17-expressing glands (data not shown). This deletion did not completely abrogate transgene activity, however, as evidenced by the presence of GFP fluorescence in the anterior column of filiform papillae in dorsal tongue epithelium (Fig. 4E and E′) and of GFP mRNA in tongue tissue (Fig. 4F). Interestingly, shh signaling is distinctly required for formation of fungiform papillae, where GFP expression is lost in the absence of the 48-bp fragment (23, 30). Residual activity in oral mucosa establishes that lack of expression in skin is not the result of transgene integration in a transcriptionally silent area of the mouse genome. While we cannot rule out that [mK17 delA 5′]-GFP exhibits an activity below the sensitivity threshold of our assays, these data establish that deletion of this internal 48-bp segment leads to a substantial loss of mK17 promoter activity in many types of epithelial appendages in vivo.
Visualizing hair cycling events in live [mK17 5′]-GFP skin tissue.
A potentially useful application of the [mK17 5′]-GFP mouse model is the visualization of the fate of K17-expressing keratinocytes during dynamic processes involving skin epithelia. The mature hair follicle, for instance, undergoes a cycle composed of three phases: anagen (growth stage), catagen (an apoptosis-driven regression stage), and telogen (resting stage) (24). In mice the first two hair cycles occur synchronously (albeit with an anterior-posterior gradient) over the initial ∼50 days following birth (9). The distribution of K17 in hair follicles (2, 40, 56) is such that variations in GFP fluorescence should reflect progression through this cycle.
Macroscopic live imaging was conducted on adult [mK17 5′]-GFP mice to test this possibility. In late-anagen-stage skin (P33), when hair follicles are fully extended within the dermis, GFP fluorescence appears as an elongated object projecting downward from the base of the protruding hair shaft at the skin surface (Fig. 5 A). By comparison, the GFP signal consists of significantly smaller dots in telogen-stage skin (P24), when resting follicles have regressed and shortened. Tissue sectioning followed by microscopic analysis confirmed that GFP fluorescence directly reflects hair follicle length, and closely mirrors that of endogenous K17, for these two stages of the hair cycle (compare Fig. 5A and B).
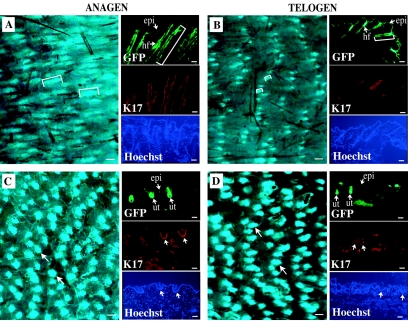
FIG. 5.
Monitoring of progression through the hair cycle and its defects in live transgenic mouse skin tissue. (A and B) [mK17 5′]-GFP transgenic mice (line 1) at P33 and P24, respectively, at which times hair follicles are in the anagen (A) and telogen (B) phases of their cycle. (C and D) P33 (C) and P24 (D) mice homozygous for the hairless allele as well as transgenic for [mK17 5′]-GFP (line 1). Panels A, B, C, and D are fluorescence recordings of live back skin tissue. Brackets depict the length of hair follicles based on GFP fluorescence. Bars, 200 μm. The small panels shown to the right of each main panel show data gathered from consecutive sections prepared from the same back skin tissues. Labeling is indicated in their lower left corners. Hoechst staining was performed to localize individual nuclei in the preparations stained for K17. Bars, 50 μm. The arrows in panels C and D point to utricles (ut). epi, epidermis; hf, hair follicles.
A similar hair staging analysis was conducted on [mK17 5′]-GFP mice that had been bred into the Hairless background (Hr). In contrast to the initial anagen, which is properly executed, pelage hair follicles fail to reenter anagen in mice homozygous for the mutant hairless allele. Progressive shedding of telogen-stage hair then gives rise to a permanent hairless phenotype in Hrhr/hr mice (6, 54). An enlargement of the infundibulum region in the uppermost region of hair follicles, which adopts a pronounced cup shape coinciding with aberrant execution of epidermal differentiation (54, 82), is the earliest morphological alteration in Hrhr/hr skin. These structures have been designated utricles. At P24, the GFP-positive dots seen in [mK17 5′]-GFP/Hrhr/hr mice (Fig. 5C) are much larger than those in the parental [mK17 5′]-GFP line (Fig. 5B). Tissue sectioning shows that these enlarged dots correspond to utricles, which also express endogenous K17 (Fig. 5C) (82). These GFP-positive dots have grown larger in size by P33 in living [mK17 5′]-GFP/Hrhr/hr skin, again reflecting corresponding changes in tissue sections (Fig. 5D). Collectively, these observations establish that progression through the hair cycle, and its defects, can be monitored through live imaging of intact [mK17 5′]-GFP mouse skin.
DISCUSSION
The mK17 gene promoter confers appendage-preferred expression in transgenic mice.
Two kilobases of 5′ upstream sequence for mK17, corresponding to the full intergenic sequence between mK17 and the telomeric mK17n (67), suffice to drive expression of a reporter sequence with the correct spatiotemporal pattern in developing and mature epithelial appendages of mouse. A 48-bp element within the mK17 promoter participates in conferring responsiveness to Gli2, as assessed in transfected keratinocytes in culture, and is required for its activity in most epithelial appendages in vivo. Visualization of [mK17 5′]-GFP reporter activity enables the tracking of placode appearance and their subsequent expansion in live mouse embryos, as well as the progression of the pelage hair follicles though their growth cycle in live adult mice. These studies thus introduce [mK17 5′]-GFP mice as a unique model whose utilization can provide, as discussed below, novel insight into the biology of epithelial appendages.
Whereas K17 is rapidly induced after a variety of insults to human and mouse skin (41), we find that neither acute injury nor the topical application of a phorbol ester leads to [mK17 5′]-GFP activation in transgenic epidermis (see Fig. S1 in the supplemental material). It follows that one or several cis-acting elements required for the manifestation of this aspect of mK17 regulation are located outside its proximal 5′ upstream sequence. They could reside in an intron and/or in the 3′ noncoding sequence of mK17 or, alternatively, elsewhere in the type I keratin locus. Of note, mK16, the gene centromeric to mK17 (40, 67), is wound inducible and phorbol ester inducible as well (53, 75). In contrast, the telomeric mK17n is not wound inducible (67). The regulatory elements responsible for induction following injury and other environmental challenges are expected to be conserved between the human and mouse K17 genes and are under investigation. Meanwhile, the lack of inducibility in vivo simplifies further study and exploitation of the mK17 gene promoter.
Implications for the molecular mechanisms regulating K17 gene expression.
Several lines of evidence point to mK17 as a likely target of shh signaling in vivo. shh-expressing cells are asymmetrically located within the hair matrix at the early anagen stage of the hair cycle (17), and these cells show higher levels of K17 antigen (39). Also, K17 is consistently expressed in basal cell carcinoma, which often arises from mutations resulting in enhanced shh signaling, in both human skin (34) and mouse models thereof (18, 19, 52). Above all, however, is the recent finding that the activity of the full-length mK17 promoter is stimulated when coexpressed alongside Gli2 in transfected human and mouse keratinocytes (7; this study). Here, we provide direct evidence that a 48-bp fragment, which is located between nucleotides −461 and −414 in the mK17 promoter and exhibits several potential Gli binding sites (7), contributes a significant fraction of the Gli responsiveness of mK17 in cultured skin keratinocytes.
Whether the loss of reporter activity in most epithelial appendages of [mK17 delA 5′]-GFP mice directly reflects decreased Gli2 responsiveness is an enticing prospect that requires further testing through more refined molecular studies. Importantly, the deleted ∼48-bp motif in [mK17 delA 5′]-GFP possesses potential binding sites for other transcription factors, such as Sp1 and AP2 (data not shown). Sp1 has been shown to modulate keratin expression in the hair follicle (4), while AP2 has defined roles in hair follicle development and cycling (55). Both Sp1 and AP2 have been shown to bind sequences located within the proximal ∼450 bp in the human K17 promoter (43). However, the binding sites for these factors are not conserved in mK17 (43). Again, more refined studies on this region of the mK17 and hK17 promoters will be required to define the role of these and other transcription factors in regulating their activity.
The significance of K17 as an shh target gene is an issue worth considering. Expression of K17 has been inferred to promote the survival of hair keratinocytes during the anagen phase of the hair cycle (42). The occurrence of mK17 transcription in keratinocytes exposed to shh, a signal leading to cell growth in hair and other epithelial appendages (27, 44, 51), therefore seems logical. shh responsiveness cannot, however, account for the onset of mK17 expression during development. Indeed, K17 expression can be detected in the early-stage epithelial placodes that form in shh null embryonic ectoderm (10, 65). The signal(s) activating mK17 transcription at the precursor placode stage thus remains to be defined.
We previously suggested, based on several arguments, that K17 could be a direct Wnt target gene during the early stages of placode formation in the embryonic ectoderm (31, 40). This notion was recently reinforced by lack of detectable K17 expression in transgenic embryos whose ectoderm ectopically expresses a soluble Wnt inhibitor (1). Surprisingly, however, we find that the activity of the mK17 gene promoter is not affected by lef-1 or tcf-4 expressed alone, or in combination with activated β-catenin, in transfected Cos1 epithelial cells (data not shown; see Fig. S1 in the supplemental material). These findings thus do not support the view that a canonical Wnt signal single-handedly underlies the onset of mK17 expression in the embryonic ectoderm. Possibly, a proximal determinant(s) conferring Wnt responsiveness could be located outside the mK17 5′ upstream sequence. Alternatively, another signal could be required alongside Wnt (29, 78), or a noncanonical Wnt signal could be involved (22).
Potential applications of the mK17 promoter and [mK17 5′]-GFP mice.
The activity profile of the mK17 promoter is distinct from those of other promoters shown to be preferentially active in transgenic mouse skin epithelia. These include, but are not limited to, K5 and K14 (46, 59, 62, 73), whose 5′ upstream sequences confer expression in the basal layer of stratified epithelia; K15, which preferentially labels the presumptive epithelial stem cells located within the hair bulge (37, 45); K1 (20, 60), which unexpectedly confers expression in all layers of the epidermis; involucrin (8, 13), which, as expected, is active in the differentiating layers of epidermis and in hair follicles; and hard keratin genes (36, 58), which are active in the hair cortex and nail plate. As such, the mK17 promoter can be exploited to drive the expression of various open reading frames, including the Cre recombinase, in transgenic mice (47). Similar K14 promoter- and K5-promoter driven “Cre” mice have been exploited for conditional gene manipulation in epidermis and other stratified epithelia (3, 72). Such K17 promoter-driven “Cre donor” mice would, upon mating to mice carrying floxed alleles, enable appendage-preferred gene manipulation.
The existing [mK17 5′]-GFP mice can be used, through fluorescence-activated cell sorting, to purify subpopulations of epithelial cells from dissected embryonic and adult tissues. Other reporter mouse models, exhibiting GFP expression in a restricted and interesting pattern within skin epithelia, have been devised (5, 35, 45, 71). In two instances, fluorescence-activated cell sorting was exploited to isolate and characterize populations of hair follicle cells enriched in epithelial stem cells (69, 71). [mK17 5′]-GFP mice could also be used as a test bed in which to test strategies aimed at modulating gene expression in live skin. Examples include small interfering RNA-mediated decreases in GFP expression in epithelial appendages or screens for small chemicals capable of enhancing or decreasing [mK17 5′]-GFP expression in skin. Either of these approaches is potentially desirable in the context of molecular therapies aimed at treating various types of skin conditions, including keratin-based genetic disorders.
Supplementary Material
Acknowledgments
We thank the Hopkins Transgenic Core Facility for production of transgenic founders; Randall Reed for providing access to his GFP imaging stereoscope; Catherine Thompson for access to a luminometer and for comments; Philip Beachy for the Gli2 expression vector; Bert Vogelstein, Harold Varmus, Elaine Fuchs, and Rudolf Grosschedl for reagents enabling us to test for Wnt-dependent transcription; and members of the Coulombe laboratory for support.
This work was supported by grant AR44232 from the National Institutes of Health to P.A.C.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andl, T., S. T. Reddy, T. Gaddapara, and S. E. Millar. 2002. WNT signals are required for the initiation of hair follicle development. Dev. Cell 2:643-653. [DOI] [PubMed] [Google Scholar]
- 2.Bernot, K. M., P. A. Coulombe, and K. M. McGowan. 2002. Keratin 16 expression defines a subset of epithelial cells during skin morphogenesis and the hair cycle. J. Investig. Dermatol. 119:1137-1149. [DOI] [PubMed] [Google Scholar]
- 3.Brakebusch, C., R. Grose, F. Quondamatteo, A. Ramirez, J. L. Jorcano, A. Pirro, M. Svensson, R. Herken, T. Sasaki, R. Timpl, S. Werner, and R. Fassler. 2000. Skin and hair follicle integrity is crucially dependent on beta 1 integrin expression on keratinocytes. EMBO J. 19:3990-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brembeck, F. H., and A. K. Rustgi. 2000. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J. Biol. Chem. 275:28230-28239. [DOI] [PubMed] [Google Scholar]
- 5.Bruen, K. J., C. A. Campbell, W. G. Schooler, S. deSerres, B. A. Cairns, C. S. Hultman, A. A. Meyer, and S. H. Randell. 2004. Real-time monitoring of keratin 5 expression during burn re-epithelialization. J. Surg. Res. 120:12-20. [DOI] [PubMed] [Google Scholar]
- 6.Cachon-Gonzalez, M. B., S. Fenner, J. M. Coffin, C. Moran, S. Best, and J. P. Stoye. 1994. Structure and expression of the hairless gene of mice. Proc. Natl. Acad. Sci. USA 91:7717-7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callahan, C. A., T. Ofstad, L. Horng, J. K. Wang, H. H. Zhen, P. A. Coulombe, and A. E. Oro. 2004. MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 18:2724-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, J. M., K. M. Albers, J. A. Garlick, R. Harrington, and L. B. Taichman. 1993. Tissue- and stratum-specific expression of the human involucrin promoter in transgenic mice. Proc. Natl. Acad. Sci. USA 90:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chase, H. B. 1954. Growth of hair. Physiol. Rev. 34:113-126. [DOI] [PubMed] [Google Scholar]
- 10.Chiang, C., R. Z. Swan, M. Grachtchouk, M. Bolinger, Y. Litingtung, E. K. Robertson, M. K. Cooper, W. Gaffield, H. Westphal, P. A. Beachy, and A. A. Dlugosz. 1999. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev. Biol. 205:1-9. [DOI] [PubMed] [Google Scholar]
- 11.Coulombe, P. A., and K. M. Bernot. 2004. Keratins and the skin, p. 497-504. In M. D. Lane and W. Lennarz (ed.), Encyclopedia of biological chemistry. Elsevier Science, New York, N.Y.
- 12.Coulombe, P. A., S. Ma, and M. Wawersik. 2001. Intermediate filaments at a glance. J. Cell Sci. 114:4345-4347. [DOI] [PubMed] [Google Scholar]
- 13.Crish, J. F., J. M. Howard, T. M. Zaim, S. Murthy, and R. L. Eckert. 1993. Tissue-specific and differentiation-appropriate expression of the human involucrin gene in transgenic mice: an abnormal epidermal phenotype. Differentiation 53:191-200. [DOI] [PubMed] [Google Scholar]
- 14.Ellis, T., I. Smyth, E. Riley, J. Bowles, C. Adolphe, J. A. Rothnagel, C. Wicking, and B. J. Wainwright. 2003. Overexpression of Sonic Hedgehog suppresses embryonic hair follicle morphogenesis. Dev. Biol. 263:203-215. [DOI] [PubMed] [Google Scholar]
- 15.Freedberg, I. M., M. Tomic-Canic, M. Komine, and M. Blumenberg. 2001. Keratins and the keratinocyte activation cycle. J. Investig. Dermatol. 116:633-640. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, E. 1995. Keratins and the skin. Annu. Rev. Cell Dev. Biol. 11:123-153. [DOI] [PubMed] [Google Scholar]
- 17.Gat, U., R. DasGupta, L. Degenstein, and E. Fuchs. 1998. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell 95:605-614. [DOI] [PubMed] [Google Scholar]
- 18.Grachtchouk, M., R. Mo, S. Yu, X. Zhang, H. Sasaki, C. C. Hui, and A. A. Dlugosz. 2000. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet. 24:216-217. [DOI] [PubMed] [Google Scholar]
- 19.Grachtchouk, V., M. Grachtchouk, L. Lowe, T. Johnson, L. Wei, A. Wang, F. de Sauvage, and A. A. Dlugosz. 2003. The magnitude of hedgehog signaling activity defines skin tumor phenotype. EMBO J. 22:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenhalgh, D. A., and D. R. Roop. 1994. Dissecting molecular carcinogenesis: development of transgenic mouse models by epidermal gene targeting. Adv. Cancer Res. 64:247-296. [DOI] [PubMed] [Google Scholar]
- 21.Grootjans, J. J., G. Reekmans, H. Ceulemans, and G. David. 2000. Syntenin-syndecan binding requires syndecan-synteny and the co-operation of both PDZ domains of syntenin. J. Biol. Chem. 275:19933-19941. [DOI] [PubMed] [Google Scholar]
- 22.Habas, R., and I. B. Dawid. 2005. Dishevelled and Wnt signaling: is the nucleus the final frontier? J. Biol. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, J. M., J. E. Hooper, and T. E. Finger. 1999. Expression of sonic hedgehog, patched, and Gli1 in developing taste papillae of the mouse. J Comp. Neurol. 406:143-155. [DOI] [PubMed] [Google Scholar]
- 24.Hardy, M. H. 1992. The secret life of the hair follicle. Trends Genet. 8:55-61. [DOI] [PubMed] [Google Scholar]
- 25.Hesse, M., T. M. Magin, and K. Weber. 2001. Genes for intermediate filament proteins and the draft sequence of the human genome: novel keratin genes and a surprisingly high number of pseudogenes related to keratin genes 8 and 18. J. Cell Sci. 114:2569-2575. [DOI] [PubMed] [Google Scholar]
- 26.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 27.Hutchin, M. E., M. S. Kariapper, M. Grachtchouk, A. Wang, L. Wei, D. Cummings, J. Liu, L. E. Michael, A. Glick, and A. A. Dlugosz. 2005. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 19:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iseki, S., A. Araga, H. Ohuchi, T. Nohno, H. Yoshioka, F. Hayashi, and S. Noji. 1996. Sonic hedgehog is expressed in epithelial cells during development of whisker, hair, and tooth. Biochem. Biophys. Res. Commun. 218:688-693. [DOI] [PubMed] [Google Scholar]
- 29.Jamora, C., R. DasGupta, P. Kocieniewski, and E. Fuchs. 2003. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung, H. S., V. Oropeza, and I. Thesleff. 1999. Shh, Bmp-2, Bmp-4 and Fgf-8 are associated with initiation and patterning of mouse tongue papillae. Mech. Dev. 81:179-182. [DOI] [PubMed] [Google Scholar]
- 31.Korinek, V., N. Barker, K. Willert, M. Molenaar, J. Roose, G. Wagenaar, M. Markman, W. Lamers, O. Destree, and H. Clevers. 1998. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 18:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 33.Kurzen, H., L. Esposito, L. Langbein, and W. Hartschuh. 2001. Cytokeratins as markers of follicular differentiation: an immunohistochemical study of trichoblastoma and basal cell carcinoma. Am. J. Dermatopathol. 23:501-509. [DOI] [PubMed] [Google Scholar]
- 34.Lacour, J. P. 2002. Carcinogenesis of basal cell carcinomas: genetics and molecular mechanisms. Br. J. Dermatol. 146(Suppl. 61):17-19. [DOI] [PubMed] [Google Scholar]
- 35.Li, L., J. Mignone, M. Yang, M. Matic, S. Penman, G. Enikolopov, and R. M. Hoffman. 2003. Nestin expression in hair follicle sheath progenitor cells. Proc. Natl. Acad. Sci. USA 100:9958-9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, M. H., C. Leimeister, M. Gessler, and R. Kopan. 2000. Activation of the Notch pathway in the hair cortex leads to aberrant differentiation of the adjacent hair-shaft layers. Development 127:2421-2432. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Y., S. Lyle, Z. Yang, and G. Cotsarelis. 2003. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J. Investig. Dermatol. 121:963-968. [DOI] [PubMed] [Google Scholar]
- 38.Mazzalupo, S., and P. A. Coulombe. 2001. A reporter transgene based on a human keratin 6 gene promoter is specifically expressed in the periderm of mouse embryos. Mech. Dev. 100:65-69. [DOI] [PubMed] [Google Scholar]
- 39.McGowan, K. M., and P. A. Coulombe. 2000. Keratin 17 expression in the hard epithelial context of the hair and nail, and its relevance for the pachyonychia congenita phenotype. J. Investig. Dermatol. 114:1101-1107. [DOI] [PubMed] [Google Scholar]
- 40.McGowan, K. M., and P. A. Coulombe. 1998. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143:469-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGowan, K. M., and P. A. Coulombe. 1998. The wound repair associated keratins 6, 16, and 17: insights into the role of intermediate filaments in specifying cytoarchitecture, p. 141-165. In J. R. Harris and H. Herrmann (ed.), Subcellular biochemistry: intermediate filaments. Plenum Publishing Co., London, United Kingdom. [PubMed]
- 42.McGowan, K. M., X. Tong, E. Colucci-Guyon, F. Langa, C. Babinet, and P. A. Coulombe. 2002. Keratin 17 null mice exhibit age- and strain-dependent alopecia. Genes Dev. 16:1412-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milisavljevic, V., I. M. Freedberg, and M. Blumenberg. 1996. Characterization of nuclear protein binding sites in the promoter of keratin K17 gene. DNA Cell Biol. 15:65-74. [DOI] [PubMed] [Google Scholar]
- 44.Mill, P., R. Mo, H. Fu, M. Grachtchouk, P. C. Kim, A. A. Dlugosz, and C. C. Hui. 2003. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 17:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris, R. J., Y. Liu, L. Marles, Z. Yang, C. Trempus, S. Li, J. S. Lin, J. A. Sawicki, and G. Cotsarelis. 2004. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22:411-417. [DOI] [PubMed] [Google Scholar]
- 46.Murillas, R., F. Larcher, C. J. Conti, M. Santos, A. Ullrich, and J. L. Jorcano. 1995. Expression of a dominant negative mutant of epidermal growth factor receptor in the epidermis of transgenic mice elicits striking alterations in hair follicle development and skin structure. EMBO J. 14:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy, A. 2000. Recombinase: the universal reagent for genome tailoring. Genesis 26:99-109. [PubMed] [Google Scholar]
- 48.Neznanov, N. S., and R. G. Oshima. 1993. cis regulation of the keratin 18 gene in transgenic mice. Mol. Cell. Biol. 13:1815-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Guin, W. M., A. Schermer, M. Lynch, and T. T. Sun. 1990. Differentiation-specific expression of keratin pairs, p. 301-334. In R. D. Goldman and P. M. Steinert (ed.), Cellular and molecular biology of intermediate filaments. Plenum Publishing Corp., New York, N.Y.
- 50.Okabe, M., M. Ikawa, K. Kominami, T. Nakanishi, and Y. Nishimune. 1997. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407:313-319. [DOI] [PubMed] [Google Scholar]
- 51.Oro, A. E., and K. Higgins. Hair cycle regulation of Hedgehog signal reception. Dev. Biol. 255:238-248. [DOI] [PubMed]
- 52.Oro, A. E., K. M. Higgins, Z. Hu, J. M. Bonifas, E. H. Epstein, Jr., and M. P. Scott. 1997. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science 276:817-821. [DOI] [PubMed] [Google Scholar]
- 53.Paladini, R. D., K. Takahashi, N. S. Bravo, and P. A. Coulombe. 1996. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J. Cell Biol. 132:381-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panteleyev, A. A., N. V. Botchkareva, J. P. Sundberg, A. M. Christiano, and R. Paus. 1999. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am. J. Pathol. 155:159-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panteleyev, A. A., P. J. Mitchell, R. Paus, and A. M. Christiano. 2003. Expression patterns of the transcription factor AP-2alpha during hair follicle morphogenesis and cycling. J. Investig. Dermatol. 121:13-19. [DOI] [PubMed] [Google Scholar]
- 56.Panteleyev, A. A., R. Paus, R. Wanner, W. Nurnberg, S. Eichmuller, R. Thiel, J. Zhang, B. M. Henz, and T. Rosenbach. 1997. Keratin 17 gene expression during the murine hair cycle. J. Investig. Dermatol. 108:324-329. [DOI] [PubMed] [Google Scholar]
- 57.Paus, R., K. Foitzik, P. Welker, S. Bulfone-Paus, and S. Eichmuller. 1997. Transforming growth factor-beta receptor type I and type II expression during murine hair follicle development and cycling. J. Investig. Dermatol. 109:518-526. [DOI] [PubMed] [Google Scholar]
- 58.Powell, B. C., and G. E. Rogers. 1990. Cyclic hair-loss and regrowth in transgenic mice overexpressing an intermediate filament gene. EMBO J. 9:1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez, A., A. Bravo, J. L. Jorcano, and M. Vidal. 1994. Sequences 5′ of the bovine keratin 5 gene direct tissue- and cell-type-specific expression of a lacZ gene in the adult and during development. Differentiation 58:53-64. [DOI] [PubMed] [Google Scholar]
- 60.Rosenthal, D. S., P. M. Steinert, S. Chung, C. A. Huff, J. Johnson, S. H. Yuspa, and D. R. Roop. 1991. A human epidermal differentiation-specific keratin gene is regulated by calcium but not negative modulators of differentiation in transgenic mouse keratinocytes. Cell Growth Differ. 2:107-113. [PubMed] [Google Scholar]
- 61.Rouabhia, M., L. Germain, F. Belanger, R. Guignard, and F. A. Auger. 1992. Optimization of murine keratinocyte culture for the production of graftable epidermal sheets. J. Dermatol. 19:325-334. [DOI] [PubMed] [Google Scholar]
- 62.Saitou, M., S. Sugai, T. Tanaka, K. Shimouchi, E. Fuchs, S. Narumiya, and A. Kakizuka. 1995. Inhibition of skin development by targeted expression of a dominant-negative retinoic acid receptor. Nature 374:159-162. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt-Ullrich, R., and R. Paus. 2005. Molecular principles of hair follicle induction and morphogenesis. Bioessays 27:247-261. [DOI] [PubMed] [Google Scholar]
- 64.Schorpp, M., T. Schlake, D. Kreamalmeyer, P. M. Allen, and T. Boehm. 2000. Genetically separable determinants of hair keratin gene expression. Dev. Dyn. 218:537-543. [DOI] [PubMed] [Google Scholar]
- 65.St. Jacques, B., H. R. Dassule, I. Karavanova, V. A. Botchkarev, J. Li, P. S. Danielian, J. A. McMahon, P. M. Lewis, R. Paus, and A. P. McMahon. 1998. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8:1058-1068. [DOI] [PubMed] [Google Scholar]
- 66.Strickland, J. E., D. A. Greenhalgh, A. Koceva-Chyla, H. Hennings, C. Restrepo, M. Balaschak, and S. H. Yuspa. 1998. Development of murine epidermal cell lines which contain an activated rasHa oncogene and form papillomas in skin grafts on athymic nude mouse hosts. Cancer Res. 48:165-169. [PubMed] [Google Scholar]
- 67.Tong, X., and P. A. Coulombe. 2004. A novel mouse type I intermediate filament gene, keratin 17n (K17n), exhibits preferred expression in nail tissue. J. Investig. Dermatol. 122:965-970. [DOI] [PubMed] [Google Scholar]
- 68.Travis, A., J. Hagman, and R. Grosschedl. 1991. Heterogeneously initiated transcription from the pre-B- and B-cell-specific mb-1 promoter: analysis of the requirement for upstream factor-binding sites and initiation site sequences. Mol. Cell. Biol. 11:5756-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trempus, C. S., R. J. Morris, C. D. Bortner, G. Cotsarelis, R. S. Faircloth, J. M. Reece, and R. W. Tennant. 2003. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J. Investig. Dermatol. 120:501-511. [DOI] [PubMed] [Google Scholar]
- 70.Troyanovsky, S. M., V. I. Guelstein, T. A. Tchipysheva, V. A. Krutovskikh, and G. A. Bannikov. 1989. Patterns of expression of keratin 17 in human epithelia: dependency on cell position. J. Cell Sci. 93:419-426. [DOI] [PubMed] [Google Scholar]
- 71.Tumbar, T., G. Guasch, V. Greco, C. Blanpain, W. E. Lowry, M. Rendl, and E. Fuchs. 2004. Defining the epithelial stem cell niche in skin. Science 303:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vasioukhin, V., L. Degenstein, B. Wise, and E. Fuchs. 1999. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA 96:8551-8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vassar, R., M. Rosenberg, S. Ross, A. Tyner, and E. Fuchs. 1989. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl. Acad. Sci. USA 86:1563-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, B., J. F. Fallon, and P. A. Beachy. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100:423-434. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Y. N., and W. C. Chang. 2003. Induction of disease-associated keratin 16 gene expression by epidermal growth factor is regulated through cooperation of transcription factors Sp1 and c-Jun. J. Biol. Chem. 278:45848-45857. [DOI] [PubMed] [Google Scholar]
- 76.Wang, Z., P. Wong, L. Langbein, J. Schweizer, and P. A. Coulombe. 2003. Type II epithelial keratin 6hf (K6hf) is expressed in the companion layer, matrix, and medulla in anagen-stage hair follicles. J. Investig. Dermatol. 121:1276-1282. [DOI] [PubMed] [Google Scholar]
- 77.Widelitz, R. B., and C. M. Chuong. 1999. Early events in skin appendage formation: induction of epithelial placodes and condensation of dermal mesenchyme. J. Investig. Dermatol. Symp. Proc. 4:302-306. [DOI] [PubMed] [Google Scholar]
- 78.Williams, B. O., G. D. Barish, M. W. Klymkowsky, and H. E. Varmus. 2000. A comparative evaluation of beta-catenin and plakoglobin signaling activity. Oncogene 19:5720-5728. [DOI] [PubMed] [Google Scholar]
- 79.Wong, P., E. Colucci-Guyon, K. Takahashi, C. Gu, C. Babinet, and P. A. Coulombe. 2000. Introducing a null mutation in the mouse K6alpha and K6beta genes reveals their essential structural role in the oral mucosa. J. Cell Biol. 150:921-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong, P., and P. A. Coulombe. 2003. Loss of keratin 6 (K6) proteins reveals a function for intermediate filaments during wound repair. J. Cell Biol. 163:327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaguchi, M., H. Saito, M. Suzuki, and K. Mori. 2000. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport 11:1991-1996. [DOI] [PubMed] [Google Scholar]
- 82.Zarach, J. M., G. M. Beaudoin III, P. A. Coulombe, and C. C. Thompson. 2004. The co-repressor hairless has a role in epithelial cell differentiation in the skin. Development 131:4189-4200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.