Abstract
The levels of replication-dependent histone mRNAs are coordinately regulated with DNA synthesis. A major regulatory step in histone mRNA metabolism is regulation of the half-life of histone mRNAs. Replication-dependent histone mRNAs are the only metazoan mRNAs that are not polyadenylated. Instead, they end with a conserved stem-loop structure, which is recognized by the stem-loop binding protein (SLBP). SLBP is required for histone mRNA processing, as well as translation. We show here, using histone mRNAs whose translation can be regulated by the iron response element, that histone mRNAs need to be actively translated for their rapid degradation following the inhibition of DNA synthesis. We also demonstrate the requirement for translation using a mutant SLBP which is inactive in translation. Histone mRNAs are not rapidly degraded when DNA synthesis is inhibited or at the end of S phase in cells expressing this mutant SLBP. Replication-dependent histone mRNAs have very short 3′ untranslated regions, with the stem-loop located 30 to 70 nucleotides downstream of the translation termination codon. We show here that the stability of histone mRNAs can be modified by altering the position of the stem-loop, thereby changing the distance from the translation termination codon.
Eukaryotic chromosomes are composed of equal amounts of DNA and histone proteins. Each time a eukaryotic cell divides, it must not only properly replicate its DNA but also synthesize large amounts of histones to package the DNA into chromatin properly. In order to coordinate the synthesis of histones and DNA, mammalian cells tightly regulate the concentrations of the replication-dependent histone mRNAs with DNA synthesis. Most of this regulation occurs at posttranscriptional levels, and in mammalian cells a major regulatory step is the regulation of the half-life of histone mRNA. The levels of replication-dependent histone mRNAs are cell cycle regulated. Histone mRNA levels increase 35-fold as cells enter S phase, and they are rapidly degraded at the end of S phase (12). In addition, replication-dependent histone mRNAs are rapidly degraded when cells are treated with inhibitors of DNA replication (29).
Replication-dependent histone mRNAs are the only metazoan mRNAs that are not polyadenylated. Instead, they end in a conserved stem-loop structure (5), which is necessary and sufficient for the rapid degradation of histone mRNAs after the inhibition of DNA replication (24). The stem-loop structure is recognized by a 31-kDa protein called the stem-loop binding protein (SLBP). SLBP is necessary for histone pre-mRNA processing (7) as well as translation of histone mRNAs, and a 15-amino-acid region of SLBP required for translational activation of histone mRNAs has been identified (27).
Histone mRNAs are rapidly degraded following treatment of cells with inhibitors of DNA synthesis. In contrast, treatment of cells with inhibitors of protein synthesis prevents rapid histone mRNA degradation after the inhibition of DNA synthesis (2) and increases histone mRNA levels in S-phase cells (31). However, it is not clear whether this is because continued protein synthesis or active translation of histone mRNAs is required for regulated histone mRNA degradation. We show here using two approaches that histone mRNAs need to be actively translated for their rapid degradation following the inhibition of DNA replication. First, we used a histone mRNA whose translation can be regulated by changing the intracellular iron concentration, due to the insertion of an iron-responsive element into the 5′ untranslated region (UTR) (8). When the translation of this mRNA was inhibited, it was not degraded after the inhibition of DNA synthesis. Second, when a mutant SLBP which cannot support efficient histone mRNA translation was expressed in HeLa cells, histone mRNAs were not rapidly degraded either following the inhibition of DNA synthesis or at the end of S phase. Therefore, the mechanisms of histone mRNA degradation at the end of S phase and following the inhibition of DNA synthesis might be similar. Taken together, these results demonstrate that translation of replication-dependent histone mRNAs is necessary for their regulated degradation in the absence of DNA synthesis.
Replication-dependent histone mRNAs have very short 3′ UTRs. The stem-loop at the 3′ end of the histone mRNAs starts 30 to 70 nucleotides (nt) from the translation termination codon in all known metazoan histone mRNAs. The fact that if the stem-loop is moved over 300 nt downstream of the translation termination codon histone mRNAs are not degraded either after the inhibition of DNA synthesis (10) or at the end of S phase (12) suggests that the stem-loop has to be located at a proper distance from the termination codon for the rapid degradation of histone mRNAs in the absence of DNA synthesis. By changing the position of the stem-loop structure with respect to the translation termination codon without changing the open reading frame (ORF), we show here that the position of the stem-loop with respect to the translation termination codon and its distance from the translation termination codon determine the half-life of the histone mRNAs. If the stem-loop is located 11 nt 3′ of the translation termination codon, histone mRNAs are constitutively unstable, whereas if it is located 11 nt 5′ of the termination codon, they are regulated normally.
MATERIALS AND METHODS
Constructs.
The starting gene for the histone H2a mRNA constructs was the mouse histone H2a gene (Hist2H2aa1; previously named H2a-614) (21). In order to construct the H2a-IRE-Wt and H2a-IRE-Mut genes, we created a KpnI site 4 nt downstream of the transcription start site using site-directed mutagenesis and inserted oligonucleotides with the following sequences into this site (Fig. 1A): IRE-WT, 5′CGATCCTGCTTCAACAGTGCTTGGACGGATCGGTAC3′; IRE-MUT, 5′CGATCCGTCCAAGCACTGTTGAAGCAGGATCGGTAC3′. These mRNAs have 5′ UTRs that are 36 nt longer than the wild-type (WT) 5′ UTR.
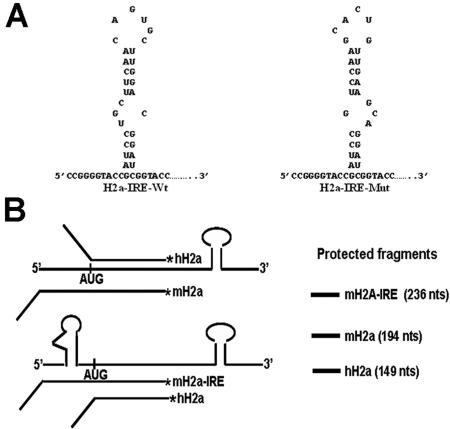
FIG. 1.
Structure of the IRE sequences and diagram of the S1 protection assays. (A) WT and mutant IRE secondary structures inserted into the 5′ UTR of the mouse H2a mRNA. (B) Schematic of the S1 nuclease assay used to simultaneously detect the endogenous human histone mRNA and the exogenous mouse histone mRNAs.
In order to construct Cls-SL, Cls-RS, and Cls-2SL genes, we created two BamHI digestion sites, the first one just 3′ of the translation termination codon and the second just 5′ of the stem-loop sequences, by site-directed mutagenesis. We digested the construct with BamHI and self-ligated the vector. Since the sequences just upstream of the stem-loop are also important for SLBP binding and all mature histone mRNAs have an ACCCA sequence just 5′ of the stem-loop, we inserted not only the stem-loop but also the sequences surrounding the stem-loop. The following sequences were inserted into the vector using the BamHI sites: Cls-SL, 5′ GATCCAAAAAGGCTCTTTTCAGAGCCACCCACG 3′; ClsRS, 5′ GATCCAAAAACCGAGATTTCTCTCGGACCCACATATAACCCCCGCGAGCTCCCAAAAAGGCTCTTTTCAGAGCCACCCACG 3′; Cls-2SL, 5′ GATCCAAAAAGGCTCTTTTCAGAGCCACCCACATATAACCCCCGCGAGCTCCCAAAAAGGCTCTTTTCAGAGCCACCCACG 3′. The stem-loop sequence is bold, the reverse stem sequence is underlined, and the sequences surrounding the stem-loop structure (in the WT mouse histone mRNA) are italic.
In order to construct the 5′ RS and 5′ SL constructs, we used the unique XmaI site present 59 nt downstream of the AUG codon. We inserted either the reverse stem sequence or the stem-loop sequence, as well as sequences surrounding the stem-loop, using the XmaI site. In order to make the 3′ RS and 3′ SL constructs, we created a BamHI site just upstream of the termination codon by site-directed mutagenesis and inserted either the reverse stem sequence or the stem-loop sequence, as well as sequences surrounding the stem-loop, using this site.
In order to create the Lng-SL and Cls-2SLLng constructs, we used the histone gene, which has a BamHI restriction site just 3′ to the stem-loop. We substituted random sequence for the stem-loop in the Lng-SL construct (below) and placed a stem-loop and histone downstream element (HDE) downstream of the stem-loop using PCR. The end of the mRNA is now 505 nt downstream of the termination codon. The random sequence inserted in place of the stem-loop was Lng-SL (5′ GGATCCAAAAAGCCAGCTAACATGCGCACCCAC 3′).
The construction of N-terminally His-tagged WT SLBP and SAVEE-SLBP has been previously described (38).
Transfections and cell culture.
HeLa cells were transfected with the indicated histone genes together with the vector expressing the neomycin resistance gene (pcDNA3) in a 10:1 ratio using Lipofectamine (Invitrogen) as the transfection reagent according to the manufacturer's procedure. Stable populations of cells expressing the transfected genes were selected. Transfection of HeLa cells stably with WT His-tagged SLBP and His-tagged SAVEE-SLBP have been described previously (38). Synchronization of HeLa cells by double-thymidine block has been described previously (36).
RNA preparation and nuclease protection assays.
Total cellular RNAs were prepared using TriZol reagent (Invitrogen) according to the manufacturer's procedure. S1 nuclease assays were performed as described previously (9), using the human H2a HIST2H2AA gene (21) labeled at the 5′ end of the AscI site, the human histone H3.3 gene labeled at the 3′ end of the NcoI site (35), or the mouse histone H2a-614 (Histh2aa1) gene labeled at the 5′ end of the AscI site (11).
RNA interference.
Synthetic 21-nt RNAs complementary to the region mutated in the SAVEE mutant SLBP were made using the Ambion small interfering RNA (siRNA) kit according to the manufacturer's protocol. The target sequence is 5′AAGTGCAGTTGAAGAAGATGA3′. HeLa cells growing in 100-mm dishes were transfected with 130 nM siRNA with Oligofectamine (Invitrogen) according to the manufacturer's protocol, and 48 h later cells were collected (single hit) or transfected again with 130 nM siRNA with Oligofectamine and collected 48 h after the second transfection (double hit) (34).
RESULTS
The iron response element (IRE) is present in the 5′ UTRs of a number of mRNAs whose translation is regulated by the intracellular iron concentration (16). The optimal distance for the IRE from the cap and the initiation codon has been determined previously (8). In order to determine whether we could alter the stability of histone mRNAs by regulating their translation, we inserted either a WT IRE or a mutant IRE into the 5′ UTR of a mouse histone H2a gene (Fig. 1A) and selected HeLa cells that expressed these mRNAs.
We used an S1 nuclease protection assay that allowed us to detect both the exogenous mouse histone H2a mRNA (Hist2h2aa1; previously named H2a-614) and the endogenous human histone H2a mRNA (HIST2H2AA mRNA derived from the orthologue of the mouse Hist2h2aa1gene) using the same mouse Hist2h2aa1 gene as a probe. This allowed us to directly compare the levels of the endogenous mRNAs with the mRNAs from the transfected genes. These mouse and human histone mRNAs have very similar ORFs but very different 5′ and 3′ UTRs. Therefore, the mouse probe protects a shorter protected fragment derived from the human histone H2a mRNA extending to the AUG codon (149 nt) and a longer fragment derived from the mouse histone H2a mRNA extending to the transcription start site (192 nt for WT mouse H2a and 230 nt for mouse H2a mRNA with the IRE sequence in the 5′ UTR) (Fig. 1B). As a loading control, we mixed an H3.3 probe with the H2a probe in some experiments. This probe protects a 411-nt fragment from the replacement variant histone, H3.3 mRNA, which is polyadenylated and not affected by inhibition of DNA replication.
First we determined whether the mouse histone H2a mRNA was regulated in parallel with the endogenous human histone H2a in HeLa cells. We treated cells stably expressing the WT mouse histone H2a mRNA with hydroxyurea (HU) to inhibit DNA synthesis and with cycloheximide (CH) to inhibit protein synthesis and analyzed the histone H2a mRNA levels by S1 assay using the mouse H2a gene as a probe. Treatment of cells with HU resulted in rapid degradation of mouse H2a mRNA very similar to endogenous human H2a mRNA (Fig. 2A, compare lanes 1 and 2), and treatment of cells with CH resulted in an approximately twofold increase in both mouse and endogenous human histone mRNA levels, as a result of stabilization of the histone mRNA (Fig. 2A, compare lanes 1 and 3) (31). Therefore, the exogenous mouse histone H2a mRNA was regulated very similarly to the endogenous human H2a mRNA in HeLa cells (Fig. 2A).
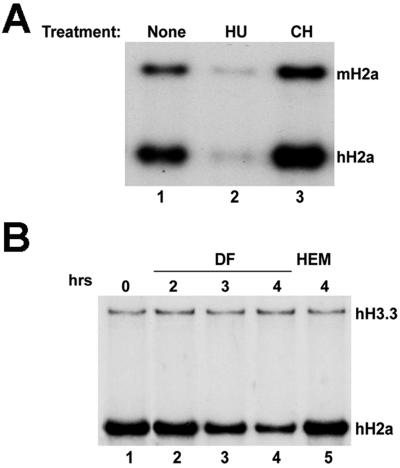
FIG. 2.
Expression of mouse histone H2a mRNA in HeLa cells. (A) Mouse histone mRNA is properly regulated in HeLa cells. Five micrograms of total RNA from untreated cells (lane 1) or cells that were treated with either HU for 45 min (lane 2) or CH for 45 min (lane 3) was analyzed by an S1 nuclease assay using the WT mouse histone H2a gene, which detects the 5′ end of histone H2a mRNAs, as a probe. The protected fragments were resolved by gel electrophoresis and detected by radioautography. The protected fragments are 192 nt for WT mouse histone H2a mRNA (mH2a) and 149 nt for the endogenous human H2a mRNA (hH2a). (B) Effects of DF and HEM treatments on histone mRNA. Five micrograms of total RNA from untreated cells (lane 1) or cells that were treated with either 5 mM DF for 2 (lane 2), 3 (lane 3), or 4 h (lane 4) or 5 mM HEM for 4 h (lane 5) was analyzed by an S1 nuclease assay using a mixture of the human replication-dependent histone H2a (hH2a) and replacement-type histone H3.3 (hH3.3) genes as a probe. The protected fragments are 198 nt for histone H2a mRNA and 411 nt for H3.3 mRNA.
The translation of mRNAs with an IRE sequence in the 5′ UTR can be regulated by altering the intracellular iron concentration. Treatment of cells with hemin (HEM) increases iron levels to the maximum level and activates translation. However, treatment of cells with desferrioxamine (Desferral; DF) reduces the iron concentration by chelating any available iron and represses the translation of IRE-containing mRNAs. There are significant amounts of iron in the medium that the cells are grown in, so a proportion of the mRNA containing the WT IRE is translated even in the absence of added HEM. We observed that treating the HeLa cells with DF resulted in a relatively rapid arrest of cell growth and loss of endogenous histone mRNA. Three hours after the addition of DF, histone mRNA levels dropped significantly (Fig. 2B, lanes 3 and 4) due to the reduction in the rate of DNA replication. However, addition of HEM for 4 h did not affect histone mRNA levels (Fig. 2B, lane 5). Therefore, we routinely used 2-h treatments with DF, which did not have a large effect on endogenous histone mRNA levels (Fig. 2B, lane 2).
The histone mRNAs containing the WT IRE and the IREMUT were expressed at similar levels in stably transfected HeLa cells. Insertion of the mutant IRE into the 5′ UTR did not affect the regulation of this mRNA compared to the endogenous human histone mRNA. The IREMUT mRNA was rapidly degraded when these cells were treated with HU for 45 min, and so was the endogenous human histone H2a mRNA (Fig. 3A, lane 2). In addition, the levels of both mRNAs were increased when these cells were treated with CH for 45 min (Fig. 3A, lane 3). Similar results were obtained when these cells were pretreated with DF for 2 h prior to the addition of either HU or CH for 45 min (Fig. 3A, lanes 4 to 6), as well as when these cells were pretreated with HEM for 2 h prior to the addition of either HU or CH for 45 min (Fig. 3A, lanes 7 to 9).
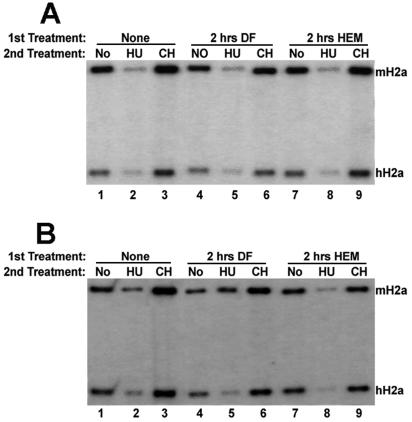
FIG. 3.
Regulation of the histone mRNAs containing an IRE in the 5′ UTR. Five micrograms of total RNA from HeLa cells stably expressing the H2a-IREMUT gene (A) or the H2a-IREWT gene (B) was analyzed by an S1 nuclease assay using the H2a-IREMUT gene (A) or the H2a-IREWT gene (B) as a probe. The cells were untreated cells (lanes 1 to 3) or cells treated for 2 h with either DF (lanes 4 to 6) or HEM (lanes 7 to 9). After the initial treatment, these cells were either not treated (lanes 1, 4, and 7) or treated for an additional 45 min with either HU (lanes 2, 5, and 8) or CH (lanes 3, 6, and 9).
In contrast, the IREWT mRNA was regulated differently from either the IREMUT mRNA or endogenous human histone H2a mRNA. When exponentially growing cells expressing the IREWT mRNA were treated with HU, the IREWT mRNA was not degraded to the same extent as the endogenous human histone H2a mRNA (Fig. 3B, lane 2). Pretreatment of cells with DF for 2 h prior to addition of HU for 45 min further stabilized IREWT mRNA (Fig. 3B, lane 5). In addition, treatment of these cells with CH increased the levels of the IREWT mRNA whether these cells were untreated or pretreated with DF prior to CH addition (Fig. 3B, lanes 3 and 6). Strikingly, when these cells were pretreated with HEM for 2 h in order to stimulate translation, prior to treatment of cells with HU, the IREWT mRNA was degraded to a similar extent as the endogenous human H2a mRNA (Fig. 3B, lane 8). Furthermore, when these cells were pretreated with HEM for 2 h, prior to treatment of cells with CH, the IREWT mRNA was stabilized (Fig. 3B, lane 9). Therefore, the stimulation of the translation of IREWT mRNA restored normal regulation of this histone mRNA, which is the rapid degradation following the inhibition of DNA replication and stabilization following the inhibition of protein synthesis. These experiments clearly show that the stability of histone mRNAs can be regulated by altering the translatability of these mRNAs without affecting the translation of other mRNAs.
SLBP is required for efficient translation of histone mRNAs both in vitro and in vivo, and a 15-amino-acid region in the N terminus of SLBP required for this translational activation has been identified (27). Therefore, a second method for selectively altering the translation of histone mRNAs is to express a mutant SLBP, which cannot support translation, in HeLa cells. We stably expressed N-terminally His-tagged WT or mutant SLBP (SAVEE-SLBP), in which the four amino acids in the core region required for histone mRNA translation were changed into alanines, in HeLa cells. Although this substitution completely abolished the ability of SLBP to activate translation of histone mRNA in either reticulocyte lysates or frog oocytes (27), it has no effect on histone pre-mRNA processing (7) or cell cycle regulation of SLBP (38).
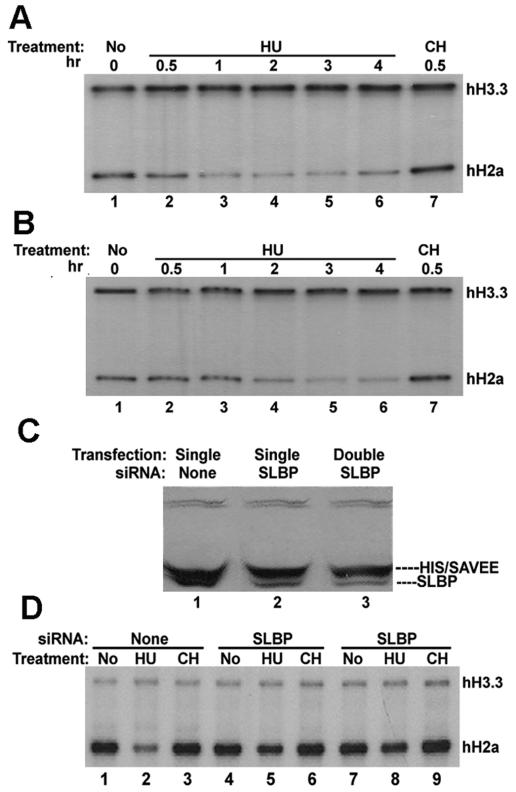
When the cells expressing His-tagged WT SLBP were treated with HU, the histone mRNAs were rapidly degraded (Fig. 4A, lanes 1 to 3). In contrast, the degradation of histone mRNAs was delayed in cells expressing His-tagged SAVEE-SLBP (Fig. 4B, lanes 1 to 4), although histone mRNAs were ultimately degraded after treatment of cells with HU for 2 or 3 h (Fig. 4B, lanes 5 and 6). The cells expressing His-tagged SAVEE-SLBP also expressed endogenous WT SLBP, which could account for the residual translation and failure to completely stabilize histone mRNAs in these cells. Therefore, we wanted to find out whether selective knockdown of endogenous WT SLBP would further stabilize histone mRNAs following the inhibition of DNA synthesis. We designed an siRNA which specifically targets the region that is mutated in SAVEE-SLBP; thereby, it will selectively reduce the expression of endogenous WT SLBP. When cells expressing SAVEE-SLBP were transfected with this siRNA either once (Fig. 4C, lanes 2) or twice (Fig. 4C, lane 3), the relative amount of endogenous SLBP was reduced drastically compared with that of mutant SLBP. Although histone mRNAs were degraded after 2 h of HU treatment in cells expressing both WT SLBP and SAVEE-SLBP (Fig. 4D, lanes 1 to 3), histone mRNAs were not degraded after 2 h of HU treatment in cells in which endogenous SLBP was selectively knocked down (Fig. 4D, lanes 4 to 9). Therefore, if the predominant SLBP in the cell is mutant SLBP which cannot support high-level translation of histone mRNAs, then the histone mRNAs are not rapidly degraded following the inhibition of DNA synthesis. These results are consistent with the suggestion that active translation of histone mRNAs is required for rapid degradation of these mRNAs following the inhibition of DNA synthesis.
FIG. 4.
Effect of expression of SAVEE-SLBP on regulation of histone mRNA. Five micrograms of total RNA from HeLa cells stably expressing His-tagged WT SLBP (A) or His-tagged SAVEE-SLBP (B) was analyzed by an S1 assay using a mixture of the human replication-dependent H2a gene and the H3.3 gene as a probe. The cells were untreated cells (lane 1) or cells that were treated with HU for 30 min (lane 2), 1 h (lane 3), 2 h (lane 4), 3 h (lane 5), or 4 h (lane 6) or with CH for 1 h (lane 7). Note that the relative intensities of the H3.3 and H2a protected fragments vary between different experiments because of different relative specific activities of the probes. (C) Lysates from HeLa cells expressing SAVEE-SLBP that were not transfected with any siRNA (lane 1) or transfected with an siRNA that specifically targets the endogenous WT SLBP once (lane 2) or twice (lane 3) were prepared 48 h after the final transfection. Ten micrograms of total protein was resolved by 10% SDS-PAGE and analyzed by Western blot assay with anti-SLBP antibody. (D) Five micrograms of total RNA from HeLa cells stably expressing His-tagged SAVEE-SLBP that were not transfected with any siRNA (lanes 1 to 3) or were transfected with SLBP siRNA either once (lanes 4 to 6) or twice (lanes 7 to 9) was analyzed by S1 nuclease assay using a mixture of histone H2a and H3.3 genes as a probe. The cells were either untreated (lanes 1, 4, and 7) or treated either with HU for 45 min (lanes 2, 5, and 8) or with CH for 45 min (lanes 3, 6, and 9) 48 h following the final transfection.
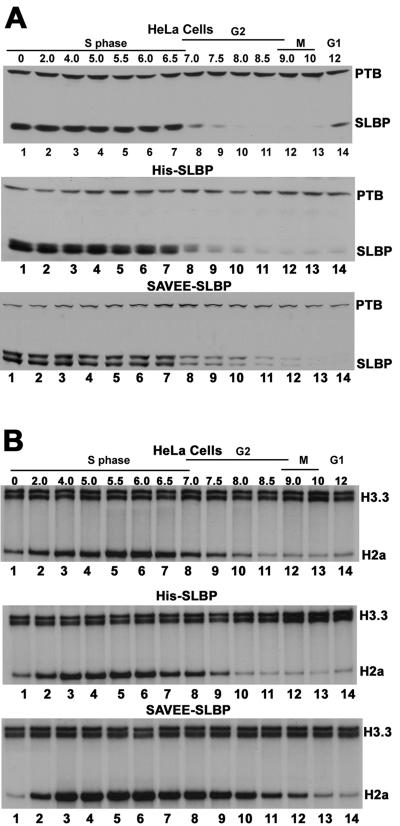
Degradation of histone mRNAs is slowed at the end of S phase in cells expressing SAVEE-SLBP.
Histone mRNAs are also rapidly degraded at the end of S phase (12). In order to determine whether regulated degradation of histone mRNAs at the end of S phase also requires active translation of these mRNAs, we synchronized HeLa cells stably expressing either His-tagged WT SLBP or His-tagged SAVEE-SLBP and analyzed the levels of histone mRNAs and SLBP every 30 min as cells exited S phase. The fact that the endogenous SLBP, the His-tagged SLBP, and the His-tagged-SAVEE-SLBP were degraded at about the same time (Fig. 5A, lanes 8 to 12) suggests that the cells expressing SAVEE-SLBP exited S phase at the same time as control HeLa cells (36, 38). The cells expressing SAVEE-SLBP degraded histone mRNA more slowly, with a 30- to 60-min delay (Fig. 5B, bottom, lanes 9 to 13) compared to degradation of the histone mRNA in normal cells or cells expressing His-tagged WT SLBP (Fig. 5B, top and middle, lanes 9 to 13). Note that as SLBP was degraded in the cells expressing SAVEE-SLBP, there was a change in the ratio of the two SLBPs at 8 and 8.5 h (Fig. 5A, bottom, lanes 10 and 11), indicating that a fraction of SAVEE-SLBP was degraded more slowly than endogenous SLBP. In addition, there was an increased amount of histone mRNA present at these same times (Fig. 5B, bottom, lanes 10 and 11) in SAVEE-SLBP-expressing cells compared to the other cells. This result is consistent with the possibility that the SAVEE-SLBP bound to histone mRNA cannot be degraded until the histone mRNA is degraded, hence releasing the SAVEE-SLBP from the histone 3′ end. These experiments show that degradation of histone mRNAs at the end of S phase also requires the active translation of these mRNAs. Moreover, these experiments suggest that the mechanisms of histone mRNA degradation at the end of S phase and after the inhibition of DNA synthesis are most likely similar.
FIG. 5.
Regulation of histone mRNA in synchronized HeLa cells stably expressing SAVEE-SLBP. HeLa cells (top) and HeLa cells stably expressing His-tagged SLBP (middle) or His-tagged SAVEE-SLBP (bottom) were synchronized by double-thymidine block, lysates were prepared at the indicated times after release from the double-thymidine block, and cell lysates and total cell RNA prepared. Equal proportions of total cell protein were resolved by 10% SDS-PAGE and analyzed by Western blot assay using both PTB and SLBP antibodies (A). The upper band of the doublet of SLBP is the His-tagged protein. Total cell RNA from equal numbers of cells was analyzed by S1 nuclease mapping for H3.3 RNA and histone H2a mRNA (B).
Translation termination is required for histone mRNA degradation.
In all metazoan histone mRNAs (65 genes in humans), the stem-loop at the 3′ end of histone mRNAs starts 30 to 75 nt from the translation termination codon (21). If the stem-loop is moved several hundred nucleotides from the stop codon, the histone mRNAs are not rapidly degraded when DNA replication is inhibited (10) or at the end of S phase (12). These results suggest that the position of the stem-loop structure with respect to the translation termination codon is important in determining the stability of histone mRNAs. The minimum distance, 30 nt, is likely sufficient to allow the ribosome to terminate translation properly without dissociating SLBP from the 3′ end of histone mRNAs.
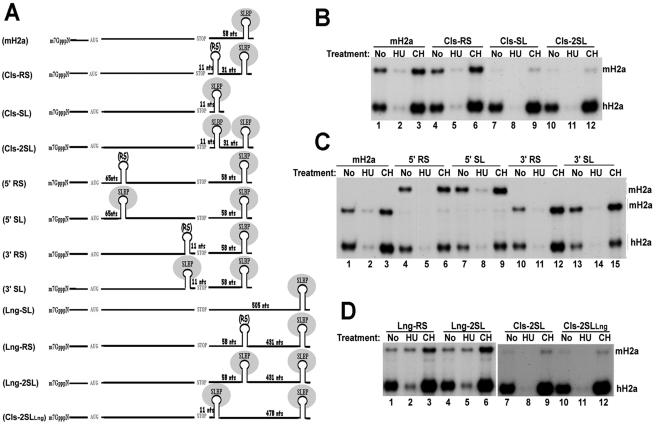
In order to further investigate the role of the 3′ end in histone mRNA degradation, we constructed mouse histone H2a genes in which we altered the position of the stem-loop. First, we tested the effect of placing the stem-loop close (11 nt downstream of the termination codon) (Cls-SL). In this case, the terminating ribosome would be expected to displace SLBP during the initial round of translation. We also constructed mouse mRNAs containing two stem-loops, the first one located 11 nt downstream of the stop codon and the second one located in the normal position (58 nt downstream of the termination codon) (Cls-2SL). As a control, we constructed a mouse gene with the stem sequences in the first stem-loop reversed, which abolishes SLBP binding (Cls-RS). The 3′ end of the Cls-2SL mRNA binds two SLBPs in vitro (data not shown). Since the histone downstream element (HDE), which binds the U7 snRNP, is absolutely required for pre-mRNA processing, the HDE was placed only after the second stem-loop, ensuring that the mRNA would only be processed after the second stem-loop. In addition, we constructed genes with an additional WT or reverse stem-loop inserted in the ORF either close to the initiation codon (59 nt downstream of the AUG codon) (5′ SL and 5′ RS) or 11 nt upstream of the termination codon (3′ SL and 3′ RS) without disrupting the ORF.
HeLa cells were stably transfected with these modified histone genes. These cells were then treated with either HU or CH, and the regulation of the modified mouse histone mRNAs was determined by an S1 nuclease assay that detected the 5′ end of the mRNAs. The Cls-SL mRNA accumulated to a very low level (Fig. 6B, lane 7), consistent with removal of SLBP during the initial round of translation destabilizing the mRNA. Furthermore, treatment of cells with CH stabilized the mRNA, suggesting that this mRNA was rapidly degraded by a pathway that requires translation (Fig. 6B, lane 9).
FIG. 6.
Expression of mouse histone mRNAs with the stem-loop in different positions. (A) The histone genes containing two stem-loops have only one HDE located downstream of the second stem-loop. The distances refer to the distance from the translation termination codon to the beginning of the first base pair of the stem-loop or reverse stem sequences. (B) Expression of histone mRNAs with a stem-loop close to the termination codon. Five micrograms of total RNA from HeLa cells stably expressing WT mouse histone H2a mRNA (lanes 1 to 3), Cls-RS mRNA (lanes 4 to 6), Cls-SL mRNA (lanes 7 to 9), or Cls-2SL mRNA (lanes 10 to 12) was analyzed by an S1 nuclease assay using the WT mouse histone gene, which detects the 5′ end of the histone H2a mRNAs, as the probe. The cells were untreated (lanes 1, 4, 7, and 10) or were treated with either HU for 45 min (lanes 2, 5, 8, and 11) or CH for 45 min (3, 6, 9, and 12). (C) Regulation of histone mRNAs with a stem-loop in the ORF. Five micrograms of total RNA from HeLa cells stably expressing WT mouse histone H2a mRNA (lanes 1 to 3), 5′ RS mRNA (lane 4 to 6), 5′ SL mRNA (lanes 7 to 9), 3′ RS mRNA (lanes 10 to 12), and 3′ SL mRNA (lanes 13 to 15) was analyzed by an S1 assay using the WT mouse histone H2a gene, which detects the 5′ end of histone H2a mRNAs, as a probe. In order to detect 5′ RS and 5′ SL histone mRNAs, either the 5′ RS gene (lanes 4 to 6) or the 5′ SL gene (lanes 7 to 9) was mixed with the WT mouse H2a gene. The protected fragments derived from the 5′ RS and 5′ SL mRNAs were longer because the probes detected the 5′ end of the histone mRNAs and these mRNAs have additional stem-loop or reverse stem sequences 59 nt downstream of the AUG codon. The cells were untreated (lanes 1, 4, 7, and 10) or were treated with either HU for 45 min (lanes 2, 5, 8, and 11) or CH for 45 min (3, 6, 9, and 12). (D) Regulation of histone mRNAs with a longer 3′ UTR. Five micrograms of total RNA from HeLa cells stably expressing Lng-RS mRNA (lanes 1 to 3), Lng-2SL mRNA (lanes 4 to 6), Cls-2SL mRNA (lanes 7 to 9), and Cls-2SLLng mRNA (lanes 10 to 12) was assayed by an S1 nuclease assay using the WT mouse histone H2a gene, which detects the 5′ end of histone H2a mRNAs, as a probe. The cells were untreated (lanes 1, 4, 7, and 10) or were treated with either HU for 45 min (lanes 2, 5, 8, and 11) or CH for 45 min (3, 6, 9, and 12).
Strikingly, the Cls-2SL mRNA also accumulated to a very low level (Fig. 6B, lane 10), suggesting that this mRNA was also constitutively unstable. In contrast, the mouse histone H2a mRNA containing a reverse stem-loop sequence 11 nt downstream of the termination codon were expressed at a level comparable to WT mouse histone mRNA (Fig. 6B, compare lanes 1 and 4). Moreover, the Cls-RS mRNA was degraded following the treatment of cells with HU (Fig. 6B, lane 5) and stabilized after the treatment of cells with CH (Fig. 6B, lane 6), like WT mouse histone H2a mRNA (Fig. 6B, lanes 1 to 3). These results demonstrate that the low-level expression of Cls-SL mRNA is not due to the presence of a secondary structure immediately after the termination codon but due to the presence of SLBP close to the termination codon.
In contrast, when we placed the stem-loop in the coding region, both 5′ SL and 3′ SL mRNAs were expressed at levels comparable to the WT mouse H2a mRNA or mouse mRNAs that have a reverse stem-loop at the same position (Fig. 6C, lanes 1, 4, 7, 10, and 13). Furthermore, all of these mRNAs were regulated normally like WT mouse H2a mRNA or endogenous human H2a mRNA. They were degraded after treatment of cells with HU (Fig. 6C, lanes 2, 5, 8, 11, and 14) and stabilized after treatment of cells with CH (Fig. 6C, lanes 3, 6, 9, 12, and 15). Therefore, having a stem-loop just before the termination codon, where the SLBP would be removed by the translating ribosome, had a very different effect from having the same structure just 3′ of the termination codon, where SLBP would be removed by the terminating ribosome. These results suggest that removal of SLBP by the terminating ribosome makes the mRNA unstable.
When the stem-loop is moved more than 300 nt from the termination codon, the mRNA is not rapidly degraded when DNA synthesis is inhibited (10). In order to further explore the effects of placing the stem-loop far downstream, we constructed genes containing two stem-loops, the first one located either at the original distance (Lng-2SL) or 11 nt downstream of the termination (Cls-2SLLng) and the second one located 505 nt downstream of the termination codon. As a control, we constructed a gene containing a reverse stem at the original position and a stem-loop located 505 nt downstream of the termination codon (Lng-RS, Fig. 6A). We stably transfected HeLa cells with these constructs, treated them with either HU or CH, and monitored the levels of these modified mRNAs using an S1 nuclease assay. Both the Lng-RS and Lng-2SL mRNAs were expressed at very similar levels (Fig. 6D, lanes 1 and 4), and neither of these mRNAs was degraded following the inhibition of DNA synthesis (Fig. 6D, lanes 2 and 5), similar to mRNAs that have only one stem-loop located 505 nt downstream of the termination codon (data not shown). These results demonstrate that the presence of a stem-loop structure at the original position does not result in histone mRNA degradation after inhibition of DNA synthesis if the histone mRNA has a long 3′ UTR with an additional stem-loop 505 nt downstream of the termination codon. In contrast, the Cls-2SLLng mRNA, which also had a long 3′ UTR with an additional stem-loop, was expressed at a very low level (Fig, 6D, lane 10), like the Cls-2SL mRNA (Fig. 6D, lane 7). Furthermore, both of these mRNAs were stabilized by treatment of cells with CH (Fig. 6D, lanes 9 and 12).
Taken together, these results suggest that SLBP bound to the stem-loop close to and 3′ of the translation termination codon is sufficient to trigger rapid degradation of histone mRNA even in the presence of another stem-loop-SLBP either 58 nt or 505 nt downstream of the termination codon. Furthermore, histone mRNAs with extended 3′ UTRs are not degraded after inhibition of DNA synthesis even when they have an additional stem-loop structure at the normal position.
DISCUSSION
The final step in mRNA metabolism is degradation of the mRNA in the cytoplasm. Each mRNA has a half-life which is determined by sequences present in the mRNA (26). Moreover, the half-life of a particular mRNA can be modulated by factors that interact with the mRNA (3). The replication-dependent histone mRNAs are an excellent example of a class of mRNAs that are coordinately regulated during the cell cycle, largely by posttranscriptional mechanisms (12, 18, 30). A major regulatory mechanism of histone mRNA metabolism is regulation of the half-lives of histone mRNAs. Histone mRNAs are rapidly degraded either after the inhibition of DNA synthesis or at the end of S phase. In contrast, treatment of cells with inhibitors of protein synthesis stabilizes histone mRNAs (2, 9, 31). Translation inhibitors also stabilize many other mRNAs (37), including mRNAs undergoing nonsense-mediated decay (4). For histone mRNAs, it has been suggested that the presence of excess histone proteins can feed back on histone mRNA levels, resulting in their rapid degradation (22, 25, 28). Thus, the requirement for translation could simply reflect a requirement for synthesis of histone protein.
Translation is required for histone mRNA degradation.
To address whether translation per se is necessary for rapid histone mRNA degradation, as opposed to the requirement for continued synthesis of a specific protein(s), we constructed a histone mRNA whose translation can be regulated in vivo by placing an IRE in the 5′ UTR. This histone mRNA was rapidly degraded when DNA synthesis was inhibited only when iron was present and this mRNA was translated efficiently. We also reduced the efficiency of histone mRNA translation by expressing a mutant SLBP which cannot translate histone mRNAs efficiently. Cells expressing the mutant SLBP together with the endogenous SLBP degraded histone mRNA at a slower rate following inhibition of DNA synthesis. Moreover, overexpression of the mutant SLBP by reducing the endogenous SLBP with a specific siRNA further stabilized histone mRNAs after the inhibition of DNA synthesis. These results are consistent with the importance of translation for histone mRNA degradation, although we cannot rule out the possibility that the region of SLBP required for translation activation is also directly involved in the activation of histone mRNA degradation. The mutant SAVEE-SLBP contains only a four-amino-acid mutation that affects translation. Even if this mutant protein also directly affects degradation, it further emphasizes the relationship between translation and histone mRNA degradation.
Taken together, these experiments rigorously demonstrate that histone mRNAs need to be translated actively for their rapid degradation after the inhibition of DNA synthesis. It is still possible that continued synthesis of histone proteins, resulting in excess soluble histone proteins, generates a signal required for degradation of histone mRNAs since the inhibition of DNA synthesis transiently increases histone protein levels.
A role for translation termination in histone mRNA degradation.
How might translation of histone mRNA contribute to histone mRNA degradation? Two requirements for the rapid degradation of histone mRNAs following the inhibition of DNA synthesis are the active translation of these mRNAs and the presence of the stem-loop structure at the 3′ end and a proper distance from the termination codon. Therefore, it is possible that after the inhibition of DNA replication, the terminating ribosome receives a signal from the 3′ end of the histone mRNP which triggers histone mRNA degradation only if the 3′ end is located at a proper distance from the termination codon.
mRNAs containing a premature termination codon are rapidly degraded by a surveillance mechanism called nonsense-mediated decay (19). Since sequences surrounding the translation termination codon can affect the efficiency of translation termination (15, 23), premature translation termination may be different from normal translation termination because the context of the termination codon is changed. In mammalian cells, recognition of the termination codon as premature requires translation and the presence of an exon-exon junction complex (EJC) more than 20 to 24 nt downstream of the translation termination codon (20). All yeast mRNAs have a short distance between the termination codon and the poly(A) tail, and the recognition of the termination codon as premature depends on the distance between the termination codon and the poly(A) tail. In yeast extracts, when a ribosome encounters a premature termination codon, it cannot terminate translation efficiently and stalls. Moreover, the tethering of poly(A)-binding protein (PABP) downstream of the premature termination codon stabilizes yeast mRNAs containing a premature termination codon (1). Since PABP interacts directly with termination factors (33), it has been proposed that the interaction between PABP and the translation termination factor helps the recycling of ribosomes to the 5′ end of the mRNA, thereby resulting in efficient reinitiation of translation following translation termination. Therefore, the presence of PABP provides a positive signal for efficient translation termination, while the presence of EJC (at least in mammalian cells) may antagonize this signal, thereby resulting in inefficient translation termination and degradation of the mRNA. Thus, the EJC bound downstream of a termination codon may affect the efficiency of translation termination.
In bacteria, there is an mRNA decay system which degrades mRNAs that are arrested in translation. If the ribosome pauses on an mRNA (e.g., an mRNA which cannot be completely translated since is has been cleaved and no longer has an in-frame stop codon), then the tmRNA (SsrA RNA) is recruited to the ribosome, resulting in trans-translation and subsequent degradation of the protein and the RNA (14). In addition, when a bacterial ribosome stalls at a termination codon, the mRNA is endonucleolytically cleaved by an unknown mechanism, allowing recruitment of tmRNA and subsequent complete degradation of the mRNA (13, 32). Thus, in bacteria there is mRNA degradation which is coupled to inefficient translation termination.
Our results show that the distance between the termination codon and SLBP affects the stability of the histone mRNA, possibly by affecting the process of translation termination. Moving the stem-loop close to the termination codon destabilizes the histone mRNA, regardless of whether there is an additional stem-loop further 3′ of the termination codon. In contrast, the presence of a stem-loop just 5′ of the termination codon does not affect the stability of histone mRNAs. The difference between these two mRNAs is that the translating ribosome will remove the SLBP prior to termination in the former case and during termination in the latter case. The interaction of SLBP with the terminating ribosome may interfere with efficient translation termination, making the histone mRNAs with a stem-loop close to the termination codon unstable. The fact that both the Cls-2SL and Cls-2SLLng mRNAs are unstable, even though these mRNAs have another stem-loop at their 3′ ends, suggests that interaction of the ribosome with the SLBP bound to the closer stem-loop likely triggers 5′-to-3′ degradation. Whether this effect is specific for SLBP or could be caused by tight binding of any protein to the 3′ UTR close to the termination codon, resulting in pausing of the terminating ribosome, cannot be determined from these experiments.
Rapid degradation of histone mRNAs after inhibition of DNA synthesis not only requires the presence of a stem-loop at a proper distance (more than 30 nt but less than 200 nt) (10) from the termination codon, but the stem-loop also has to be located at the 3′ end of the mRNA. The distance constraint suggests there is an interaction between the 3′ end of the histone mRNP and the terminating ribosome, which is likely prevented by lengthening the 3′ UTR. Histone mRNAs with the stem-loop at the proper position but with a longer 3′ UTR, due to the presence of either a poly(A) tail (17) or another stem-loop at the end of the mRNA (505 nt downstream of the termination codon), are not degraded after the inhibition of DNA synthesis (Fig. 6C). This result suggests that histone mRNA degradation is probably initiated by exonucleolytic degradation rather than endonucleolytic cleavage. An exonuclease which can specifically bind the stem-loop-SLBP complex has been identified recently (6). Perhaps the role of the initial exonucleolytic degradation is disruption of the stem-loop-SLBP interaction, which then may lead to activation of 5′-to-3′ degradation in the same way that deadenylation is followed by decapping and activation of 5′-to-3′ degradation (3).
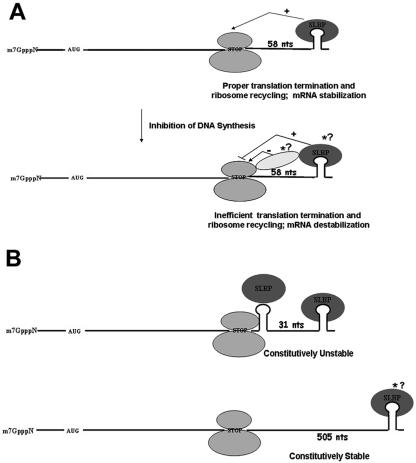
In all metazoan histone mRNAs, the distance between the termination codon and the stem-loop has been highly conserved. The stem-loop is located 30 to 70 nt downstream of the termination codon, which is just beyond the distance a terminating ribosome will cover. We postulate that this distance constraint ensures efficient translation of histone mRNAs, as well as appropriate regulation of the histone mRNA half-life. At least in yeast, efficient translation termination requires the presence of PABP close to the termination codon (1). We propose that, similar to PABP, SLBP is normally involved in not only efficient translation (27) but also translation termination of histone mRNA. Following the inhibition of DNA replication, a signal is generated which prevents efficient translation termination of histone mRNAs (Fig. 7). For SLBP to interact with the terminating ribosome when DNA synthesis is inhibited, it must be sufficiently close to the termination codon, accounting for the observed dependence of the distance of the stem-loop from the termination codon. When the stem-loop is close to the termination codon, the interaction of the terminating ribosome with SLBP triggers rapid histone mRNA degradation, resulting in a constitutively unstable mRNA. Since there is also a requirement that the stem-loop be at the 3′ end of the mRNA, either the initial degradation step involves an exonuclease (6) or the 3′ end of the mRNA is required for productive interaction of SLBP with the ribosome. It is possible that there is regulation of the efficiency of translation termination of other mRNAs and that this is yet another mechanism by which cells can regulate gene expression.
FIG. 7.
Model for histone mRNA degradation. (A) During S phase, histone mRNAs are efficiently translated with the help of SLBP. We propose that SLBP positively stimulates translation termination and ribosome recycling, leading to stabilization of histone mRNAs. However, following the inhibition of DNA synthesis, translation termination becomes inefficient, leading to degradation of histone mRNAs. This can be achieved by preventing positive signaling from SLBP, by modifying SLBP, or by creating a negative signal from a ribosome associated factor. (B) Histone mRNAs containing a stem-loop close to the termination codon are constitutively unstable, since removal of SLBP by the terminating ribosome might lead to inefficient translation termination. On the other hand, histone mRNAs with a longer 3′ UTR are constitutively stable because the distal SLBP cannot interact with the terminating ribosome.
Acknowledgments
This work was supported by NIH grant GM29832 to W.F.M.
We thank Bob Duronio for critical comments on the manuscript. We thank Mariano Garcia-Blanco (Duke University) for the PTB antibody.
REFERENCES
- 1.Amrani, N., R. Ganesan, S. Kervestin, D. A. Mangus, S. Ghosh, and A. Jacobson. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112-118. [DOI] [PubMed] [Google Scholar]
- 2.Baumbach, L. L., F. Marashi, M. Plumb, G. Stein, and J. Stein. 1984. Inhibition of DNA replication coordinately reduces cellular levels of core and H1 histone mRNAs: requirement for protein synthesis. Biochemistry 23:1618-1625. [DOI] [PubMed] [Google Scholar]
- 3.Caponigro, G., and R. Parker. 1996. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60:233-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, J., M. Fogel-Petrovic, and L. E. Maquat. 1990. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol. Cell. Biol. 10:5215-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 6.Dominski, Z., X. Yang, H. Kaygun, and W. F. Marzluff. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12:295-305. [DOI] [PubMed] [Google Scholar]
- 7.Dominski, Z., L.-X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goossen, B., and M. W. Hentze. 1992. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol. Cell. Biol. 12:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graves, R. A., and W. F. Marzluff. 1984. Rapid, reversible alterations in histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol. Cell. Biol. 4:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graves, R. A., N. B. Pandey, N. Chodchoy, and W. F. Marzluff. 1987. Translation is required for regulation of histone mRNA degradation. Cell 48:615-626. [DOI] [PubMed] [Google Scholar]
- 11.Graves, R. A., S. E. Wellman, I.-M. Chiu, and W. F. Marzluff. 1985. Differential expression of two clusters of mouse histone genes. J. Mol. Biol. 183:179-194. [DOI] [PubMed] [Google Scholar]
- 12.Harris, M. E., R. Böhni, M. H. Schneiderman, L. Ramamurthy, D. Schümperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes, C. S., and R. T. Sauer. 2003. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12:903-911. [DOI] [PubMed] [Google Scholar]
- 14.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 15.Kisselev, L. L., and R. H. Buckingham. 2000. Translational termination comes of age. Trends Biochem. Sci. 25:561-566. [DOI] [PubMed] [Google Scholar]
- 16.Klausner, R. D., T. A. Rouault, and J. B. Harford. 1993. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell 72:19-28. [DOI] [PubMed] [Google Scholar]
- 17.Levine, B. J., N. Chodchoy, W. F. Marzluff, and A. I. Skoultchi. 1987. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3′ end of the mRNA. Proc. Natl. Acad. Sci. USA 84:6189-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lüscher, B., C. Stauber, R. Schindler, and D. Schümperli. 1985. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3′-terminal part of the gene. Proc. Natl. Acad. Sci. USA 82:4389-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maquat, L. E. 2002. Nonsense-mediated mRNA decay. Curr. Biol. 12:R196-R197. [DOI] [PubMed] [Google Scholar]
- 20.Maquat, L. E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5:89-99. [DOI] [PubMed] [Google Scholar]
- 21.Marzluff, W. F., P. Gongidi, K. R. Woods, J. P. Jin, and L. Maltais. 2002. The human and mouse replication-dependent histone genes. Genomics 80:487-498. [PubMed] [Google Scholar]
- 22.McLaren, R. S., and J. Ross. 1993. Individual purified core and linker histones induce histone H4 mRNA destabilization in vitro. J. Biol. Chem. 268:14637-14644. [PubMed] [Google Scholar]
- 23.Nakamura, Y., K. Ito, and L. A. Isaksson. 1996. Emerging understanding of translation termination. Cell 87:147-150. [DOI] [PubMed] [Google Scholar]
- 24.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltz, S. W., and J. Ross. 1987. Autogenous regulation of histone mRNA decay by histone proteins in a cell-free system. Mol. Cell. Biol. 7:4345-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pesole, G., F. Mignone, C. Gissi, G. Grillo, F. Licciulli, and S. Liuni. 2001. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276:73-81. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez, R., and W. F. Marzluff. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22:7093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sariban, E., R. S. Wu, L. C. Erickson, and W. M. Bonner. 1985. Interrelationships of protein and DNA syntheses during replication of mammalian cells. Mol. Cell. Biol. 5:1279-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sittman, D. B., R. A. Graves, and W. F. Marzluff. 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. USA 80:1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stauber, C., and D. Schümperli. 1988. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 16:9399-9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stimac, E., V. E. Groppi, Jr., and P. Coffino. 1984. Inhibition of protein synthesis stabilizes histone mRNA. Mol. Cell. Biol. 4:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunohara, T., K. Jojima, Y. Yamamoto, T. Inada, and H. Aiba. 2004. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10:378-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchida, N., S. I. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 277:50286-50292. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 35.Wellman, S. E., P. J. Casano, D. R. Pilch, W. F. Marzluff, and D. B. Sittman. 1987. Characterization of mouse H3.3-like histone genes. Gene 59:29-39. [DOI] [PubMed] [Google Scholar]
- 36.Whitfield, M. L., L.-X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen, T. J., P. S. Machlin, and D. W. Cleveland. 1988. Autoregulated instability of β-tubulin mRNAs by recognition of the nascent amino terminus of β-tubulin. Nature 334:580-585. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, L.-X., Z. Dominski, X. Yang, P. Elms, C. S. Raska, C. H. Borchers, and W. F. Marzluff. 2003. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 23:1590-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]