Abstract
We have previously formulated a list of approximately 2,000 RNA octamers as putative exonic splicing enhancers (PESEs) based on a statistical comparison of human exonic and nonexonic sequences (X. H. Zhang and L. A. Chasin, Genes Dev. 18:1241-1250, 2004). When inserted into a poorly spliced test exon, all eight tested octamers stimulated splicing, a result consistent with their identification as exonic splicing enhancers (ESEs). Here we present a much more stringent test of the validity of this list of PESEs. Twenty-two naturally occurring examples of nonoverlapping PESEs or PESE clusters were identified in six mammalian exons; five of the six exons tested are constitutively spliced. Each of the 22 individual PESEs or PESE clusters was disrupted by site-directed mutagenesis, usually by a single-base substitution. Eighteen of the 22 disruptions (82%) resulted in decreased splicing efficiency. In contrast, 24 control mutations had little or no effect on splicing. This high rate of success suggests that most PESEs function as ESEs in their natural context. Like most exons, these exons contain several PESEs. Since knocking out any one of several could produce a severalfold decrease in splicing efficiency, we conclude that there is little redundancy among ESEs in an exon and that they must work in concert to optimize splicing.
The splicing together of exons during the maturation of mRNA from a primary transcript represents a step that is fundamental in the transfer of information from DNA to protein for most genes in higher eukaryotes. This process is catalyzed by a supramolecular particle known as the spliceosome, which serves as the site for the two transesterification reactions that remove an intron and connect two adjacent exons. The spliceosome is composed of five small nuclear RNA molecules and perhaps hundreds of proteins (20, 23, 33, 55). Despite this complexity, much has been learned about the identity of these components, their function, and the sequence of events that culminates in exon joining (2, 5, 16, 21). Less understood are the earliest events that initiate splicing, in which the boundaries of the intron that must be removed are identified. The absolute accuracy of this molecular recognition is critical to gene function and must depend on signals comprised of sequence and/or structural elements in the RNA substrate.
In higher eukaryotes, several classes of splicing sequence elements can be distinguished based on their function and location; the splice sites themselves constitute one such class (31, 39). The 5′ splice site is comprised of a 9-base consensus sequence that includes an almost universally conserved GU surrounded by additional nucleotides that are less well conserved. Similarly, the 3′ splice contains a highly conserved AG surrounded by a less conserved C and G and an upstream tract of about 10 bases that is rich in pyrimidines. Splice site sequences from tens of thousands of introns have been used to define these two consensus sequences and to construct position-specific scoring matrices to evaluate concordance to these consensuses (43). However, the great majority of splice sites do not conform to the consensus; for example, less than 5% match one of the four consensus sequences (MAG/GTRAGT, where M is A or C and R is A or G) for the 5′ splice site, and more than a quarter include three or more mismatches among the seven variable bases. Moreover, sequences that match the consensuses as well as or better than real splice sites can be easily found within introns. Even if such false 3′ and 5′ sites are paired to define false exons of typical size, the pseudo exons so defined outnumber the real exons by at least an order of magnitude (39, 44).
A third element required for splicing is the branch point. The first step in splicing involves an attack of the 5′ exon-intron boundary by a nucleotide near the 3′ end of the intron. The result is cleavage at the exon-intron boundary and the formation of a lariat structure in which a new 5′-2′ phosphodiester linkage is formed at the so-called branch point. The branch point base is usually A, and there is a loose consensus of YNYURAY that surrounds this A (underlined). However, the consensus sequence in this case is very poorly conserved and its position is variable, being usually between 15 and 35 bases upstream of the 3′ end of the intron (11, 17, 19, 21). As a result of this sequence and positional flexibility, cryptic branch points can be readily found upstream of most pseudo exons as well as exons. However, relatively few branch points have been experimentally identified, and there is evidence that the branch point is recognized early in splicing not only by the specific splicing factor SF1 (25) but also by exon-specific splicing factors (41). Thus, it is possible that the exact location and sequence of the branch point play a role in the recognition of 3′ splice sites.
The demonstration that a downstream 5′ splice site can greatly stimulate splicing at an upstream 3′ splice site led Robberson and colleagues to propose the exon definition model of splicing. According to this model, interactions of splicing factors across the exon precede the interaction between exons that constitutes the splicing reaction (35). This model has received ample support from genetic studies showing that mutation at either end of an exon usually leads to exon skipping rather than intron retention (6, 26, 34).
Additional information for the recognition of splice sites comes in the form of enhancer and silencer elements, which are categorized according to their location and effect: exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs). It is clear that there are interactions between these different types of elements (2, 13, 24, 36), but how this splicing information is integrated to produce a particular splicing outcome is not yet understood.
ESEs have been the most intensively studied of these elements. In most cases studied, ESEs bind and respond to particular SR proteins, a family so named because they contain one or more arginine-serine repeat domains (46). Although most particular examples have been examined in the context of alternative splicing (2, 3, 7, 54), it is likely that ESEs are also important for constitutive splicing (37) and thus for splicing in general. In addition to the many cases detecting individual ESEs associated with specific transcripts, attempts have been made to identify a broad range of sequences that can function as ESEs in a particular context. In these experiments, random oligomers were inserted into a poorly splicing exon and the effective sequences were then collected from spliced molecules after iterative selection (9, 12, 27, 28, 38). In several of these studies, splicing was assayed so as to depend on the presence of a single SR protein. In this way it was possible to identify not only the ESE sequences but also the SR proteins (SRp40, SRp55, SC35, ASF/SF2, 9G8) that targeted those sequences. As a result, different families of sequences have been defined, each with a consensus associated with a particular SR protein. Although distinct from each other, these sequence families typically display considerable degeneracy, such that a search algorithm based on agreement to the consensuses for just four SR proteins (8) finds a high frequency of candidate ESEs in both exons and introns.
Global definitions of ESEs have been sought using computational methods based on the overrepresentation of such sequences in exons or exon classes. Fairbrother et al. (15) identified 238 hexamer sequences that occurred more frequently in exons with weak splice sites than in exons with strong splice sites. When tested by insertion into a test exon, almost all of the hexamers (termed RESCUE-ESEs) enhanced splicing as predicted. We previously described a set of RNA octamers that were found more frequently in exons than in two regions with no requirement for such sequences: pseudo exons and intronless genes (51). In this computation, we restricted the analysis to exons occurring in 5′-untranslated regions (5′UTRs) so as to avoid detecting differences based on protein coding information. A statistical index was computed for each possible octamer based on these two comparisons. Octamers whose frequency difference surpassed a threshold for both the pseudo exon comparison score (P-score) and the 5′UTR of intronless genes score (I-score) were designated as putative ESEs (PESEs). Similarly, octamers with a lower prevalence in exons were designated as PESSs (putative ESSs). The predicted positive or negative effects of representative octamers of each class were confirmed by their insertion into test minigenes. From these results two lists of 2,069 PESEs and 974 PESSs were generated.
In both of these computational predictions, a high proportion of all possible sequences was found to be capable of acting as ESEs: 5.8% of hexamers for the RESCUE-ESEs and 3.2% of octamers. Moreover, given the high success rates of the tests (>90%), these proportions must be considered as minimums. Thus, ESEs seem to be omnipresent in exons, a conclusion that was implied by the high degree of degeneracy found in the earlier sequence selection experiments. However, this conclusion is critically dependent on the assumption that the activity of these nonnative oligomers in a small number of heterologous test systems reflects the action of natural splicing signals in natural exons. That is, although it has been shown that these sequences can act as ESEs, the question remains as to whether they do act as such when found within their natural context.
To address these questions we have performed site-directed mutagenesis to knock out PESEs found in a series of natural exons and tested the effect on splicing. In particular we asked the following questions. (i) Do the predicted PESE sequences reflect ESEs that function within their natural context? (ii) Do most exons require ESEs for splicing? (iii) When several functional ESEs are present, do they act redundantly or in concert?
More than two-thirds of these mutational disruptions reduced splicing, supporting the validity of the PESE scoring system and indicating the widespread use of ESEs to define splice sites. Most individual ESE disruptions produced a severalfold effect on splicing, suggesting that ESEs must work together to enhance splicing.
MATERIALS AND METHODS
Exon cloning.
Five human exons with or without their flanking intronic sequences beyond the splice site consensus sequences were amplified from human genomic DNA (placental DNA; Sigma) using primers tailed with EagI restriction sites (primer sequences are available on request). The amplified fragments were cut by EagI and inserted into the NotI site in pDCH1P12D (51). The insertion results in a 3-exon minigene in which the test exon is bounded by hamster dhfr exon 1 and the joined dhfr exons 4, 5, and 6. The sixth exon, dhfr-2-3, is composed of the joined hamster dhfr exons 2 and 3, found in pDCH1P12 (51). Among these six exons, five are constitutively spliced, and one (wt1-5) is alternatively spliced (18). Exons are considered constitutively spliced if no instance of their exclusion is present in the literature or expressed sequence tag databases.
Mutagenesis.
We used PCR ligation-based mutagenesis described by Ali and Steinkasserer (1). All mutant constructs were sequenced to confirm the mutations.
Splicing assay.
HEK 293 cells cultured in 35-mm dishes were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer. The cells were then incubated for 24 h. Total RNA was extracted using RNAwiz (Ambion) following the manufacturer's protocol. The extracted RNA was treated with DNase I, and half the RNA was reverse transcribed (RT) using Omniscript and random hexamers from QIAGEN. One-tenth of the RT product served as template in the following PCR labeled with [α-32P]dATP (10): forward primer, CGCCAAACUUGGGGGAAGCA; reverse primer, CGGAACUGCCUCCAACUAUC; initial denaturation, 93°C for 5 min; denaturation, 93°C, 30 s; annealing, 61°C, 30 s; extension, 72°C, 1 min; 28 cycles; final extension, 72°C, 7 min. Results of polyacrylamide gel electrophoresis were quantified with a PhosphorImager. At least three independent transfections were carried out for each of the six wild-type exons. A mutation was considered as effective if it reduced the splicing efficiency (spliced/[spliced + skipped]) to less than half compared to the wild type. Two exceptions were the second and third mutations of wt1-5; in this case no exon skipping was ever detected in wild-type transfections, and so any detectable exon skipping (>2%) was considered noteworthy.
Comparison with RESCUE-ESEs and ESEfinder.
The 238 RESCUE-ESE hexamers (14) were located in both wild-type and mutated exons. A mutation is said to disrupt a RESCUE-ESE if it reduced the number of RESCUE-ESEs in the sequence. For ESEfinder, we used default thresholds to identify sites respon-sive to the four SR proteins ASF/SF2, SC35, SRp40, and SRp55 (8). We analyzed the occurrences and locations of these responsive sites in the six exons and compared them with our PESEs and RESCUE-ESEs. A mutation is said to disrupt an ESEfinder ESE if it reduced the number of such sites in a sequence.
RESULTS
Exons.
We selected six exons to conduct our tests; five are spliced constitutively (chuk-8, clcn7-3, dhfr-2-3, hbb-2, thbs4-12) and one (wt1-5) is alternatively spliced with ∼70% inclusion (18). All are human except dhfr-2-3, a composite exon that originally resided in the hamster dhfr minigene that was used as a host context. To provide a sensitive assay for the detection of a splicing deficit, we wanted to start with wild-type exon sequences that already exhibited some exon skipping. When inserted along with about 60 nucleotides (nt) of flanking sequence, only chuk-8 and clcn7-3 had this attribute, yielding 11% and 34% exon skipping, respectively. The other four exons yielded close to 100% splicing. In order to encourage some skipping of the remaining exons, we deleted about 50 nt from both intronic flanks, retaining the regions of the consensus splice sites from 14 nt upstream to 6 nt downstream of the exon borders. In the case of wt1-5 the flank deletions abolished splicing, so the flanked version was used. Deletion of dhfr-2-3 flanks was not possible since the cloning site in the host sequence is made up of the dhfr-2-3 flanks. The flank deletions produced the desired effect in hbb-2 and thbs4-12, leading to 52% and 35% skipping, respectively, and so these were used in subsequent experiments. Thus, for two of the six exons tested, we used versions deprived of their natural intronic flanks. A more complete examination of the effects of exon flanks on the splicing of these exons is described elsewhere (53).
The mutations.
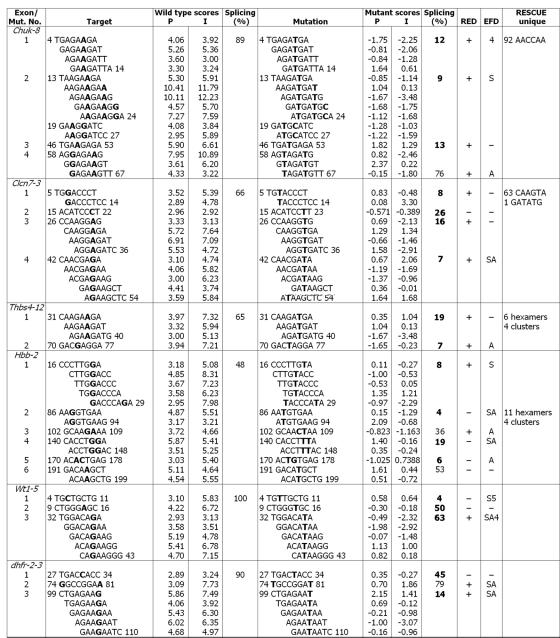
To identify PESEs for mutagenesis, we assigned two z-scores to each octamer in each exon, calculated as described previously (51). A P-score measures overrepresentation of an octamer in exons as compared to pseudo exons, and an I-score is based on a similar comparison to the 5′UTR of intronless genes. An octamer was designated as a PESE if it exceeded a value of 2.88 (P < 0.002) in both comparisons (51). The P- and I-scores were usually, but not always, in good agreement (Fig. 1, columns P and I). These exons contained between 4 and 14 PESEs, which fell into two to seven clusters. Profiles of the average of the two scores for all exons are presented in Fig. 2 (black curves). In addition to the PESE peaks, a lesser number of PESS valleys can also be seen.
FIG. 1.
Characteristics of PESE mutations. Site-directed mutagenesis was used to create point mutations within the indicated 6 exons positioned as the central exon in a 3-exon minigene. The sequence changes accompanying the 22 mutations created in PESEs are shown, with the mutated bases in bold. PESE scores for 8-mers overlapping the mutated sites are given as calculated previously (51) either on the basis of comparing exons with pseudo exons (P) or with the 5′UTRs of intronless genes (I). Only those overlapping 8-mers with PESE scores >2.88 based on both criteria are shown. The numbers indicate the first and last nucleotide of an octamer or cluster. The percent splicing (100 × spliced/[spliced + skipped]) is based on RT-PCR of transfected 293 cells. The boldface entries indicate a reduction in splicing to less than half that of the wild type (16 cases) or an increase in skipping of at least 10-fold (2 cases in wt1-5). The last three columns show the changes with respect to other methods of ESE prediction. The column labeled RED indicates whether the mutation also resulted in a RESCUE-ESE disruption. In the column labeled EFD, a letter or numeral indicates a disruption of an ESE predicted by ESEfinder; here the splicing factor specificity is denoted as follows: A, ASF/SF2; S, SC35; 4, SRp40; 5, SRp55. In the last column RESCUE-ESEs that do not fall within PESE sequences are noted. Mut. No., mutation number.
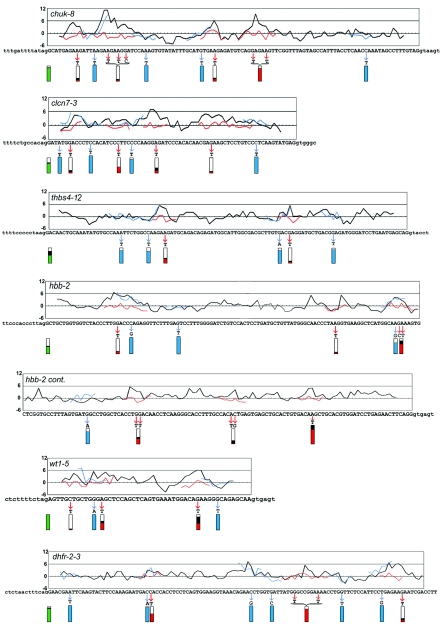
FIG.2.
Characteristics of PESE and control mutants. The black curves in the graphs show the PESE score profile across the exon. Successive windows of 8 nt were scored as described in Zhang and Chasin (51); the average of two scoring systems (P and I) is plotted for simplicity (separate profiles for each scoring system can be seen in Fig. 4). The score for each 8-mer is plotted near the middle of the 8-mer, at position 4. Point mutations in 22 predicted ESE clusters are indicated by the red arrows. The scores for the mutant sequences are shown by the red curves where they differ from the wild type. Twenty-four control mutations were of two types, both indicated by blue arrows and curves: 12 control mutations in PESE clusters were designed to not substantially change the PESE score and 12 control mutations outside PESE or PESS clusters also had little effect on the PESE score. Splicing phenotypes are indicated by the extent of filling of the rectangles below each mutated region. The green rectangles at the left show the extent of splicing (spliced/[spliced + skipped]) for the wild-type exons. The red and blue rectangles show the extent of splicing of the experimental and control mutants, respectively. To point out the effect of mutations on splicing, the colored areas represent the extent of splicing relative to that of the wild type set equal to 1; the black regions above the red or green areas indicate the range of values from two or three replicate transfections. Note that the splicing data presented in Fig. 1 and Table 1 have not been normalized in this way. The last 40 nt of the dhfr-2-3 exon were not mutated and are not shown. cont., continued.
Mutations were designed according to the following criteria: (i) to introduce as few base substitutions as possible; (ii) to reduce the P- and I-scores to less than 2.88 and ideally to as close to 0 as possible; (iii) to avoid reducing both scores to less than −2.88, which could be considered a PESS (51); and (iv) if more than one PESE clustered to form a single peak, the mutation(s) had to disrupt the entire cluster. We made 22 disruptions, individually knocking out every PESE or PESE cluster in the five human exons and three of seven in dhfr-2-3. Pursuance of the competing criteria described above involved some trade offs. We were able to effect the desired changes with single-base substitutions in 16 of the 22 cases; there were five double-base substitutions and one triple. The 22 disruptions affected 58 individual PESE octamers; their average score changed from 5.1 for the wild types to −0.3 for the mutants. None of the mutations produced a PESS, but in five cases one of the two comparison scores (the P-score) did fall below −2.88. Among the 116 mutant scores (58 octamers, 2 scores each), only one remained above 2.4. All specific sequence changes and the associated scores are shown in Fig. 1. The effect of the mutations on the average score profiles is shown by the red curves in Fig. 2.
The splicing phenotypes.
The effects of these mutations on splicing were assessed by transient transfection into HEK 293 cells followed by quantitative analysis of radioactive RT-PCR products (Fig. 3). The results are summarized in Fig. 1 in terms of absolute splicing efficiency and more usefully in Fig. 2 as splicing efficiency relative to that of the wild type (red rectangles). Of the 22 PESE disruptions, 16 (73%) produced a strong effect on splicing efficiency, defined here as a decrease to less than half that of the wild type. This success rate rose to 18 (82%) when two wt1-5 mutations were included; these mutants produced 15% and 43% exon skipping compared to no detectable skipping in the wild type.
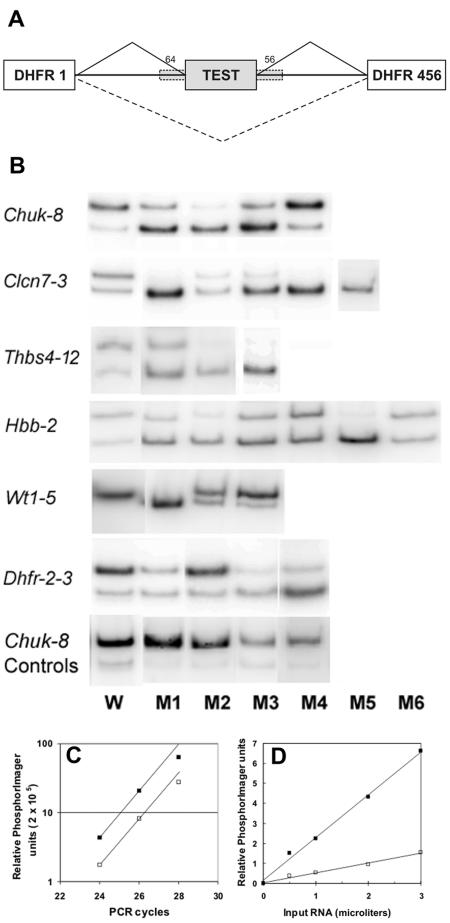
FIG. 3.
Splicing patterns in PESE mutants. A. General map of the constructs used. The dashed boxes indicate that exon flanks (∼50 nt) were included for most exons (chuk-8, clcn7-3, wt1-5, dhfr-2-3) and excluded for hbb-2 and thbs4-12 to generate a partial splicing phenotype for the wild-type exon in these cases. B. HEK 293 cells were transfected with mutant plasmids, and the resulting mRNA was amplified by RT-PCR using radioactive dATP, separated in polyacrylamide gels and subjected to phosphorimaging. The lane numbers reflect the order of the PESE disruption mutations within each exon, shown in Fig. 2. The results of one of duplicate experiment are shown. Also shown are control mutations in chuk-8 that do not disrupt a PESE. Control mutations in the other exons are not shown. The PhosphorImager data was converted to a tiff file and formatted using Corel PhotoPaint. C. Dependence of PhosphorImager signal intensity on PCR cycle number. D. Dependence of PhosphorImager signal intensity on the amount of RNA input. Two microliters, corresponding to the total RNA from approximately 15,000 cells, were routinely used for the RT reaction. W, wild type.
This high success rate raised the question of whether almost any change in the exon sequence would decrease splicing. We therefore similarly created and assayed 24 control mutations, divided into two categories of 12 each. In category 1, single-base substitutions were designed to make a minimal change in the P- and I-scores of a PESE. Category 2 mutations were made in regions containing neither PESEs nor PESSs and also made minimal changes in the scores. These base changes and their effect on the average scores are shown in Fig. 2 (blue arrows) and Table 1. The blue curves in Fig. 2 show the changes in the profiles brought about by these control mutations; it can be seen that their neutrality extends over all the overlapping sequences affected. The control mutations had little effect on splicing: efficiencies ranged from 77% to 115% compared to the wild type (Table 1 and blue rectangles in Fig. 2). RT-PCR results for the control mutations in one exon (chuk-8) are shown in Fig. 3.
TABLE 1.
Characteristics of control mutations
| Exon | Wild-type splicing (%)a | Positionb | Change | Mutant splicing (%)a | REDc | EFDd | RESCUE uniquee |
|---|---|---|---|---|---|---|---|
| Mutations that turn enhancers into other enhancers | |||||||
| chuk-8 | 89 | 15 | A→T | 95 | − | − | |
| 45 | G→T | 88 | − | − | |||
| clcn7-3 | 66 | 4 | A→T | 70 | + | 5 | GATATG |
| 25 | C→T | 60 | − | − | |||
| thbs4-12 | 65 | 31 | C→T | 50 | − | − | |
| 70 | G→A | 55 | − | AS | |||
| hbb-2 | 48 | 26 | C→G | 52 | + | − | CAGAGG |
| 105 | A→G | 35 | + | − | AAGAAA AGAAAG | ||
| wt1-5 | 100 | 12 | G→A | 100 | − | − | |
| dfhr-23 | 90 | 30 | C→A | 85 | − | 4 | |
| 61 | C→G | 93 | − | − | |||
| 100 | T→G | 95 | − | 4S | |||
| Mutations that turn neutral sequences to other neutral sequences | |||||||
| chuk-8 | 89 | 29 | A→T | 92 | + | − | |
| 92 | C→T | 85 | − | 4 | |||
| clcn7-3 | 66 | 13 | C→T | 75 | − | − | |
| 61 | C→T | 70 | − | AS | |||
| thbs4-12 | 65 | 23 | A→T | 54 | − | − | |
| 86 | G→T | 58 | + | A | GGAGATGAGATG | ||
| hbb-2 | 48 | 40 | G→T | 55 | − | 5 | |
| 130 | T→A | 39 | − | − | |||
| wt1-5 | 100 | 43 | G→T | 100 | − | − | |
| dfhr-23 | 90 | 7 | A→T | 95 | + | − | AACGAA |
| 67 | G→C | 92 | − | − | |||
| 88 | G→T | 88 | − | − |
Exon inclusion out of all species.
From the 5′ end of exon.
RED, disruption of an ESE predicted by RESCUE-ESE.
EFD, disruption of an ESE predicted by ESEfinder (5, SRp55; 4, SRp40; A, ASF/SF2; S, SC35).
Disrupted ESE predicted by RESCUE-ESE but not in the PESE list.
A mutation that decreases splicing is not necessarily evidence for an enhancer; the possibility that a silencer has been created must always be considered. In these experiments we were careful not to create PESSs when disrupting PESEs. In three additional cases, we purposely designed mutations that created PESSs (both P- and I-scores less than −2.88) concomitant with disrupting a PESE. The splicing phenotypes of these three mutants seemed to be more severe than those of the mutants that simply had a PESE disrupted; the former exhibited an average of 10% splicing relative to the wild type (Fig. 3, clcn7-3 M5, thbs4-12 M3, and dhfr-2-3 M4) compared to 26% for the 18 effective disruptions in Fig. 1.
As mentioned above, we removed the natural flanking sequences from two of our test constructs, thbs4-12 and hbb-2, so that the wild type exhibited some initial exon skipping (35 to 52%). To see if this sensitizing strategy was necessary, we examined the effects of this flank removal in the most extreme case, hbb-2, where it caused a decrease from 100% to 48% splicing efficiency. All six PESE disruptions were reproduced in the flanked version of this exon. None of the mutations affected splicing in the presence of the intronic flanks (data not shown), indicating that intronic flanks here contain redundant splicing enhancers that can compensate for the loss of an exonic enhancer.
A PESE/PESS prediction website.
We have established a web server that allows users to identify PESEs and PESSs in their own sequences: http://cubweb.biology.columbia.edu/pesx.
DISCUSSION
Predicted ESEs function in natural exons.
One goal of this work was to determine whether PESEs predicted on the basis of computed statistical features actually function as ESEs in situ. A negative result here would have been inconclusive, as one could readily imagine ESEs to be arranged as redundant signals. The results turned out to be strongly positive, with 18 of 22 PESE disruptions either decreasing splicing efficiency by at least a factor of 2 (16 of 22) or by increasing skipping from an undetectable level to a substantial fraction (2 of 22). One conclusion that can be drawn is that our method of prediction is fairly robust; most PESEs are functioning as ESEs in their natural context.
Although our long-range goal is to identify the earliest exon definition signals and the SR proteins that bind to most ESEs are thought to act early in exon recognition, our assay method does not differentiate between a role for ESEs in recognition and a role in the biochemical process of splicing, which are not mutually exclusive in any case. An examination of the effects of these mutations on cell-free assembly of RNP complexes will be necessary to resolve this issue.
Single-base substitutions that disrupt ESEs.
The mutational disruptions were designed to bring the P- and I-scores below 2.88. This arbitrarily set z-score threshold corresponds to a probability of less than 0.002 that an octamer with such a score would occur by chance. Over 2,000 PESE sequences were defined in this way, representing over 3% of all possible octamers (2,069 out of 65,536). Moreover, they are overrepresented in exons, the average internal exon having 10.5 such octamers (51). As might be expected, different PESEs can be grouped into families of overlapping or related sequences; as a result, they tend to occur in clusters. Since any single-base substitution would change eight overlapping octamers, it was not always easy to find a single change that did not leave an overlapping PESE with a high score. A transversion of a purine to thymine was by far the most common change that allowed this parsimony. The 22 disruptions comprised 29 single-base substitutions; 26 of these involved a change to a T, 23 being transversions. The effectiveness of T is not surprising, since T is underrepresented among PESEs (16% versus 25% to 30% for the other three bases). It is interesting to note that a change to T is also the most common change among random missense mutations that disrupt splicing: 47% compared to 21% for a control set of mutations that did not affect splicing (51). In contrast, T is highly overrepresented among PESS sequences (48%).
Inefficient splicing phenotypes are not the result of NMD.
Mutations that generate premature termination codons can often reduce the steady-state level of mRNA via nonsense-mediated decay (NMD) (29, 47). In such cases NMD could lead to an underestimate of exon inclusion and an exaggeration of the skipping phenotype. However, NMD is not affecting the conclusions here. The T substitutions in mutated PESEs generated nonsense codon triplets in 13 of the 22 disruptions, but the triplet constituted an in-frame stop codon in only three of these cases. Moreover, in two of these three cases the stop codon was less than 50 nt from the 3′ end of the central exon; premature chain terminations within this region in penultimate exons do not usually give rise to NMD (29, 32). The one remaining case produced only a 30% decrease in splicing relative to the wild type (hbb-2, third PESE disruption) and was not counted as an effective disruption. Moreover, it appears that NMD is not operating in this system, as two of the control mutations produced in-frame stop codons beyond the 50-nt limit yet did not reduce the proportion of spliced RNA. We have previously argued that NMD does not operate for the chuk-8 and thbs4-12 exons based on their response to insertions (51). The lack of correlation between exon skipping and in-frame stop codons also does not support the operation of nonsense-mediated aberrant splicing (30, 49) for these exons.
Relation of PESEs to ESEs predicted by other methods.
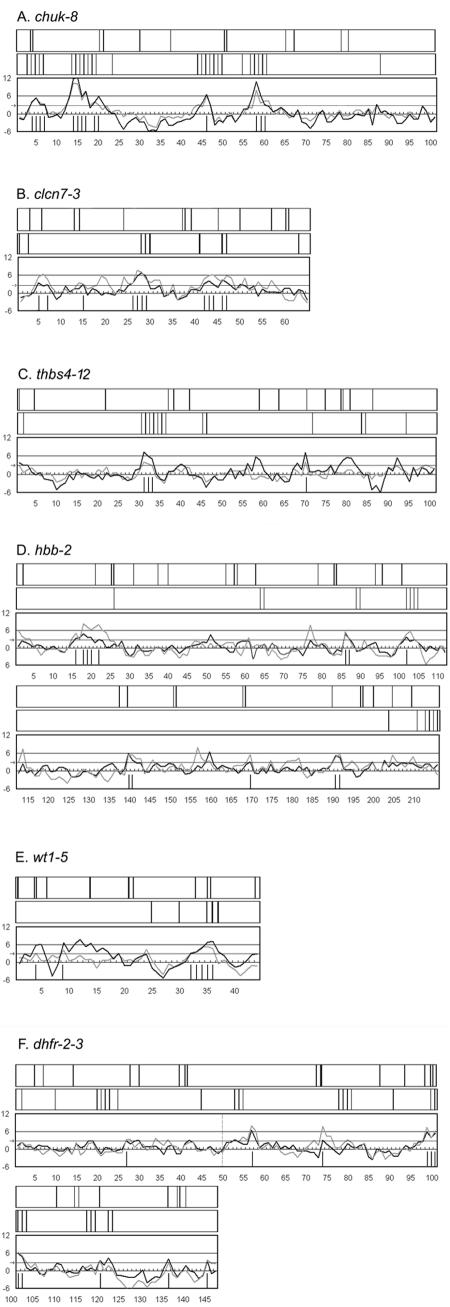
The validation of most of the PESEs as ESEs prompted us to analyze these six test exon sequences for the presence of ESEs predicted by other methods: RESCUE-ESEs identified by a different computational strategy (15) and those predicted by ESEfinder based on functional selection (8, 27, 28). These comparisons are presented in Fig. 4, which also displays the separate P- and I-scores for PESEs in profiles for each exon. PESEs are predicted when both scores exceed 2.88; the positions of the other ESEs are indicated by vertical lines above the profiles.
FIG. 4.
Comparison of different ESE prediction methods. A to F. The graphs show PESE scores for the six tested exons. Unlike the graphs in Fig. 2, two curves are shown here: the dark curves depict scores based on a comparison of real exons to pseudo exons and the light curve is based on a comparison to the 5′UTRs of intronless genes (51). The vertical lines under the curves denote the starting positions of individual PESEs. The dashed vertical line in panel F indicates the dhfr exons 2 and 3 joint. The rectangle directly above each graph depict the positions of ESE 6-mers predicted by the RESCUE method (15). The top rectangle in each panel depicts the positions ESEs predicted by ESEfinder (8), based on the consensuses of sequences selected for function from among random oligomers (27, 28).
There is good agreement between RESCUE-ESEs and PESEs here. Nevertheless, there are some notable differences, which are documented in columns 10 and 12 of Fig. 1. RESCUE-ESE did not identify 7 of the 18 PESEs validated here (39%). Conversely, there were 16 RESCUE-ESE hexamers or hexamer clusters in these exons that we did not assign as PESEs (Fig. 1, last column). That not all of these RESCUE-ESEs are functional is indicated by our control experiments. Of 24 control mutations, 6 happened to disrupt RESCUE-ESEs yet had essentially no effect on splicing (Table 1, column 6).
ESEfinder also predicted ESEs that overlapped with 11 of the 18 validated PESEs, the same number as RESCUE-ESE, albeit in mostly different sequences. There is less overall agreement between ESEfinder ESEs and PESEs (Fig. 4), mainly because ESEfinder predicted about twice as many clusters of sequences (an average of 11 per exon) compared to PESEs (4.3) or RESCUE-ESEs (6.3). The different results with ESEfinder may be attributable to base compositional differences between ESEfinder ESEs and RESCUE-ESEs or PESEs, the most striking of which is CpG content. The sequences upon which ESEfinder is based (kindly provided by Adrian Krainer) contain 9.3% CpG dinucleotides, which is high compared to exons in general (4%) and to PESEs (4%) and RESCUE-ESEs (3%) in particular. It could be that CpGs happen to be a common feature of the binding sites of the four SR proteins (ASF/SF2, SRp40, SRp55 and SC35) upon which ESEfinder is based (8, 27, 28). However, a high CpG content was also evident in the ESE sequences isolated by Schaal and Maniatis (38) based on responsiveness to two additional SR proteins (9G8 and SRp20 in addition to ASF/SF2 and SC35) and in the AC-rich sequences selected by Coulter et al. in vivo (12). Moreover, CpGs may be underrepresented in the computationally defined sequences, since RESCUE-ESEs are very high in A+T (14) and the strategy used to identify PESEs may have selected against CpGs (51).
On the other hand, there may be biases in functional selections as well. Most functional selections insert sequences into the terminal exon of a 2-exon construct rather than an internal exon, they are usually carried out in vitro, they assess activity in one particular context, and through iterations they select for a tighter binding subset of functional sequences. The identification of ESE sequences using SELEX to isolate oligomers that simply bind to SR proteins may be less subject to context biases and have resulted in consensus sequences that are significantly different from most of those identified by functional selection (4, 9, 45).
In summary, these comparisons show some overlaps and some differences. It is likely that each of these three methods will identify ESEs missed by the other two. Thus, the true number of ESEs will be even greater than that predicted and verified by any individual method. The availability of a Web site (http://cubweb.biology.columbia.edu/pesx) for identifying PESEs will allow researchers to take advantage of the strong predictor performance seen here.
In designing these mutations, we were careful not to create any PESSs so as to focus on ESE disruption per se. However, after these experiments were completed, Wang et al. described a list of 103 hexamers (the FAS-hex-3 set) identified as ESS candidates by genetic selection; upon testing, two-thirds of these hexamers decreased splicing (50). Two of the 18 PESE disruptions described here created such an ESS candidate (wt1-5-1 and wt1-5-2), so it is possible that in these two cases silencing may have contributed to the phenotype.
Prevalence of ESEs in exons.
In addition to providing a validation of the PESE sequences, these results have implications about the mode of action of ESEs. First, the high success rate of the mutagenesis disclosed a remarkable lack of redundancy among the multiple ESEs in these exons. All but one of the exons tested are constitutively spliced and as such should represent the great majority of exons. It follows that most exons require several ESEs to effect efficient splicing.
Second, in most cases each of several individual disruptions abolished most splicing, meaning that two or more ESEs must be acting in concert to achieve efficient splicing. In all five constitutive exons there were two to four disruptions that reduced splicing to less than 50% and one to three that reduced splicing to less than 25%. For instance, in the large hbb-2 exon, disruption of each of four separated ESEs reduced splicing to 10%, 19%, 23%, and 32% of the wild-type control. If enhancement were simply additive, an average decrease to 75% would be expected for each of the four individual ESEs. These results stand in contrast to the experiments of Hertel and Maniatis, who found that enhancers of dsx splicing acted in an additive manner (22). Those experiments focused on the activation of a 3′ splice site of a terminal exon, perhaps a simpler situation that required only a single enhancer complex. The concerted action of ESEs seen here fits well with a picture of SR proteins binding to ESE sequences to form a bridge between the two ends of an internal exon as part of the exon definition process (2, 16, 35, 40). A requirement for more than one ESE for spliceosome formation has been proposed by Shen et al. (41). Thus, part of the concerted action could be reflecting the two intron definitions ultimately required of all internal exons; i.e., one ESE per exon would promote spliceosome formation across the upstream intron and another across the downstream intron.
Interestingly, the greatest effect on splicing brought about by an ESE disruption was seen in the alternatively spliced wt1-5 exon where a single-base substitution reduced splicing 50-fold. This exon was also completely dependent on its natural flanks, implying a reliance on ISEs as well. These results are consistent with the view of alternatively spliced exons as being “weaker” in general and thus more vulnerable to perturbation of splicing elements. As a class, alternatively spliced exons exhibit a greater conservation of flanking sequences than do constitutively spliced exons (42) and include a greater number of ESSs (51).
Relationship between ESEs and ISEs.
The lack of redundancy among ESEs is not seen when considering the action of other elements, namely the ISEs that must reside in the near flanks of some of these exons. ISEs may generally provide an alternative means for the recruitment of splicing factors. We have previously described pentamer sequences that are overrepresented in exon flanks; this overrepresentation is limited to about 50 nt beyond the splice site consensus sequences (52, 53). These candidate ISE sequences (52, 53) are quite distinct from PESEs (51). For hbb-2 the decreases in splicing brought about by the disruption of single ESEs were completely suppressed if the near flanks of these exons were also provided. We have previously shown that a low splicing phenotype produced by depriving chuk-8 of its flanks could be substantially reversed by the insertion of any of eight different PESE octamers (51), again pointing to ISEs and ESEs as alternatives. Moreover, the mutational disruption of a PESS in flank-deprived chuk-8 also restored splicing (unpublished results). It remains to be seen whether ISEs can provide all the enhancement necessary for efficient splicing, i.e., whether they can suppress multiple ESE knockouts.
A redundancy with ISEs may explain why mutational screens have not revealed a strong dependence of splicing on ESEs. Despite the fact that exonic changes are being increasingly recognized among mutations resulting in human genetic disease (7, 16), by far the larger number of characterized splicing mutations have been located in the splice sites. Similarly, splicing mutants of cultured cells have mostly been affected in splice sites (6, 10, 34, 48). The failure to detect ESE mutations could be due to the leaky nature of these mutations; a low proportion of correctly spliced mRNA may well allow a wild-type phenotype at the cellular or organismic level. Moreover, an expectation that multiple ESEs would provide a large mutational target compared to splice sites may not be justified, as most single-base substitution mutations may not compromise ESE function; the mutations created here were specifically tailored to knock out the ESE character (score) of these sequences.
Acknowledgments
L.C. was supported by funds from Columbia University. X.Z. is a predoctoral Faculty Fellow of Columbia University.
REFERENCES
- 1.Ali, S. A., and A. Steinkasserer. 1995. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques 18:746-750. [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeois, C. F., F. Lejeune, and J. Stevenin. 2004. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 78:37-88. [DOI] [PubMed] [Google Scholar]
- 5.Burge, C. B., T. H. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 6.Carothers, A. M., G. Urlaub, D. Grunberger, and L. A. Chasin. 1993. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol. Cell. Biol. 13:5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 8.Cartegni, L., J. Wang, Z. Zhu, M. Q. Zhang, and A. R. Krainer. 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31:3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavaloc, Y., C. F. Bourgeois, L. Kister, and J. Stevenin. 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5:468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, I. T., and L. A. Chasin. 1993. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol. Cell. Biol. 13:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua, K., and R. Reed. 2001. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 21:1509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulter, L. R., M. A. Landree, and T. A. Cooper. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Expert-Bezancon, A., A. Sureau, P. Durosay, R. Salesse, H. Groeneveld, J. P. Lecaer, and J. Marie. 2004. hnRNP A1 and SR proteins, ASF/SF2 and SC35 have antagonistic functions in splicing of beta-tropomyosin exon 6B. J. Biol. Chem. 279:38249-38259. [DOI] [PubMed] [Google Scholar]
- 14.Fairbrother, W. G., D. Holste, C. B. Burge, and P. A. Sharp. 2004. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2:E268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairbrother, W. G., R. F. Yeh, P. A. Sharp, and C. B. Burge. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297:1007-1013. [DOI] [PubMed] [Google Scholar]
- 16.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 17.Green, M. R. 1986. Pre-mRNA splicing. Annu. Rev. Genet. 20:671-708. [DOI] [PubMed] [Google Scholar]
- 18.Haber, D. A., R. L. Sohn, A. J. Buckler, J. Pelletier, K. M. Call, and D. E. Housman. 1991. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc. Natl. Acad. Sci. USA 88:9618-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, N. L., and P. Senapathy. 1990. Distribution and consensus of branch point signals in eukaryotic genes: a computerized statistical analysis. Nucleic Acids Res. 18:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 22.Hertel, K. J., and T. Maniatis. 1998. The function of multisite splicing enhancers. Mol. Cell 1:449-455. [DOI] [PubMed] [Google Scholar]
- 23.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 24.Kan, J. L., and M. R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent, O. A., D. B. Ritchie, and A. M. Macmillan. 2005. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol. Cell. Biol. 25:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krawczak, M., J. Reiss, and D. N. Cooper. 1992. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90:41-54. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H. X., S. L. Chew, L. Cartegni, M. Q. Zhang, and A. R. Krainer. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, H. X., M. Zhang, and A. R. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maquat, L. E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev. Mol. Cell Biol. 5:89-99. [DOI] [PubMed] [Google Scholar]
- 30.Mendell, J. T., C. M. ap Rhys, and H. C. Dietz. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298:419-422. [DOI] [PubMed] [Google Scholar]
- 31.Mount, S. M. 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10:459-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neu-Yilik, G., N. H. Gehring, M. W. Hentze, and A. E. Kulozik. 2004. Nonsense-mediated mRNA decay: from vacuum cleaner to Swiss army knife. Genome Biol. 5:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25:1147-1149. [DOI] [PubMed] [Google Scholar]
- 34.O'Neill, J. P., P. K. Rogan, N. Cariello, and J. A. Nicklas. 1998. Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat. Res. 411:179-214. [DOI] [PubMed] [Google Scholar]
- 35.Robberson, B. L., G. J. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rooke, N., V. Markovtsov, E. Cagavi, and D. L. Black. 2003. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaal, T. D., and T. Maniatis. 1999. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaal, T. D., and T. Maniatis. 1999. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19:1705-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senapathy, P., M. B. Shapiro, and N. L. Harris. 1990. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183:252-278. [DOI] [PubMed] [Google Scholar]
- 40.Shen, H., and M. R. Green. 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16:363-373. [DOI] [PubMed] [Google Scholar]
- 41.Shen, H., J. L. Kan, and M. R. Green. 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13:367-376. [DOI] [PubMed] [Google Scholar]
- 42.Sorek, R., and G. Ast. 2003. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 13:1631-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens, R. M., and T. D. Schneider. 1992. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol. 228:1124-1136. [DOI] [PubMed] [Google Scholar]
- 44.Sun, H., and L. A. Chasin. 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 20:6414-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tacke, R., and J. L. Manley. 1999. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11:358-362. [DOI] [PubMed] [Google Scholar]
- 46.Tacke, R., and J. L. Manley. 1999. Functions of SR and Tra2 proteins in pre-mRNA splicing regulation. Proc. Soc. Exp. Biol. Med. 220:59-63. [DOI] [PubMed] [Google Scholar]
- 47.Urlaub, G., P. J. Mitchell, C. J. Ciudad, and L. A. Chasin. 1989. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol. Cell. Biol. 9:2868-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valentine, C. R. 1998. The association of nonsense codons with exon skipping. Mutat. Res. 411:87-117. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J., Y. F. Chang, J. I. Hamilton, and M. F. Wilkinson. 2002. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10:951-957. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Z., M. E. Rolish, G. Yeo, V. Tung, M. Mawson, and C. B. Burge. 2004. System identification and analysis of exonic splicing silencers. Cell 119:831-845. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, X. H., and L. A. Chasin. 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 18:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, X. H., K. A. Heller, I. Hefter, C. S. Leslie, and L. A. Chasin. 2003. Sequence information for the splicing of human pre-mRNA identified by support vector machine classification. Genome Res. 13:2637-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, X. H., C. Leslie, and L. A. Chasin. 2005. Dichotomous splicing signals in exon flanks. Genome Res. 15:768-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, Z. M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11:278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]