Abstract
Protein-coding genes of trypanosomes are mainly transcribed polycistronically and cleaved into functional mRNAs in a process that requires trans splicing of a capped 39-nucleotide RNA derived from a short transcript, the spliced-leader (SL) RNA. SL RNA genes are individually transcribed from the only identified trypanosome RNA polymerase II promoter. We have purified and characterized a sequence-specific SL RNA promoter-binding complex, tSNAPc, from the pathogenic parasite Trypanosoma brucei, which induces robust transcriptional activity within the SL RNA gene. Two tSNAPc subunits resemble essential components of the metazoan transcription factor SNAPc, which directs small nuclear RNA transcription. A third subunit is unrelated to any eukaryotic protein and identifies tSNAPc as a unique trypanosomal transcription factor. Intriguingly, the unusual trypanosome TATA-binding protein (TBP) tightly associates with tSNAPc and is essential for SL RNA gene transcription. These findings provide the first view of the architecture of a transcriptional complex that assembles at an RNA polymerase II-dependent gene promoter in a highly divergent eukaryote.
Small nuclear RNAs (snRNAs) play an essential role in the production of functional mRNAs in all eukaryotic cells. In human cells, a preinitiation complex containing RNA polymerase II is recruited to those snRNA gene promoters that contain a single DNA element located 50 to 70 bp upstream from the transcription start site (15). This 20-bp core promoter element, called the proximal sequence element (PSE), is recognized by a transcription factor known as the snRNA-activating protein complex (SNAPc; also known as PTF) (15, 35). In Drosophila melanogaster, snRNA production requires a bipartite PSE, divided into PSEA and PSEB regions (19). Drosophila RNA polymerase II is recruited to snRNA gene promoters only when the PSEA contains an invariant trinucleotide.
The trypanosome family of unicellular parasitic organisms includes the medically important genera Leishmania and Trypanosoma and the insect-infective genus Leptomonas. In contrast to most eukaryotic organisms, trypanosomes express a novel snRNA called the spliced-leader (SL) RNA (for reviews, see references 5 and 6). Trypanosome SL RNA is the sole eukaryotic snRNA that contains a complex hypermethylated cap (3) and functions as an exon donor in the trans-splicing reaction that is required for the production of every mRNA (18, 33). The SL RNA gene also is the only trypanosome gene for which an RNA polymerase II promoter has been defined (10).
In trypanosomes, RNA polymerase II functions in at least three distinct ways. First, it must produce large amounts of the capped SL RNA, which donates its 5′ 39-nucleotide SL to every mRNA. Second, it must selectively transcribe most protein-coding genes (in Trypanosoma brucei, RNA polymerase I is responsible for transcribing genes encoding the major surface proteins) (12). Finally, trypanosome RNA polymerase II transverses many genes in a single polymerization reaction, relying on cotranscriptional RNA processing reactions to fragment the polycistronic pre-mRNAs into capped and polyadenylated mRNAs.
The largest subunit of trypanosome RNA polymerase II is unusual among eukaryotic RNA polymerase II enzymes in lacking the standard heptapeptide repeat at its carboxyl end and containing multiple di-serines (8, 10, 32). Typical RNA polymerase II-associated basal transcription factors that have been studied extensively in yeasts and metazoans lack recognizable orthologs in trypanosomes. For example, transcription factor IIB (TFIIB) and most subunits of TFIIH and TFIIF cannot be readily identified in the completed trypanosome genome sequences. A protein considered to be the trypanosome ortholog of the highly conserved TATA-binding protein, TBP (16), has been identified using bioinformatics and can be modeled onto the classical TBP structure (6; A. Deaconescu, S. K. Burley, and G. A. M. Cross, unpublished observations). This protein, also referred to as TRF4, plays a vital role in transcription by all three RNA polymerases (28). However, as no TATA elements have been identified in any trypanosome gene promoter in the limited studies so far reported, the mechanisms by which TBP functions in trypanosome gene expression are a tantalizing open question.
The SL RNA gene promoter likely recruits RNA polymerase II through protein-DNA interactions that occur within the three elements PBP-1E, PBP-2E, and the initiator element (IR), which constitute the SL RNA gene promoter in most of the well-studied trypanosomatids (1, 5, 13, 23). Detailed studies in Leptomonas seymouri showed that PBP-1E, located 60 to 80 bp upstream from the transcription start site, is essential for SL RNA gene expression and recruits the SNAP complex-like transcription factor PBP-1 (now designated tSNAPc), which is a 122-kDa sequence-specific DNA-binding protein complex containing three subunits of 57 kDa, 46 kDa, and 36 kDa (22). The PBP-1E shares functional but not sequence homology to metazoan PSEs. The two downstream elements PBP-2E and IR, in concert with PBP-1E, coordinate the assembly of an RNA polymerase II-containing preinitiation complex that directs SL RNA expression (7).
This report presents data on T. brucei SNAPc, reveals that it is tightly associated with TBP, and demonstrates that both proteins are essential for SL RNA gene transcription. Two of the tSNAPc polypeptides, tSNAP50 and tSNAP26, are similar to subunits of SNAPc, whereas a third polypeptide, tSNAP42, is divergent from other SNAP subunits and may be unique to trypanosomes.
MATERIALS AND METHODS
Trypanosome cell lines and transfections.
Procyclic (tsetse midgut form) parasites of the wild-type T. brucei Lister 427 strain were previously transfected to generate a strain that contains and constitutively expresses T7 RNA polymerase and tetracycline repressor, coupled to drug resistance markers. This cell line, 29-13, was cultured at 27°C in SDM-79 supplemented with 10% fetal bovine serum and containing 15 μg/ml G418 and 25 μg/ml hygromycin to maintain the foreign genes. Stably transfected cell lines were produced by transfecting pLEW111 (pLEW82 − the luciferase gene) derivatives into 29-13, using electroporation (34). Transfectants were selected using 2.5 μg/ml phleomycin in the presence of 20 ng/ml tetracycline.
Plasmid constructions.
pAD50, which contains a tandem affinity protein (TAP)-tagged tSNAP50, was constructed by introducing the tSNAP50 open reading frame (ORF) with a carboxyl-terminal TAP cassette into pLEW111. This gene was amplified from wild-type T. brucei genomic DNA by PCR using two primers: one corresponded to the amino terminus of the ORF and contained a 5′ NruI site, and the second corresponded to the carboxyl terminus and contained a 5′ XhoI site. The carboxyl-terminal TAP cassette, along with the flanking XhoI and BamHI sites, was PCR amplified from pBS1761c and ligated to the ORF after XhoI digestion. This DNA was reamplified using primers that corresponded to the amino terminus of tSNAP50 and the carboxyl terminus of the TAP tag. This ∼1,900-bp DNA fragment was NruI and BamHI digested and inserted into pLew111.
pJM26, which contains TAP-tagged tSNAP26, was constructed by initially inserting the ∼780-bp tSNAP26 ORF between the HindIII and BamHI sites of pLew111. The ORF was amplified from genomic DNA by PCR, using primers that corresponded to the amino and carboxyl termini of this gene with flanking HindIII and BamHI sites. The carboxyl-terminal TAP tag was inserted into the BamHI site 3′ to the ORF.
The Ty-1 epitope (Ty tag) (EVHTNQDPLD), which reacts with the BB2 monoclonal antibody (a kind gift from Keith Gull), was introduced at the amino terminus of T. brucei TBP by PCR of a chromosome X shotgun sequencing clone from T. brucei TREU 927, the official genome project strain (4). The 5′ primer contained a HindIII site followed by a sequence encoding an initiator Met, the Ty tag, and the 5′ end of the TBP ORF. The 3′ primer was complementary to the 3′ end of the TBP ORF and terminated in an NruI site. pLEW82 was digested with BamHI, and the 5′ overhang was filled in using Klenow fragment and then digested with HindIII. The TBP PCR product was NruI and HindIII digested and ligated into the vector.
In each case, the DNA sequence of the recombinant plasmid was verified. Each construct was transfected into 29-13 cells, as described above.
Antibodies.
Antibody reagents were generated from recombinant proteins produced as maltose binding protein fusions. After IPTG (isopropyl-β-d-thiogalactopyranoside) induction, bacterial cells were sonicated and proteins were purified according to the manufacturer's protocol (New England Biolabs). Soluble tSNAP42, tSNAP26, and TBP recombinant proteins were used to raise polyclonal antibodies in rabbits. Anti-tSNAP50 antibodies were raised against the amino-terminal half of the protein in chickens. All antibodies were produced at Lampire Corporation, Pipersville, Pa.
In vitro transcription and electrophoretic mobility shift analysis.
Transcription extracts were made from wild-type T. brucei parasites following the method described elsewhere (17). Extract protein concentrations were routinely 6 to 8 μg/μl.
pJP10 was generated using recombinant PCR, starting with two partially overlapping PCR products that extended across the bp −125 to +120 region of the wild-type SL RNA gene. During the initial PCRs, a 20-bp tag was inserted at position +63 within the SL RNA coding region. The recombinant ∼265-bp PCR product was cloned into pBSIISK(+) (Stratagene), and supercoiled DNA was used to program transcription reactions. Transcripts were detected by primer extension using a 32P (5′ end)-labeled oligonucleotide primer that hybridized to the 20-nt RNA tag.
For gel shift analysis, the wild-type (WT) SL RNA gene promoter, spanning the region from bp −115 to +2 of the SL RNA gene, was produced by PCR amplification using pJP10 as template. The DNA was end labeled using [32P]ATP and T4 kinase and purified using Probequant G50 spin columns (Amersham). In DNA competition experiments, mutant promoter competitor was generated by recombinant PCR followed by gel purification.
Protein purification.
The TAP purification protocol was adapted from Rigaut et al. (27). Nuclei were isolated as described previously (17) from 4 × 1010 transgenic T. brucei organisms. Prior to immunoglobulin G (IgG) chromatography, nuclei were lysed using 0.4 M KCl in TX buffer (7, 17), nucleic acid was separated from protein by two ammonium sulfate steps or DEAE chromatography, and extract was loaded onto an IgG column. After rigorous washing, proteins were eluted from the IgG column by tobacco etch virus (TEV) protease digestion. Proteins bound in the subsequent calmodulin affinity step were eluted with 2 mM EGTA. For mass spectrometry analysis, purified proteins were concentrated using a Centricon 10 (Millipore) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For velocity sedimentation, purified proteins were concentrated, loaded onto 5-ml 5 to 20% (wt/vol) sucrose gradients in 150 mM potassium glutamate-TX buffer, and centrifuged at 100,000 × g for 18 h. Highly purified protein from calmodulin chromatography or velocity sedimentation steps were used for transcription add-back reactions.
Ty-tagged TBP was purified from nuclear extracts from transgenic parasites. Monoclonal antibody BB2 (4) against the Ty epitope was bound to protein A Sepharose and used for affinity chromatography. Proteins were incubated with beads in IP buffer (20 mM HEPES, pH 7.9, 150 mM sucrose, 2.5 mM MgCl2, 1 mM EDTA, 2.5 mM dithiothreitol, 0.1% Nonidet P-40, and 1 μM each pepstatin, leupeptin, and phenylmethylsulfonyl fluoride) containing 200 mM KCl for 3 h. Following stringent washing, protein was eluted from antibody-containing beads using a synthetic Ty peptide (100 μg/ml). Purified Ty-TBP was verified by silver staining of SDS-PAGE gels and by Western blot analysis.
Coimmunoprecipitations.
Preimmune, anti-tSNAP42, anti-tSNAP26, and anti-TBP antibodies were cross-linked to protein A Sepharose beads (14). Twenty microliters of antibody-bound beads was washed in IP buffer containing either 200 mM or 400 mM KCl and incubated for 4 to 6 h with 1 mg of nuclear extract protein in 400 μl IP buffer. Precipitates were washed five times, boiled in SDS-PAGE sample buffer, and analyzed by immunoblotting, using the ECL enhanced chemiluminescence detection system (Amersham).
Immunodepletions.
Preimmune, anti-tSNAP42, and anti-TBP antibodies covalently attached to protein A Sepharose were used for immunodepletions. Two hundred microliters (1.5 mg protein) of nuclear extract was incubated with 100 μl of beads for two subsequent rounds. Depletions using anti-TBP antibodies were performed in IP buffer with 400 mM potassium glutamate. Depletions using anti-tSNAP42 were performed using 250 mM salt. Thirty to 50 μg of protein of depleted extract was used for transcription reactions.
Protein alignments.
Multiple alignments of tSNAP42 proteins from T. brucei, Trypanosoma cruzi, Leishmania major, and Leptomonas seymouri were performed using CLUSTALW. The ProFit program, a sequence-to-structure search algorithm for protein fold recognition in Proceryon's ProHit package, was used to perform fold recognition searches for all four trypanosome tSNAP42 sequences (http://www.proceryon.com/solutions/prohit_pro.html). The search was performed under default parameters against known structures in SCOP40, a minimally redundant library of PDB structures. In each case the query sequence was analyzed for the energy potential of the forced alignment with every known structure and then a ranking was produced. Thdldx scores greater than 40 were considered significant. The resulting sequence-to-structure alignments were then manually inspected.
RESULTS
Purification of the T. brucei SNAP complex.
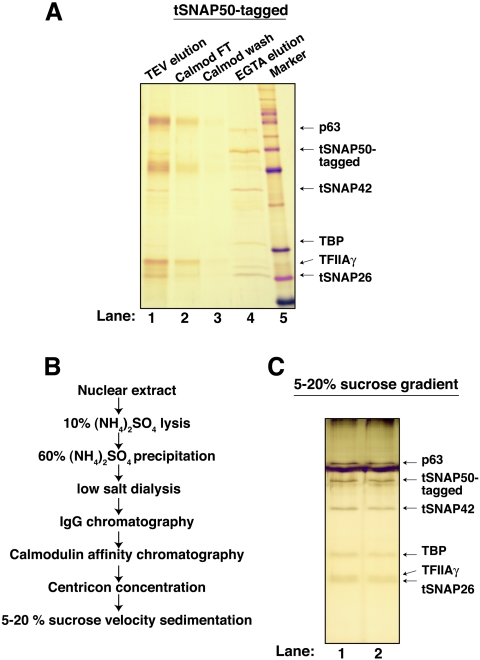
To study the pathogenic trypanosome T. brucei, we affinity tagged the trypanosome ortholog of Leptomonas p57 (Lsp57). Since the calculated molecular mass of the T. brucei protein is ∼50 kDa, we refer to this protein as tSNAP50. Procyclic T. brucei cells, designed for regulated expression of the protein, were stably transfected with the TAP-tagged construct. Nuclear extracts were prepared and protein was purified using the purification scheme outlined in Fig. 1B. The final purification step, Fig. 1A, lane 4, indicated that few polypeptides cofractionated with the TAP-tagged tSNAP50. Each major band was eluted and analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry and quadrupole time of flight analyses. The ∼50-kDa protein band was confirmed to be tSNAP50. Two other prominent polypeptides were identified. The 42-kDa protein (tSNAP42) was orthologous to the Leptomonas tSNAP subunit, p46. The smallest protein, which migrated at ∼26 kDa, was anticipated to be related to the p36 subunit of Leptomonas tSNAP, but the corresponding gene had never been cloned. Edman sequencing data derived from the Leptomonas protein (our unpublished data) suggested that T. brucei p26 is orthologous to Leptomonas p36. This smallest subunit of tSNAPc is now named tSNAP26. The copurification of TAP-tagged tSNAP50 with tSNAP42 and tSNAP26 confirms that we have isolated the T. brucei ortholog of the Leptomonas tSNAPc transcription factor.
FIG. 1.
Composition of tSNAPc and its associated TBP. (A) Protein fractions from IgG chromatography (lane 1), calmodulin flowthrough (lane 2), calmodulin wash (lane 3), EGTA elution (lane 4), and markers (BenchMark; Invitrogen) (lane 5) are shown on an SDS-10% polyacrylamide gel. The three subunits of tSNAPc (tSNAPP50, tSNAP42, and tSNAP26) and TBP, as well as p63 and TFIIA-γ, are identified by arrows. (B) Purification scheme of TAP-tSNAPc and TBP. (C) Silver-stained SDS-10% polyacrylamide gel of 5 to 20% sucrose gradient fractions with peak transcription factor activity.
A cofractionating band that migrated at ∼30 kDa was identified as TBP (Fig. 1A). Subsequent sucrose gradient sedimentation eliminated the possibility that TBP coeluted as a minor contaminant in the purification. Figure 1C shows adjacent fractions collected from the sedimentation-based fractionation. TBP remained associated with tSNAPc through this purification step. Two minor bands copurified with tSNAPc and TBP. Mass spectrometric and database analyses suggested that the smaller polypeptide, which migrates at ∼27 kDa, is the trypanosome ortholog of eukaryotic TFIIA-γ. The larger polypeptide is a 63-kDa hypothetical protein (Fig. 1A and C). The function of these two polypeptides in SL RNA transcription is currently under study.
Trypanosome SNAP26 is a truncated metazoan SNAP43.
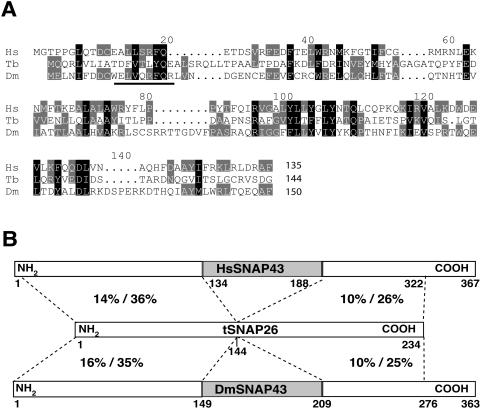
We utilized a combination of mass spectrometry, genome database analysis, and gene cloning to analyze the smallest polypeptide in the purified tSNAPc. A comparison of the L. seymouri, T. brucei, Leishmania major, and Trypanosoma cruzi orthologs showed that the smallest subunits of T. brucei and T. cruzi are both ∼10 kDa smaller than both the Leishmania and Leptomonas proteins. Using T. brucei SNAP26 as our reference sequence, we compared this trypanosome SNAPc subunit with all five human SNAPc subunits and the three reported Drosophila SNAPc subunits (15, 19). T. brucei SNAP26 and its T. cruzi and L. major orthologs appear to be internally deleted versions of the metazoan SNAP43 protein (Fig. 2). SNAP43 is an essential subunit of human SNAPc and is highly conserved between humans and flies. The amino acid sequence comparisons demonstrate significant conservation between the termini of T. brucei SNAP26 and the metazoan SNAP43 proteins (Fig. 2). The central region of the metazoan proteins, indicated by a gray box in the human and Drosophila proteins in Fig. 2, is absent from the trypanosome protein and might serve a metazoan-specific role in snRNA gene transcription. The trypanosome SNAP26 protein may represent a more primitive version of this important SNAPc subunit.
FIG. 2.
Trypanosome SNAP26 resembles an internally deleted metazoan SNAP43. (A) The sequence alignment shows the similarity of the amino-terminal half of T. brucei (Tb) SNAP26 to human (Homo sapiens [Hs]) (NP_003073) and Drosophila (Dm) (NP_650499) SNAP43. The alignment was obtained using the CLUSTALW program and refined manually. Dark shading indicates identical or conserved residues in all three proteins. Medium shading highlights similarity between two proteins. The underlined region shows the T. brucei SNAP26 sequence that corresponds to Edman-derived data from the purified L. seymouri p36 subunit of tSNAPc (our unpublished data). (B) Schematic of proteins. Dashed lines indicate regions of similarity among the proteins. Percent identities and similarities are shown. The central region of the metazoan proteins (indicated by a gray box) is absent from the trypanosome protein.
tSNAP42 is a novel protein and contains potential DNA binding domains.
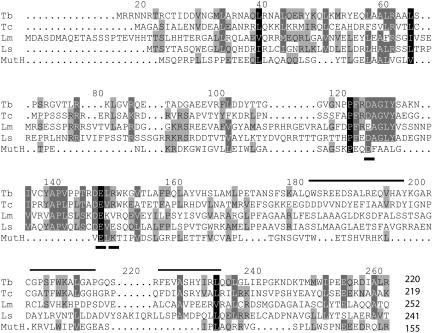
T. brucei and T. cruzi SNAP42 proteins align over their full length, as do the L. major and L. seymouri proteins (Fig. 3). Using ProFit analysis, which performs sequence-to-structure searches, the majority of the top hits for all four trypanosome proteins were protein domains that bind phosphate, usually in the context of a nucleotide or a nucleic acid polymer. These findings suggest that tSNAP42 may bind to the phosphate backbone of DNA in a sequence-independent manner.
FIG. 3.
Multiple sequence alignment of T. brucei (Tb) SNAP42 with its orthologs from T. cruzi (Tc), L. major (Lm), and L. seymouri (Ls) using CLUSTALW. Dark shading indicates identical or conserved amino acids in all five proteins. Medium shading with white letter overlays shows amino acid similarity among four proteins. Medium shading alone indicates amino acid similarity between three sequences. The signature triplet helices, present in a single Myb domain, are overlined. The Escherichia coli MutH (P06722) alignment shown was obtained by ProFit analysis. The three active site amino acids (D, E, and K) in MutH that aligned in structure and sequence to the trypanosome proteins are underlined.
To gain more insight into the function of tSNAP42, we further inspected the sequence-to-structure alignments between T. brucei SNAP42 and T. cruzi SNAP42 against their top three hits from ProFit. We focused on the highly conserved amino acid residues and the known active sites of the reported structures. Bacterial double-stranded DNA binding protein MutH showed the most striking similarity in both its overall fold and active-site regions (2). Interestingly, a local BLAST search revealed homology among all four trypanosome SNAP42 proteins in the regions structurally aligned with MutH active sites (Fig. 3, underlined amino acids).
Protein motif searches revealed a single Myb-like domain in T. brucei SNAP42, although this domain was less apparent in T. cruzi, L. major, and L. seymouri orthologs. Further inspection of all four tSNAP42 proteins uncovered conserved, closely spaced, triplet alpha helices reminiscent of Myb and Myb-related SANT domains, which often signal protein-DNA interactions (Fig. 3, overlined amino acids). In summary, both motif searches and ProFit analyses suggest that tSNAP42 is a sequence-independent DNA binding component of tSNAPc. This idea is consistent with our initial modeling of the Leptomonas SNAP42 (7). The specificity of tSNAPc recruitment to the SL RNA gene promoter is likely conferred by either the tSNAP26 or tSNAP50 subunits of the protein complex.
TBP is tightly associated with tSNAPc.
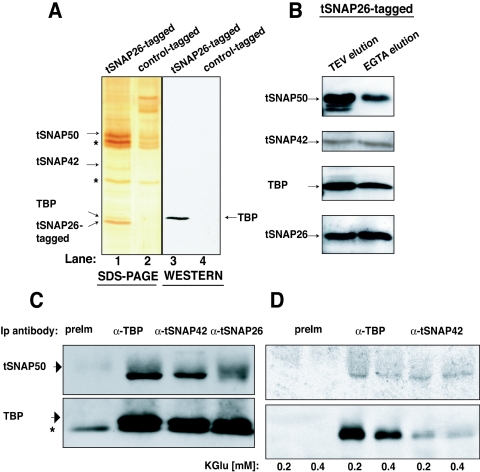
Copurification of TBP with tSNAPc from the TAP-tagged tSNAP50 strain indicated an association among the proteins. To verify this interaction, we designed two sets of experiments. First, we produced TAP-tagged tSNAP26 transgenic parasites. The cofractionation of tSNAP50, tSNAP42, and TBP with TAP-tagged tSNAP26 was confirmed by SDS-polyacrylamide gel electrophoresis (Fig. 4A, lane 1). The presence of TBP was further confirmed by Western blot analysis (Fig. 4A, lane 3). Polyclonal antibodies were also used to verify that these proteins cofractionated throughout affinity purification (Fig. 4B). A parallel purification of a control TAP-tagged RNA processing protein (J. Milone and V. Bellofatto, unpublished) in the same genetic background ruled out the possibility that TBP was copurifying as a nonspecific contaminant (Fig. 4A, lanes 2 and 4). The major bands that copurified with both TAP-tagged proteins are marked by asterisks.
FIG. 4.
TBP is associated with tSNAPc. (A) A silver-stained SDS-10% polyacrylamide gel of affinity-purified proteins from a TAP-tagged tSNAP26 (lane 1) or a control TAP-tagged (lane 2) strain. Nonspecific proteins observed in both lanes are marked by asterisks. Lanes 3 and 4 show a Western blot of the corresponding proteins, probed with anti-TBP antibody. (B) Western analysis of proteins from TAP-tSNAP26 extracts after sequential IgG (lane 1) and calmodulin affinity (lane 2) purification, probed with the indicated antibodies. (C) Western analysis of tSNAP50 and TBP after immunoprecipitation (Ip) from wild-type T. brucei extracts by antisera (α-) as indicated above each lane. The asterisk indicates antibody light chain. (D) Western analysis of tSNAP50 and TBP after immunoprecipitation from wild-type T. brucei extracts with the antisera at different salt concentrations. Total amounts of precipitated protein are less at 0.4 M salt than at 0.2 M, probably reflecting reduced antibody-antigen affinity. PreIm, preimmune.
In a second set of experiments, we used wild-type T. brucei for coimmunoprecipitation assays to rule out the possibility of a concentration-dependent association of overexpressed TAP-tagged tSNAPc subunits with TBP. Anti-TBP, anti-tSNAP42, and anti-tSNAP26 antibodies were incubated with nuclear extracts. Antibodies against tSNAP42 or tSNAP26 coimmunoprecipitated tSNAP50, indicating the expected strong association among the tSNAPc subunits (Fig. 4C). Importantly, both anti-tSNAP42 and anti-tSNAP26 also precipitated TBP. In a reciprocal immunoprecipitation, anti-TBP antibody efficiently coprecipitated tSNAPc, as judged by the presence of tSNAP50. To investigate the strength of the TBP-tSNAPc association, we challenged the stability of the interaction at a higher salt concentration (Fig. 4D). Anti-TBP antibody still coprecipitated tSNAPc at 0.4 M salt. Anti-tSNAP42 antibodies also detected the tight TBP interaction with tSNAPc. Taken together, these data suggest that a subpopulation of TBP interacts stably with tSNAPc.
tSNAPc specifically interacts with the SL RNA gene promoter.
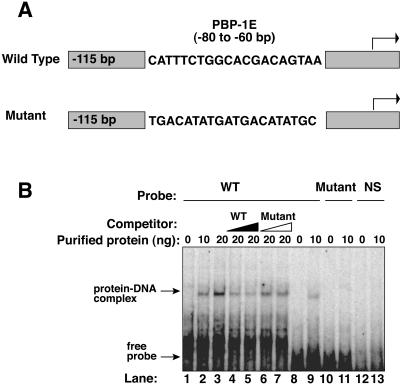
To test if tSNAPc binds to the SL RNA gene promoter, we performed electrophoretic mobility shift assays. tSNAPc formed a specific complex with a 117-bp DNA (Fig. 5A), as detected by a slowly migrating protein-DNA complex in a neutral polyacrylamide gel (Fig. 5B, lanes 2, 3, and 9). This protein-DNA complex was destabilized by adding increasing amounts of unlabeled wild-type DNA (lanes 4 and 5) but was resistant to similar amounts of a DNA mutated in PBP-1E (lanes 6 and 7). Neither the mutant SL RNA gene promoter probe nor a completely unrelated DNA sequence bound tSNAPc (lanes 11 and 13).
FIG. 5.
tSNAPc binds specifically to PBP-1E in the SL RNA gene promoter. (A) Wild-type and mutant SL RNA gene promoter sequences. (B) Formation of a protein-DNA complex with purified tSNAPc. Lanes 1 to 7 contain the wild-type (WT) DNA probe shown in A. Protein amounts used are indicated above each lane. Ten-fold and 20-fold molar excesses of wild-type (lanes 4 and 5) and mutant DNA used as competitors (lanes 6 and 7) are indicated. A slightly shorter wild-type SL RNA promoter probe, corresponding to bp −97 to −17, was used in lanes 8 and 9. A mutant SL RNA promoter of similar size (lanes 10 and 11) and a nonspecific probe (lanes 12 and 13) did not form protein-DNA complexes.
tSNAPc promotes SL RNA transcription in vitro.
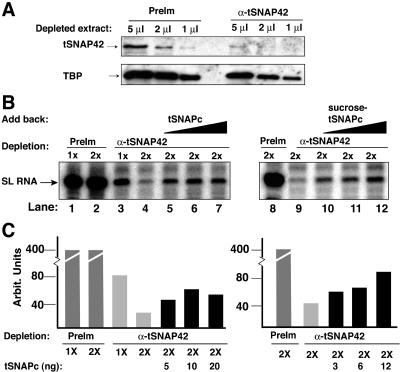
To test the function of tSNAPc in SL RNA gene transcription, nuclear extracts were incubated with either preimmune or anti-tSNAP42 IgG beads. Anti-tSNAP42 antibodies efficiently depleted tSNAPc, as indicated by immunoblot analysis of tSNAP42 (Fig. 6A). The immune sera also removed a small portion of TBP from the extract. This was expected, as we predict that only a small percentage of TBP is associated with tSNAPc. Immunodepletion of tSNAPc abrogated SL RNA transcription (Fig. 6B, lanes 3, 4, and 9). Add-back experiments, using affinity-purified tSNAPc (Fig. 1A), restored SL RNA transcription (lanes 5 to 7), as did even more highly purified (Fig. 1C) protein (lanes 10 to 12). These results, which are quantified in Fig. 6C, demonstrated that tSNAPc is required for SL RNA gene transcription.
FIG. 6.
tSNAPc is required for SL RNA gene transcription. (A) Western blot analysis of preimmune-depleted (PreIm) and anti-tSNAP42-depleted (α-tSNAP42) nuclear extracts. Only a portion of TBP was depleted from the extracts. (B) Transcription activity was lost in the tSNAPc-depleted extract (compare lanes 3, 4, and 9 with 1, 2, and 8). Depleted extracts were reactivated when supplemented with 5, 10, and 20 ng of affinity-purified tSNAPc (lanes 5 to 7) or 3, 6, and 12 ng of sucrose velocity-purified tSNAPc (lanes 10 to 12). (C) Quantification, in arbitrary units, of transcription activities shown in panel B.
TBP is essential for SL RNA transcription.
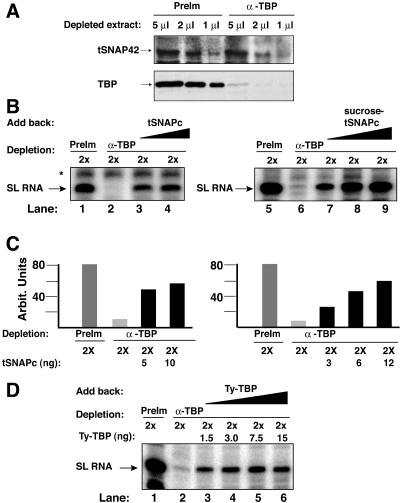
The association of TBP with tSNAPc led us to investigate the function of TBP in SL RNA transcription. We incubated transcription-competent extracts with either preimmune or anti-TBP beads. The immune sera efficiently depleted TBP (Fig. 7A). Neither mock-depleted nor TBP-depleted extracts were deficient in tSNAPc, as indicated by Western blot analysis of tSNAP42. The TBP-depleted extracts lost transcriptional activity (Fig. 7B, lanes 2 and 6), whereas mock-depleted extracts maintained transcriptional competency (lanes 1 and 5). Addition of small amounts of either affinity-purified or sedimentation velocity-purified tSNAPc, containing TBP, reestablished transcription in a concentration-dependent manner (lanes 3 and 4 and 7 to 9). Quantification confirmed that tSNAPc restoration resulted in robust gene transcription (Fig. 7C).
FIG. 7.
TBP is essential for SL RNA gene transcription. (A) Western blot analysis of preimmune-depleted (PreIm) and anti-TBP-depleted (α-TBP) nuclear extracts. A small amount of tSNAPc was depleted by anti-TBP; TBP was efficiently removed. (B) Anti-TBP-depleted extracts lost their transcription activity (compare lanes 2 and 6 with 1 and 5). Addition of either TAP-purified tSNAPc, containing TBP (lanes 3 and 4), or sucrose velocity-sedimented tSNAPc, containing TBP (lanes 7 to 9), complemented SL RNA transcription. (C) Quantification, in arbitrary units, of transcription activities shown in panel B. (D) Addition of a purified Ty-tagged TBP (amounts shown above each lane) restored SL RNA gene transcription (lanes 3 to 6).
To address directly the TBP requirement in SL RNA transcription, we immunopurified TBP from a trypanosome line expressing Ty-tagged TBP. The isolated TBP did not contain detectable levels of any tSNAPc subunits, as judged by Western analysis (data not shown), but it efficiently reestablished SL RNA gene expression in TBP-depleted extracts (Fig. 7D, lanes 3 to 6). This observation and the inability of a tSNAPc-depleted extract to drive SL RNA transcription, even though it retains significant amounts of TBP, leads to the conclusion that SL RNA synthesis is dependent on a functional association of tSNAPc and TBP.
DISCUSSION
This biochemical study establishes the molecular structure and demonstrates the essential role of the unique trypanosomal transcription factor tSNAPc in SL RNA gene transcription. Production of SL RNA is essential for gene expression in these evolutionarily distant and pathogenic protozoa. In this study the evidence shows that the trypanosomal SNAPc is comprised of at least three subunits. One of these subunits, tSNAP50, has been previously reported, and two subunits, tSNAP42 and tSNAP26, have been newly identified. Furthermore, we have also discovered that tSNAPc interacts tightly with trypanosomal TBP.
Our characterization of tSNAPc demonstrates that this factor is related to human and Drosophila SNAPc. This relationship between tSNAPc and metazoan SNAPc is based on the sequence homology among the various subunits. Specifically, our findings indicate that the SNAP50 polypeptides in human and trypanosome SNAPc are orthologous. In addition, the tSNAP26 subunit of tSNAPc is homologous to human and Drosophila SNAP43, with the highest similarity in the amino termini of the three orthologs (19, 31). Finally, tSNAP42 and metazoan SNAP190 share an unusual motif. Metazoan SNAP190 proteins have 4.5 clustered Myb repeats, whereas tSNAP42 has a single set of triple helices that constitute a single Myb domain (21). These observations demonstrate that even a primitive eukaryote such as T. brucei utilizes a related set of polypeptides to achieve transcription of RNA polymerase II-dependent essential small nuclear RNAs.
A comparison of the overall composition of tSNAPc with metazoan SNAPc implies that the metazoan protein is an embellished version of a simpler factor. In human cells, SNAPc contains five polypeptides. The largest subunit is SNAP190. This protein interacts in a regulatory fashion with the transcriptional activator Oct-1 through its carboxyl-terminal region (9, 24). Specifically, in the absence of the Oct-1 protein, the carboxyl end of SNAP190 seems to block SNAPc binding to its promoter. Interestingly, neither the Drosophila SNAP190 protein nor any region of the three tSNAPc subunits contains a sequence similar to the Oct-1 binding region of mammalian SNAP190 (19). It logically follows that neither the trypanosome SL RNA gene promoter nor the Drosophila U1 snRNA gene promoter contains an Oct-1 binding site (called the distal sequence element in the mammalian U1 snRNA promoter) (15). Indeed, neither of these two promoters seems to contain an upstream enhancer-type distal sequence element that would recruit a transcriptional activator. It is possible that in trypanosomes sufficient output of SL RNA is achieved in the absence of classic transcriptional activators and relies on a cooperative transcriptional effect imparted by the highly reiterated organization of the SL RNA genes. In all trypanosomes, SL RNA genes are arranged in the genome as closely spaced genes repeated, often without interruption, about a hundred times.
The amino-terminal regions of metazoan SNAP43 and tSNAP26 represent the most conserved regions of all three proteins (fly to human regions are 26% identical and 42% similar; T. brucei comparisons are shown in Fig. 2) (19). Coimmunoprecipitation experiments that tested interactions among the various human SNAPc polypeptides demonstrated that the amino-terminal region of SNAP43 is sufficient for its interaction with SNAP50 (31). The SNAP50-SNAP43 association is likely essential for SNAPc function as it occurs between two essential SNAPc subunits. Our observation that trypanosomes contain orthologs to SNAP50 and SNAP43 argues that the functionally important human SNAP50-SNAP43 interaction is retained down the evolutionary tree.
Furthermore, a comparison of tSNAP26 with human and Drosophila SNAP43 highlights our contention that tSNAPc is related to, but distinct from, metazoan SNAPc. The trypanosome protein is far smaller than the metazoan one because it is missing the carboxyl-terminal extensions of the metazoan protein as well as an internal stretch of ∼50 amino acids. This internal region is the precise portion of human SNAP43, and presumably Drosophila SNAP43, that interacts with SNAP190 (31). Therefore, in the absence of a SNAP190 ortholog in trypanosomes, this specific internal region of tSNAP26 need not exist. Based on these findings, it appears that tSNAPc is a streamlined version of the metazoan factor.
The abridged version of trypanosomal SNAPc requires us to ask the question why is there a structural difference in SNAPc composition between trypanosomes and metazoans? A simple explanation is that SNAPc can have distinct roles in each eukaryote. The function of tSNAPc in trypanosome snRNA transcription may be different from the role SNAPc plays in metazoan snRNA expression. Specifically, in human cells, identical SNAPc factors are involved in both RNA polymerase II- and RNA polymerase III-dependent snRNA gene transcription (15). This also seems to be the case in the Drosophila and sea urchin systems, although these studies are less complete (19, 20). In these organisms, a common proximal sequence element recruits SNAPc, which then likely interacts with additional factors, probably stabilized through additional protein-DNA and protein-protein contacts, to determine polymerase specificity. Trypanosome snRNA genes are clearly divided into two classes: the SL RNA genes are a single class, requiring RNA polymerase II and containing a tSNAPc binding element and two additional elements that likely recruit additional proteins. Other snRNA genes, including U2 and U6, contain adjacent tRNA or tRNA-like genes and are expressed using RNA polymerase III (25). Therefore, we argue that tSNAPc may be essential only for SL RNA expression and may play a minor role, if any, in the expression of trypanosome RNA polymerase III-dependent snRNA genes. In support of this suggestion, recent in vitro DNA binding studies, using T. brucei U2 and U6 snRNA genes, did not find any tSNAP50-DNA associations (29). However, the function of tSNAPc in U2 and U6 snRNA transcription may not be that simple, as chromatin immunoprecipitation studies performed in Leptomonas and using antibodies against the tSNAP50 subunit of Leptomonas tSNAPc suggest that this factor is positioned near these two snRNA genes (11).
tSNAP42 has a single Myb domain, which suggests that this subunit may function in DNA binding. Indeed, earlier studies of Leptomonas tSNAPc showed that this protein could be cross-linked to the SL RNA gene promoter (22). However, a single Myb domain is insufficient for stable DNA binding; other protein structures are necessary. These may be found in other regions of tSNAP42, as computer modeling clearly suggests that both the T. brucei and T. cruzi polypeptides have characteristics of several nucleic acid binding proteins. This finding is consistent with our earlier molecular threading analysis of L. seymouri SNAP42 (7). The high overall structural correlation, as well as the alignment between the three key active site residues in bacterial MutH and tSNAP42 suggests that this unique trypanosome polypeptide may mimic MutH in its ability to interact with DNA in the context of other proteins. This situation is reminiscent of the way MutH is recruited to DNA in the context of MutL and MutS (2, 30). Possibly tSNAP42 needs its other tSNAPc partners, tSNAP50 and tSNAP26, to be recruited to the SL RNA gene promoter.
It was interesting to discover that trypanosome TBP is associated with tSNAPc. In humans, TBP exists alone or as part of a variety of complexes, including SL1, TFIID, and TFIIIB (26). Studies using human cell extracts and recombinant proteins demonstrated that TBP is essential for both RNA polymerase II- and RNA polymerase III-dependent snRNA transcription. In addition, TBP associated with SNAPc to form a salt-stable complex and TBP was immunoprecipitated with antibodies that recognized SNAP43 (15). Recently chromatin immunoprecipitation studies demonstrated the presence of trypanosome TBP at multiple genetic loci, including the SL RNA gene-proximal region (28). Our findings suggest a precise role for TBP in SL RNA gene expression. We envision that TBP associates with tSNAPc in the absence of DNA to form a complex that is then recruited, via the sequence-specific binding capacity of tSNAPc, 60 to 80 bp upstream from the SL RNA transcription start site. This “complex preformation followed by binding” argument is based on several observations. First, transcription extracts lose transcriptional activity when depleted of TBP and regain their activity when either free TBP or TBP complexed with tSNAPc is used in reconstitution experiments. Second, tSNAPc associates with TBP in coimmunprecipitation studies. Third, TBP cofractionates with tSNAPc during tSNAPc purifications. Collectively, these points illustrate that tSNAPc and TBP are assembled in a DNA-independent complex prior to preinitiation complex formation. It is likely that this complex is further stabilized in part by direct TBP-DNA contacts.
Most TBP proteins contain two phenylalanine pairs that make close contact with TATA elements in DNA. The trypanosome TBP lacks the first of these phenylalanine pairs and is thus expected to inefficiently bind TATA sequences. This is not surprising as no known gene promoters in trypanosomes contain TATA elements. However, the second phenylalanine pair (positions 288 and 305 in human TBP; positions 206 and 229 in T. brucei TBP) is present in all trypanosome TBPs. We predict that this amino acid pair is important in SL RNA gene promoter binding, albeit in an unknown manner. This view is supported by experiments using human TBP in which two phenylalanine residues (F228 and F305) were mutated. These alterations significantly reduced RNA polymerase II-dependent U1 snRNA transcription in HeLa extracts (36). In summary, we conclude that there is a functional conservation of TBP in its role in RNA polymerase II-dependent snRNA gene expression in trypanosomes.
Unfortunately, we do not yet know which subunit of tSNAPc interacts with TBP, nor do we know what other factors may be involved in preinitiation complex formation at the SL RNA gene promoter. Toward solving this problem, we are identifying additional proteins that associate either with the SL RNA gene promoter or directly with tSNAPc. Two other tSNAPc-associated proteins copurified with tSNAPc. One of these proteins is the trypanosome ortholog of TFIIA-γ, a known basal transcription factor that functions in human snRNA gene expression. The other, p63, is a novel protein that does not resemble any metazoan SNAPc subunits (unpublished observations). In addition, we have previously characterized a Leptomonas protein, PBP-2, as a DNA-binding protein that associates with the tSNAPc/DNA complex (22). These proteins may all play specific roles in directing SL RNA gene expression in trypanosomes.
Acknowledgments
We thank all Bellofatto lab members for comments on the manuscript; Chris Utter for help with antibody production; Joseph Milone for the control cell line; Martin Marinus for helpful discussions; Hong Li for mass spectrometry analysis; Douglass Lamont, David Martin, and Mike Ferguson for making available the TrypPEP database; Sara Melville for providing chromosome X shotgun sequencing clones; and the Tri-Trypanosome Genome Projects for their accessible data.
This work was supported by NIH grants AI29478 to V.B. and AI21729 to G.A.M.C. and American Heart Association Postdoctoral Fellowship 0425791T to J.B.P.
REFERENCES
- 1.Agami, R., R. Aly, S. Halman, and M. Shapira. 1994. Functional analysis of cis-acting DNA elements required for expression of the SL RNA gene in the parasitic protozoan Leishmania amazonensis. Nucleic Acids Res. 22:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban, C., and W. Yang. 1998. Structural basis for MutH activation in E. coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 17:1526-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. [PubMed] [Google Scholar]
- 4.Bastin, P., Z. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235-239. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, D. A., S. Thomas, and N. R. Sturm. 2003. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 5:1231-1240. [DOI] [PubMed] [Google Scholar]
- 6.Das, A., and V. Bellofatto. 2004. Genetic regulation of protein synthesis in trypanosomes. Curr. Mol. Med. 4:577-584. [DOI] [PubMed] [Google Scholar]
- 7.Das, A., and V. Bellofatto. 2003. RNA polymerase II-dependent transcription in trypanosomes is associated with a SNAP complex-like transcription factor. Proc. Natl. Acad. Sci. USA 100:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evers, R., A. Hammer, J. Kock, W. Jess, P. Borst, S. Memet, and A. W. Cornelissen. 1989. Trypanosoma brucei contains two RNA polymerase II largest subunit genes with an altered C-terminal domain. Cell 56:585-597. (Erratum, 58:following 419.) [DOI] [PubMed] [Google Scholar]
- 9.Ford, E., M. Strubin, and N. Hernandez. 1998. The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes Dev. 12:3528-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilinger, G., H. Luo, and V. Bellofatto. 2004. In vivo transcription analysis utilizing chromatin immunoprecipitation reveals a role for trypanosome transcription factor PBP-1 in RNA polymerase III-dependent transcription. Mol. Biochem. Parasitol. 134:169-173. [DOI] [PubMed] [Google Scholar]
- 12.Gunzl, A., T. Bruderer, G. Laufer, B. Schimanski, L. C. Tu, H. M. Chung, P. T. Lee, and M. G. Lee. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2:542-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunzl, A., E. Ullu, M. Dorner, S. P. Fragoso, K. F. Hoffmann, J. D. Milner, Y. Morita, E. K. Nguu, S. Vanacova, S. Wunsch, A. O. Dare, H. Kwon, and C. Tschudi. 1997. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67-76. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane (ed.). 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 17.Huie, J., P. He, and V. Bellofatto. 1997. In vitro transcription of the Leptomonas seymouri SL RNA gene using homologous cell extracts. Mol. Biochem. Parasitol. 90:183-192. [DOI] [PubMed] [Google Scholar]
- 18.LeBowitz, J. H., H. Smith, L. Rushce, and S. M. Beverley. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7:996-1007. [DOI] [PubMed] [Google Scholar]
- 19.Li, C., G. A. Harding, J. Parise, K. J. McNamara-Schroeder, and W. E. Stumph. 2004. Architectural arrangement of cloned proximal sequence element-binding protein subunits on Drosophila U1 and U6 snRNA gene promoters. Mol. Cell. Biol. 24:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, J.-M., R. P. Haberman, and W. F. Marzluff. 1996. Common factors direct transcription through the proximal sequence elements (PSEs) of the embryonic sea urchin U1, U2, and U6 genes despite minimal sequence similarity among the PSEs. Mol. Cell. Biol. 16:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipsick, J. S. 1996. One billion years of Myb. Oncogene 13:223-235. [PubMed] [Google Scholar]
- 22.Luo, H., and V. Bellofatto. 1997. Characterization of two protein activities that interact at the promoter of the trypanosomatid spliced leader RNA. J. Biol. Chem. 272:33344-33352. [DOI] [PubMed] [Google Scholar]
- 23.Luo, H., G. Gilinger, D. Mukherjee, and V. Bellofatto. 1999. Transcription initiation at the TATA-less spliced leader RNA gene promoter requires at least two DNA-binding proteins and a tripartite architecture that includes an initiator element. J. Biol. Chem. 274:31947-31954. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, S., J.-B. Yoon, T. Gerster, and R. G. Roeder. 1992. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 12:3247-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakaar, V., A. O. Dare, D. Hong, E. Ullu, and C. Tschudi. 1994. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 14:6736-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 27.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 28.Ruan, J.-P., G. K. Arhin, E. Ullu, and C. Tschudi. 2004. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol. Cell. Biol. 24:9610-9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schimanski, B., L. Ma, and A. Gunzl. 2004. Failure to detect binding of Trypanosoma brucei SNAPc to U2 and U6 snRNA gene sequences by in vitro transcription competition and pull-down assays. Mol. Biochem. Parasitol. 137:293-296. [DOI] [PubMed] [Google Scholar]
- 30.Schofield, M. J., and P. Hsieh. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579-608. [DOI] [PubMed] [Google Scholar]
- 31.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593-2620. [DOI] [PubMed] [Google Scholar]
- 32.Smith, J. L., J. R. Levin, C. J. Ingles, and N. Agabian. 1989. In trypanosomes the homolog of the largest subunit of RNA polymerase II is encoded by two genes and has a highly unusual C-terminal domain structure. Cell 56:815-827. [DOI] [PubMed] [Google Scholar]
- 33.Ullu, E., K. R. Matthews, and C. Tschudi. 1993. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol. 13:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 35.Yoon, J.-B., and R. G. Roeder. 1996. Cloning of two proximal sequence element-binding transcription factor subunits (γ and δ) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol. Cell. Biol. 16:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, X., L. Schramm, N. Hernandez, and W. Herr. 2003. A shared surface of TBP directs RNA polymerase II and III transcription via association with different TFIIB family members. Mol. Cell 11:151-161. [DOI] [PubMed] [Google Scholar]