Abstract
Translocations, deletions, and chromosome fusions are frequent events seen in cancers with genome instability. Here we analyzed 358 genome rearrangements generated in Saccharomyces cerevisiae selected by the loss of the nonessential terminal segment of chromosome V. The rearrangements appeared to be generated by both nonhomologous end joining and homologous recombination and targeted all chromosomes. Fifteen percent of the rearrangements occurred independently more than once. High levels of specific classes of rearrangements were isolated from strains with specific mutations: translocations to Ty elements were increased in telomerase-defective mutants, potential dicentric translocations and dicentric isochromosomes were associated with cell cycle checkpoint defects, chromosome fusions were frequent in strains with both telomerase and cell cycle checkpoint defects, and translocations to homolog genes were seen in strains with defects allowing homoeologous recombination. An analysis of human cancer-associated rearrangements revealed parallels to the effects that strain genotypes have on classes of rearrangement in S. cerevisiae.
The development and progression of cancer are correlated with genetic instability. These genomic changes are associated with either a microsatellite instability (MSI) phenotype, involving defects in mismatch repair and a dramatic increase in base substitution and insertion/deletion mutation rates, or a chromosomal instability (CIN) phenotype, involving changes in chromosome number and structure (reviewed in reference 55); however, some tumors have been observed with both MSI and CIN. A causal role for CIN in tumorigenesis is still debated; however, most cancers are associated with dramatic changes to the chromosomal complement (68), and several hereditary cancer predisposition syndromes are closely linked to CIN (49). Using the mutator hypothesis (61), it has been argued that changes in gene dosage, a loss of heterozygosity, a deregulation of gene expression, and the generation of gain-of-function protein chimeras due to CIN are sufficient to drive tumorigenesis in many cases, even without mutations in oncogenes or tumor suppressor genes (29). Thus, understanding CIN is likely to provide some insight into the mechanisms of tumorigenesis.
One of the major barriers to understanding the CIN phenotype, even for model organisms such as the yeast Saccharomyces cerevisiae, is that there are very few genetic systems capable of systematic characterizations of genome rearrangements (49, 105). In principle, rearrangements can arise through both homologous recombination (HR) and nonhomologous end-joining (NHEJ) pathways. In practice, the lack of a detailed understanding of the genetic and biochemical mechanisms that underlie these types of rearrangements in vivo has remained a bottleneck to understanding the generation of genome rearrangements in cancer and other human diseases.
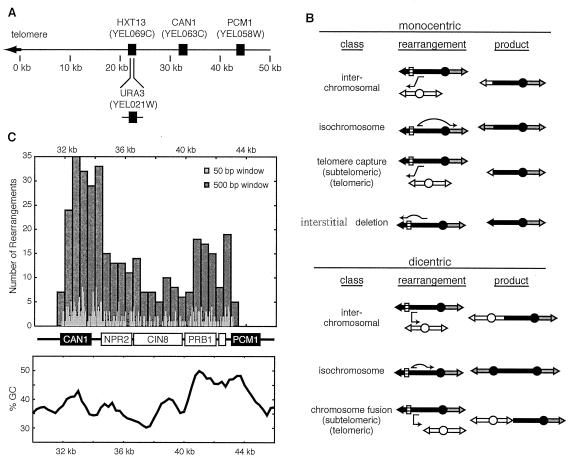
An assay designed to analyze the formation of translocations and other gross chromosomal rearrangements (GCRs) was developed with haploid strains of S. cerevisiae (17). Using the CAN1 and URA3 genes as counterselectable markers for a nonessential portion of the left arm of chromosome V (Fig. 1A), both translocations (Fig. 1B) and other types of GCRs, including broken chromosomes healed by de novo telomere addition, can be selected. For translocations, one of the breakpoints defining such rearrangements must occur in the 12-kilobase region between the most distal essential gene, PCM1 (YEL058), and the telomeric end of the most central marker, CAN1 (YEL063), whereas the other breakpoint can lie anywhere else in the genome. Numerous breakpoint sequences have been determined for GCRs arising in both wild-type and mutant strains (17, 44, 69-74, 79). These data are notable in that the GCR assay minimally influences the diversity of rearrangements formed. Here we use a large number of breakpoints previously isolated in numerous genetic backgrounds, but not described in detail, to explore the mechanistic features of the pathways that give rise to these rearrangements. Remarkably, as described below, many of the correlations observed between the genetic background and the recovered rearrangements match observations for tumors, demonstrating that yeast can provide substantial insight into CIN in cancer.
FIG. 1.
Rearrangements selected by the GCR assay can target multiple chromosomal sites. (A) Positions of the genes important to the GCR assay on the left arm of chromosome V. HXT13 was replaced by URA3. Rearrangements are selected by simultaneous counterselection against CAN1 and URA3. (B) The GCRs recovered included a variety of nonreciprocal translocations that generate mono- and dicentric products. Chromosome fusions result from dicentric translocations into telomeres or subtelomeric repeats of other chromosomes. (C) Distribution of breakpoints on chromosome V for the GCR assay. The breakpoints for all GCR isolates are plotted along the 15-kb region between the most centromeric marker (CAN1) and the most telomeric essential gene (PCM1). The 358 rearrangements are displayed at a 50 (light gray)- and a 500 (dark gray)-nucleotide resolution. The relative GC content of the chromosomal arm, calculated for a sliding 100-bp window, is displayed below the genes.
MATERIALS AND METHODS
Isolation, mapping, and breakpoint sequencing of individual GCR mutants.
All breakpoints analyzed in this paper were determined previously and were summarized but not reported in detail (17, 44, 69-74, 79). Briefly, yeast cultures were grown nonselectively and plated on plates containing canavanine (Can) and 5-fluoroorotic acid (5FOA). Single Canr 5FOAr colonies arising from separate cultures were analyzed to avoid obtaining multiple isolates of individual rearrangement events. The positions of the breakpoints were mapped by overlapping PCRs specific for chromosome V between PCM1 and CAN1. Breakpoints were amplified using nested arbitrarily primed PCR products amplified with one primer within the region of the last successful mapping reaction and then were sequenced.
Breakpoint analysis.
Breakpoints were analyzed using custom software that identified direct matches to the chromosome V sequence (or sequences of human genes involved in specific translocations for human translocations). Statistics for breakpoint features were then derived using the reference Saccharomyces genome sequence and the positions of mapped breakpoints. In all cases, breakpoint positions determined by this method matched those resulting from BLAST searches. For illustrative purposes, subtelomeric repeats, rRNA gene repeats, and transposons were merged into a representative repeat that superimposed all breakpoints from sequence-diverged copies. The categorization of translocations into coding sequences of essential genes cross-referenced the gene definitions and phenotypes downloaded from the Saccharomyces Genome Database (http://www.yeastgenome.org).
RESULTS
Rearrangements recovered in the GCR assay.
Observed rearrangements (see Table S1 in the supplemental material) are predicted to fall into several classes on the basis of sequences at the breakpoint junction (Fig. 1B). The most common are the predicted monocentric translocations (Table 1); however, the classes observed are strongly influenced by genotype (discussed below). Thus, the distribution of rearrangement classes is biased by which strains have been studied.
TABLE 1.
Average identity length at junction breakpoints depends on the rearrangement formed and the strain genotype
| Rearrangement class | Subtype or gene | No. of rearrangements | Avg identity (range) | Theoretical avg (range)a |
|---|---|---|---|---|
| All | 358 | 3.7 (0-15) | 0.46 (0-21) | |
| Monocentric | Interchromosomal | 122 | 3.6 (0-15) | 0.46 (0-21) |
| Isochromosome | 11 | 4.4 (2-10) | 0.46 (0-18) | |
| Subtelomeric capture | 15 | 3.8 (0-13) | 0.44 (0-13) | |
| Telomeric capture | 0 | NAb | NAb | |
| Interstitial deletion | 43 | 3.1 (0-14) | 0.46 (0-15) | |
| Dicentricc | Interchromosomal | 21 | 4.3 (0-11) | 0.46 (0-21) |
| Isochromosome | 19 | 7.1 (1-14) | 0.46 (0-18) | |
| Subtelomeric fusion | 42 | 3.2 (0-14) | 0.43 (0-13) | |
| Telomeric fusion | 24 | NAb | NAb | |
| Repetitive | Ty elements | 46 | 2.5 (0-10) | |
| rRNA gene | 9 | 3.7 (0-8) | ||
| Complexd | 6 | 4.0 (0-9) | ||
| NHEJ mutationse | yku70/hdf1 | 7 | 7.4 (3-14) | |
| lig4 | 24 | 5.8 (0-14) | ||
| HR mutationse | rad52 | 21 | 2.4 (0-6) | |
| rad54f | 12 | 1.8 (0-4) | ||
| rad51 | 36 | 3.4 (0-12) | ||
| rad55 | 15 | 3.3 (0-13) | ||
| rad59 | 37 | 3.3 (0-13) | ||
| rdh54/tid1 | 14 | 3.9 (0-10) | ||
| rad51 rad59 | 7 | 2.7 (0-5) |
Analyzed from a computational generation of all translocations involving breakpoint regions of chromosome V and appropriate regions of the rest of the genome (over 1.3 × 1011 translocations were generated and analyzed for the monocentric interchromosomal translocations alone).
NA, not applicable.
Dicentric products are predicted on the basis of the sequence orientation at the breakpoint.
Rearrangements involving chromosome V that would be predicted to generate direct repeats (n = 4) or circular chromosomes (n = 2).
For strains containing mutations, breakpoints are included whenever the mutation occurs so that, for example, breakpoints in rad52 rad51 and rad52 rad51 rad59 strains would be included in both the rad52 and rad51 entries.
Five rad54 rearrangements were from a rad54 tlc1 strain five other rad54 tlc1 isolates were telomeric fusions and are not considered here), and seven were from a rad54 rad52 strain.
One of the breakpoints for each of the 358 rearrangements studied occurred within the 12-kb region between PCM1 and the telomeric end of CAN1 (Fig. 1C). The distribution of these breakpoints appears to be roughly correlated with the relative GC content of the region. An important deviation from this trend is at the AT-rich intergenic region between the CAN1 and NPR2 genes, which is associated with a large number of breakpoints. A similar distribution is observed for de novo telomere additions (see Fig. S1 in the supplemental material) that target short TG-rich regions during the healing of broken chromosomes (83).
Sequence identities at rearrangement breakpoints.
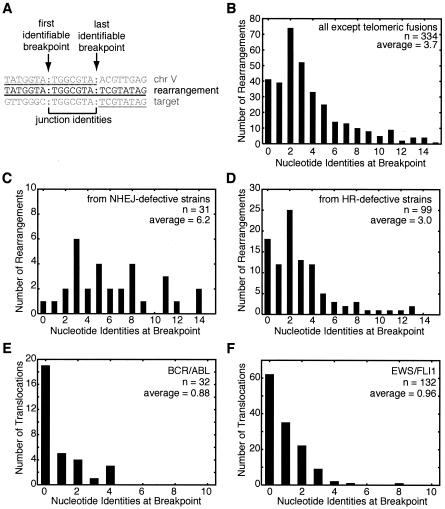
Almost all of the recovered rearrangements appear to be created solely from sequences from chromosome V and the target chromosome (Fig. 2A). These rearrangements frequently occur between short regions of sequence identity on chromosome V and the target DNA. The lengths of these perfect identities at the rearrangement range from 0 to 15 bases, with an average of 3.7 bases for the 334 rearrangements that do not involve the heterogeneous telomeric repeats (Fig. 2B; Table 1). These observed identities are longer than would be predicted by chance (Table 1). Moreover, breakpoints with more than five bases of identity tend to have adjacent regions of homology (see Fig. S2 in the supplemental material).
FIG. 2.
Rearrangement structure and distribution of identities at the junctions. (A) The rearrangement breakpoint positions can be ambiguous if there are regions of identity between chromosome V and the target sequence. The example rearrangement sequence (middle) is aligned with the chromosome V (top) and target (bottom) sequences. Contiguous matches to the rearrangement are underlined. Colons indicate breakpoint positions defined by two systematic methods. The last identifiable chromosome V breakpoint is the last nucleotide that matches the chromosome V sequence from the database; this method forces the nucleotide after the last identifiable breakpoint to never match the chromosome V sequence. The first identifiable target breakpoint is at the first nucleotide which could have come from the target; hence, there is no identity of the target with the nucleotide before the first identifiable breakpoint. (B) Distribution of breakpoints by the number of nucleotide identities between chromosome V and the target chromosome for the 334 rearrangements not involving fusions to the heterogeneous telomeric sequences. (C) Distribution of identity lengths for breakpoints from strains with mutations in genes involved in NHEJ (lig4 or yku70). (D) Distribution of identity lengths for breakpoints from strains with mutations in genes involved in HR (rad51, rad52, rad54, rad55, rad59, or rdh54). (E) Analysis of breakpoint identities at the genomic junctions of BCR/ABL fusions from leukemias suggesting that these translocations are formed by NHEJ. Breakpoints for both products of the reciprocal translocations, the der(9) and der(22) chromosomes, were combined from previous published studies (20, 21, 25, 41, 57, 98, 108, 113). (F) Analysis of EWS fusions to FLI1 (n = 113) and other targets, including ERG (n = 3), E1AF (n = 2), ZSG (n = 1), and CHOP (n = 1), from Ewing's sarcomas and other cancers suggesting that these translocations were formed by NHEJ. Breakpoints from both derivative chromosomes were included when available. Breakpoint sequence data were taken from previous reports (7, 30, 46, 66, 75, 77, 81, 114).
We have previously documented some cases where rearrangements are dependent on NHEJ (eliminated by a lig4 mutation) (69) and one case where rearrangements are dependent on HR (dramatically reduced by rad51 and rad59 mutations) (79). Thus, we analyzed this data set of rearrangements to more comprehensively examine the roles of both NHEJ and HR in rearrangement formations. Remarkably, we discovered that rearrangements recovered from strains with mutations in LIG4 or a gene encoding one of the Ku subunits (yku70/hdf1) showed longer breakpoint junction sequence identities than the average (Table 1). On the other hand, strains with HR defects that eliminated both the Rad51- and Rad59-dependent pathways (e.g., rad52 single mutants or rad51 rad59 double mutants) led to shorter junction identities (Table 1). This effect was not observed when only a single HR pathway was compromised (e.g., rad51 or rad59 single mutants), which is consistent with studies suggesting these genes are parts of distinct recombination pathways (19, 96). Surprisingly, a single mutation in rad54 was associated with rearrangements with identity lengths similar to those in rad52 single mutants or rad51 rad59 double mutants (Table 1), although Rad54 has been implicated primarily in the Rad51-dependent pathway (86). Thus, this result may suggest a key role for Rad54 in HR-mediated rearrangements or in the ability of HR to compete with NHEJ for repairing DNA damage.
Importantly, only the NHEJ-deficient strains had identity lengths that were statistically different from the average (P < 0.005) (Fig. 2C and D), suggesting that NHEJ generated most rearrangements in the total data set. However, the effect of specific HR mutations on identity length (Table 1) suggests that HR does contribute to the formation of some rearrangements, even in NHEJ-proficient strains. Even with NHEJ-deficient strains, though, the observed 6- to 7-bp breakpoint identities are shorter than the minimum homology lengths required for efficient Rad51- and Rad59-dependent recombination (19, 45). These facts suggest that both the Rad51- and Rad59-dependent pathways can promote rearrangements at short regions of sequence identity, albeit at suboptimal rates. Consistent with this, the maximal rates of translocations and deletions seen are very low, even for mutants with GCR rates that are 5 orders of magnitude higher than the wild-type rate (∼4.5 × 10−6/cell generation). Moreover, rearrangements into essential genes are less common in HR-defective strains (see Table S2 in the supplemental material), and detailed analyses of selected rearrangements have shown that translocations are nonreciprocal, as would be predicted for translocations into essential genes in haploid strains (17, 79). Thus, the apparent predominance of NHEJ and the use of low-efficiency HR targets in the formation of GCRs may reflect the fact that highly efficient HR involving longer regions of homology is likely to result in the formation of an intact chromosome V using genetic information from another copy of chromosome V.
Some of the longest-identity translocation breakpoints that could be formed would result from rearrangements targeting paralogs of the genes in the PCM1-CAN1 region. However, only 3 of 88 rearrangements involving CAN1 were with paralogs such as ALP1 and LYP1, whereas none involving either CIN8 or PRB1 were translocations to their paralogs. One CAN1/ALP1 translocation was found in a rad51 tlc1 strain, and individual CAN1/ALP1 and CAN1/LYP1 translocations were found in an sgs1 strain which is defective in suppressing homoeologous recombination (70, 99, 101). These translocations involved long homoeologous stretches; however, the rearrangement breakpoints did not occur at the regions of maximum identity (the average identity length was only 7 bases). These rearrangements are similar to selected fusions between the pma1-105 and PMA2 H+-ATPases (40).
A similar analysis of junction identities in translocations from human cancers also suggests an NHEJ-based mechanism. We selected 32 BCR/ABL translocations and 132 EWS/FLI1 translocations from leukemia and Ewing's tumors, respectively (95), because of the number of such breakpoint sequences available. We did not analyze rearrangements that could not be mimicked by the yeast assay, such as translocations involving V(D)J recombination or rearrangements involving long palindromic sequences (36, 52). The observed junction identities were short, with an average of 0.88 and 0.96 bases, respectively (Fig. 2E and F). These rearrangements mostly involved introns (see Fig. S3 in the supplemental material), and a computational analysis showed that millions of rearrangements with larger identities could be generated that would result in identical protein chimeras (the maximum identities are 26 and 31 bases for BCR/ABL and EWS/FLI, respectively). Thus, homologies do not drive these rearrangements. These breakpoints are similar to smaller numbers of other sequenced breakpoints from humans that have 0 to 5 bases of identity. These cancer-associated rearrangements show less breakpoint homology than breakpoints resulting from the rejoining of linearized simian virus 40 DNA in monkey cells (88) and show shorter homology than rearrangements resulting from NHEJ in yeast (Table 1) (51), possibly due to the (at least partial) dependence of NHEJ in budding yeast, but not in mammals, on end processing by the Mre11 complex (27); indeed, defects in the Mre11 complex lead to reduced homology at translocation breakpoints (17).
Nonrepetitive chromosomal target sequences.
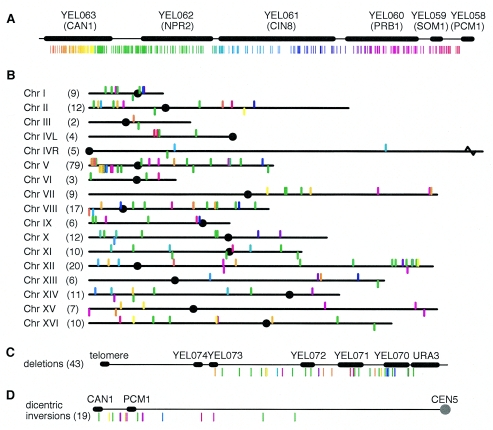
Of the 358 rearrangements (Fig. 3A), 222 involved nonrepetitive target sequences and included translocations to every chromosome (Fig. 3B). Both coding and noncoding regions were targeted (see Table S2 in the supplemental material); however, there were no rearrangements involving centromeres. There was no clear correlation between the position of the breakpoint between CAN1 and PCM1 and the target site, even for rearrangements that only involved chromosome V.
FIG. 3.
Distribution of rearrangements in the nonrepetitive region of the genome. (A) Vertical lines indicate the nucleotide coordinates of rearrangement breakpoints along chromosome V. Lines are colored by hue according to their location. (B) Vertical lines indicate rearrangements in each of the chromosomes. Lines above the chromosome are predicted to generate monocentric products; lines below the chromosome are predicted to be dicentric on the basis of the orientation of sequences at the junction. Line colors are based on their positions on chromosome V as in panel A. Circles are centromeres. (C) Distribution of target sites for interstitial deletions on chromosome V displayed as described above for panel B. (D) Distribution of target sites for dicentric isochromosomes on chromosome V displayed as described above for panel B.
Chromosome V was the most frequent target, with 79 breakpoints. Interstitial deletions, which comprised 12% of the rearrangements, were overrepresented relative to the length of the target region, which is only 0.17% of the genomic DNA (Fig. 3C). The deletions had short average junction identities (3.2 nucleotides) and were more frequent in HR-defective strains (24 of 43) than in NHEJ-defective strains (2 of 43, with 6 and 12 nucleotides of identity at the junctions), consistent with their being generated primarily by NHEJ (Fig. 4). It should be noted that the formation of interstitial deletions by NHEJ requires only one resected break on chromosome V rather than multiple independent events. The midpoints between the two recovered ends for these deletions were uniformly distributed on chromosome V (data not shown). The predicted dicentric isochromosomes (Fig. 3D) had the longest average breakpoint junction identities of any translocation class (7.1 nucleotides) (Table 1). These events comprised 7.8% of the rearrangements, even though the relevant target region only comprised 0.9% of the total chromosomal DNA. Most of these (9 of 19) were identified in strains with mutations causing defects in both telomerase and cell cycle checkpoints, such as the mec1 sml1 lig4 tlc1 and tel1 lig4 tlc1 mutants (Fig. 4). Dicentric isochromosomes in telomerase-deficient strains were previously found to be nonreciprocal, dependent on Rad51, and partially dependent on Rad59 and were suggested to result from a specific HR-dependent mechanism (79). The fact that the isochromosome breakpoint junctions had longer average sequence identities (Table 1) is consistent with these conclusions. In contrast, the 11 predicted monocentric isochromosomes resulting from rearrangements involving the right arm of chromosome V did not occur more frequently than translocations to other chromosomes (Fig. 3B). Additionally, these monocentric isochromosomes did not show a bias for a particular genotypic background, unlike dicentric isochromosomes (Fig. 4). Thus, the frequency of targeting of the left arm of chromosome V in rearrangements is due primarily to specific rearrangement mechanisms based on the positioning of the assay region rather than to some special property of chromosome V.
FIG. 4.
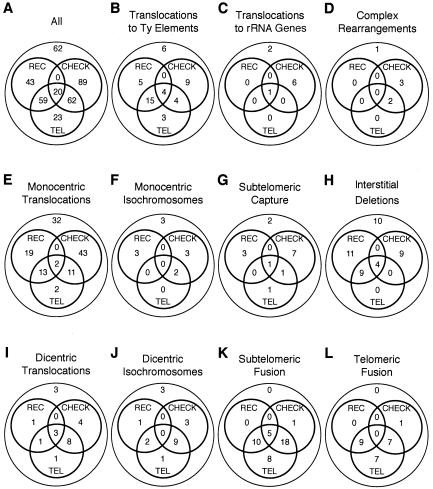
Genotype dependence of recovering specific classes of rearranged chromosomes. (A) The numbers of rearrangements recovered from strains containing mutations in genes responsible for recombination (REC; rad51, rad52, rad54, rad55, rad59, and/or rdh54), telomerase (TEL; tlc1 or est2), or cell cycle checkpoints (CHECK; mec1, rad53, tel1, rad9, dun1, chk1, pds1, and/or mec3) are displayed as Venn diagrams. Numbers at the places where two or three circles overlap represent the numbers of double or triple mutants, respectively. The number in the outer circle represents the number of rearrangements that do not possess any of the mutations listed above. (B to L) Similar breakdown of each rearrangement class, as defined in Table 1. Note that the subtelomeric (K) and telomeric fusions (L) that do not fall into the circle representing telomerase mutations possess a mutation in TEL1, which is known to be involved in telomere maintenance.
The 122 predicted monocentric translocations between chromosome V and other chromosomes were the most common type of translocation (Table 1) and were observed in the greatest diversity of mutant backgrounds (Fig. 4). The average of 3.7 bases of identity at the translocation breakpoints is similar to that for the collection of all rearrangements. In contrast, the 21 predicted dicentric interchromosomal translocations and dicentric isochromosomes were primarily observed in strains containing either checkpoint mutations (mec1, rad53, and tel1) or telomerase mutations (tlc1 and est2; 17 of 21 for dicentric translocations and 15 of 19 for dicentric isochromosomes) (Fig. 4). Random chromosome breakage and rejoining by either NHEJ or HR predict equal numbers of monocentric and dicentric translocations. However, monocentric rearrangements outnumbered potential dicentric rearrangements, suggesting that cell cycle checkpoint-proficient strains do not tolerate dicentric translocations and/or do not permit them to undergo subsequent rearrangements to yield stably inherited products.
Chromosomal rearrangements in human cancers.
We searched the 47,800 karyotypes in the Mitelman database (http://cgap.nci.nih.gov/Chromosomes/Mitelman) to attempt to identify correlations between rearrangement classes and human cancer types similar to those seen with genetic backgrounds in the yeast studies described here. The frequencies of karyotypes with terminal and interstitial deletions showed a rough correlation with those of most other rearrangements (including large-scale chromosomal duplications and insertions [data not shown]), suggesting similar rates of formation in CIN cancers.
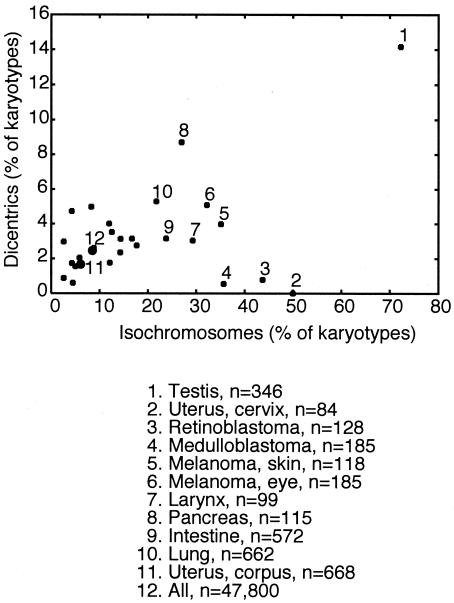
On the other hand, isochromosomes and dicentric translocations were more poorly correlated with deletions and showed a tendency to be associated with specific cancer types. Overall, dicentric chromosomes from human cancers were rare (2.5% [n = 47,800]), with a higher prevalence in testicular cancer (14% [n = 346]), pancreatic cancer (10% [n = 115]), and squamous cell carcinomas from various sources (7.0% [n = 572]) (Fig. 5). The general infrequency of dicentric translocations may reflect their potential instability. We also specifically searched the database for complex translocations that may have arisen from rearrangements of dicentric chromosomes into more stable monocentric products; however, most karyotypic data do not have a sufficient resolution to allow this type of analysis.
FIG. 5.
Frequencies of isochromosomes and dicentric chromosomes in human cancers. The 47,800 karyotypes from the Mitelman database (cgap.nci.nih.gov/Chromosomes/Mitelman) were screened for the propensity to form different types of chromosomal rearrangements. Subsets of the karyotypes grouped by cancer location, such as the lungs, or type, such as medulloblastoma, were also screened for the percentage of karyotypes containing at least one dicentric chromosome or isochromosome. Cancers with high percentages of these rearrangements are labeled. Unlabeled cancer groups lacking a clear bias included Ewing's tumors (n = 333), rhabdomyosarcomas (n = 192), chrondrosarcomas (n = 181), neuroblastomas (n = 232), Wilms's tumors (n = 427), liposarcomas (n = 259), hepatoblastomas (n = 50), adenosarcomas (n = 3,253), leukemias (n = 25,156), lymphomas (n = 4,237), and cancers of the skin (n = 328), breast (n = 961), ovary (n = 598), brain (n = 1,588), prostate (n = 230), and thyroid (n = 304). Karyotypes for these groups were not necessarily kept distinct; for example, the skin melanoma group samples were also included in the skin cancer group.
Like dicentric translocations, isochromosomes also show strong cancer type specificity. Intriguingly, recent analyses showed that many cancer-associated isochromosomes identified by karyotypic analysis are also dicentric (31, 112), and some of them have been shown to be more consistent with break-induced replication and other recombination models than with classic centromere misdivision models (111). Isochromosomes are infrequent in all cancers (8.6% [n = 47,800]) (Fig. 5). However, these aberrant chromosomes are far more frequent in retinoblastomas (44% [n = 128]), testicular cancers (including seminomas and teratomas; 72% [n = 250]), and cervical cancers (50% [n = 84]) (Fig. 5). Although inactivation of the retinoblastoma (Rb) pathway is very common in most cancers (4, 94), each of the three cancers with the highest frequencies of isochromosomes is known to be associated with characteristic isochromosomes (see Fig. S4 in the supplemental material) and with pRB deficiency as an early, if not initial, event. Both sporadic and hereditary retinoblastomas are associated with mutations in the gene encoding pRB (62); the cells believed to give rise to most testicular germ cell tumors normally do not express pRB (5); and many cervical cancers are linked to infection by human papillomavirus, which encodes the E6 and E7 oncoproteins that inactivate p53 and pRB, respectively (28). In contrast, endometrial cancers, which are not caused by human papillomavirus, have no bias for isochromosomes (6.1% [n = 668]) (Fig. 5). These defects are reminiscent of the checkpoint-defective S. cerevisiae backgrounds that frequently form similar chromosomal rearrangements, i.e., predicted dicentric translocations and isochromosomes.
Repeated chromosomal target sequences.
The S. cerevisiae genome contains a number of repeated sequences, including rRNA genes, telomeric and subtelomeric sequences, Ty elements, and 2μm plasmid DNA. Repeated sequences were the target sites of 136 of the 358 rearrangements observed, including rRNA genes, telomeric and subtelomeric sequences, and Ty1 and Ty2 elements (Fig. 6). No translocations to Ty3, Ty4, or Ty5 elements or 2μm plasmid DNA were observed.
FIG. 6.
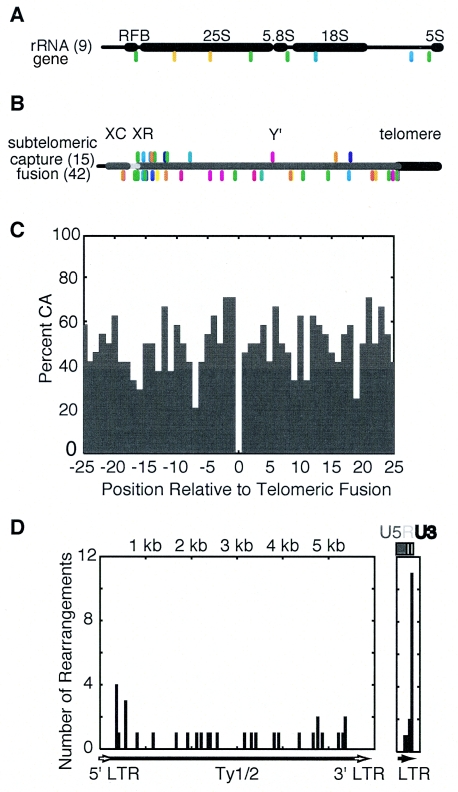
Distribution of rearrangements targeting repetitive elements in the genome. (A) A small number of rearrangements target the rRNA gene repeats and are fairly evenly distributed across the repeats. The 25S, 5.8S, and 18S rRNA genes are spliced from a single transcript, whereas the 5S rRNA gene is transcribed separately. The replication fork-blocking sequence, which is also the position of the HOT1 recombination site, is indicated by RFB. (B) Positions of breakpoints that fall into the subtelomeric repeat sequences. Above the horizontal line are subtelomeric captures, and below the horizontal line are subtelomeric fusions. Since the sequences of the subtelomeric repeats vary, all breakpoints were mapped by sequence alignment with the right arm of chromosome V for graphing purposes. (C) Chromosome fusions to the heterogeneous telomeric sequences are most frequent in telomerase-defective strains and show no sequence bias towards C or A nucleotides before the junction (positions −1 and before) or after the junction (positions +1 and after). (D) Distribution of breakpoints in Ty1 and Ty2 transposons at a 50-bp resolution. Rearrangement positions were mapped onto the YERCTy1-1 transposon. Breakpoints that did not match the sequence of this transposon exactly were mapped onto it by sequence homology. Due to the ambiguity of breakpoints mapping to LTR sequences, which could be either 5′ or 3′ LTRs in transposons or isolated delta sequences, LTR breakpoints are displayed separately. Remarkably, most rearrangements in the LTR occurred within the terminal U3 element (12 into a 45-base region) rather than the R element (2 into a 49-base region) or U5 element (1 into a 243-base region).
Rearrangements targeting the rRNA genes were underrepresented relative to the proportion of rRNA genes in the genome. The tandem rRNA gene repeats make up ∼10% of the genome but only accounted for 2.5% of the rearrangements. This is reminiscent of the suppression of meiotic recombination at the rRNA gene locus within the nucleolus (24), although it may be due to the efficient repair of breaks in the rRNA genes by single-stranded annealing. Rearrangements in the rRNA genes did not involve the HOT1 rRNA gene recombinational hotspot (110), and the recovered junction sequences do not reveal if the rearrangements targeted the rRNA gene repeats on chromosome XII or extrachromosomal rRNA gene circles (97). Remarkably, the majority (six of nine) of rearrangements involving the rRNA genes in this data set were isolated from strains containing a tel1 mutation (Fig. 4; see Table S1 in the supplemental material).
In contrast, rearrangements involving the Ty elements of the related Ty1 and Ty2 families were overrepresented; Ty1/2 elements account for 3% of the genome and 13% of all rearrangement targets, despite the lack of a Ty element in the chromosome V breakpoint region. Most (26 of 46) were from strains with a tlc1 or est2 mutation, and of the remaining 20, 7 were from strains containing a tel1 mutation that also affects telomere maintenance (Fig. 4). Most rearrangements targeted the long terminal repeat sequences (Fig. 6D), which can be isolated or located within a functional Ty element, and typically had short junction sequence identities (Table 1), suggesting that these rearrangements tended to be formed by NHEJ.
Fusions of a broken chromosome V to both subtelomeric (Fig. 6B) and telomeric (Fig. 6C) repeats were found in strains with telomerase or telomerase maintenance defects (64 of 66 had a tlc1 or est2 mutation, and 66 of 66 had tlc1, est2, and/or tel1 mutations) (Fig. 4). These subtelomeric fusions and telomeric fusions to the poly(CA)1-3 strand lacked a bias towards the target sequence (Table 1: Fig. 6C), unlike the strong G/T bias observed in the breakpoints of de novo telomere additions (83). In contrast, 12 of the 15 monocentric subtelomeric capture events (Fig. 6B) were observed in strains with a functional telomerase (Fig. 4). No telomere capture events were observed in 164 translocations from telomerase-defective strains, suggesting that telomeres are poor targets for capture by NHEJ or HR due to their short length or sequestration by protein factors (109). This observation is also consistent with previous observations that the majority of telomere additions are due to telomerase-dependent de novo telomere addition (83).
Identical rearrangement products.
Twenty-five rearrangement products were observed more than once and accounted for 52 of the 358 rearrangements studied (see Table S3 in the supplemental material). These identical rearrangement products involved the same breakpoints on chromosome V and on the non-chromosome-V target. The products were isolated from independent cultures and usually from different strains, although frequently these strains had similar genetic defects. This group of identical rearrangement products accounts for 91% of the multiply observed non-chromosome-V target breakpoints but for only 74% of the multiply observed chromosome V breakpoints. These multiply used breakpoints are clearly nonrandom (Table 2); this frequency could only be attributed to chance if there were only ∼2,200 targets in the genome rather than the millions that would be predicted based on the size of the genome. Remarkably, even breakpoint sites that were within a few nucleotides of each other on chromosome V did not translocate to adjacent locations; this suggests that either the sequence at the end of the chromosome fragments is critical for targeting or that target site selection influences which bases are retained at the junction.
TABLE 2.
Multiply used rearrangement breakpoints are observed more frequently than expected
| Breakpoint usagea | No. of source breakpointsb (avg identity length)
|
No. of target breakpointsb (avg identity length)
|
||||
|---|---|---|---|---|---|---|
| Last IDc | First IDc | Randomd | Last IDc | First IDc | Randomd | |
| 1 | 286 (3.3) | 299 (3.3) | 348 | 281 (3.7) | 282 (3.7) | 334 |
| 2 | 31 (3.7) | 25 (3.4) | 10 | 24 (4.5) | 23 (4.7) | 0 |
| 3 | 2 (2.2) | 3 (6.0) | 0 | 3 (4.9) | 2 (8.5) | 0 |
| 4 | 1 (8.25) | 0 | 0 | 0 | 0 | 0 |
Number of times that any single nucleotide was at a rearrangement.
Number of nucleotide positions that are rearrangements.
Distributions calculated using the first and last identifiable definitions for breakpoints (see Fig. 2A).
Number of identical breakpoints that would be predicted by a Poisson distribution of 358 events in 12 kb (for the chromosome V source) or 334 events in 12 Mb (for the unique genome targets).
The presence of multiple identical rearrangement products could reflect either an increased rate of occurrence or selection for specific rearrangements. However, we were unable to determine any single factor that explained the selection of either breakpoint in the identical rearrangement products or in rearrangements that were only observed once. First, the average junction identity length for the identical rearrangements was only slightly longer than the average and was not unique in the genome. Second, breakpoints did not appear to be biased by potential non-B-DNA structures suggested by theoretical folding with MFOLD (115), nor did the breakpoint distributions correlate with the best free folding energies calculated for overlapping 80-mers along chromosome V. This is in contrast to proposals for human deletions and translocations (23, 114), although we observed potential structures in some source, target, and rearrangement sequences. Third, we observed no correlation with the average replication time of a wild-type strain (85) (see Fig. S5 in the supplemental material) or the positions of replication origins or termination zones, as suggested for rearrangements that change the gene dosage (50) (see Fig. S6 in the supplemental material). Fourth, breakpoints were not correlated with transcriptional activity, based on microarray data (22), or with positions relative to genes (data not shown). Fifth, there was no clear correlation of chromosome V or target breakpoints with sister-chromatid attachment sites, as defined by Mcd1 binding sites (34). Sixth, and finally, we observed no special correlation with evolutionary breakpoints derived from chromosomal alignments of Ashyba gossypii or Kluyveromyces waltii that diverged from S. cerevisiae prior to the duplication of its genome (26, 47, 111; data not shown), unlike proposals for biases of human translocations towards segmental duplications identified by using the mouse genome (3).
Alternatively, the identical rearrangements could indicate a role for selection, such as with translocations observed in wine-producing yeast strains (8, 78, 80). However, only three of the identical rearrangement products, including one chimeric gene fusion, had any potential for a selective advantage over the original strain by disregulating gene expression (see Table S4 in the supplemental material). The identical rearrangement products could also be selected because of the duplication of a portion of the target chromosome by nonreciprocal translocation mechanisms; however, there is no significant difference in the chromosomal regions targeted by the identical versus unique rearrangements.
DISCUSSION
The analysis of genome rearrangement breakpoints described here and the results of previous genetic studies describe a number of features of genome rearrangements in S. cerevisiae. First, genome rearrangements promoted by NHEJ and HR each have a characteristic homology signature, and most rearrangements selected in our studies using a chromosome V-based assay appear to be generated by NHEJ. Second, virtually any chromosomal region can be targeted by genome rearrangements, yet identical rearrangements are surprisingly frequent. Third, the genetic background strongly influences the type of genome rearrangements that can be recovered, both for repetitive and for nonrepetitive element targets. And fourth, the rearrangements observed in yeast have characteristics in common with those observed in human cancers. Together, these data suggest that rearrangements are formed through competition between alternative pathways and are dependent upon the strain genotype. Similarly, an evolving genotype during cancer progression may dramatically affect the spectrum of rearrangements seen in individual cancers.
The isolates studied here must have escaped efficient HR repair, since selection occurs against any events that restore the intact chromosome V structure. However, both HR and NHEJ could, in principle, generate rearrangements, and both have been shown to be important in specific cases (69, 79). Our analysis shows that both NHEJ and HR generate GCRs and that these mechanisms have different breakpoint homology signatures (6.1 versus 2.5 bases of identity at the junctions [on average]). For the 358 rearrangements in this data set, these signatures suggest that NHEJ is the major mechanism for the formation of recovered rearrangements, even in strains in which HR is functional. Some HR-mediated rearrangements do occur, as revealed by the effects of rad52 single mutants and rad51 rad59 double mutants, and these had increased average homology lengths at their breakpoints (Table 1). When both NHEJ and HR are inactivated, such in a rad52 lig4 double mutant strain, no translocations are seen (69).
Even though HR did mediate the formation of some rearrangements, the largest homologies in the rest of the genome were not used (Table 1), and the homologies used by both Rad51- and Rad59-dependent recombination machineries were shorter than their minimal efficient pairing lengths (19, 45). These results suggest that these rearrangements involve competition between NHEJ of other breaks in the genome and low-efficiency HR that is unable to scan much of the genome for targets. Thus, the relative proficiency of NHEJ in most genetic backgrounds and the limited homologies in HR-mediated rearrangements are consistent with the results of recent studies suggesting that chromosomal breaks throughout the nucleus are brought to central repair foci (56). Similarly, the proposed colocalization of chromosomal breaks for repair is also consistent with the fact that all chromosomes are involved. Perhaps one of the more striking results is that there are no clear hotspots for rearrangements involving similar chromosomal regions or repetitive elements once the left arm of chromosome V is omitted from consideration. Our data are less consistent with proposals that recurrent rearrangements, such as those observed during carcinogenesis, are dependent on adjacent nonrandom positioning of interphase chromosomes (43, 91), unless the chromosomes proximal to chromosome V vary from cell to cell even within the same genetic background.
Despite the extensive characterization of dynamic features of the yeast genome, we were unable to identify any single factor explaining the distribution of rearrangement sites for either the unique or identical rearrangements isolated. This suggests that rearrangements and, presumably, initiating breaks are generated by a number of independent and competing mechanisms. This mirrors the difficulty in assigning rearrangements in human cancers due to specific genomic features, including VDJ recombination heptamer and nonamer signals, eukaryotic topoisomerase II cleavage sites, chi-like octamers, translin binding sequences, purine-pyrimidine tracts, and homopolymeric tracts, as well as repetitive elements such as Alu and SINE repeats and long terminal repeats (LTRs) (reviewed in reference 58). The fact that adjacent breakpoints on chromosome V do not select similar targets is reminiscent of the fact that over 38 different partner genes on a large number of different chromosomes are fused to breaks induced in the human MLL gene (90), which is involved in most cases of infant acute lymphoblastic leukemia (89). Despite this fact, 70 of 358 rearrangements involve repeated targeting of identical sites in the chromosome V source, and 52 of 358 involve repeated targeting of the same nucleotides in both the source and the target, which is far more common than would be expected by chance. Intriguingly, the S. cerevisiae identical rearrangements were frequently recovered from strains with similar genotypes (see Table S3 in the supplemental material), which suggests that the genetic background may have an important role in forming or selecting the recovered rearrangements. Rather remarkably, identical rearrangements have also been observed in human cancers; 2 of 19 API2/MALT1 fusion breakpoints isolated from MALT lymphomas were also identical, and another 2 of the 19 share the MALT1 breakpoint (58).
The genetic background of the yeast strains studied dramatically influenced the types of rearrangements seen (Fig. 4). Telomerase-defective strains showed a dramatic increase in chromosome fusions with both telomeres and subtelomeric elements as well as rearrangements with Ty elements. The high frequency of chromosome fusions in telomerase-deficient strains suggests either that telomeres maintained by HR are structured differently than those maintained by telomerase or that NHEJ competes more effectively with the HR-based maintenance of telomeres than with telomerase-based maintenance of telomeres (79). On the other hand, translocations to the subtelomeric elements resulting in telomere capture events can occur in telomerase-proficient strains, suggesting that the proteins that prevent fusion reactions do not prevent other types of recombination, consistent with frequent mitotic recombination between Y′ subtelomeric elements (63). Rearrangements involving Ty1 and Ty2 elements were also more frequent in telomerase-defective strains. However, these rearrangements involving Ty elements are not HR events between repeated Ty elements, as observed in laboratory strains, natural isolates, the chromosomal evolution of closely related species of the Saccharomyces sensu stricto group, and recent studies of fragile sites due to the suppression of DNA polymerases (14, 33, 54, 84, 107). Rather, these rearrangements may be due to the Ty1 mobilization observed in strains with telomere dysfunction, possibly leading to the transient production of some type of rearrangement target such as a double-strand break (92, 93); telomerase defects could cause Ty elements to become fragile, similar to the suppression of polymerases (54); or telomerase defects could alter the closed chromatin structure of Ty elements (6).
Additionally, translocations involving paralogs of CAN1 (LYP1 and ALP1) are more frequently observed in strains containing an sgs1 mutation. SGS1 has previously been shown to suppress homoeologous recombination, similar to some of the mismatch repair (MMR) genes (70, 99, 101). Defects in SGS1 cause increased recombination and defects in the human homolog BLM which are associated with a predisposition to many forms of cancer, and they also cause increased sister-chromatid exchanges (15, 53, 76, 101). These observations suggest mechanisms by which MSI and CIN genetic instability phenotypes in cancer can be formed. In addition to the allelic loss of MMR genes by chromosomal instability and the secondary mutational inactivation of genes that suppress genomic instability by defects in mismatch repair, CIN phenotypes might arise due to defects in mechanisms, possibly including MMR, that suppress translocations to homoeologous sequences. In support of this view, colorectal cancers with combined MSI and CIN phenotypes have been observed (1), and more recent data suggest that in some cases these phenotypes may be dependent on each other (35, 103).
Predicted dicentric chromosomes and isochromosomes were predominantly seen in checkpoint-defective backgrounds. A likely explanation for this is that dicentric chromosomes are unstable. These products can comigrate to the same pole during mitosis (38, 87, 102), inactivate one of the centromeres (2, 12, 32, 39, 102), or undergo additional cycles of rearrangements during mitosis (59, 60, 65, 67). Checkpoint defects would allow cells to progress through the cell cycle in the presence of such broken chromosomes. The mechanism proposed for the formation of dicentric isochromosomes in checkpoint-deficient strains (79) is consistent with those for many isochromosomes in humans (112) and correlates with the accumulation of isochromosomes in cancers with an early loss of Rb (Fig. 4). During retinoblastoma progression, only 0.6 megabase of the p arm of chromosome 6 needs to be amplified (18), and multiple types of chromosomal rearrangements have been observed to increase the copy number of this region of the genome (13). Despite this, isochromosome 6p is the most frequently observed mechanism of amplification (100), suggesting that isochromosomes readily form in Rb-defective cells. Rb controls the G1/S transition and the progression through S phase, inhibits apoptosis, and prevents aneuploidy through the suppression of Mad2 overexpression (10, 16, 42, 48); consistent with this, checkpoint defects in yeast often cause an increased accumulation of genome rearrangements and chromosome copy number changes.
Moreover, the karyotypes of tumors and cell lines derived from patients with specific cancer predisposition syndromes also suggest similar relationships between chromosomal products and checkpoint defects. Most patients with the Li-Fraumeni cancer syndrome possess germ line defects in p53 (64). Fibroblasts from Li-Fraumeni patients showed a dramatic accumulation of dicentric chromosomes and a relative increase in acentric fragments, double minutes, ring chromosomes, and gaps as cells were passaged in culture or after immortalization (9, 11, 106). Karyotypes from patients with BRCA1- and BRCA2-defective cancers resemble those from Li-Fraumeni syndrome patients; this could reflect the early loss of BRCA1 and BRCA2 or the fact that >90% of these tumors accumulated mutations in p53 (reviewed in reference 82). On the other hand, mutations in ataxia telangiectasia, A-T-like diseases, and Nijmegen breakage syndrome, which also cause checkpoint defects, tend to be associated with the development of B- and T-cell lymphomas. These lymphomas possess characteristic VDJ recombination-associated translocations between chromosomes 7 and 14 (104) and do not clearly correlate with the accumulation of other specific types of chromosomal aberrations. This may reflect selection for specific rearrangements that occur during the development of lymphomas and drive the carcinogenesis process.
The best-known example of genotype effects on genetic instability in cancer is the MSI phenotype associated with mismatch repair defects (37); however, substantially less is known about the CIN phenotype due to the low resolution of cytogenetic data, the lack of sequence information available for rearrangements that do not drive tumorigenesis, and the lack of information about genetic defects in CIN tumors. The dramatic influence of the strain genotype on the types of rearrangements recovered from S. cerevisiae demonstrated here raises the possibility that the characterization of the types of genome rearrangements seen in human cancers can provide insights into their mechanisms of formation and the presence of genetic defects in these cancers. Moreover, these results suggest that the evolving genotype of cancer cells influences which genes are readily misregulated by genomic instability and thereby may control carcinogenesis.
Supplementary Material
Acknowledgments
We thank Karen Arden and Meng-Er Huang for helpful discussions and critical readings of the manuscript.
This work was supported by National Institutes of Health grant GM26017 (to R.D.K.) and by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation and the Robert Black Charitable Trust (to C.D.P.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdel-Rahman, W. M., K. Katsura, W. Rens, P. A. Gorman, D. Sheer, D. Bicknell, W. F. Bodmer, M. J. Arends, A. H. Wyllie, and P. A. W. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA 98:2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agudo, M., J. P. Abad, I. Molina, A. Losada, P. Ripoll, and A. Villasante. 2000. A dicentric chromosome of Drosophila melanogaster showing alternate centromere inactivation. Chromosoma 109:190-196. [DOI] [PubMed] [Google Scholar]
- 3.Armengol, L., M. A. Pujana, J. Cheung, S. W. Scherer, and X. Estivill. 2003. Enrichment of segmental duplications in regions of breaks of synteny between human and mouse genomes suggests their involvement in evolutionary rearrangements. Hum. Mol. Genet. 12:2001-2008. [DOI] [PubMed] [Google Scholar]
- 4.Bartek, J., J. Bartkova, and J. Lukas. 1997. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 237:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova, J., C. Lukas, C. S. Sorensen, E. R. Meyts, N. E. Skakkebaek, J. Lukas, and J. Bartek. 2003. Deregulation of the RB pathway in human testicular germ cell tumours. J. Pathol. 200:149-156. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Aroya, S., P. A. Mieczkowski, T. D. Petes, and M. Kupiec. 2004. The compact chromatin structure of a Ty repeated sequence suppresses recombination hotspot activity in Saccharomyces cerevisiae. Mol. Cell 15:221-231. [DOI] [PubMed] [Google Scholar]
- 7.Bhagirath, T., S. Abe, T. Nojima, and M. C. Yoshida. 1995. Molecular analysis of a t(11;22) translocation junction in a case of Ewing's sarcoma. Genes Chromosomes Cancer 13:126-132. [DOI] [PubMed] [Google Scholar]
- 8.Bidenne, C., B. Blondin, S. Dequin, and F. Vezinhet. 1992. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr. Genet. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 9.Bischoff, F. Z., S. O. Yim, S. Pathak, G. Grant, M. J. Siciliano, B. C. Giovanella, L. C. Strong, and M. A. Tainsky. 1990. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer Res. 50:7979-7984. [PubMed] [Google Scholar]
- 10.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 11.Boyle, J. M., E. L. D. Mitchell, M. J. Greaves, S. A. Roberts, K. Tricker, E. Burt, J. M. Varley, J. M. Birch, and D. Scott. 1998. Chromosome instability is a predominant trait of fibroblasts from Li-Fraumeni families. Br. J. Cancer 77:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brock, J. A., and K. Bloom. 1994. A chromosome breakage assay to monitor mitotic forces in budding yeast. J. Cell Sci. 107:891-902. [DOI] [PubMed] [Google Scholar]
- 13.Cano, J., O. Oliveros, and E. Yunis. 1994. Phenotype variants, malignancy, and additional copies of 6p in retinoblastoma. Cancer Genet. Cytogenet. 76:112-115. [DOI] [PubMed] [Google Scholar]
- 14.Casaregola, S., H. V. Nguyen, A. Lepingle, P. Brignon, F. Gendre, and C. Gaillardin. 1998. A family of laboratory strains of Saccharomyces cerevisiae carry rearrangements involving chromosomes I and III. Yeast 14:551-564. [DOI] [PubMed] [Google Scholar]
- 15.Chaganti, R. S. K., S. Schonberg, and J. German. 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71:4508-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chau, B. N., and J. Y. Wang. 2003. Coordinated regulation of life and death by RB. Nat. Rev. Cancer 3:130-138. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 18.Chen, D., S. Pajovic, A. Duckett, V. D. Brown, J. A. Squire, and B. L. Gallie. 2002. Genomic amplification in retinoblastoma narrowed to 0.6 megabase on chromosome 6p containing a kinesin-like gene, RKIN. Cancer Res. 62:967-971. [PubMed] [Google Scholar]
- 19.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, S. J., Z. Chen, L. d'Auriol, M. Le Coniat, D. Grausz, and R. Berger. 1989. Ph1+ bcr− acute leukemias: implication of Alu sequences in a chromosomal translocation occurring in the new cluster region within the BCR gene. Oncogene 4:195-202. [PubMed] [Google Scholar]
- 21.Chen, S. J., Z. Chen, M. P. Font, L. d'Auriol, C. J. Larson, and R. Berger. 1989. Structural alterations of the BCR and ABL genes in Ph1 positive acute leukemias with rearrangements in the BCR gene first intron: further evidence implicating Alu sequences in the chromosome translocation. Nucleic Acids Res. 17:7631-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1997. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 23.Chuzhanova, N., S. S. Abeysinghe, M. Krawczak, and D. N. Cooper. 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer. II. Potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum. Mutat. 22:245-251. [DOI] [PubMed] [Google Scholar]
- 24.Davis, E. S., B. K. Shafer, and J. N. Strathern. 2000. The Saccharomyces cerevisiae RDN1 locus is sequestered from interchromosomal meiotic ectopic recombination in a SIR2-dependent manner. Genetics 155:1019-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Klein, A., T. van Agthoven, C. Groffen, N. Heisterkamp, J. Groffen, and G. Grosveld. 1986. Molecular analysis of both translocation products of a Philadelphia-positive CML patient. Nucleic Acids Res. 14:7071-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wang, A. Flavier, T. D. Gaffrey, and P. Phillipsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 27.Dudasova, Z., A. Dudas, and M. Chovanec. 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28:581-601. [DOI] [PubMed] [Google Scholar]
- 28.Duensing, S., and K. Munger. 2004. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int. J. Cancer 109:157-162. [DOI] [PubMed] [Google Scholar]
- 29.Duesberg, P., and R. Li. 2003. Multistep carcinogensis: a chain reaction of aneuploidizations. Cell Cycle 2:202-210. [PubMed] [Google Scholar]
- 30.Dunn, T., L. Praissman, N. Hagag, and M. V. Viola. 1994. ERG gene is translocated in an Ewing's sarcoma cell line. Cancer Genet. Cytogenet. 76:19-22. [DOI] [PubMed] [Google Scholar]
- 31.Fioretos, T., B. Stroembeck, T. Sandberg, B. Johansson, R. Billstrom, A. Borg, P. G. Nilsson, H. van den Berghe, A. Hagemeijer, F. Mitelman, and M. Hoglund. 1999. Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood 94:225-232. [PubMed] [Google Scholar]
- 32.Fischer, A. M., L. Al-Gazali, T. Pramathan, R. Quaife, A. E. Cockwell, J. C. Barber, W. C. E. A. J. Axelman, B. R. Migeon, and C. Tyler-Smith. 1997. Centromeric inactivation in a dicentric human Y:21 translocation chromosome. Chromosoma 106:199-206. [DOI] [PubMed] [Google Scholar]
- 33.Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver, and E. J. Louis. 2000. Chromosomal evolution in Saccharomyces. Nature 405:451-454. [DOI] [PubMed] [Google Scholar]
- 34.Glynn, E. F., P. C. Megee, H. G. Yu, C. Mistrot, E. Unal, D. E. Koshland, J. L. DeRisi, and J. L. Gerton. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goel, A., C. N. Arnold, D. Niedzwiecki, D. K. Chang, L. Ricciardiello, J. M. Carethers, J. M. Dowell, L. Wasserman, C. Compton, R. J. Mayer, M. M. Bertagnolli, and C. R. Boland. 2003. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 63:1608-1614. [PubMed] [Google Scholar]
- 36.Gotter, A. L., T. H. Shaikh, M. L. Budarf, C. H. Rhodes, and B. S. Emanuel. 2004. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 13:103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grady, W. M. 2004. Genomic instability and colon cancer. Cancer Metastasis Rev. 23:11-27. [DOI] [PubMed] [Google Scholar]
- 38.Haber, J. E., P. C. Thorburn, and D. Rogers. 1984. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics 106:185-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hande, M. P., E. Sampler, P. Landsdorp, and M. A. Blasco. 1999. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 144:589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris, S., K. S. Rudnicki, and J. E. Haber. 1993. Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics 135:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heisterkamp, N., K. Stam, J. Groffen, A. de Klein, and G. Grosveld. 1985. Structural organization of the bcr gene and its role in the Ph′ translocation. Nature 315:758-761. [DOI] [PubMed] [Google Scholar]
- 42.Hernando, E., Z. Nahle, G. Juan, E. Diaz-Rodriguez, M. Alaminos, M. Hemann, L. Michel, V. Mittal, W. Gerald, R. Benezra, S. W. Lowe, and C. Cordon-Cardo. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430:797-802. [DOI] [PubMed] [Google Scholar]
- 43.Hilliker, A. J. 1985. Assaying chromosome arrangements in embryonic interphase nuclei of D. melanogaster by radiation induced interchanges. Genet. Res. 47:13-18. [Google Scholar]
- 44.Huang, M. E., A. G. Rio, A. Nicolas, and R. D. Kolodner. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100:11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishida, S., K. Yoshida, Y. Kaneko, Y. Tanaka, Y. Sasaki, F. Urano, A. Umezawa, J. Hata, and K. Fujinaga. 1998. The genomic breakpoint and chimeric transcripts in the EWSR1-ETV4/E1AF gene fusion in Ewing sarcoma. Cytogenet. Cell Genet. 82:278-283. [DOI] [PubMed] [Google Scholar]
- 47.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617-624. [DOI] [PubMed] [Google Scholar]
- 48.Knudsen, E. S., C. Buckmaster, T. T. Chen, J. R. Fermisco, and J. Y. Wang. 1998. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 12:2278-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 50.Koszul, R., S. Caburet, B. Dujon, and G. Fischer. 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23:234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kramer, K. M., J. A. Brock, K. Bloom, J. K. Moore, and J. E. Haber. 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 14:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kueppers, R., and R. Dalla-Favera. 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20:5580-5594. [DOI] [PubMed] [Google Scholar]
- 53.Langois, R. G., W. L. Bigbee, R. H. Jensen, and J. German. 1989. Evidence for increased in vivo mutation and somatic recombination in Bloom's syndrome. Proc. Natl. Acad. Sci. USA 86:670-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemoire, F. J., N. P. Destyareva, K. Lobachev, and T. D. Petes. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosomal fragile sites. Cell 120:587-598. [DOI] [PubMed] [Google Scholar]
- 55.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 56.Lisby, M., and R. Rothstein. 2004. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 16:328-334. [DOI] [PubMed] [Google Scholar]
- 57.Litz, C. E., J. S. McClure, C. M. Copenhaver, and R. D. Brunning. 1993. Duplication of small segments within the major breakpoint cluster region in chronic myelogenous leukemia. Blood 81:1567-1572. [PubMed] [Google Scholar]
- 58.Liu, H., R. A. Hamoudi, H. Ye, A. Ruskone-Fourmestraux, A. Dogan, P. G. Issacson, and M. Q. Du. 2004. t(11;18)(q21;q21) of mucosa-associated lymphoid tissue lymphoma results from illegitimate non-homologous end joining following double strand breaks. Br. J. Haematol. 125:318-329. [DOI] [PubMed] [Google Scholar]
- 59.Lo, A. W., L. Sabatier, B. Fouladi, G. Pottier, M. Ricoul, and J. P. Murnane. 2002. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia 4:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lo, A. W., C. N. Sprung, B. Fouladi, M. Pedram, L. Sabatier, M. Ricoul, G. E. Reynolds, and J. P. Murnane. 2002. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol. Cell. Biol. 22:4836-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loeb, L. A. 2001. A mutator phenotype in cancer. Cancer Res. 61:3230-3239. [PubMed] [Google Scholar]
- 62.Lohmann, D. R. 1999. RB1 gene mutations in retinoblastoma. Hum. Mutat. 14:283-288. [DOI] [PubMed] [Google Scholar]
- 63.Louis, E. J., and J. E. Haber. 1990. Mitotic recombination among subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 124:547-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malkin, D., F. P. Li, L. C. Strong, J. F. Fraumeni, Jr., C. E. Nelson, D. H. Kim, J. Kassel, M. A. Gryka, F. Z. Bischoff, M. A. Tainsky, and S. H. Friend. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250:1233-1238. [DOI] [PubMed] [Google Scholar]
- 65.Maringele, L., and D. Lydall. 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18:2663-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mastrangelo, T., M. Piergiorgio, S. Tornielli, F. Bullrich, M. A. Testi, A. Mezzelani, P. Radice, A. Azzarelli, S. Pilotti, C. M. Croce, M. A. Pierotti, and G. Sozzi. 2000. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene 19:3799-3804. [DOI] [PubMed] [Google Scholar]
- 67.McClintock, B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26:234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitelman, F. 1991. Catalog of chromosome aberrations in cancer. Wiley Liss, New York, N.Y.
- 69.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411:1073-1076. [DOI] [PubMed] [Google Scholar]
- 70.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 71.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint function in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 72.Myung, K., and R. D. Kolodner. 2003. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair 2:243-258. [DOI] [PubMed] [Google Scholar]
- 73.Myung, K., and R. D. Kolodner. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Myung, K., V. Pennaneach, E. S. Kats, and R. D. Kolodner. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Obata, K., H. Hiraga, T. Nojima, M. C. Yoshida, and S. Abe. 1999. Molecular characterization of the genomic breakpoint junction in a t(11;22) translocation in Ewing sarcoma. Gene Chromosomes Cancer 25:6-15. [PubMed] [Google Scholar]
- 76.Onoda, F., M. Seki, A. Miyajima, and T. Enomoto. 2000. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 459:203-209. [DOI] [PubMed] [Google Scholar]
- 77.Panagopoulos, I., C. Lassen, M. Isaksson, F. Mitelman, N. Mandahl, and P. Aman. 1997. Characteristic sequence motifs at the breakpoints of the hybrid genes FUS/CHOP, EWS/CHOP, and FUS/ERG in myxoid liposarcoma and acute myeloid leukemia. Oncogene 15:1357-1362. [DOI] [PubMed] [Google Scholar]
- 78.Park, H., and A. T. Bakalinsky. 2000. SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16:881-888. [DOI] [PubMed] [Google Scholar]
- 79.Pennaneach, V., and R. D. Kolodner. 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36:612-617. [DOI] [PubMed] [Google Scholar]
- 80.Perez-Ortin, J. E., A. Querol, S. Puig, and E. Barrio. 2002. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 12:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peter, M., F. Mugneret, A. Aurias, G. Thomas, H. Magdelenat, and O. Delattre. 1996. An EWS/ERG fusion with a truncated N-terminal domain of EWS in a Ewing's tumor. Int. J. Cancer 67:339-342. [DOI] [PubMed] [Google Scholar]
- 82.Powell, S. N., and L. A. Kachnic. 2003. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 22:5784-5791. [DOI] [PubMed] [Google Scholar]
- 83.Putnam, C. D., V. Pennaneach, and R. D. Kolodner. 2004. Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:13262-13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rachidi, N., P. Barre, and B. Blodin. 1999. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol. Gen. Genet. 261:841-850. [DOI] [PubMed] [Google Scholar]
- 85.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 86.Rattray, A. J., and L. S. Symington. 1995. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics 139:45-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rhoades, M. M. 1953. Preferential segregation in maize, p. 66-81. In J. W. Gowen (ed.), Heterosis. Iowa State Press, Ames, Iowa.
- 88.Roth, D. B., and J. H. Wilson. 1986. Nonhomologous recombination in mammalian cells: role for short sequence homologies in joining reactions. Mol. Cell. Biol. 6:4295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rowley, J. D. 1998. The critical role of chromosome translocation in human leukemia. Annu. Rev. Genet. 32:495-519. [DOI] [PubMed] [Google Scholar]
- 90.Rowley, J. D. 2000. Molecular genetics in acute leukemia. Leukemia 14:513-517. [DOI] [PubMed] [Google Scholar]
- 91.Sachs, R. K., L. R. Hlatky, and B. J. Trask. 2002. Radiation-produced chromosomal aberrations: colorful clues. Trends Genet. 16:143-146. [DOI] [PubMed] [Google Scholar]
- 92.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curico. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scholes, D. T., A. E. Kenny, E. R. Gamache, Z. Mou, and M. J. Curico. 2003. Activation of a LTR-retrotransposon by telomere erosion. Proc. Natl. Acad. Sci. USA 100:15736-15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sherr, C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60:3689-3695. [PubMed] [Google Scholar]
- 95.Shtivelman, E., B. Lifshitz, R. P. Gale, and E. Canaani. 1985. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature 315:550-554. [DOI] [PubMed] [Google Scholar]
- 96.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 98.Soekarman, D., J. van Denderen, L. Hoefsloot, M. Moret, T. Meeuwsen, J. van Baal, A. Hagemeijer, and G. Grosveld. 1990. A novel variant of the bcr-abl fusion product in Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia 4:397-403. [PubMed] [Google Scholar]
- 99.Spell, R. M., and S. Jinks-Robertson. 2004. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 168:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Squire, J., R. A. Phillips, S. Boyce, R. Godbout, B. Rogers, and B. L. Gallie. 1984. Isochromosome 6p, a unique chromosomal abnormality in retinoblastoma: verification by standard staining techniques, new densitometric methods, and somatic cell hybridization. Hum. Genet. 66:46-53. [DOI] [PubMed] [Google Scholar]
- 101.Sugawara, N., T. Goldfarb, B. Studamire, E. Alani, and J. E. Haber. 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101:9315-9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sullivan, B. A., and H. F. Williard. 1998. Stable dicentric chromosomes with two functional centromeres. Nat. Genet. 20:227-228. [DOI] [PubMed] [Google Scholar]
- 103.Tang, R., C. R. Changchien, M. C. Wu, C. W. Fan, K. W. Liu, J. S. Chen, H. T. Chien, and L. L. Hsieh. 2004. Colorectal cancer without high microsatellite instability and chromosomal instability—an alternative genetic pathway to human colorectal cancer. Carcinogenesis 25:841-846. [DOI] [PubMed] [Google Scholar]
- 104.Taylor, A. M., J. A. Metcalfe, J. Thick, and F. Mak. 1996. Leukemia and lymphoma in ataxia telangiectasia. Blood 6:423-438. [PubMed] [Google Scholar]
- 105.Tennyson, R. B., N. Ebran, A. E. Herrera, and J. E. Lindsley. 2002. A novel selection system for chromosome translocations in Saccharomyces cerevisiae. Genetics 160:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsutsui, T., S. Kumakura, T. Tamura, T. W. Tsutsui, M. Sekiguichi, T. Higuchi, and J. C. Barrett. 2003. Immortal, telomerase-negative cell lines derived from a Li-Fraumeni patient exhibit telomere length variability and chromosomal and minisatellite instabilities. Carcinogenesis 24:953-965. [DOI] [PubMed] [Google Scholar]
- 107.Umezu, K., M. Hiraoka, M. Mori, and H. Maki. 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160:97-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van der Feltz, M. J. M., M. K. K. Shivji, P. B. Allen, N. Heisterkamp, J. Groffen, and L. M. Wiedermann. 1989. Nucleotide sequence of both reciprocal translocation junction regions in a patient with Ph positive acute lymphoblastic leukaemia, with a breakpoint within the first intron of the BCR gene. Nucleic Acids Res. 17:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vega, L. R., M. K. Mateyak, and V. A. Zakian. 2003. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 4:948-959. [DOI] [PubMed] [Google Scholar]
- 110.Voelkel-Meiman, K., R. L. Keil, and G. S. Roeder. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48:1071-1079. [DOI] [PubMed] [Google Scholar]
- 111.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 112.Wolff, D. J., A. P. Miller, D. L. Van Dyke, S. Schwartz, and H. F. Willard. 1996. Molecular definition of breakpoints associated with human Xq isochromosomes: implications for mechanisms of formation. Am. J. Hum. Genet. 58:154-160. [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang, J. G., J. M. Goldman, and N. C. P. Cross. 1995. Characterization of genomic BCR-ABL breakpoints in chronic myeloid leukaemia by PCR. Br. J. Haematol. 90:138-146. [DOI] [PubMed] [Google Scholar]
- 114.Zucman-Rossi, J., P. Legoix, J. M. Victor, B. Lopez, and G. Thomas. 1998. Chromosome translocation based on illegitimate recombination in human tumors. Proc. Natl. Acad. Sci. USA 95:11786-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zuker, M., D. H. Matthews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.