Abstract
The ESC-E(Z) complex of Drosophila melanogaster Polycomb group (PcG) repressors is a histone H3 methyltransferase (HMTase). This complex silences fly Hox genes, and related HMTases control germ line development in worms, flowering in plants, and X inactivation in mammals. The fly complex contains a catalytic SET domain subunit, E(Z), plus three noncatalytic subunits, SU(Z)12, ESC, and NURF-55. The four-subunit complex is >1,000-fold more active than E(Z) alone. Here we show that ESC and SU(Z)12 play key roles in potentiating E(Z) HMTase activity. We also show that loss of ESC disrupts global methylation of histone H3-lysine 27 in fly embryos. Subunit mutations identify domains required for catalytic activity and/or binding to specific partners. We describe missense mutations in surface loops of ESC, in the CXC domain of E(Z), and in the conserved VEFS domain of SU(Z)12, which each disrupt HMTase activity but preserve complex assembly. Thus, the E(Z) SET domain requires multiple partner inputs to produce active HMTase. We also find that a recombinant worm complex containing the E(Z) homolog, MES-2, has robust HMTase activity, which depends upon both MES-6, an ESC homolog, and MES-3, a pioneer protein. Thus, although the fly and mammalian PcG complexes absolutely require SU(Z)12, the worm complex generates HMTase activity from a distinct partner set.
The Polycomb group (PcG) proteins of Drosophila melanogaster form chromatin complexes that repress transcription during development (see references 9, 44, and 53 for reviews). The most intensively studied targets of PcG repression are the fly Hox genes, which are differentially activated along the anterior-posterior axis in early embryos. Once initial Hox activity patterns are established, PcG complexes maintain Hox-repressed states during subsequent embryonic, larval, and pupal stages. There are approximately 15 fly PcG repressors, and they are implicated in silencing many other targets, besides Hox genes, in the fly genome (11, 31, 33, 45). Thus, PcG proteins are general repressors that provide a key model for understanding chromatin mechanisms that propagate transcriptional off states during development.
Fly PcG complexes have been defined by fractionation and purification from embryo extracts (7, 34, 36, 48, 50, 59). The two best-characterized PcG complexes are the ESC-E(Z) complex and Polycomb repressive complex 1 (PRC1). These complexes are biochemically separable and contain distinct sets of PcG subunits. The ESC-E(Z) complex has histone methyltransferase (HMTase) activity which depends upon the SET domain of the catalytic subunit, E(Z) (34). The primary target of ESC-E(Z) HMTase activity is lysine 27 of histone H3 (7, 34). This H3-K27 methylation is thought to help recruit the second PcG complex, PRC1, to specific chromatin sites; the Polycomb (PC) subunit of PRC1 binds to trimethyl-H3-K27 in vitro (3, 7, 8, 32), and E(Z) function is required for PRC1 localization to Polycomb response elements (PREs) in vivo (3, 42, 67). Thus, a stepwise model has been proposed whereby the ESC-E(Z) complex marks local chromatin with methyl-H3-K27, and this in turn recruits PRC1 to target sites where it directly executes PcG silencing (3, 52, 67). The original recruitment of the ESC-E(Z) complex to PREs depends upon the DNA-binding PcG proteins PHO and PHO-Like (67).
The ESC-E(Z) complex contains four core subunits: extra sex combs (ESC), Enhancer of zeste [E(Z)], Suppressor of zeste-12 [SU(Z)12], and nucleosome-remodeling factor 55 (NURF-55) (7, 34). Though the E(Z) SET domain provides the catalytic active site, the noncatalytic subunits also make critical contributions to function. This is apparent from HMTase assays which show that a recombinant four-subunit ESC-E(Z) complex is greater than 1,000-fold more active than E(Z) alone (34). In agreement with this, genetic studies show that loss of either ESC or SU(Z)12 yields embryonic Hox phenotypes that are as severe as those caused by loss of E(Z) function (2, 20, 56). Clearly, enzyme activity in vitro and PcG repression in vivo depend upon tight partnership of E(Z) with its noncatalytic cohorts. The molecular roles of these subunits and their mechanisms for potentiating activity are not yet clear. Indeed, in the absence of a three-dimensional structure of the complex, much remains to be determined about how the complex is built from its constituent parts and how it functions.
The ESC-E(Z) complex is strikingly conserved from flies to mammals. The corresponding complexes isolated from HeLa cells, which have been called Polycomb repressive complexes 2 and 3 (PRC2/3), contain homologs of the four fly subunits and have similar HMTase activities that target H3-K27 (3, 4, 26, 27). In addition to Hox gene regulation, mammalian ESC-E(Z) complexes are implicated in X chromosome inactivation (41, 51) and in progression of prostate and breast cancers (23, 62). Even more evolutionarily striking, plants contain clear homologs of ESC, E(Z), SU(Z)12, and NURF-55, which function together in developmental processes, including seed differentiation and control of flowering (15, 29, 54). A particularly fascinating role for the plant PcG homologs is in the process of vernalization (14, 24). In this case, flowering time is influenced by temperatures experienced weeks earlier, with the PcG proteins thought to provide memory by maintaining chromatin states for the long intervening period. This conservation across kingdoms implies that the ESC-E(Z) complex supplies an ancient function in chromatin modification.
The biological functions of ESC-E(Z) homologs in Caenorhabditis elegans have also been intensively studied. The E(Z) homolog, MES-2, and the ESC homolog, MES-6, are subunits in a worm MES complex that is required for germ line development and gene silencing (13, 17, 21, 25, 68) and also plays a role in Hox gene repression in the soma (46). Besides MES-2 and MES-6, the MES complex contains MES-3, which is a novel protein unrelated to PcG proteins from other species. In addition, the worm genome lacks a recognizable Su(z)12 homolog. Nevertheless, like the fly and mammalian complexes, the MES complex methylates H3-K27 in worms (1). Thus, there are functional parallels between the worm and fly complexes but there are also major differences in subunit compositions.
Here we analyze recombinant ESC-E(Z) complexes to define contributions of individual subunits and roles of their functional domains. We assess assembly and HMTase activities of complexes lacking particular subunits or bearing mutations in single subunits. Many of the subunit mutations mimic existing missense alleles, so in vitro properties can be compared to genetic behavior in vivo. Our analysis of stable subunit interactions implies that E(Z) and SU(Z)12 occupy central positions in the complex, whereas ESC and NURF-55 associate more peripherally. We find that SU(Z)12 and ESC, which directly contact E(Z), are each needed for HMTase function in vitro. In agreement with this, we also find that ESC is critically required for global H3-K27 methylation in fly embryos. Among the missense alleles analyzed, mutations in the cysteine-rich CXC domain of E(Z), in conserved surface loops of ESC, and in the VEFS domain of SU(Z)12 are notable for compromising HMTase activity but not assembly of the complex. Thus, these domains are implicated in facilitating E(Z) catalytic function. Finally, we find that a recombinant worm MES complex, built from three subunits, has robust HMTase activity which requires both noncatalytic subunits. We discuss how worms and flies may have evolved distinct strategies to build active HMTase complexes.
MATERIALS AND METHODS
Expression and purification of protein complexes.
Baculovirus expression of recombinant proteins was performed using the Bac-to-Bac system (Invitrogen). Anti-FLAG immunoaffinity purification of complexes was performed essentially as described previously (34), except washes in BC buffer were performed up to 1.2 M KCl and elutions were performed in batch for 1 h with 0.8 mg/ml FLAG peptide. Mutant complexes were prepared in parallel with a wild-type control and purified at least twice independently for each complex. Protein complexes were purified from insect Sf9 cells grown at 28 to 29°C, which corresponds to the loss-of-function restrictive temperature for E(z)32 (C545Y), E(z)61 (C603Y), and E(z)28 (C363Y).
Baculovirus constructs and site-directed mutagenesis.
Full-length cDNAs encoding FLAG-ESC, E(Z), SU(Z)12, NURF-55, and FLAG-E(Z) inserted into pFastBac1 were described previously (34). Additional FLAG-SU(Z)12 and hemagglutinin (HA)-ESC constructs were produced by inserting tag-encoding oligonucleotides at the N termini of the respective untagged expression constructs. pFastBac1 derivatives for MES protein expression were generated using the Gateway recombination system (Invitrogen). Full-length mes-2, mes-3, and mes-6 cDNAs were inserted into pDONR201 (Invitrogen), generating mes entry vectors. Destination vectors were generated by engineering pFastBac1 to contain either an attL cassette alone or a FLAG tag sequence followed by an attL cassette. Recombination between mes entry vectors and destination vectors generated FLAG-tagged and untagged versions of MES-2, MES-3, and MES-6. Site-directed mutations were generated using the Quick Change mutagenesis kit (Stratagene) or the GeneTailor mutagenesis system (Invitrogen). In-frame deletions of E(Z) or SU(Z)12 domains were constructed using a PCR-based strategy followed by sequencing to confirm intact open reading frames. Deletion of E(Z) domain II spans residues 259 to 370, and deletion of the SU(Z)12 VEFS domain spans residues 527 to 603.
HMTase assays and substrates.
Histone methyltransferase assays were performed as described previously (34). HMTase assays were repeated at least twice using independently prepared complexes. Relative HMTase levels are based upon visual comparison of signals obtained with various concentrations of complexes. The estimate of threefold HMTase reduction resulting from loss of NURF-55 (Fig. 1A) is derived from four independent tests, two using FLAG-ESC complexes and two using FLAG-SU(Z)12 complexes.
FIG. 1.
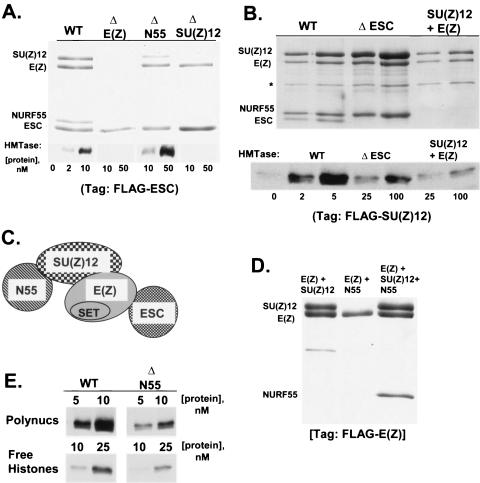
Assembly and activity of recombinant subcomplexes missing individual subunits of the ESC-E(Z) complex. (A) Subunit compositions (top) and HMTase activities (bottom) of subcomplexes obtained after omission of indicated subunits. Wild-type (WT) four-subunit complex (left lane) and subcomplexes were affinity purified using FLAG-ESC. (B) As in panel A, except complexes purified using FLAG-SU(Z)12. The second lane of each pair shows twice as much material loaded. The rightmost lanes show subcomplex obtained after coexpression of only SU(Z)12 plus E(Z). An asterisk here, and in subsequent figures, denotes hsp70, which often contaminates baculovirus-produced complexes (10, 34). (C) Model for the four-subunit complex deduced from stable interactions detected in panels A and B. The catalytic subunit, E(Z), occupies a central position and can bind stably and independently to ESC and SU(Z)12. (D) Subunit compositions of subcomplexes obtained after coexpression of the indicated subunits and purified using FLAG-E(Z). (E) HMTase activities of wild-type four-subunit complex versus trimeric complex lacking NURF-55. Activities were compared on polynucleosome (Polynucs; top) and free histone (bottom) substrates.
Polynucleosome substrates, consisting of 8- to 12-mers prepared from HeLa cells, were used throughout this work except for experiments in Fig. 1E and 6C, which used bovine free histones (Roche) as a substrate. Polynucleosomes were prepared essentially as described previously (16), using nuclei extracted from HeLa cells (National Cell Culture Center). After final sucrose gradient separation of polynucleosome arrays, fractions containing 8- to 12-mers were dialyzed in 20 mM HEPES, pH 7.5, 0.1 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, and 0.5 mM phenylmethylsulfonyl fluoride, and polynucleosomes were concentrated using Amicon Ultra centrifugal filter devices.
FIG. 6.
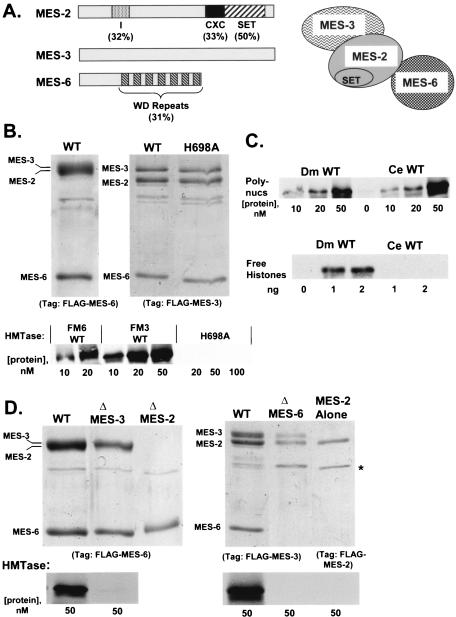
Properties and subunit contributions of recombinant worm MES complexes. (A) Domain organizations of the three worm MES proteins. Percent identities shown are between worm MES-2 and fly E(Z) or between worm MES-6 and fly ESC (17, 25). Boundaries of the CXC and SET domains are as defined (28), whereas domain I similarity spans residues 101 to 175 of fly E(Z). The illustration on the right shows a model of the MES complex based upon stable interactions detected in panels B and D below and in reference 68. (B) Assembly (top) and HMTase activities (bottom) of wild-type trimeric MES complexes or a trimeric complex bearing MES-2-H698A. Complexes were purified via FLAG-MES-6 (FM6) or FLAG-MES-3 (FM3) as indicated. (C) HMTase activities of wild-type four-subunit fly (D. melanogaster [Dm]) complex versus wild-type three-subunit worm (C. elegans [Ce]) complex. Activities were compared on polynucleosome (Polynucs; top) and free histone (bottom) substrates. Numbers in the top panel indicate concentrations of complexes (in nM), and numbers in the bottom panel denote free histone amounts (in ng) in reactions with the indicated complex at 50 nM. (D) Assembly and activity of recombinant subcomplexes missing individual subunits of the worm MES complex. Complexes on the left were purified via FLAG-MES-6, and complexes on the right were purified via FLAG-MES-3.
Generation of esc and E(z) mutant embryos and Western blot analysis.
Wild-type embryos were collected from stock y Df(1)w67c2 at 25°C (0 to 16 h). E(z) mutant embryos were collected from a homozygous E(z)61 stock at 18°C (0 to 24 h) or at 29°C (0 to 12 h). esc mutant embryos were collected as the progeny of esc10 b pr/esc2 Cy0 adults at 25°C (0 to 16 h). esc10 is a 380-kb deficiency that removes the esc locus, and esc2 is an apparent null allele resulting from a frameshift (12, 47, 56). Embryos were dechorionated in 50% bleach and flash frozen in liquid nitrogen. Protein extracts were prepared by resuspending the embryos in an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer containing 0.4 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml chymostatin, 1 μg/ml pepstatin A, and 1 μg/ml antipain; crushing with a pestle; and then incubating at 95°C for 10 min. Proteins were fractionated on 15% SDS-polyacrylamide gel electrophoresis (PAGE) gels and transferred to Immobilon-P (Millipore) (Fig. 2A) or on 10% gels followed by transfer to Protran (Schleicher and Schuell) (Fig. 2B). The blots were blocked for at least 30 min in 5% nonfat dry milk and incubated in primary antibody overnight at 4°C. The trimethyl-H3K27 antibody was used at 1:1,000, and the dimethyl-H3K27 and dimethyl-H3K9 antibodies were used at 1:500. These antibodies have been described previously (40) and were kindly provided by Thomas Jenuwein. The antibody against unmodified histone H3 (residues 1 to 20; Upstate Biotech) was used at 1:250. Antibodies against SU(Z)12 and E(Z) have been described previously (6, 34) and were used at 1:500 and 1:100, respectively. A monoclonal antibody against α-tubulin (DM-1A; Sigma) was used at 1:2,000. Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies (Jackson Immuno Laboratories) were used at 1:10,000. Signals were developed using an ECL chemiluminescence detection kit (Amersham).
FIG. 2.
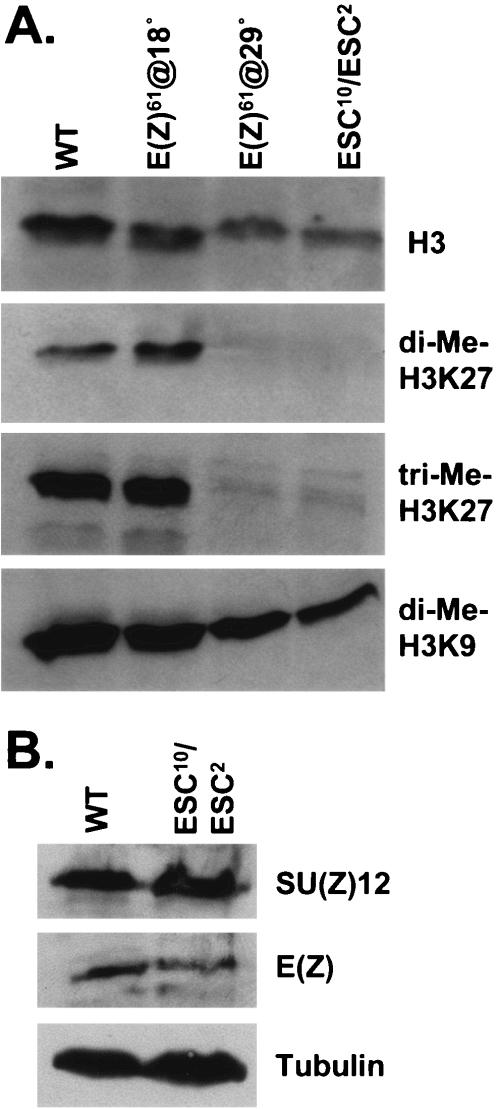
Levels of methylated histone H3 in fly embryos bearing an ESC or E(Z) loss-of-function mutation. (A) Western blots were performed on fly embryo extracts using antibodies that detect either unmodified histone H3 (top row) or the indicated forms of methylated H3 (bottom rows). Extracts were prepared from wild-type (WT) embryos (lane 1), E(z)61 embryos collected at permissive temperature (18°C, lane 2), E(z)61 embryos at restrictive temperature (29°C, lane 3), or embryos collected as progeny of esc10/esc2 parents (lane 4). (B) As in panel A, except blots were incubated with antibodies against SU(Z)12, E(Z), or tubulin (loading control) as indicated.
RESULTS
SU(Z)12 and ESC are required for histone methyltransferase activity.
We used a baculovirus expression system to produce, purify, and test recombinant forms of the fly ESC-E(Z) complex for assembly and HMTase activity in vitro. As we described previously (34), the four-subunit ESC-E(Z) complex prepared this way methylates histone H3 at lysine 27 with an approximately fivefold substrate preference for polynucleosome arrays versus free histones. Polynucleosomes were used as a substrate throughout this study, except where specifically noted otherwise.
To determine which noncatalytic subunits are required for HMTase activity, we compared enzyme activities of the four-subunit complex to subcomplexes obtained when one or two subunits were omitted from the coexpression mix. Figure 1A shows subunit dropout experiments performed using FLAG-tagged ESC for purification. Single loss of E(Z) yields FLAG-ESC alone (lane 2), which indicates that ESC primarily interacts with E(Z) in the normal complex. In contrast, single loss of NURF-55 yields a trimeric FLAG-ESC/E(Z)/SU(Z)12 complex (lane 3), which is catalytically active at levels about threefold reduced from wild type. Single loss of SU(Z)12 leads to loss of NURF-55 as well, suggesting that NURF-55 association is primarily through SU(Z)12; the resulting FLAG-ESC/E(Z) dimer (lane 4) is catalytically inactive. These results show that SU(Z)12 is vital for HMTase function, whereas NURF-55 makes only a minimal contribution to activity of the complex.
To assess contributions of ESC, we purified complexes with the FLAG tag instead placed at the N terminus of SU(Z)12. Figure 1B shows that the four-subunit complex prepared this way has comparably robust activity. If ESC is singly deleted, a stable subcomplex containing the other three subunits is obtained (lanes 3 and 4). This complex has detectable HMTase activity, but it is 25- to 50-fold reduced relative to the wild type. Finally, coexpression of only FLAG-SU(Z)12 plus E(Z) yields a stable dimer which is even further reduced in activity (lanes 5 and 6). Taken together, the subunit dropout experiments identify SU(Z)12 and ESC as most critical for potentiating E(Z) HMTase activity. In addition, these tests define the stable subunit interactions that support complex assembly, which are summarized by the model in Fig. 1C.
In addition to these results, several studies have described subunit interactions of fly or mammalian versions of the ESC-E(Z) complex (5, 35, 39, 59, 69). Two issues appear unresolved after comparing the data sets. First, there are different views on the location of E(Z) in the complex. One study of the mammalian complex suggests that EZH2 is located peripherally and assembles solely through contact with the ESC homolog, EED (5). Two other studies of mammalian subunits also detect stable EZH2-SU(Z)12 interactions (39, 69) and suggest a more central location for EZH2. Our results (Fig. 1A to C) are consistent with another study of the fly complex (35), which detected stable contacts of E(Z) with both ESC and SU(Z)12, and support a central location for E(Z). Second, there are different results on whether NURF-55 binds stably to E(Z). In agreement with our SU(Z)12 dropout result (Fig. 1A), two studies on the mammalian complex find that the NURF-55 homolog RbAp48 cannot stably associate with EZH2-EED in the absence of SU(Z)12 (5, 39). However, studies of the fly subunits have reported pairwise binding of NURF-55 to E(Z) (35, 59). To directly assess this using our assay conditions, we compared stable subcomplexes obtained after coexpression of FLAG-E(Z) with NURF-55 plus or minus SU(Z)12. As shown in Fig. 1D, the stable E(Z)-SU(Z)12 dimer and E(Z)-SU(Z)12-NURF-55 trimer were obtained (lanes 1 and 3), but we failed to detect a stable E(Z)-NURF-55 dimer (lane 2) in our system.
Since the mammalian homolog of NURF-55 has histone-binding activity (64), it has been hypothesized to help mediate substrate interactions. However, NURF-55 binds primarily to a portion of histone H4 that is inaccessible in the nucleosome context and it fails to bind nucleosome arrays on its own (30, 64). Thus, in addition to polynucleosome substrate, we also tested ESC-E(Z) complexes lacking NURF-55 for activity upon free histones. Figure 1E shows that, as with polynucleosomes, NURF-55 loss leads to a modest reduction in HMTase activity on free histones.
ESC is required for H3-K27 methylation in fly embryos.
Previous work using a temperature-sensitive E(z) allele has shown that H3-K27 methylation of bulk chromatin in fly embryos depends upon E(Z) function (3). Thus, we consider E(Z) to be the predominant sponsor of H3-K27 modification in flies. If the ESC and SU(Z)12 contributions to E(Z) HMTase activity (Fig. 1) are fundamentally required, then we would expect these subunits to be similarly key for H3-K27 methylation in vivo. Figure 2A shows tests to directly assess the in vivo requirement for ESC. Western blots were performed on fly embryo extracts using antibodies that recognize methylated forms of histone H3 (40). As expected (3), levels of methylated H3-K27 are dramatically reduced in extracts from E(z)61 embryos collected at restrictive temperature, which serves as a positive control. In the test sample (lane 4), we find that extracts from esc-null mutant embryos show similar bulk loss of methylated H3-K27. These dramatic reductions are observed with antibodies that detect either dimethyl- or trimethyl-H3-K27, but no significant differences are seen with an antibody that detects dimethyl-H3-K9. Figure 2B also shows that SU(Z)12 and E(Z) persist at similar levels in these esc mutant embryos. We conclude that the ESC noncatalytic subunit is an obligate functional partner in E(Z) complexes in fly embryos.
Mutational analysis of E(Z) domains.
As depicted in Fig. 3A, several E(Z) functional domains have been identified based upon established in vitro roles and/or high homology among E(Z) homologs in other species (19, 28). In agreement with its assumed role as the core catalytic domain (43), we have previously shown that the E(Z) SET domain is required for HMTase activity in vitro and Hox gene repression in vivo (34). In addition, pairwise binding studies have identified an N-terminal ESC-interacting domain (EID) and homology domain I contains a binding site for another PcG protein, Polycomb-like (PCL), which may associate with the core ESC-E(Z) complex in vivo (18, 38, 58, 60). However, molecular roles for the highly conserved cysteine-rich CXC domain and homology domain II have not been defined.
FIG. 3.
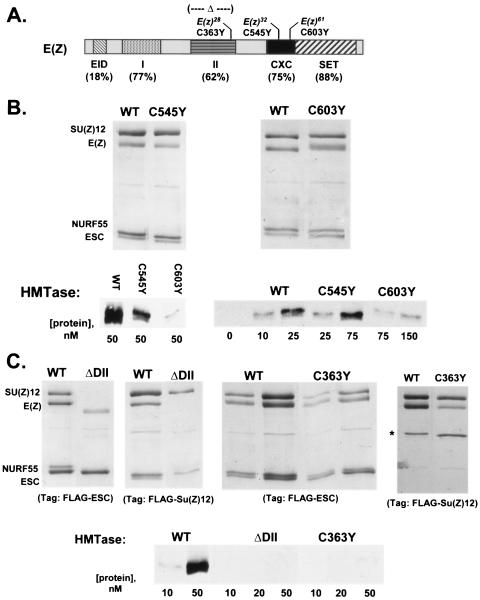
Effects of E(Z) mutations on assembly and HMTase activity of ESC-E(Z) complexes. (A) Domain organization of E(Z) as defined by comparison to mammalian homologs (19, 28). Percent identities between fly E(Z) and human EZH2 are shown for each domain. EID represents the ESC-interacting domain (18, 58). Mutations analyzed in this study are indicated. WT, wild type. (B) Assembly (top) and HMTase activities (bottom) of complexes containing E(Z) CXC domain mutations, purified using FLAG-SU(Z)12. (C) Assembly (top) and HMTase activities (bottom) of complexes containing E(Z) domain II mutations. For Δdomain II analysis (top left), subcomplexes obtained using FLAG-ESC or FLAG-SU(Z)12 are shown. For C363Y analysis (top right), either the four-subunit complex with FLAG-ESC or just pairwise binding to FLAG-SU(Z)12 is shown. The second lane of each pair shows twice as much material loaded. HMTase assays (bottom) were performed on subcomplexes obtained with FLAG-ESC.
To address contributions of the CXC domain, we analyzed two missense mutations, C545Y and C603Y, for effects upon assembly and HMTase activity of the recombinant ESC-E(Z) complex. These mutations correspond to E(z) loss-of-function alleles, E(z)32 and E(z)61, respectively (6). Figure 3B shows that both CXC mutant forms of E(Z) assemble into the four-subunit complex comparably to the wild type. However, both mutant complexes show reduced levels of HMTase activity. Based upon the concentration ranges employed (Fig. 3B, bottom right), we estimate that the C545Y complex is reduced about 3-fold and the C603Y complex shows a more dramatic loss of at least 8- to 10-fold. Thus, one role of the CXC domain is to augment HMTase activity of E(Z).
To analyze function of domain II, we tested mutant forms of E(Z) bearing either an in-frame deletion of domain II (ΔII) or the C363Y missense mutation, which corresponds to the E(z)28 loss-of-function allele (6). Western blots showed that the E(Z)ΔII protein is stably expressed in Sf9 cells after baculovirus infection (data not shown). Assembly of E(Z)ΔII into ESC-E(Z) complexes was assessed independently using FLAG tags on either ESC or SU(Z)12. Purification with the former yielded a FLAG-ESC/E(Z)ΔII dimer, whereas purification with the latter yielded a FLAG-SU(Z)12/NURF-55 dimer (Fig. 3C, left). Both results suggest that domain II is required for stable binding of E(Z) to SU(Z)12.
A more subtle assembly defect is observed with the C363Y mutant. Figure 3C (middle panel) shows that four-subunit complexes are obtained with E(Z)C363Y; however, the subunit ratios appeared slightly altered, relative to the wild type prepared in parallel, suggesting that the mutant complexes are not as stable. Specifically, FLAG-ESC appeared slightly overrepresented compared to other subunits. Given that complete deletion of domain II disrupts binding to SU(Z)12, we tested for direct pairwise binding of E(Z)C363Y to FLAG-SU(Z)12. Figure 3C (right panel) shows that some dimer is recovered but that SU(Z)12 binding to E(Z)C363Y is less efficient than that to the wild type. Although assembly of the four-subunit complex appears only partially affected, we find that C363Y-containing complexes nevertheless show a dramatic reduction in HMTase activity (Fig. 3C, bottom).
Mutations in ESC.
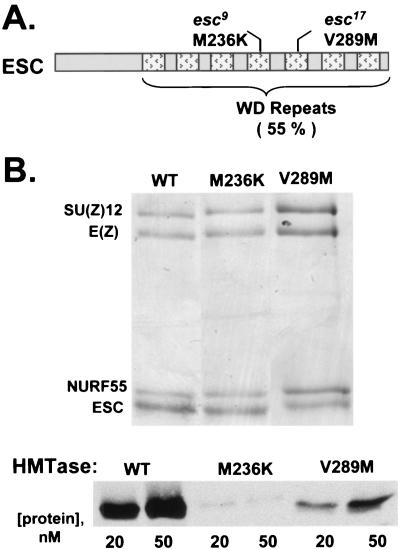
The domain organization of ESC is depicted in Fig. 4A. Most of ESC is occupied by seven WD repeats which together fold into a β-propeller (37, 58). This structure provides a scaffold for protein interactions, and several of the surface loops that emanate from the propeller core have been implicated in binding to E(Z) (18, 58). Aside from the highly conserved WD repeat region, there is a relatively unconserved N-terminal tail of about 70 amino acids.
FIG. 4.
Effects of ESC mutations on assembly and HMTase activity of ESC-E(Z) complexes. (A) Domain organization of ESC (37, 58). Percent identity between fly ESC and human EED is shown for the WD repeat region. Mutations analyzed in this study are indicated. WT, wild type. (B) Assembly (top) and HMTase activities (bottom) of complexes containing indicated ESC mutations. Complexes were purified using FLAG-ESC bearing the mutations.
Several of the characterized esc mutant alleles are missense mutations that alter surface loops in close proximity on one side of the β-propeller. We chose to analyze M236K (esc9) and V289M (esc17) in the context of recombinant ESC-E(Z) complexes, since they preserve the structure of the ESC β-propeller and maintain binding to E(Z) in vitro (58). Thus, they seemed most likely to create ESC loss of function without simply destabilizing the protein or eliminating complex assembly. Indeed, we find that complexes harboring either mutant form of ESC are able to assemble comparably to the wild type (Fig. 4B). However, both mutants cause a significant reduction of HMTase activity (Fig. 4B, bottom). V289M appears to be at least fivefold reduced compared to the wild type, and M236K shows an even more dramatic loss of activity. These results suggest that these mutations produce loss of function in vivo by compromising enzyme activity rather than the stability of ESC-E(Z) complexes.
Mutational analysis of SU(Z)12 domains.
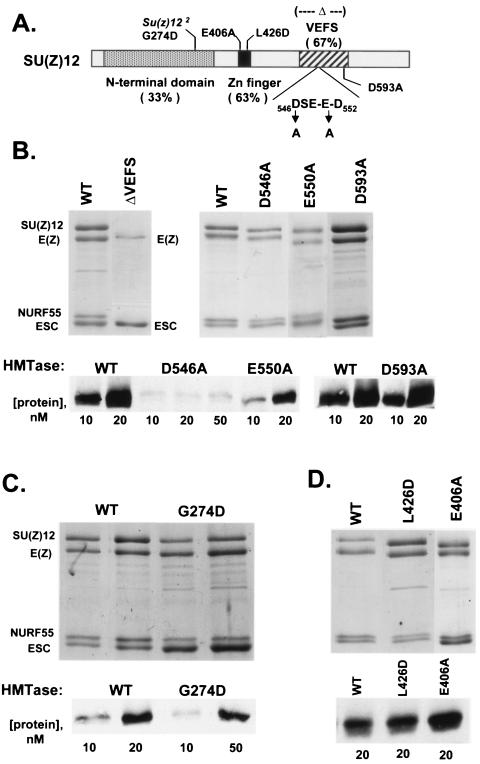
Based upon sequence similarity among fly, plant, and mammalian homologs, two putative functional domains were identified in SU(Z)12 (2). There is a C-terminal domain of about 80 amino acids, termed the VEFS domain, plus a single Cys2-His2 zinc finger (Fig. 5A). The zinc finger appears insufficient for DNA binding (2). The molecular roles of these domains are not known. Besides these two domains, there is also high sequence similarity throughout the N-terminal halves of fly and human SU(Z)12 (2). The sole characterized Su(z)12 missense allele is located in this region (Fig. 5A).
FIG. 5.
Effects of SU(Z)12 mutations on assembly and HMTase activity of ESC-E(Z) complexes. (A) Domain organization of SU(Z)12 as defined by comparison to its plant and mammalian homologs (2). Percent identities between fly and human SU(Z)12 are shown for each domain. Mutations analyzed in this study are indicated. (B) Assembly (top) and HMTase activities (bottom) of complexes containing SU(Z)12 VEFS domain mutations. WT, wild type. (C) Assembly (top) and HMTase activities (bottom) of complexes containing the SU(Z)12-G274D mutation. The second lane of each pair shows twice as much material loaded. (D) Assembly (top) and HMTase activities (bottom) of complexes containing SU(Z)12 zinc finger mutations. All complexes shown in this figure were purified via FLAG-ESC.
To address roles of the VEFS homology region, we first analyzed an in-frame deletion of the entire domain. Western blots showed that this shortened version of SU(Z)12 accumulates stably in the Sf9 cells used for complex production (data not shown). Figure 5B shows that a FLAG-ESC/E(Z) dimer is obtained upon coexpression of SU(Z)12ΔVEFS with the other three subunits. This result is similar to that obtained when SU(Z)12 is omitted from the expression mix (Fig. 1A) and implies that the VEFS domain is required for stable binding of SU(Z)12 to E(Z).
We next analyzed site-directed missense mutations of conserved residues within the VEFS domain. Two of these, D546A and E550A, reside within a negatively charged DSE-E-D motif located in the N-terminal half of the domain (Fig. 5A). The third mutation, D593A, targets a charged residue in the C-terminal half. Figure 5B shows that all three of these SU(Z)12 mutants support assembly of four-subunit ESC-E(Z) complexes. Thus, while loss of the entire VEFS domain disrupts E(Z) contact, apparently none of the three single VEFS changes has that effect. Intriguingly, the three VEFS mutations differ widely in their effects upon HMTase activity: D593A retains wild-type levels of activity, E550A is modestly reduced, and D546A shows at least a 10-fold reduction (Fig. 5B). These results suggest that the DSE-E-D motif of the VEFS domain plays a role in potentiating E(Z) HMTase activity.
The G274D mutation is a Su(z)12 loss-of-function allele that retains some function in vivo (2). Given this genetic behavior, we investigated if there was a discernible defect in G274D recombinant complexes. Figure 5C (left panel) shows that there is a modest assembly defect; although G274D four-subunit complexes are obtained, the stoichiometry appears altered, with ESC and E(Z) overrepresented compared to the ratios in a parallel wild-type control. The simplest interpretation is that G274D binding to E(Z) is somewhat compromised. Similar in degree to the assembly defect, a modest reduction in HMTase activity is also observed with G274D (Fig. 5C, bottom). Thus, the G274D partial loss of function in vivo correlates with compromised assembly and reduced HMTase activity in vitro.
To address possible roles of the zinc finger, we generated two missense changes at conserved positions outside of the two C and two H residues. Both mutations, L426D and E406A, produce charge alterations in the zinc finger region in the hopes of compromising the finger without disrupting its overall structure, which presumably requires the Cys2-His2 backbone. Figure 5D shows that both missense mutants are comparable to the wild type in complex assembly and HMTase activity. Thus, either these mutations are inadequate to compromise domain function or the role of the zinc finger is not revealed by these in vitro assays. For example, the SU(Z)12 zinc finger could be needed to specifically target the ESC-E(Z) complex to PREs in vivo.
Requirements for subunits in the worm MES complex.
We initially determined whether the three identified subunits of the worm MES complex (depicted in Fig. 6A) are sufficient to assemble into a stable complex with robust HMTase activity. Additional subunits could be required since the MES complex, though well characterized (1, 68), has not been extensively purified from worm extracts. Baculovirus constructs were generated to express each MES protein in either untagged form or with a FLAG tag placed at the extreme N terminus. Figure 6B shows complexes purified independently using FLAG tags on MES-6 (left) or on MES-3 (right). A stable trimeric complex was obtained in both cases. In the case of the FLAG-MES-6 complex, untagged MES-2 and MES-3 comigrate as a broad doublet, which was confirmed by Western blot (not shown). The addition of a FLAG tag to MES-3 alters its mobility so that all three subunits are resolved in this version of the complex (right panel). Both of these wild-type MES complexes have robust HMTase activity (Fig. 6B, bottom). To confirm that this activity is intrinsic to recombinant purified complex, a mutant complex bearing an H698A mutation in MES-2 was tested in parallel. This histidine residue (underlined) is in the highly conserved (H/R)XXNHS motif of the SET domain and is required for HMTase activity in E(Z) and other SET domain proteins (34, 43, 55, 66). Figure 6B shows that the H698A complex assembles normally but lacks detectable HMTase activity.
The worm MES complex, like the corresponding fly complex, targets H3-K27 in vivo (1). However, there is also evidence that the fly and worm complexes differ with respect to substrate specificities; the fly complex can methylate either nucleosomes or free histones (34), whereas the worm complex immunoprecipitated from embryo extracts can methylate nucleosomes but not free histones (1). To determine if the recombinant worm complex shares this specificity, we compared its HMTase activities on both types of substrate (Fig. 6C). Although the recombinant worm and fly complexes show comparable HMTase levels on polynucleosomes, we find that the worm complex cannot methylate free histones.
To determine how the worm complex is built and to define contributions of the subunits to HMTase function, we performed subunit dropout experiments similar to our analysis of the fly complex (Fig. 1). Experiments using FLAG-MES-6 for purification are shown in the left panel of Fig. 6D. If MES-3 is omitted from the trimeric mix, then a stable FLAG-MES-6/MES-2 dimer is obtained (middle lane) which lacks HMTase activity. If MES-2 is omitted, then no complex forms and only FLAG-MES-6 is purified (lane 3). The right panel shows proteins purified using FLAG-MES-3. If MES-6 is omitted, then a FLAG-MES-3/MES-2 dimer is recovered (middle lane), which lacks HMTase activity. In agreement with these results, we find that the MES-2 SET domain subunit alone lacks HMTase activity (Fig. 6D, rightmost lane). We conclude that both MES-3 and MES-6 are required to partner with MES-2 to produce robust HMTase activity. Furthermore, these results imply a model for the worm complex where MES-2 is located centrally and forms independent stable contacts with MES-3 and MES-6 (Fig. 6A). Thus, the ∼255-kDa MES complex detected in worm embryos (68), which is close in size to the sum of the three MES proteins (∼230 kDa), likely represents an enzymatically active complex consisting solely of these components.
DISCUSSION
Critical role for ESC.
Our in vitro and in vivo data indicate that the noncatalytic ESC subunit makes a critical contribution to HMTase function of the ESC-E(Z) complex (Fig. 1, 2, and 4). In particular, since global levels of H3-K27 methylation are similarly reduced by genetic loss of ESC or E(Z) (Fig. 2), ESC appears to be an obligate functional partner for E(Z) HMTase activity.
We can envision two main molecular explanations for the ESC requirement. First, ESC could potentiate HMTase activity through direct interaction with E(Z). ESC binding could trigger a conformational change in E(Z) that improves catalytic efficiency, and/or ESC residues could directly interact with and influence the E(Z) active site. Alternatively, the main role of ESC could be to bind nucleosomes. In this scenario, ESC would boost HMTase activity by facilitating interaction of the enzyme complex with its substrate. Based on several lines of evidence, we favor a mechanism that works through direct ESC-E(Z) contact. First, the ESC M236K and V289M mutations, which significantly reduce HMTase activity (Fig. 4), are located in surface loops previously shown to mediate direct ESC contact with E(Z) (18, 58). Furthermore, M236K displays dominant-negative properties in vivo (58). This genetic behavior is consistent with an enzyme complex that assembles normally (Fig. 4) but is compromised in catalytic function. Second, a recent report documents that ESC lacks nucleosome-binding activity on its own and that addition of ESC to a trimeric NURF-55/SU(Z)12/E(Z) complex has little additive effect on ability to bind nucleosomes (35). Finally, ESC potentiation through direct E(Z) binding is supported by evolutionary considerations. Every organism examined that has an E(Z) homolog, ranging from plants to worms, flies, and humans, has at least one ESC homolog. In addition, 28 residues within the ESC surface loops implicated in E(Z) binding are identical from flies to humans (37, 49). This conservation may reflect a tight functional requirement wherein direct ESC-E(Z) partnership, combining to produce HMTase activity, is maintained by evolutionary pressure. Future studies will be needed to define the precise biochemical mechanism by which ESC potentiates HMTase activity, including tests for binding-induced conformational changes in E(Z).
Studies on the mammalian homolog of ESC, called EED, have also highlighted its important role as a regulatory subunit. In human cells, multiple EED isoforms are expressed, which differ in the extents of their N-terminal tails (26). These isoforms are generated by alternative start codon usage of the same EED mRNA. Intriguingly, incorporation of particular EED isoforms into EZH2 complexes can shift the enzyme specificity so that K26 of histone H1 is methylated in addition to H3-K27 (26). Thus, it appears that the ESC/EED subunit can influence both catalytic efficiency and lysine substrate preference. In the fly system, ESC isoforms produced from the same mRNA have not been detected. Instead, alternative ESC isoforms could be supplied by an esc-related gene (CG5202) located about 150 kb proximal to esc. Since mutations in this second esc gene, called esc-like (escl), have not yet been reported, its in vivo contributions remain to be assessed. However, since genetic loss of ESC alone dramatically reduces global methylation of H3-K27 in fly embryos (Fig. 2), we conclude that ESC is the predominant functional E(Z) partner during embryonic stages.
Functional contribution of SU(Z)12 and its conserved VEFS domain.
Our studies on recombinant complexes show that fly SU(Z)12 is absolutely required for HMTase activity of the ESC-E(Z) complex (Fig. 1 and 5). A key requirement for SU(Z)12 in mammalian EZH2 complexes has also been established based upon in vitro tests and loss-of-function studies in vivo (5, 22, 39). How does SU(Z)12 contribute molecularly to HMTase activity? Again, we can envision two main possibilities: influence through direct contact with E(Z) or by mediating nucleosome binding. To address this, it is instructive to consider our SU(Z)12 mutants affecting the conserved VEFS domain (Fig. 5B). Deletion of the entire VEFS domain eliminates assembly of the fly complex by disrupting SU(Z)12-E(Z) binding. Pairwise binding assays with mammalian SU(Z)12 have similarly shown that the VEFS domain is needed for binding to EZH2 in vitro (69). Thus, a conserved function of this domain is to contact E(Z). However, we also have missense mutations within the VEFS domain, D546A and E550A, which preserve full complex assembly yet have reduced levels of HMTase (Fig. 5B). Taken together, these results implicate the VEFS domain in both binding to E(Z) and potentiating its enzyme activity, which suggests a connection between these two functions. On the other hand, a recent report provides evidence that SU(Z)12 contributes to affinity for nucleosomes (35). Although SU(Z)12 cannot bind to nucleosomes by itself, the SU(Z)12/NURF-55 dimer has nucleosome-binding properties that are similar to those of the four-subunit complex. Thus, as also suggested for the human PRC2 complex (5), at least one role of SU(Z)12 is to mediate nucleosome binding. Further work will be needed to define the SU(Z)12 functional domains required for interactions with NURF-55 and with nucleosomes. Based on the available data, SU(Z)12 potentiation of E(Z) HMTase activity may involve both direct E(Z) contact and facilitated binding to nucleosome substrate.
Requirements for E(Z) domains besides the SET domain.
Although the SET domain is the most well-characterized functional domain of E(Z), the adjacent cysteine-rich CXC domain is also remarkably conserved from flies to humans (28) (Fig. 3A). To address CXC function, we analyzed in vitro properties of two missense mutants, C545Y and C603Y. Both mutations correspond to E(z) loss-of-function alleles; in particular, in vivo effects of the E(z)61 mutation (C603Y) have been well documented. This mutation disrupts global H3-K27 methylation in embryos and causes loss of methyl-H3-K27 from a Hox target gene in imaginal disks (3, 67) (Fig. 2). We find that mutant complexes bearing E(Z)-C603Y can assemble normally but show an approximately 10-fold reduction in HMTase levels (Fig. 3B). C545Y causes a more modest HMTase reduction, which parallels results obtained with the analogous substitution (C588Y) in human EZH2 (27). These results suggest that the CXC domain interfaces with the SET domain to produce robust HMTase activity. In this regard, the CXC domain could be considered similar to cysteine-rich “preSET” domains required for robust HMTase activity in other SET domain proteins (43, 70). Another effect of these CXC mutations in vivo is that they dislodge E(Z) from target sites in chromatin (3, 6, 67). Although the molecular basis for this dissociation is not known, our in vitro assembly results (Fig. 3B) suggest that it is not due to wholesale destabilization of the ESC-E(Z) complex. The dissociation may reflect another proposed role for the CXC domain, which is to interact with the PcG targeting factor PHO (67).
We also performed in vitro tests to investigate the role of E(Z) domain II (Fig. 3C). Both the complete domain deletion and the C363Y missense mutation show that domain II is required for stable association of E(Z) with SU(Z)12. Thus, the composite domain organization of E(Z) (Fig. 3A) reflects division of labor among catalytic functions and requirements for complex assembly. In addition, it appears that none of the E(Z) domains are specifically built for nucleosome interactions, as E(Z) plays little or no role itself in stable binding of the complex to nucleosomes (35).
Possible roles for NURF-55.
The NURF-55 subunit is distinct from the other three subunits in several ways. First, it makes only minimal contributions to in vitro HMTase activity in both the fly (Fig. 1) and mammalian (5) complexes. Second, whereas the other three subunits appear dedicated to PcG function, NURF-55 is present in diverse chromatin-modifying complexes, including NURF, chromatin assembly factor 1 (CAF-1), and histone deacetylase complexes (30, 57, 61, 65). The ability of the mammalian NURF-55 homologs RbAp46 and RbAp48 to bind to free histone H4 (64) has led to the suggestion that NURF-55 may help chromatin complexes interact with substrate. Indeed, the absence of a NURF-55-related protein from the trimeric worm MES complex could help explain its inability to methylate free histones (Fig. 6C). The free histone-binding property of NURF-55 has also prompted the intriguing suggestion that silencing by the ESC-E(Z) complex in vivo could involve methylation of histones prior to nucleosome assembly (63). Since NURF-55 loss-of-function alleles have not been described in flies, many questions about roles of NURF-55 remain to be addressed. Even with alleles available, the multiplicity of NURF-55-containing complexes will likely complicate in vivo dissection of its PcG functions.
Divergence of worm and fly PcG complexes.
The basic enzymatic function of the ESC-E(Z) complex, to methylate H3-K27, is shared between the worm, fly, and mammalian versions. Another similarity revealed from our study of recombinant worm complexes is that robust HMTase activity depends critically upon the two noncatalytic subunits. Although MES-6 and MES-3 can each individually bind to the catalytic subunit, MES-2, all three subunits are required together to produce enzyme activity (Fig. 6). Since worm MES-6 is a WD repeat protein related to fly ESC (25), it seems likely that MES-6 and ESC potentiate HMTase activity through similar mechanisms. As discussed above, we suggest this mechanism entails direct subunit interactions rather than an influence upon affinity for nucleosomes. However, a major puzzle is presented by the dissimilarity between worm MES-3 and fly SU(Z)12. Though each is required for HMTase activity in their respective complexes, we are unable to recognize any relatedness between these two proteins in primary sequence or predicted secondary structure arrangement. From an evolutionary standpoint, it appears that SU(Z)12 represents the more ancient partner, as it is functionally conserved across plant and animal kingdoms (2, 14, 22). MES-3, a novel protein, may have evolved more recently to replace SU(Z)12 in the worm complex. Since a molecular role attributed to SU(Z)12 in the fly complex is nucleosome binding (35), we speculate that MES-3 may supply this function for the worm complex. There are many strategies for building nucleosome contact domains, as represented among divergent chromatin proteins, so MES-3 could have acquired functional similarity without overt sequence similarity to SU(Z)12. In this view, MES-3 function in the worm complex would require, at minimum, affinity for nucleosomes and ability to bind MES-2. In this regard, it is interesting that MES-2 appears to lack domain II, which is needed in E(Z) for stable binding to SU(Z)12 (Fig. 3C). Presumably, MES-2 has instead acquired a site for stable MES-3 interaction (Fig. 6). In summary, we suggest that the E(Z)/ESC and MES-2/MES-6 dimers have been conserved as core subunits of the HMTase complex, whereas the additional required partners in each complex, SU(Z)12 and MES-3, have been allowed to diverge. Future studies will be needed, including functional tests of chimeric worm and fly proteins, to address such a model.
Acknowledgments
We thank Nicole Francis and Dan Mallin for advice and help with Sf9 cell culture and the baculovirus system. We are grateful to Julio Herrera for advice on preparation of polynucleosomes and to Thomas Jenuwein for providing antibodies. We also thank Ellen Miller and Sarah Malmquist for help in generating several of the mutant constructs used in this work and Wenchao Wang for providing Gateway plasmid constructs.
This work was supported by National Institutes of Health (NIH) grants GM49850 to J.A.S. and GM34059 to S.S. C.S.K. was supported in part by NIH training grant HD07480.
REFERENCES
- 1.Bender, L. B., R. Cao, Y. Zhang, and S. Strome. 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14:1639-1643. [DOI] [PubMed] [Google Scholar]
- 2.Birve, A., A. K. Sengupta, D. Beuchle, J. Larsson, J. A. Kennison, A. Rasmuson-Lestander, and J. Muller. 2001. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128:3371-3379. [DOI] [PubMed] [Google Scholar]
- 3.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 4.Cao, R., and Y. Zhang. 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14:155-164. [DOI] [PubMed] [Google Scholar]
- 5.Cao, R., and Y. Zhang. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15:57-67. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, E. A., and R. S. Jones. 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development 122:4073-4083. [DOI] [PubMed] [Google Scholar]
- 7.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 8.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 10.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 11.Franke, A., M. DeCamillis, D. Zink, N. Cheng, H. W. Brock, and R. Paro. 1992. Polycomb and Polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 11:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frei, E., D. Bopp, M. Burri, S. Baumgartner, J. E. Edström, and M. Noll. 1985. Isolation and structural analysis of the extra sex combs gene of Drosophila. Cold Spring Harbor Symp. Quant. Biol. 50:127-134. [DOI] [PubMed] [Google Scholar]
- 13.Garvin, C., R. Holdeman, and S. Strome. 1998. The phenotype of mes-2, mes-3, mes-4 and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics 148:167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendall, A. R., Y. Y. Levy, A. Wilson, and C. Dean. 2001. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107:525-535. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich, J., P. Puangsomlee, M. Martin, D. Long, E. M. Meyerowitz, and G. Coupland. 1997. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386:44-51. [DOI] [PubMed] [Google Scholar]
- 16.Herrera, J. E., K. L. West, R. L. Schiltz, Y. Nakatani, and M. Bustin. 2000. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol. Cell. Biol. 20:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holdeman, R., S. Nehrt, and S. Strome. 1998. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development 125:2457-2467. [DOI] [PubMed] [Google Scholar]
- 18.Jones, C. A., J. Ng, A. J. Peterson, K. Morgan, J. Simon, and R. S. Jones. 1998. The Drosophila esc and E(z) proteins are direct partners in Polycomb group-mediated repression. Mol. Cell. Biol. 18:2825-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, R. S., and W. M. Gelbart. 1993. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol. Cell. Biol. 13:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, R. S., and W. M. Gelbart. 1990. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126:185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, W. G., and A. Fire. 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125:2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18:1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleer, C. G., Q. Cao, S. Varambally, R. Shen, I. Ota, S. A. Tomlins, D. Ghosh, R. G. Sewalt, A. P. Otte, D. F. Hayes, M. S. Sabel, D. Livant, S. J. Weiss, M. A. Rubin, and A. M. Chinnaiyan. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 100:11606-11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohler, C., and U. Grossniklaus. 2002. Epigenetics: the flowers that come in from the cold. Curr. Biol. 12:R129-R131. [DOI] [PubMed] [Google Scholar]
- 25.Korf, I., Y. Fan, and S. Strome. 1998. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development 125:2469-2478. [DOI] [PubMed] [Google Scholar]
- 26.Kuzmichev, A., T. Jenuwein, P. Tempst, and D. Reinberg. 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14:183-193. [DOI] [PubMed] [Google Scholar]
- 27.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laible, G., A. Wolf, R. Dorn, G. Reuter, C. Nislow, A. Lebersorger, D. Popkin, L. Pillus, and T. Jenuwein. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, M., P. Bilodeau, E. S. Dennis, W. J. Peacock, and A. Chaudhury. 2000. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97:10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Balbas, M. A., T. Tsukiyama, D. Gdula, and C. Wu. 1998. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. USA 95:132-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurange, C., and R. Paro. 2002. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 16:2672-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min, J., Y. Zhang, and R. M. Xu. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moazed, D., and P. H. O'Farrell. 1992. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development 116:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 35.Nekrasov, M., B. Wild, and J. Muller. 2005. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 6:348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, J., C. M. Hart, K. Morgan, and J. A. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, J., R. Li, K. Morgan, and J. Simon. 1997. Evolutionary conservation and predicted structure of the Drosophila extra sex combs repressor protein. Mol. Cell. Biol. 17:6663-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell, S., L. Wang, S. Robert, C. A. Jones, R. Saint, and R. S. Jones. 2001. Polycomblike PHD fingers mediate conserved interaction with Enhancer of zeste protein. J. Biol. Chem. 276:43065-43073. [DOI] [PubMed] [Google Scholar]
- 39.Pasini, D., A. P. Bracken, M. R. Jensen, E. L. Denchi, and K. Helin. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23:4061-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577-1589. [DOI] [PubMed] [Google Scholar]
- 41.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 42.Rastelli, L., C. S. Chan, and V. Pirrotta. 1993. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 12:1513-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 44.Ringrose, L., and R. Paro. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38:413-443. [DOI] [PubMed] [Google Scholar]
- 45.Ringrose, L., M. Rehmsmeier, J. M. Dura, and R. Paro. 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell 5:759-771. [DOI] [PubMed] [Google Scholar]
- 46.Ross, J. M., and D. Zarkower. 2003. Polycomb group regulation of Hox gene expression in C. elegans. Dev. Cell 4:891-901. [DOI] [PubMed] [Google Scholar]
- 47.Sathe, S. S., and P. J. Harte. 1995. The Drosophila extra sex combs protein contains WD motifs essential for its function as a repressor of homeotic genes. Mech. Dev. 52:77-87. [DOI] [PubMed] [Google Scholar]
- 48.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 49.Schumacher, A., O. Lichtarge, S. Schwartz, and T. Magnuson. 1998. The murine Polycomb-group gene eed and its human orthologue: functional implications of evolutionary conservation. Genomics 54:79-88. [DOI] [PubMed] [Google Scholar]
- 50.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 51.Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4:481-495. [DOI] [PubMed] [Google Scholar]
- 52.Simon, J. A. 2003. Polycomb group proteins. Curr. Biol. 13:R79-R80. [DOI] [PubMed] [Google Scholar]
- 53.Simon, J. A., and J. W. Tamkun. 2002. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12:210-218. [DOI] [PubMed] [Google Scholar]
- 54.Spillane, C., C. MacDougall, C. Stock, C. Kohler, J. P. Vielle-Calzada, S. M. Nunes, U. Grossniklaus, and J. Goodrich. 2000. Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 10:1535-1538. [DOI] [PubMed] [Google Scholar]
- 55.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Struhl, G. 1981. A gene product required for correct initiation of segmental determination in Drosophila. Nature 293:36-41. [DOI] [PubMed] [Google Scholar]
- 57.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 58.Tie, F., T. Furuyama, and P. J. Harte. 1998. The Drosophila Polycomb Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 125:3483-3496. [DOI] [PubMed] [Google Scholar]
- 59.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 60.Tie, F., J. Prasad-Sinha, A. Birve, A. Rasmuson-Lestander, and P. J. Harte. 2003. A 1-megadalton ESC/E(Z) complex from Drosophila that contains Polycomblike and RPD3. Mol. Cell. Biol. 23:3352-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyler, J. K., M. Bulger, R. T. Kamakaka, R. Kobayashi, and J. T. Kadonaga. 1996. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 16:6149-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varambally, S., S. M. Dhanasekaran, M. Zhou, T. R. Barrette, C. Kumar-Sinha, M. G. Sanda, D. Ghosh, K. J. Pienta, R. G. Sewalt, A. P. Otte, M. A. Rubin, and A. M. Chinnaiyan. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624-629. [DOI] [PubMed] [Google Scholar]
- 63.Vermaak, D., K. Ahmad, and S. Henikoff. 2003. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 15:266-274. [DOI] [PubMed] [Google Scholar]
- 64.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1997. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8:96-108. [DOI] [PubMed] [Google Scholar]
- 65.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 66.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 67.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]
- 68.Xu, L., Y. Fong, and S. Strome. 2001. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc. Natl. Acad. Sci. USA 98:5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto, K., M. Sonoda, J. Inokuchi, S. Shirasawa, and T. Sasazuki. 2004. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J. Biol. Chem. 279:401-406. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]