Abstract
WW domains are protein modules that mediate protein-protein interactions through recognition of proline-rich peptide motifs and phosphorylated serine/threonine-proline sites. To pursue the functional properties of WW domains, we employed mass spectrometry to identify 148 proteins that associate with 10 human WW domains. Many of these proteins represent novel WW domain-binding partners and are components of multiprotein complexes involved in molecular processes, such as transcription, RNA processing, and cytoskeletal regulation. We validated one complex in detail, showing that WW domains of the AIP4 E3 protein-ubiquitin ligase bind directly to a PPXY motif in the p68 subunit of pre-mRNA cleavage and polyadenylation factor Im in a manner that promotes p68 ubiquitylation. The tested WW domains fall into three broad groups on the basis of hierarchical clustering with respect to their associated proteins; each such cluster of bound proteins displayed a distinct set of WW domain-binding motifs. We also found that separate WW domains from the same protein or closely related proteins can have different specificities for protein ligands and also demonstrated that a single polypeptide can bind multiple classes of WW domains through separate proline-rich motifs. These data suggest that WW domains provide a versatile platform to link individual proteins into physiologically important networks.
Many signaling proteins contain modular domains that mediate specific protein-protein interactions, frequently through the recognition of short peptide motifs in their binding partners (56). In many cases these interactions are regulated by posttranslational modifications, such as phosphorylation. Interaction domains can thereby control the subcellular localization, enzymatic activity, and substrate specificity of regulatory proteins and the assembly of multiprotein complexes, and thus the flow of information through signaling pathways.
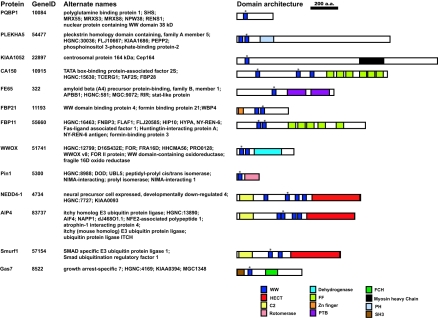
WW domains comprise a family of protein-protein interaction modules that are found in many eukaryotes and are present in approximately 50 human proteins (6; see Fig. 1). Within these polypeptides, WW domains are joined to a number of distinct interaction modules, including phosphotyrosine-binding domains (i.e., in the FE65 protein) and FF domains (CA150 and FBP11), as well as protein localization domains, such as C2 (NEDD4 family proteins) and pleckstrin homology domains (PLEKHA5). WW domains are also linked to a variety of catalytic domains, including HECT E3 protein-ubiquitin ligase domains (in NEDD4 family proteins), rotomerase/peptidyl prolyisomerase domains (Pin1), and Rho GTPase-activating protein domains. Consequently, WW domain-containing proteins are involved in a variety of cellular processes, including transcription, RNA processing, protein trafficking, receptor signaling, and control of the cytoskeleton (32, 33, 68). WW domain-mediated interactions have been implicated in cancer (4, 75), in hereditary disorders, such as Liddle's syndrome (66) and Rett's syndrome (8), as well as in Alzheimer's (46, 48) and Huntington's (20, 55) diseases.
FIG. 1.
Domain organization and alternate names of WW domain-containing proteins analyzed in this study. WW domain-containing proteins examined in this study, their unique GeneIDs, as well as alternate names are indicated. The relative domain organization of each of the proteins was determined by searching the National Center for Biotechnology Information conserved domain database (CDD) (51) and is indicated. WW domains specifically analyzed in this study are indicated with an asterisk. a.a., amino acids; PTB, phosphotyrosine binding; PH, pleckstrin homology; SH3, Src homology 3.
WW domains are typically 35 to 40 amino acids in length (6) and fold into a three-stranded, antiparallel β sheet with two ligand-binding grooves (30, 38, 50, 72). WW domains bind a variety of distinct peptide ligands including motifs with core proline-rich sequences, such as PPXY (amino acid single-letter code; X is any amino acid) (PY) (14), PPLP (2, 18), as well as proline/arginine-containing (PR) sequences (3) and phosphorylated serine/threonine-proline sites [p(S/T)P] (49, 76). WW domains have been classified into four groups on the basis of their binding to peptide ligands (3, 19, 67). Group I WW domains, which include the NEDD4 family proteins and the 65-kDa Yes-associated protein (YAP65), recognize PY motifs (14, 66). Group II WW domains, such as the FE65 WW domain and the first WW domain of FBP11, recognize PPLP motifs (2, 18), and group III WW domains (FBP30 WW1) recognize PR motifs (3). Group IV WW domains, such as those from Pin1 (49) and PDX-1 C-terminus-interacting factor 1 (21), recognize p(S/T)P motifs (76). Group II and III WW domains can be rather versatile in their binding properties, since they not only recognize both PPLP- and PR-containing peptides (with varied affinities) but also polyproline stretches often containing glycine, methionine, or arginine (29, 39, 40, 54), and therefore could be viewed as a single set.
A number of groups have identified WW domain-binding proteins through screening cDNA expression libraries and yeast two-hybrid analysis (2, 14, 18, 36, 53). These studies, coupled with probing of peptide libraries, have suggested that consensus sequences are recognized by individual WW domains (29, 54). To obtain a more comprehensive view of the functions of WW domain-containing proteins, as well as the ability of individual WW domains to recognize cellular proteins, we have used tandem mass spectrometry (MS) to identify human polypeptides that associate with a range of WW domains. This approach has several advantages. (i) It is not biased towards previously determined peptide ligands and provides orthogonal results regarding WW domain-binding properties that can be overlaid on peptide-binding data. (ii) It allows the identification of proteins that interact indirectly with WW domains, providing new information regarding the cellular complexes and machinery engaged by WW domains. (iii) It facilitates the identification of phosphorylated ligands for group IV WW domains.
Here, we identify 148 T-cell proteins that selectively associate with 10 human WW domains. Interestingly, hierarchical clustering of this data set organized the WW domains into three groups on the basis of their protein-binding properties, which correlates with the presence of specific proline-rich or phosphorylated peptide motifs in these associated proteins. The screen identified novel, direct, and biologically relevant binding partners for WW domain-containing proteins, as well as associated proteins that bind WW domains indirectly as components of multiprotein complexes. Finally, we show that WW domains from the same protein or protein families can have distinct binding preferences and that individual proteins can potentially serve as scaffolds to recruit multiple WW domains of different classes to distinct binding sites. These results argue that the reiterated use of a small interaction domain can yield a versatile network of protein interactions that influences many facets of cellular behavior.
MATERIALS AND METHODS
Antibodies and cDNAs.
The polyclonal antisera to AIP4 (74) and p68 and p25 antisera (60) have been previously described. Antibodies were purchased as follows: anti-Diaphanous 1 polyclonal antisera (ImmunoGlobe, Himmelstadt, Germany); anti-FLAG M2 monoclonal antibody (MAb) (Sigma, St. Louis, MO); anti-Myc 9E10 MAb, antibody to the large subunit of RNA polymerase II (anti-RNA Pol II LS), antihemagglutinin (anti-HA), and anti-EWSR1 (anti-EWS) polyclonal antisera (Santa Cruz, Santa Cruz, CA); anti-CA150 polyclonal antisera (Abcam, Cambridge, MA); and anti-Pin1 polyclonal antisera (Upstate Biotechnology, Inc., Upstate, NY). The HA-tagged ubiquitin construct was a gift from Ivan Dikic (Goethe University Medical School, Frankfurt, Germany). The doubly Myc-tagged AIP4 constructs were generated by PCR of our previously published constructs (74) with the amino terminus of the published AIP4 sequence (31) replacing the amino-terminal Itch portion of the fusion used in our earlier study. These constructs were cloned into the pCDNA3.1A expression vector (Invitrogen, San Diego, CA). The Flag-tagged p68 (Flag-p68) and KIAA0144 and the corresponding tyrosine-to-alanine (Y/A) mutation in the PY motif, were generated by PCR and cloned into the pCMVFlac eukaryotic expression vector (Sigma). All PCR products were sequence verified. Production of the His-tagged, recombinant pre-mRNA cleavage and polyadenylation factor Im (CFIm) p68 and p25 has been previously described (16).
Construction of glutathione S-transferase (GST) fusion proteins.
Oligonucleotides for the WW domains including four or five amino acids N and C terminal to the conserved tryptophan residues of each of the WW domains (see Table S1 in the supplemental material) were purchased from Sigma. Complementary oligonucleotides (approximately 2 mM of each) were annealed and cloned into BamHI/EcoRI-cut pGEXKT (Amersham Biosciences AB, Uppsala, Sweden). All constructs were sequence verified. The AIP4 WW domain fusion proteins have been previously described (74).
Purification of GST fusion proteins.
GST fusion proteins were purified from BL21 Escherichia coli bacterial lysate with glutathione-Sepharose beads (Amersham). Fusion protein-containing beads were stored at 4°C as a 10% slurry in phosphate-buffered saline containing 0.02% NaN3. Fusion protein used for SPOTS blotting was eluted from the beads with 50 mM Tris base and 25 mM glutathione (Sigma), and the eluates were dialyzed overnight at 4°C in phosphate-buffered saline.
Cell culture.
Jurkat cell pellets were purchased from the National Cell Culture Center (Minneapolis, MN) where cells were grown in RPMI 1640 medium supplemented with 0.1 mM sodium pyruvate and 10% normal calf serum. 293T cells were grown in Dulbecco modified Eagle medium (Gibco BRL, Cambridge, Ontario, Canada) supplemented with 10% fetal calf serum (Gibco BRL).
GST-WW domain pulldowns for MS analysis.
Jurkat cells were resuspended in 1% NP-40 lysis buffer supplemented with 1 mM NaVO4, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin at 1.0 × 108 cells/ml and incubated on a nutator in the cold for 30 min. Detergent-insoluble material was removed by centrifugation at ∼50,000 × g for 30 min at 4°C. One milliliter of lysate (1.0 × 108 cell equivalents) was then added to 80 μg of GST fusion protein precoupled to glutathione-Sepharose beads and allowed to incubate for 6 h on a nutator at 4°C. The beads were washed four times with 1% NP-40 lysis buffer with inhibitors, and proteins were removed from the beads by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Running and staining of SDS-PAGs.
Samples were run on 10% SDS-PAGs and washed a minimum of three times for 10 min each time with distilled, deionized water before being stained with GelCode colloidal Coomassie blue reagent (MJS BioLynx, Inc., Brockville, Ontario, Canada) overnight at room temperature. Gels were destained the next day with distilled, deionized water.
Excision of bands for MS.
Individual bands were excised using an Investigator ProPic Gel Cutting Robotic workstation (Genomic Solutions, Ann Arbor, MI) and delivered to a single well of a 96-well microtiter plate with prepunched holes at the bottom of each well (Genomic Solutions).
Reduction, alkylation, and tryptic digestion of samples.
The PCR microtiter plate was transferred to a Genomic Solutions ProGest Digestion robot for “in-gel” trypsin digestion where protein bands were washed, reduced with dithiothreitol, alkylated with iodoacetamide, before being digested with sequence-grade, modified trypsin (Promega, Madison, WI) as previously described (28). Tryptic peptides were then extracted from the gel for analysis by MS.
Liquid chromatography-MS (LC-MS).
Tryptic peptides were analyzed by liquid chromatography-tandem MS (LC-MS/MS) using two systems. The first was an Ultimate high-performance liquid chromatography system equipped with a FAMOS 96-well autosampler (LC Packings-Dionex, Sunnyvale, CA) linked to a Q-TRAP mass spectrometer (MDS Sciex, Concord, Ontario, Canada). The second was an HP 1100 high-performance liquid chromatography system (Palo Alto, CA) connected to an LCQ-Deca mass spectrometer (Thermo Electron, San Jose, CA). Peptides were separated on custom-made 75-μm-inner-diameter PicoTip columns packed with 5-μm C-18 beads. The gradient was 3 to 60% of acetonitrile during 25 min with a total run time of 45 min. Data were analyzed in batch using the Mascot search engine (57), and proteins were considered “hits” if two independent peptides or a single peptide with a Mascot score of 50 or higher was found. Protein hits were converted to gene identifiers (GeneIDs) for further analysis. Any “hit” that was not seen in two of three independent experiments or was seen binding to GST alone was eliminated. Also excluded were all actins, myosins, keratins, spectrins, and heat shock proteins. The cellular process for each “hit” was assigned on the basis of published work and GeneOntology information provided by the Gene Ontology Consortium (10). For the identification of phosphopeptides, the data were analyzed using the Spectrum Mill software (Agilent Technologies, Palo Alto, CA). The validity of phosphorylation sites was manually verified by examination of the MS/MS fragmentation spectra.
Transfection of 293T cells, GST pulldowns, and immunoprecipitation experiments.
293T cells were transfected in 10-cm plates using CaPO4 as previously described (42). Transfected cells were lysed in 1 ml 1% NP-40 lysis buffer supplemented with inhibitors (see above) for 10 min on ice. Detergent-insoluble material was removed by spinning at ∼18,000 × g in a microcentrifuge, and the concentration of the cleared lysate was determined by bicinchoninic acid assay (Pierce, Rockford, IL). An equivalent amount of cleared lysate was incubated with 2 μg of the indicated antibodies and either protein A or G-Sepharose or anti-mouse immunoglobulin G-agarose for 1 h on a rocker at 4°C. For GST fusion protein experiments, equivalent amounts of lysate were incubated for 1 h on a rocker at 4°C with ∼8 μg of GST fusion protein preabsorbed on glutathione-Sepharose beads. Immunoprecipitates and GST pulldowns were then washed three times with 1% NP-40 lysis buffer with inhibitors, and bound proteins were eluted by boiling in SDS-PAGE sample buffer.
Western blotting.
Cell lysate, immunoprecipitations, and GST pulldowns were separated on SDS-PAGs, transferred to Polyscreen polyvinylidene fluoride (PVDF) membranes (Perkin-Elmer), and blocked for 1 h in Tris-buffered saline containing 0.5% Tween 20 (TBST) supplemented with 5% skim milk powder. Membranes were probed with the indicated antibodies (1 μg/ml in TBST) as previously described (34), and proteins were visualized using enhanced chemiluminescence reagent. Blots were stripped by incubating the membrane for 1 h in TBST, pH 2, and reprobed as described above.
Hierarchical clustering.
Pearson correlation coefficients were computed to reflect the similarities between the different WW domains with respect to the proteins they precipitated and vice versa. The x and y axes were hierarchically clustered independently using an average linkage method, and the matrix was reorganized to reflect the hierarchically clustered ordering (17). The tree shows the relatedness between items based on the clustering, with items closer together being more related (65).
Ubiquitylation experiments.
For ubiquitylation experiments, transfected cells were treated for 1 h with 50 μM of the proteasome inhibitor MG132 (BACHEM, King of Prussia, PA) prior to lysis. Cells were lysed and lysates processed as indicated above with the exception that 50 μM MG132 and 20 mM N-ethylmaleimide (Sigma) were added to the lysis buffer.
Synthesis and blotting of SPOTS membranes.
SPOTS blots (23) were synthesized, using standard 9-fluorenylmethoxycarbonyl (Fmoc) chemistry, in 30 × 20 spot arrays using a Multipep peptide synthesizer adapted for SPOTS synthesis (Intavis AG, Cologne, Germany). Membranes were blocked for at least 1 h in TBST with 5% skim milk powder before being probed overnight with the indicated GST fusion protein (1 μM in TBST) in the cold. The following day, blots were washed three times for 10 min each time in TBST, incubated with a horseradish peroxidase-conjugated, anti-GST MAb (B14) (Santa Cruz) for 45 min at room temperature, and then washed with TBST three more times for 10 min each time before visualizing SPOTS by exposing the membranes to enhanced chemiluminescence reagent.
RESULTS
Cluster analysis groups WW domains according to their binding partners.
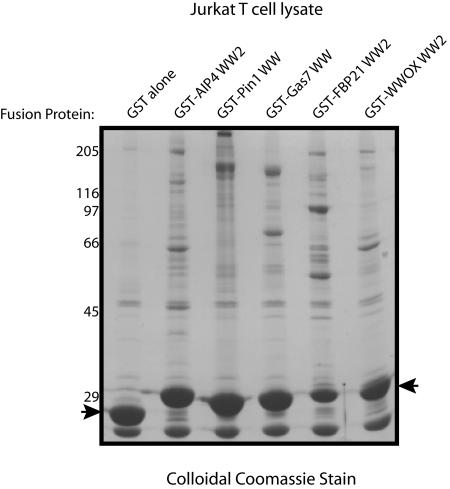
To identify cellular proteins that interact with individual WW domains, we constructed GST fusion proteins of 15 WW domains from 13 proteins containing WW domains (Fig. 1). This set includes WW domains that have different binding preferences for peptide ligands and are derived from proteins with diverse molecular functions. Distinct WW domains from the same protein (FBP21 or CA150) and from proteins with similar domain organizations (i.e., NEDD4-1/AIP4/Smurf1 and CA150/FBP11) were also part of the test set. Fusion proteins were expressed in bacteria, purified, and used to precipitate proteins from Jurkat T-cell lysates as described in Materials and Methods. A representative gel stained with colloidal Coomassie blue shows that the GST-WW domain fusion proteins can precipitate proteins from Jurkat cell lysates not seen with GST alone (Fig. 2). Ten of the tested WW domains associated with specific proteins and showed distinct binding profiles (Fig. 2), whereas five (Smurf1 WW1, CA150 WW1, PQBP1, PLEKHA5, and KIAA1052) did not bind specific proteins in this assay (data not shown), potentially because they require additional sequences either to fold into functional modules or to recognize ligands (15, 29, 30, 50, 54). Proteins that were precipitated by the 10 WW domains were then excised from the relevant gel and analyzed by LC-MS/MS. Precipitations with each GST-WW domain were repeated three times, and only proteins identified in two of three independent experiments with a minimum score of confidence (see Materials and Methods) were included for subsequent analysis. Using these criteria, a total of 148 proteins were identified as being precipitated by one or more of the 10 WW domains but not by GST alone (see Table S2 in the supplemental material). The majority of these proteins represent novel WW domain-associated proteins and are involved in processes such as transcription, RNA processing, and regulation of the cytoskeleton.
FIG. 2.
Purification of WW domain-binding proteins. Proteins were precipitated from Jurkat cell lysate with the indicated fusion proteins as outlined in Materials and Methods. Precipitated proteins were separated on SDS-PAGs, and gels were stained with colloidal Coomassie blue to visualize proteins. One representative gel is shown here. The positions of molecular mass markers (in kilodaltons) are indicated to the left. The positions of bait proteins are indicated by arrows.
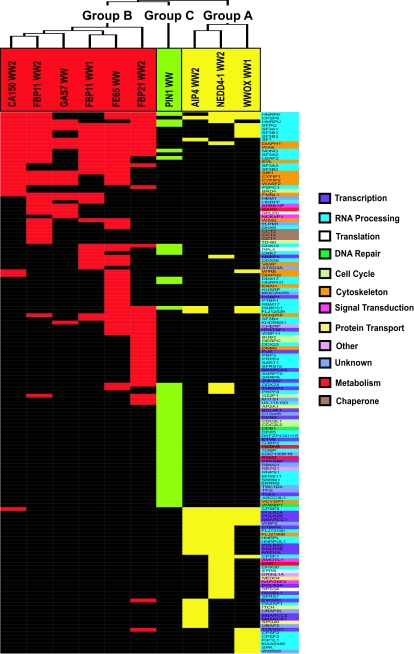
In order to compare the binding properties of the different WW domains, we employed hierarchical clustering to group the WW domains on the basis of the similarity of the proteins they recognized. This clustering separated the WW domains into three classes (Fig. 3). One cluster (group A [shown in yellow]) includes the NEDD4-1 WW2, AIP4 WW2, and WWOX WW1 domains. A second group comprises the FE65 WW, Gas7 WW, FBP21 WW2, FBP11 WW1 and WW2, and CA150 WW2 domains (group B [shown in red]). The WW domain of Pin1 clustered independently of the other WW domains and thus forms its own group (group C [shown in green]). Thus, despite the fact that these WW domain complexes may contain both direct and indirect interaction partners, data based exclusively on the analysis of full-length proteins isolated by WW domains identified three groups of domains, raising the possibility that these might correspond to groups previously distinguished by their selectivity for short peptide motifs.
FIG.3.
Hierarchical clustering of WW domains with precipitated proteins. The GST-WW domains (abscissa) and precipitated proteins (ordinate) were hierarchically clustered as outlined in Materials and Methods. The tree at the top indicates the relatedness of the WW domains with respect to precipitated proteins. This clustering distinguishes three groups of WW domains: group A (AIP4 WW2, NEDD4-1 WW2, and WWOX WW1) in yellow, group B (FBP11 WW1 and WW2, FBP21 WW2, FE65 WW, CA150 WW2, and Gas7 WW) in red, and group C (Pin1 WW) in green. The color of the precipitated protein indicates cellular function and is shown to the right.
Clusters of WW domain ligands are enriched for specific WW domain-binding motifs.
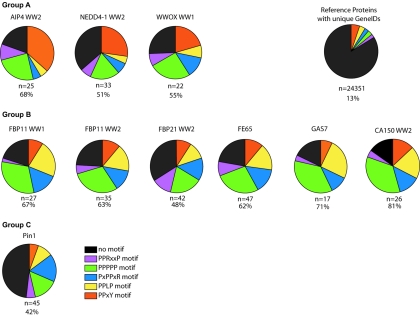
WW domains bind a number of proline-based peptide ligands including PY, PPLP, PR, and p(S/T)P motifs as well as polyproline stretches (2, 3, 14, 18, 29, 39, 40, 49, 54, 76). Therefore, we examined whether PPXY, PPLP, and PPPPP motifs are present in the WW-binding proteins from Jurkat cells (Fig. 4). Since a clear consensus sequence does not exist for PR peptide ligands, we searched for the PPRXP and PXPPXR motifs described by Otte et al. (54). Analysis of over 24,000 reference proteins with unique GeneIDs showed that only 13% of these polypeptides contained at least one such motif. In contrast, over 50% of the proteins isolated in association with the analyzed WW domains possess at least one of these peptide sequences, with the exception of Pin1 WW domain-binding proteins. Furthermore, WW domains assigned to group A by hierarchical clustering were enriched for precipitated proteins with PPXY motifs, while group B WW domains preferentially precipitated proteins containing PPLP, PPPPP, and PXPPXR motifs (Fig. 4). This relationship is highlighted by superimposing the presence of these motifs on the clustered data of Fig. 3 (see Fig. S1 in the supplemental material).
FIG. 4.
Examination of precipitated proteins for known proline-based WW domain-binding motifs. The proteins precipitated with each of the GST-WW pulldowns were analyzed for the presence of the indicated proline-containing motifs. Note that the pie charts do not equal 100%, since individual proteins can have more than one motif. The number of proteins precipitated by each WW domain and the percentage of proteins with at least one of the proline-rich motifs are indicated below the corresponding pie charts. The ∼24,000 reference proteins with unique GeneIDs were downloaded from the National Center for Biotechnology Information database and searched locally for the indicated motifs.
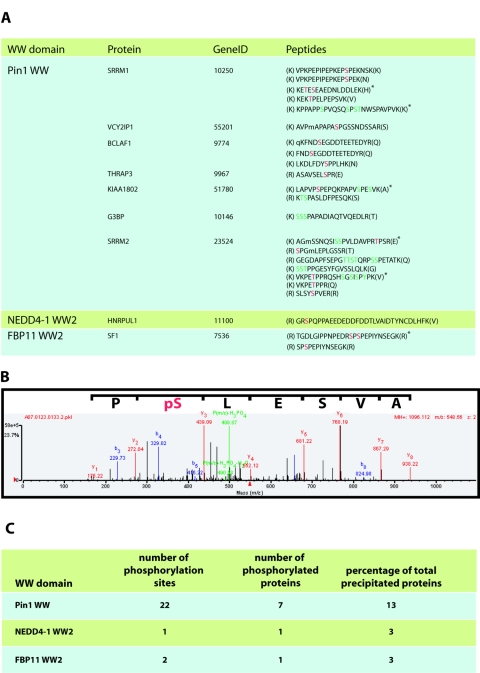
The majority of the proteins precipitated by the WW domain of Pin1 lacked these proline-rich motifs, and there was no enrichment for any specific proline-rich sequence (Fig. 4; see Fig. S1 in the supplemental material). This is consistent with the ability of this domain to recognize p(S/T)P ligands. We therefore examined the MS data for tryptic peptides containing phosphorylated serine or threonine and compared Pin1 phosphorylation data with results obtained for representatives of group A (NEDD4-1 WW2) and group B (FBP11 WW2) WW domains (Fig. 5A). Twenty-two distinct serine/threonine phosphorylation sites were found in 7 of the 45 proteins precipitated by the Pin1 WW domain (Fig. 5C), and the majority of these phosphorylation sites conform to the Pin1 consensus, p(S/T)P. A single phosphorylation site from one protein was identified in the NEDD4-1 WW2 pulldowns, while two phosphorylation sites were identified for the single phosphoprotein precipitated by the FBP11 WW2 (Fig. 5C). While this analysis is not comprehensive, the GST-Pin1 WW domain pulldowns appear to be enriched for phosphorylated-serine/threonine peptides which fit the consensus for Pin1 WW domain binding.
FIG. 5.
Analysis of GST-WW domain pulldowns for phosphopeptides. (A) The MS data from the GST-Pin1 WW, NEDD4-1 WW2, and FBP11 WW2 pulldowns were analyzed for the presence of phosphopeptides as described in Materials and Methods. Identified phosphopeptides and their host proteins are indicated. Definitively phosphorylated residues are indicated in red, and possible phosphorylation sites where the identification was ambiguous are indicated in green. A lowercase “m” denotes an oxidized methionine, while a lowercase “q” denotes a pyroglutamine residue. Amino acids in parentheses are residues N and C terminal to the peptide in the protein sequence. Peptides indicated by an asterisk are doubly phosphorylated. (B) The MS/MS spectra for the THRAP3 phosphopeptide is shown. Note that only b (blue), y (red), and parental ion minus phosphoric acid (P(m/z) − H3PO4) or minus phosphoric acid and water (P(m/z) − H3PO4-H2O) (green) are indicated. The number associated with each ion is the mass/charge ratio (m/z) of that ion. (C) Statistics showing the number of phosphorylation sites, number of phosphorylated proteins, and number of phosphorylated proteins as a percentage of total proteins precipitated with each WW domain (from Fig. 3).
Taken together, these results indicate that the binding properties of individual WW domains for full-length cellular proteins can be integrated with their selective recognition of peptide ligands to provide a more comprehensive view of their physiological interactions. This approach may be of general utility in exploring the functions of interaction domains.
Domain-based screen identifies novel, physiologically relevant WW domain-associated proteins.
Several of the proteins identified as binding to specific WW domains have been previously shown to coimmunoprecipitate with the corresponding WW domain-containing protein. For example, transfected FE65 coprecipitates with endogenous Mena from Cos-7 cells (18), and CA150 coprecipitates with the SF1 splicing factor through its second WW domain (45). We identified both of these interactions in our screen (ENAH [GeneID 55740] and SF1 [GeneID 7536]). In addition, NEDD4-1 and its yeast orthologue, Rsp5p, directly interact with the RNA Pol II LS (12) and induce its ubiquitylation in response to UV-induced DNA damage (1). We found RNA Pol II LS in association with NEDD4-1 WW2 and also with WW domains from AIP4 and WWOX (POL2RA, GeneID 5430).
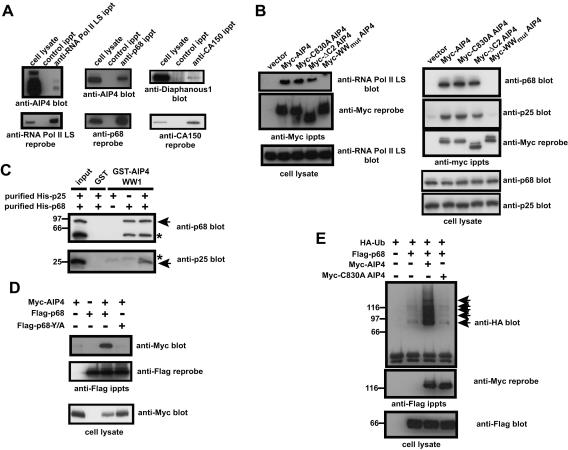
However, the majority of the proteins we identified have not been previously shown to interact with the relevant WW domain-containing proteins. Therefore, we assessed whether selected proteins identified as binding isolated WW domains in vitro could associate with the cognate full-length proteins in cells (Fig. 6A). In a fashion similar to that of NEDD4-1, endogenous AIP4 coimmunoprecipitated with the RNA Pol II LS. More interestingly, we tested whether AIP4 associates in vivo with the p68 subunit of CFIm (p68) (CPSF6, GeneID 11052), which precipitated with the AIP4 WW2 domain in vitro. The CFIm complex is involved in the earliest events of pre-mRNA cleavage prior to the addition of the poly(A) tail (59, 60) and is formed through the heterodimerization of p68 with a p25 subunit (CPSF5, GeneID 11051) which was also precipitated with the AIP4 WW2 domain (Fig. 3). Indeed, the p68 subunit of CFIm coprecipitated with AIP4 from Jurkat T-cell lysate. We also found that CA150, which has been reported to regulate transcription and splicing (25, 45, 63, 69) and to bind the Huntingtin protein (27), coprecipitated with Diaphanous 1 (DIAPH1, GeneID 1729) (Fig. 6A), a protein with links to the cytoskeleton (73), but which has also been shown to be present in the nucleus in NIH 3T3 cells (70).
FIG. 6.
Identified proteins are direct, in vivo, binding partners. (A) Proteins were precipitated from 293T cell lysate (anti-RNA Pol II LS immunoprecipitate [ippt]) or Jurkat cell lysate (anti-p68 ippts) with the indicated antibodies, separated on SDS-PAGs, transferred to PVDF membranes, and blotted with antiserum against AIP4 or Diaphanous 1 (top blots). Blots were stripped and reprobed with the indicated antibodies (bottom blots). (B) 293T cells were transfected with cDNAs coding for a doubly Myc-tagged AIP4 (Myc-AIP4), enzymatically inactive AIP4 mutant (Myc-C830A AIP4), an AIP4 construct lacking the entire C2 domain (Myc-ΔC2 AIP4), or a construct in which the second tryptophan (tyrosine in the case of the fourth WW domain) residue in each WW domain had been mutated to alanine (Myc-WWmut AIP4). Cell lysates were immunoprecipitated with the anti-Myc MAb 9E10, and immunoprecipitates were separated on SDS-PAGs, transferred to PVDF membranes, and blotted for the large subunit of RNA polymerase II (anti-RNA Pol II LS) or the p68 and p25 subunits of CFIm (anti-p68 and anti-p25). The blots were stripped and reprobed with the 9E10 anti-Myc MAb (bottom blots). Cell lysates are included to show that the levels of RNA Pol II LS, p68, and p25 are equivalent in each of the lysates. Note that the dot in the top blot of the Myc-WWmut AIP4 pulldown is an artifact. (C) Baculovirus-expressed, purified recombinant His-tagged p68 or p25 protein was incubated with GST alone or a GST fusion protein of the first WW domain of AIP4 (GST-AIP4 WW1). GST precipitates were separated on SDS-PAGs, transferred to nitrocellulose, and blotted with either anti-p68 antiserum (top blot) or anti-p25 antiserum (bottom blot). The positions of molecular mass markers (in kilodaltons) are indicated to the left. The positions of p68 and p25 proteins are indicated by the arrows. The asterisk in the top blot indicates a p68 degradation product, while the asterisk in the bottom blot indicates the GST-AIP4 WW1 fusion protein. The input is one/fifth of the material used for the pulldown experiment. (D) 293T cells were transfected as indicated with cDNAs coding for a doubly Myc-tagged AIP4 (Myc-AIP4), Flag-tagged p68 protein (FLAG-p68), or Flag-tagged p68 protein in which the tyrosine residue in the PY motif had been mutated to alanine (FLAG-p68 Y/A). Cell lysates were immunoprecipitated with the anti-FLAG M2 MAb, and immunoprecipitates were separated on SDS-PAGs, transferred to PVDF membranes, and blotted with the anti-Myc MAb (top blot). The blot was stripped and reprobed with the anti-Flag M2 MAb (middle blot). Cell lysate is included to show that equivalent amounts of Myc-tagged AIP4 were expressed in the appropriate transfections (bottom blot). (E) 293T cells were transfected as indicated with cDNAs coding for a Flag-tagged p68 protein (FLAG-p68), a doubly Myc-tagged AIP4 (Myc-AIP4), an enzymatically inactive AIP4 mutant (Myc-C830A AIP4), or an HA-tagged ubiquitin (HA-Ub) construct. Cell lysates were immunoprecipitated with the anti-FLAG M2 MAb, and immunoprecipitates were separated on SDS-PAGs, transferred to PVDF membranes, and blotted with an anti-HA antiserum (top blot). The blot was stripped and reprobed with the anti-Myc 9E10 MAb (middle blot). Cell lysate is included to show that equivalent amounts of FLAG protein were expressed in the appropriate transfections (bottom blot). The positions of ubiquitylated p68 proteins are indicated by arrows. The positions of molecular mass standards (in kilodaltons) are indicated to the left.
We next tested whether these in vivo interactions are indeed WW domain dependent. Figure 6B shows that Myc-tagged AIP4 coprecipitates with the RNA Pol II LS and the p68 and p25 subunits of CFIm from 293T cell lysate. Similar results were obtained with Myc-tagged AIP4 variants lacking the entire C2 domain (ΔC2) or rendered enzymatically inactive by substitution of a critical catalytic cysteine within the HECT domain (C830A). However, collectively inactivating the four WW domains in AIP4 by mutating the second conserved tryptophan residue in each case (tyrosine in the case of the fourth WW domain) (13, 50) abolished AIP4 interaction with all three of these binding partners (Fig. 6B).
While these coprecipitation experiments identify proteins that associate with AIP4 in cells, they do not prove that these interactions are direct. To explore this point, we tested whether baculovirus-expressed, purified, recombinant His-tagged p68 and p25 subunits of CFIm could be precipitated with a GST fusion protein containing the first WW domain of AIP4. Although both the p68 and p25 subunits possess a single PY motif and therefore could potentially directly interact with AIP4, only the purified p68 protein on its own precipitated with GST-AIP4 WW1, whereas p25 did not (Fig. 6C). However, the p25 subunit was precipitated with the GST-AIP4 WW1 fusion protein when coincubated with the p68 subunit, likely through heterodimerizing with the p68 protein.
To test whether the single PY motif of p68 is required for its WW domain-dependent interaction with AIP4, we coexpressed Myc-tagged AIP4 in 293T cells with either Flag-p68 or Flag-p68 Y/A, which contains a mutation that disrupts WW domain binding (14, 74). Unlike wild-type Flag-p68, the Flag-p68 Y/A protein did not coprecipitate the Myc-tagged AIP4 (Fig. 6D), indicating that the interaction between AIP4 and p68 depends on both the AIP4 WW domains and the p68 PY motif.
Since AIP4 is an E3 protein-ubiquitin ligase, we examined whether the interaction between p68 and AIP4 might promote p68 ubiquitylation. Figure 6E shows that when Flag-tagged p68 is coexpressed with an HA-tagged ubiquitin construct in 293T cells, the HA-tagged ubiquitin is incorporated into p68, forming a characteristic ubiquitin ladder. Moreover, the further expression of Myc-tagged AIP4, but not the enzymatically inactive C830A mutant, strongly enhanced p68 ubiquitylation. Taken together, these data suggest that proteins identified in this domain-based screen can serve as direct physiological binding partners for WW domain-containing polypeptides. Furthermore, the screen can identify a protein such as the p25 subunit of CFIm, which is recruited into a complex with a WW domain protein through an indirect association. In addition, complexes such as CFIm/AIP4 identified in the WW domain screen can be functional in vivo, as indicated by the AIP4-mediated ubiquitylation of p68.
WW domain screen isolates multiprotein complexes involved in transcription, splicing, chromatin remodeling, and cytoskeletal organization.
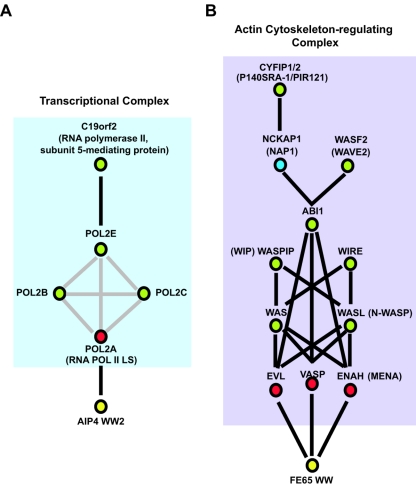
The data for CFIm suggest that the WW domains can bind proteins that are themselves components of stable multiprotein complexes. In fact, analysis of the WW domain-binding proteins identified by LC-MS/MS suggests that several such complexes are recruited to WW domains. For instance, several studies have shown that the WW domains of NEDD4 family proteins bind the RNA Pol II LS (POLR2A) (1, 12). Indeed, we found the RNA Pol II LS associated with the AIP4 WW2, NEDD4 WW2, and WWOX WW1 domains. Moreover, we identified additional RNA Pol II subunits, as well as proteins reported to bind RNA Pol II subunits (RNA polymerase II, subunit 5-mediating protein [C19orf2] [GeneID 8725]) (Fig. 7A). These data argue that the WW domain pulldowns precipitate larger transcriptional complexes. Additionally, we identified proteins of the ENA/VASP family (ENAH [Mena], VASP, and EVL) in pulldowns with the FE65 WW domain and other group B (II/III) WW domains (Fig. 7B). The FE65 WW domain reportedly binds a PPLP motif in Mena (18) and EVL (44), and the related VASP protein also has this motif. Collectively, these proteins control cell motility through regulation of the actin cytoskeleton (61). The ENA/VASP proteins associate with WASP/WAVE family proteins (11), which also regulate the actin cytoskeleton through the Arp2/3 complex, and we found several family members (WAS [WASP], WASL [N-WASP], and WASF2 [WAVE2]) in group B (II/III) WW domain pulldowns (Fig. 7B and Fig. 3) (5). These proteins can recruit additional polypeptides, such as WASPIP (WIP), WIRE, ABI1, and CYFIP1/2, which were also identified in the group B (II/III) WW domain precipitations. It will be of interest to determine how WW domain interactions may regulate this actin polymerizing complex; in one instance, a WW domain protein, FBP11, has been shown to interfere with N-WASP function by sequestering it in the nucleus (52).
FIG. 7.
Protein complexes precipitated by GST-WW domains. (A) The transcriptional complex precipitated by the AIP4 WW2 domain is shown. The WW domain is indicated in yellow, direct WW domain-binding proteins in red, and likely indirectly precipitating proteins in green. Gene symbols and common names (in parentheses) are provided. Black lines indicate a direct physical interaction, while gray lines indicate a physical association, but not necessarily direct. (B) Actin cytoskeleton-regulating complex precipitated by the FE65 WW domain (as well as other group B [II/III] WW domains) is shown and labeled as described above for panel A. Although not found in the FE65 pulldowns, NAP1 (NCKAP1) was included (in blue) as it has been shown to bridge ABI1 with CYFIP1/2 (24, 35) and was found in association with other group B (II/III) WW domains (Fig. 3). Note that this model represents one possible cytoskeleton-regulating complex based on published work. Since the WASP proteins (WAS and WASL), WIPIP, WIRE, WASF2, and ABI1 are extremely proline rich and contain motifs that are predicted to interact with group B WW domains, they may interact directly with group B WW domains.
Other cellular complexes were also seen in WW domain pulldowns. We purified components that make up the U2 splicing complex (SF3A1, -2, and -3 and SF3B1, -2, -3, and -4) with group B (II/III) WW domains, subunits of the chaperonin-containing t-complex polypeptide 1 (CCT) that facilitate the folding of actin, tubulin, and other cytosolic proteins (CCT2, CCT4, CCT7) (71) with FBP11 WW2 and several components of the chromatin remodeling SWI/SNF complex (ARID1A, SMARCC1, SMARCC2, and SMARCE1) with the AIP4 WW2 domain. These results are consistent with the notion that WW domain proteins interact with larger multiprotein complexes involved in multiple facets of cellular regulation.
Specificity within the WW domains of AIP4 and between the related WW domains of AIP4 and NEDD4-1.
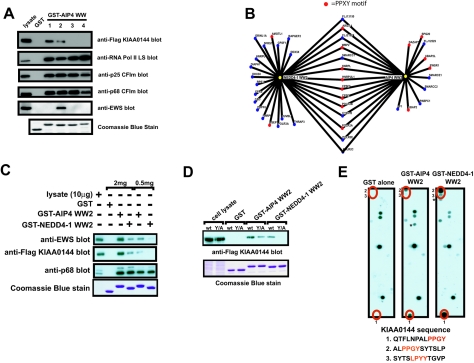
In the MS-based analysis, we used the second WW domain of AIP4 to precipitate proteins from Jurkat cell lysate. However, AIP4 possesses four WW domains, which may differ in their ability to bind ligands (22, 26, 47, 64). Therefore, we investigated whether proteins identified by MS as precipitating with the second WW domain could also bind to the other WW domains of AIP4. Figure 8A shows that the RNA Pol II LS and the p68 and p25 subunits of CFIm were precipitated by each of four WW domains, whereas EWS (EWSR1, GeneID 2130), a protein with an RNA recognition motif that is frequently rearranged with Ets family transcription factors in cancers such as Ewing's sarcoma (41), was specifically precipitated by AIP4 WW2. Similarly, a Flag-tagged KIAA0144 protein (UBAP2L, GeneID 9898), a protein of unknown function with a ubiquitin-associated domain, was selectively precipitated by the first and second WW domains. Thus, the isolated domains of tandem WW domain arrays can show both common and selective binding, suggesting that the tandem WW domains of AIP4 may act synergistically to bind common partners or alternatively may act as a scaffold to recruit specific proteins to individual WW domains.
FIG. 8.
Specificity of binding between the different WW domains of AIP4 and the second WW domains of AIP4 and NEDD4-1. (A) 293T cells were transfected with a cDNA coding for a Flag-tagged KIAA0144 protein (FLAG-KIAA0144 blot) or left untransfected (other blots). Cell lysates were prepared, and an equivalent amount of lysate was precipitated with GST alone or with the indicated GST-AIP4 WW domain. Half of each precipitate was separated on an SDS-PAG, transferred to a PVDF membrane, and blotted with the indicated antibody. The other half of the GST pulldown was separated on an SDS-PAG and stained with Coomassie blue to show that equivalent amounts of GST fusion protein were used for the pulldowns (bottom blot). Cell lysate is included to show the electrophoretic mobilities of the indicated proteins. (B) Osprey diagram (7) showing the proteins identified by MS to precipitate with the second WW domains of NEDD4-1 and AIP4. Proteins with PY motifs are indicated in red. Note that the large subunit of RNA polymerase II (POL2A) does not have a PY motif but does possess several copies of the YSPTSPS heptamer sequence in its C-terminal domain (CTD), which has been shown to bind the WW domains of NEDD4 family proteins (12). (C) 293T cells were transfected with a cDNA coding for a Flag-tagged KIAA0144 protein (FLAG-KIAA0144 blot) or left untransfected (other blots). Cell lysates were prepared, and proteins were precipitated with the indicated GST fusion proteins from either 2 mg or 500 μg of cell lysate. Half of each precipitate was separated on an SDS-PAG, transferred to a PVDF membrane, and blotted with the indicated antibody. The other half of the GST pulldown was separated on an SDS-PAG and stained with Coomassie blue to show that equivalent amounts of GST fusion protein were used for the pulldowns (bottom blot). Cell lysate (10 μg) was included to show the electrophoretic mobility of the immunoblotted protein and to give an indication of the amount of protein precipitated by the GST fusion proteins. (D) 293T cells were transfected with a cDNA coding for a Flag-tagged KIAA0144 protein (wild type [wt]) or a Flag-tagged KIAA0144 protein in which the tyrosine residue in the PY motif had been mutated to alanine (Y/A). Cell lysates were prepared, and proteins were precipitated with the indicated fusion proteins from equivalent amounts of cell lysate. Half of each precipitate was separated on an SDS-PAG, transferred to a PVDF membrane, and blotted with the anti-FLAG M2 MAb. The other half of the GST pulldown was separated on an SDS-PAG and stained with Coomassie blue to show that equivalent amounts of GST fusion protein were used for the pulldowns (bottom blot). Cell lysate was included to show the electrophoretic mobility of the FLAG-KIAA0144 protein. (E) SPOTS membranes were prepared as outlined in Materials and Methods. Proteins (12-mers) comprising the entire coding sequence of KIAA0144 were generated, with a moving window of four amino acids. SPOTS membranes were probed with 1 μΜ of either GST-AIP4 WW2, GST-NEDD4-1 WW2, or GST alone. Spots specific to GST-AIP4 WW2 and NEDD4-1 WW2 are numbered and indicated in red. The sequences of these peptides (with the PY motif and LPXY motifs in red) are shown below the blots. Spots marked by an asterisk were recognized by GST alone in other experiments.
Many of the proteins precipitated by the second WW domains of AIP4 and NEDD4-1 possess at least one PY motif (Fig. 8B and Fig. 4). While many of these proteins were precipitated by WW domains from both proteins, several were selectively precipitated by only one of the WW domains (e.g., UBAP2L [KIAA0144] and EWSR1 [EWS]). Direct analysis by immunoblotting of proteins isolated from various amounts of 293T cell lysate showed that the second WW domains of NEDD4-1 and AIP4 were equivalent in their abilities to precipitate the p68 subunit of CFIm, whereas the KIAA0144 and EWS proteins were preferentially precipitated by the AIP4 WW2 domain (Fig. 8C). This preferential binding of EWS and KIAA0144 likely explains why the EWS and KIAA0144 proteins were not identified as NEDD4-1 WW2-binding proteins by MS (Fig. 3; see Table S2 in the supplemental material).
To confirm that binding of the AIP4 and NEDD4-1 WW domains to the KIAA0144 protein was through the PY motif in KIAA0144, we mutated the tyrosine residue in the PY motif. Figure 8D shows this substitution greatly decreased KIAA0144 precipitation by both the AIP4 and NEDD4-1 WW2 domains. The mutation did not completely abolish binding and had a more severe effect on precipitation by the NEDD4-1 WW2 domain than AIP4 WW2 (Fig. 8D). To probe whether the PY motif is the main site of interaction for the WW domains of AIP4 and NEDD4-1 and determine whether there may be additional binding sites, as suggested by the PY mutants, we generated an array of overlapping peptides corresponding to the entire sequence of KIAA0144 and determined which peptides were recognized by GST-AIP4 WW2 and GST-NEDD4-1 WW2 fusion proteins. A representative array (Fig. 8E) shows that three peptides were reproducibly and specifically recognized by GST-AIP4 WW2 and GST-NEDD4-1 WW2 (two strongly and one weakly, indicated in red) but not GST alone. The two strong-intensity spots (spots 1 and 2) include the PY motif, while the weak-intensity spot (spot 3) contains an LPXY motif, a motif that has been shown to bind the WW domains of NEDD4 family proteins (13, 62). Taken together, these data show that recognition of KIAA0144 by the second WW domains of AIP4 and NEDD4-1 is largely dependent on the KIAA0144 PY motif and that individual group A (I) WW domains show preference for specific PY motifs.
An individual protein with distinct binding motifs for different classes of WW domains.
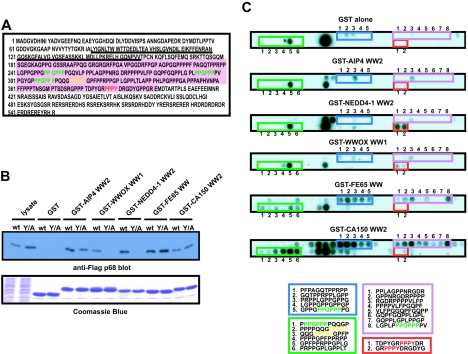
A few proteins identified in the MS screen were precipitated by WW domains that typically recognize different classes of proline-rich ligands. For example, the p68 protein of CFIm was precipitated by seven of the WW domains analyzed (AIP4 WW2, NEDD4-1 WW2, WWOX WW, CA150 WW2, FBP21 WW2, and FBP11 WW2). This raised the question as to whether p68 possessed a single “generic” WW domain recognition motif, or alternatively, distinct recognition motifs recognized by different classes of WW domains. The sequence of p68 (Fig. 9A) shows that the C terminus possesses numerous RS repeats, a characteristic of SR proteins (9). The p68 protein is also extremely proline rich and has a single PY motif, a PPPPP stretch, and three PPGPPP motifs. Since the PY motif of p68 is required for association with AIP4 (Fig. 6), we tested whether it is also important for association with other WW domains that recognize p68. Mutation of the p68 PY motif abolished binding to the WW domains of WWOX and NEDD4-1. In contrast, the AIP4 WW2 domain showed reduced but still significant binding to the p68 Y/A mutant, while the FE65 WW domain and CA150 WW2 domains were equally able to precipitate wild-type p68 and the p68 Y/A mutant (Fig. 9B). To locate the recognition motifs for these latter WW domains, we synthesized a peptide SPOTS array corresponding to the proline-rich region of p68 (Fig. 9A, pink region) and probed this with GST-WW fusion proteins (Fig. 9C). As expected, the NEDD4-1 WW2, WWOX WW1, and AIP4 WW2 domains recognized two overlapping peptides containing the PY motif (Fig. 9C, red box). The fact that the AIP4 WW2 domain recognized no additional peptides suggests that the binding of this WW domain to the p68 Y/A mutant still involves the p68 PY motif (Fig. 9B). The FE65 WW domain and CA150 WW2 domain collectively bound a distinct set of proline-rich peptides that were not recognized by the NEDD4-1, WWOX, or AIP4 WW domain or GST alone (Fig. 9C, blue, pink, and green boxes). These included peptides comprising the PPPPP stretch and the three PPGPPP motifs, as well as several other peptides rich in proline and proline/arginine (Fig. 9C). These results illustrate the specificity of group A (I) WW domains for PY motifs and show the more promiscuous nature of the group B (II/III) WW domains. Furthermore, the data suggest that WW domains may interact with p68 at multiple sites, with specific classes of WW domains recruited to separate sites. The p68 protein may therefore exemplify a scaffold with the ability to recruit multiple classes of WW domains and WW domain proteins through distinct proline-rich sequences.
FIG. 9.
Recognition of the p68 CFIm protein by multiple WW domains. (A) The coding sequence of the p68 protein is shown. The proline-rich region used for the SPOTS blots in panel C is indicated in pink. The PY (red), PPPPP stretch (yellow), and three PPGPPP motifs (green) are also indicated. The RNA recognition motif is underlined. (B) 293T cells were transfected with cDNAs coding for either a Flag-tagged p68 protein (wild type [wt]) or a Flag-tagged p68 protein in which the tyrosine residue in the PY motif had been mutated to alanine (Y/A). Cell lysates were prepared, and an equivalent amount of lysate was precipitated with GST alone or the indicated GST-WW domain fusion protein. Half of each precipitate was separated on an SDS-PAG, transferred to a PVDF membrane, and blotted with the anti-FLAG M2 MAb. The other half of the GST pulldown was separated on an SDS-PAG and stained with Coomassie blue to show that equivalent amounts of GST fusion protein were used for the pulldowns (bottom blot). Cell lysate is included to show the electrophoretic mobilities of the indicated proteins; note that the p68 Y/A was consistently expressed at higher levels than the wt protein. (C) Peptides (12-mers), with a moving window of five amino acids, were generated for the proline-rich region of p68 (panel A, pink region), and SPOTS membranes were probed with 1 μM of GST alone or the indicated GST-WW domain fusion protein. Peptides recognized by one group of WW domains over another are boxed and numbered, and the peptide sequences are provided below the blots. In the boxed sequences, the PY (red), PPPPP (yellow), and PPGPPP (green) motifs in the peptides are highlighted.
DISCUSSION
To explore the range of cellular processes and ligand-binding preferences exhibited by individual WW domains, we have undertaken an MS-based screen to identify proteins from Jurkat T cells that associate in vitro with GST-WW domain fusion proteins taken from a broad range of WW-containing proteins. We have used unsupervised hierarchical clustering to group WW domains on the basis of their similarity of precipitated protein ligands in this assay. This approach yielded three subsets of WW domains that cluster on the basis of the similarities of their protein-binding partners. Many of these associated proteins contain proline-rich sequences typical of peptide ligands for WW domains. Indeed, we found that proteins in one cluster (A) are enriched for PY motifs and are recognized by WW domains with a known predilection for PY peptides (i.e., group I WW domains). In a similar fashion, proteins in a second cluster (B) possess PPLP and proline/arginine-containing motifs, as well as polyproline stretches containing glycine and methionine, and associate with WW domains previously classified as group II/III on the basis of their peptide-binding properties. The final cluster (C) contains proteins that are recognized by the Pin1 WW domain (group IV); these show no preference for the specific proline-rich motifs analyzed but are enriched in phosphorylated-Ser/Thr-Pro sites. Although we have included the FBP21 WW2 in group II/III (cluster B), it has a fair number of specific binding partners (Fig. 3) and has previously been suggested to lie outside the conventional WW domain groups (29). Hierarchical clustering therefore reveals a striking relationship between the full-length proteins recognized by a particular subset of WW domains in cell lysates and the peptide-binding specificities of these domains. This suggests that significant information regarding the molecular recognition properties of interaction domains can be inferred from proteomic-scale analysis of their protein-binding partners. Furthermore, these data argue that the WW domain-binding motifs identified by the analysis of synthetic peptides are used extensively in the context of intact proteins to determine binding specificity.
While some of the polypeptides precipitated by WW domains in this analysis have been described as binding partners for specific WW domain proteins in cells, most of these interactions have not been previously identified. We therefore used the p68/p25 subunits of the CFIm complex to address whether a novel interaction detected in this in vitro screen might be relevant in cells, whether it involves the anticipated recognition of a specific proline-rich motif by one or more WW domains, whether WW domains can recruit multiple components of a larger protein complex, and whether such interactions are functional in vivo. The p68 protein contains RS repeats and a RNA recognition motif, which together with p25 binds pre-mRNA and aids in the recruitment of other 3′ processing factors, such as cleavage and polyadenylation specificity factor (16, 59, 60). Indeed, we have found that the AIP4 E3 protein-ubiquitin ligase binds directly to the p68 CFIm subunit in a fashion that requires functional AIP4 WW domains and a PY motif in p68. Although p25 has a PY motif, it does not interact directly with AIP4 WW domains but rather binds p68, which acts as a bridge to AIP4. Furthermore, we identified an SR protein, SFRS7 (9G8), which interacts with the RS domain of p68 (16) in the NEDD4-1 WW2 pulldowns (Fig. 3), providing further support for the notion that WW domain pulldowns can precipitate cellular complexes. From a functional point of view, AIP4 stimulates the selective ubiquitylation of p68 (but not p25) in cells. Ubiquitylation has many functions, including degradation via the 26S proteasome, but can also positively and negatively regulate molecular interactions independent of degradation (37, 43). It will be of interest to explore whether ubiquitylation of p68 may regulate its binding to RNA or its ability to recruit other factors important for pre-mRNA cleavage.
Taken together, these data suggest that WW domains may regulate an extensive network of protein-protein interactions in vivo. Two additional points support this view. First, the WW domains tested in this screen isolated a number of multiprotein complexes with central functions in cellular regulation, including assemblies involved in transcription, splicing, chromatin remodeling, and actin polymerization. WW domain proteins may therefore serve to bridge or regulate such cellular machinery, for example in the coupling of transcription to splicing (25, 45, 63). In support of this notion, we have used peptide arrays to show that the p68 CFIm subunit contains multiple proline-rich sequences that are able to bind WW domains of different classes in vitro. Thus, WW domains can potentially function to establish networks of interactions, and this may be especially true of proteins with tandem WW domains.
In this regard, the screen compared the second WW domains of the related NEDD4 family E3 ubiquitin ligases NEDD4-1 and AIP4. While many similarities were seen in the proteins precipitated by the second WW domains of AIP4 and NEDD4-1, there were some proteins specifically precipitated with one WW domain (Fig. 8B). Further investigation of two such selective proteins, KIAA0144 and EWS, confirmed that these proteins were indeed precipitated much more efficiently by the AIP4 WW2 domain than by NEDD4-1 WW2 (Fig. 8C). Since humans have nine NEDD4 family proteins (33), an important question is whether these proteins are functionally overlapping or whether they have specific targets. Our data, although limited to one WW domain for both AIP4 and NEDD4-1, suggest that both these paradigms may apply, as some proteins are recognized equivalently by the two domains, while others are selective for one. Of interest, a chromosomal inversion that disrupts the promoter of the Itch/AIP4 gene in mice leads to a hyperactivation of cells of the immune system and an autoimmune-like phenotype; this also suggests that these proteins can have nonoverlapping biological functions (58).
In conclusion, our experiments show that WW domains, and by extension WW domain-containing proteins, associate with multiprotein cellular complexes, many of which have not been previously described. Thus, WW domains provide a versatile module with which to assemble regulatory protein networks.
Supplementary Material
Acknowledgments
R.J.I. was a recipient of fellowships from the National Cancer Institute of Canada (supported by Funds from the Terry Fox Run) and the Leukemia & Lymphoma Society. T.P. is a Distinguished Scientist of the Canadian Institutes for Health Research (CIHR). This work was supported by grants from the CIHR, the Ontario R&D Research Fund, and Genome Canada.
We also thank the National Cell Culture Center for tissue culture services.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Beaudenon, S. L., M. R. Huacani, G. Wang, D. P. McDonnell, and J. M. Huibregtse. 1999. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford, M. T., D. C. Chan, and P. Leder. 1997. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 16:2376-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford, M. T., D. Sarbassova, J. Xu, P. Leder, and M. B. Yaffe. 2000. A novel pro-Arg motif recognized by WW domains. J. Biol. Chem. 275:10359-10369. [DOI] [PubMed] [Google Scholar]
- 4.Bednarek, A. K., K. J. Laflin, R. L. Daniel, Q. Liao, K. A. Hawkins, and C. M. Aldaz. 2000. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 60:2140-2145. [PubMed] [Google Scholar]
- 5.Bompard, G., and E. Caron. 2004. Regulation of WASP/WAVE proteins: making a long story short. J. Cell Biol. 166:957-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork, P., and M. Sudol. 1994. The WW domain: a signalling site in dystrophin? Trends Biochem. Sci. 19:531-533. [DOI] [PubMed] [Google Scholar]
- 7.Breitkreutz, B. J., C. Stark, and M. Tyers. 27February2003, posting date. Osprey: a network visualization system. Genome Biol. 4:R22. [Online.] http://genomebiology.com/2003/4/3/R22. [DOI] [PMC free article] [PubMed]
- 8.Buschdorf, J. P., and W. H. Stratling. 2004. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J. Mol. Med. 82:135-143. [DOI] [PubMed] [Google Scholar]
- 9.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 10.Camon, E., M. Magrane, D. Barrell, D. Binns, W. Fleischmann, P. Kersey, N. Mulder, T. Oinn, J. Maslen, A. Cox, and R. Apweiler. 2003. The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 13:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellano, F., C. Le Clainche, D. Patin, M. F. Carlier, and P. Chavrier. 2001. A WASp-VASP complex regulates actin polymerization at the plasma membrane. EMBO J. 20:5603-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A., S. Cheang, X. Espanel, and M. Sudol. 2000. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:20562-20571. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H. I., A. Einbond, S. J. Kwak, H. Linn, E. Koepf, S. Peterson, J. W. Kelly, and M. Sudol. 1997. Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J. Biol. Chem. 272:17070-17077. [DOI] [PubMed] [Google Scholar]
- 14.Chen, H. I., and M. Sudol. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 92:7819-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, W., and J. T. Campanelli. 1999. WW and EF hand domains of dystrophin-family proteins mediate dystroglycan binding. Mol. Cell Biol. Res. Commun. 2:162-171. [DOI] [PubMed] [Google Scholar]
- 16.Dettwiler, S., C. Aringhieri, S. Cardinale, W. Keller, and S. M. L. Barabino. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 279:35788-35797. [DOI] [PubMed] [Google Scholar]
- 17.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ermekova, K. S., N. Zambrano, H. Linn, G. Minopoli, F. Gertler, T. Russo, and M. Sudol. 1997. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila Enabled. J. Biol. Chem. 272:32869-32877. [DOI] [PubMed] [Google Scholar]
- 19.Espanel, X., and M. Sudol. 1999. A single point mutation in a group I WW domain shifts its specificity to that of group II WW domains. J. Biol. Chem. 274:17284-17289. [DOI] [PubMed] [Google Scholar]
- 20.Faber, P. W., G. T. Barnes, J. Srinidhi, J. Chen, J. F. Gusella, and M. E. MacDonald. 1998. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 7:1463-1474. [DOI] [PubMed] [Google Scholar]
- 21.Fan, H., K. Sakuraba, A. Komuro, S. Kato, F. Harada, and Y. Hirose. 2003. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 301:378-385. [DOI] [PubMed] [Google Scholar]
- 22.Fotia, A. B., A. Dinudom, K. E. Shearwin, J. P. Koch, C. Korbmacher, D. I. Cook, and S. Kumar. 2003. The role of individual Nedd4-2 (KIAA0439) WW domains in binding and regulating epithelial sodium channels. FASEB J. 17:70-72. [DOI] [PubMed] [Google Scholar]
- 23.Frank, R. 2002. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—principles and applications. J. Immunol. Methods 267:13-26. [DOI] [PubMed] [Google Scholar]
- 24.Gautreau, A., H. Y. Ho, J. Li, H. Steen, S. P. Gygi, and M. W. Kirschner. 2004. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA 101:4379-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstrohm, A. C., T. R. Albrecht, C. Sune, M. T. Bedford, and M. A. Garcia-Blanco. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21:7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey, K. F., A. Dinudom, P. Komwatana, C. N. Jolliffe, M. L. Day, G. Parasivam, D. I. Cook, and S. Kumar. 1999. All three WW domains of murine Nedd4 are involved in the regulation of epithelial sodium channels by intracellular Na+. J. Biol. Chem. 274:12525-12530. [DOI] [PubMed] [Google Scholar]
- 27.Holbert, S., I. Denghien, T. Kiechle, A. Rosenblatt, C. Wellington, M. R. Hayden, R. L. Margolis, C. A. Ross, J. Dausset, R. J. Ferrante, and C. Neri. 2001. The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington's disease pathogenesis. Proc. Natl. Acad. Sci. USA 98:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houthaeve, T., H. Gausepohl, M. Mann, and K. Ashman. 1995. Automation of micro-preparation and enzymatic cleavage of gel electrophoretically separated proteins. FEBS Lett. 376:91-94. [DOI] [PubMed] [Google Scholar]
- 29.Hu, H., J. Columbus, Y. Zhang, D. Wu, L. Lian, S. Yang, J. Goodwin, C. Luczak, M. Carter, L. Chen, M. James, R. Davis, M. Sudol, J. Rodwell, and J. J. Herrero. 2004. A map of WW domain family interactions. Proteomics 4:643-655. [DOI] [PubMed] [Google Scholar]
- 30.Huang, X., F. Poy, R. Zhang, A. Joachimiak, M. Sudol, and M. J. Eck. 2000. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat. Struct. Biol. 7:634-638. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda, M., A. Ikeda, L. C. Longan, and R. Longnecker. 2000. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 268:178-191. [DOI] [PubMed] [Google Scholar]
- 32.Ilsley, J. L., M. Sudol, and S. J. Winder. 2002. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell Signal 14:183-189. [DOI] [PubMed] [Google Scholar]
- 33.Ingham, R. J., G. Gish, and T. Pawson. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23:1972-1984. [DOI] [PubMed] [Google Scholar]
- 34.Ingham, R. J., L. Santos, M. Dang-Lawson, M. Holgado-Madruga, P. Dudek, C. R. Maroun, A. J. Wong, L. Matsuuchi, and M. R. Gold. 2001. The Gab1 docking protein links the B cell antigen receptor to the phosphatidylinositol 3-kinase/Akt signaling pathway and to the SHP2 tyrosine phosphatase. J. Biol. Chem. 276:12257-12265. [DOI] [PubMed] [Google Scholar]
- 35.Innocenti, M., A. Zucconi, A. Disanza, E. Frittoli, L. B. Areces, A. Steffen, T. E. Stradal, P. P. Di Fiore, M. F. Carlier, and G. Scita. 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6:319-327. [DOI] [PubMed] [Google Scholar]
- 36.Jolliffe, C. N., K. F. Harvey, B. P. Haines, G. Parasivam, and S. Kumar. 2000. Identification of multiple proteins expressed in murine embryos as binding partners for the WW domains of the ubiquitin-protein ligase Nedd4. Biochem. J. 351:557-565. [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser, P., K. Flick, C. Wittenberg, and S. I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303-314. [DOI] [PubMed] [Google Scholar]
- 38.Kanelis, V., D. Rotin, and J. D. Forman-Kay. 2001. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 8:407-412. [DOI] [PubMed] [Google Scholar]
- 39.Kato, Y., K. Nagata, M. Takahashi, L. Lian, J. J. Herrero, M. Sudol, and M. Tanokura. 2004. Common mechanism of ligand recognition by group II/III WW domains: redefining their functional classification. J. Biol. Chem. 279:31833-31841. [DOI] [PubMed] [Google Scholar]
- 40.Komuro, A., M. Saeki, and S. Kato. 1999. Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J. Biol. Chem. 274:36513-36519. [DOI] [PubMed] [Google Scholar]
- 41.Kovar, H. 2003. Ewing tumor biology: perspectives for innovative treatment approaches. Adv. Exp. Med. Biol. 532:27-37. [DOI] [PubMed] [Google Scholar]
- 42.Krebs, D. L., Y. Yang, M. Dang, J. Haussmann, and M. R. Gold. 1999. Rapid and efficient retrovirus-mediated gene transfer into B cell lines. Methods Cell Sci. 21:57-68. [DOI] [PubMed] [Google Scholar]
- 43.Kuras, L., A. Rouillon, T. Lee, R. Barbey, M. Tyers, and D. Thomas. 2002. Dual regulation of the Met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell 10:69-80. [DOI] [PubMed] [Google Scholar]
- 44.Lambrechts, A., A. V. Kwiatkowski, L. M. Lanier, J. E. Bear, J. Vandekerckhove, C. Ampe, and F. B. Gertler. 2000. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J. Biol. Chem. 275:36143-36151. [DOI] [PubMed] [Google Scholar]
- 45.Lin, K. T., R. M. Lu, and W. Y. Tarn. 2004. The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 24:9176-9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liou, Y. C., A. Sun, A. Ryo, X. Z. Zhou, Z. X. Yu, H. K. Huang, T. Uchida, R. Bronson, G. Bing, X. Li, T. Hunter, and K. P. Lu. 2003. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature 424:556-561. [DOI] [PubMed] [Google Scholar]
- 47.Lott, J. S., S. J. Coddington-Lawson, P. H. Teesdale-Spittle, and F. J. McDonald. 2002. A single WW domain is the predominant mediator of the interaction between the human ubiquitin-protein ligase Nedd4 and the human epithelial sodium channel. Biochem. J. 361:481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, P. J., G. G. Wulf, X. Z. Zhou, P. Davies, and K. P. Lu. 1999. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature 399:784-788. [DOI] [PubMed] [Google Scholar]
- 49.Lu, P. J., X. Z. Zhou, M. Shen, and K. P. Lu. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283:1325-1328. [DOI] [PubMed] [Google Scholar]
- 50.Macias, M. J., M. Hyvonen, E. Baraldi, J. Schultz, M. Sudol, M. Saraste, and H. Oschkinat. 1996. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382:646-649. [DOI] [PubMed] [Google Scholar]
- 51.Marchler-Bauer, A., J. B. Anderson, P. F. D. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizutani, K., S. Suetsugu, and T. Takenawa. 2004. FBP11 regulates nuclear localization of N-WASP and inhibits N-WASP-dependent microspike formation. Biochem. Biophys. Res. Commun. 313:468-474. [DOI] [PubMed] [Google Scholar]
- 53.Murillas, R., K. S. Simms, S. Hatakeyama, A. M. Weissman, and M. R. Kuehn. 2002. Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J. Biol. Chem. 277:2897-2907. [DOI] [PubMed] [Google Scholar]
- 54.Otte, L., U. Wiedemann, B. Schlegel, J. R. Pires, M. Beyermann, P. Schmieder, G. Krause, R. Volkmer-Engert, J. Schneider-Mergener, and H. Oschkinat. 2003. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci. 12:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Passani, L. A., M. T. Bedford, P. W. Faber, K. M. McGinnis, A. H. Sharp, J. F. Gusella, J. P. Vonsattel, and M. E. MacDonald. 2000. Huntingtin's WW domain partners in Huntington's disease post-mortem brain fulfill genetic criteria for direct involvement in Huntington's disease pathogenesis. Hum. Mol. Genet. 9:2175-2182. [DOI] [PubMed] [Google Scholar]
- 56.Pawson, T., and P. Nash. 2003. Assembly of cell regulatory systems through protein interaction domains. Science 300:445-452. [DOI] [PubMed] [Google Scholar]
- 57.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 58.Perry, W. L., C. M. Hustad, D. A. Swing, T. N. O'Sullivan, N. A. Jenkins, and N. G. Copeland. 1998. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 18:143-146. [DOI] [PubMed] [Google Scholar]
- 59.Ruegsegger, U., K. Beyer, and W. Keller. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 271:6107-6113. [DOI] [PubMed] [Google Scholar]
- 60.Ruegsegger, U., D. Blank, and W. Keller. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243-253. [DOI] [PubMed] [Google Scholar]
- 61.Sechi, A. S., and J. Wehland. 2004. ENA/VASP proteins: multifunctional regulators of actin cytoskeleton dynamics. Front. Biosci. 9:1294-1310. [DOI] [PubMed] [Google Scholar]
- 62.Shcherbik, N., Y. Kee, N. Lyon, J. M. Huibregtse, and D. S. Haines. 2004. A single PXY motif located within the carboxyl terminus of Spt23p and Mga2p mediates a physical and functional interaction with ubiquitin ligase Rsp5p. J. Biol. Chem. 279:53892-53898. [DOI] [PubMed] [Google Scholar]
- 63.Smith, M. J., S. Kulkarni, and T. Pawson. 2004. FF domains of CA150 bind transcription and splicing factors through multiple weak interactions. Mol. Cell. Biol. 24:9274-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder, P. M., D. R. Olson, F. J. McDonald, and D. B. Bucher. 2001. Multiple WW domains, but not the C2 domain, are required for inhibition of the epithelial Na+ channel by human Nedd4. J. Biol. Chem. 276:28321-28326. [DOI] [PubMed] [Google Scholar]
- 65.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38:1409-1438. [Google Scholar]
- 66.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 67.Sudol, M., and T. Hunter. 2000. NeW wrinkles for an old domain. Cell 103:1001-1004. [DOI] [PubMed] [Google Scholar]
- 68.Sudol, M., K. Sliwa, and T. Russo. 2001. Functions of WW domains in the nucleus. FEBS Lett. 490:190-195. [DOI] [PubMed] [Google Scholar]
- 69.Sune, C., T. Hayashi, Y. C. Liu, W. S. Lane, R. A. Young, and M. A. Garcia-Blanco. 1997. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 17:6029-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tominaga, T., W. Meng, K. Togashi, H. Urano, A. S. Alberts, and M. Tominaga. 2002. The Rho GTPase effector protein, mDia, inhibits the DNA binding ability of the transcription factor Pax6 and changes the pattern of neurite extension in cerebellar granule cells through its binding to Pax6. J. Biol. Chem. 277:47686-47691. [DOI] [PubMed] [Google Scholar]
- 71.Valpuesta, J. M., J. Martin-Benito, P. Gomez-Puertas, J. L. Carrascosa, and K. R. Willison. 2002. Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 529:11-16. [DOI] [PubMed] [Google Scholar]
- 72.Verdecia, M. A., M. E. Bowman, K. P. Lu, T. Hunter, and J. P. Noel. 2000. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 7:639-643. [DOI] [PubMed] [Google Scholar]
- 73.Wallar, B. J., and A. S. Alberts. 2003. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13:435-446. [DOI] [PubMed] [Google Scholar]
- 74.Winberg, G., L. Matskova, F. Chen, P. Plant, D. Rotin, G. Gish, R. Ingham, I. Ernberg, and T. Pawson. 2000. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol. Cell. Biol. 20:8526-8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wulf, G. M., A. Ryo, G. G. Wulf, S. W. Lee, T. Niu, V. Petkova, and K. P. Lu. 2001. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20:3459-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yaffe, M. B., M. Schutkowski, M. Shen, X. Z. Zhou, P. T. Stukenberg, J. U. Rahfeld, J. Xu, J. Kuang, M. W. Kirschner, G. Fischer, L. C. Cantley, and K. P. Lu. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957-1960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.