Abstract
Fundamentally different recombination defects cause apoptosis of mouse spermatocytes at the same stage in development, stage IV of the seminiferous epithelium cycle, equivalent to mid-pachynema in normal males. To understand the cellular response(s) that triggers apoptosis, we examined markers of spermatocyte development in mice with different recombination defects. In Spo11−/− mutants, which lack the double-strand breaks (DSBs) that initiate recombination, spermatocytes express markers of early to mid-pachynema, forming chromatin domains that contain sex body-associated proteins but that rarely encompass the sex chromosomes. Dmc1−/− spermatocytes, impaired in DSB repair, appear to arrest at or about late zygonema. Epistasis analysis reveals that this earlier arrest is a response to unrepaired DSBs, and cytological analysis implicates the BRCT-containing checkpoint protein TOPBP1. Atm−/− spermatocytes show similarities to Dmc1−/− spermatocytes, suggesting that ATM promotes meiotic DSB repair. Msh5−/− mutants display a set of characteristics distinct from these other mutants. Thus, despite equivalent stages of spermatocyte elimination, different recombination-defective mutants manifest distinct responses, providing insight into surveillance mechanisms in male meiosis.
Recombination promotes accurate segregation of homologous chromosomes during the first meiotic division and is initiated by the formation of DNA double-strand breaks (DSBs) by the evolutionarily conserved SPO11 protein. Many factors, including the DNA strand exchange protein DMC1 and the meiosis-specific MutS homolog MSH5, act on these DSBs to catalyze recombination with a homologous, nonsister chromatid (see references 23 and 24 for reviews). Many mutations that affect the formation or repair of DSBs result in arrest and/or programmed cell death during prophase of meiosis I in mice, as in many other organisms (11, 21, 38). This response reveals that cellular systems monitor the recombination process, presumably to prevent the formation of gametes with damaged or aneuploid genomes. However, it is not always clear what molecular defect is responsible for triggering cell death. By examining oocyte development in several mouse mutants, we recently demonstrated that distinct DNA damage-dependent and independent responses drive the loss of recombination-defective meiocytes in the female germ line (13). Specifically, the absence of DSB formation (in a Spo11−/− mutant) caused only a partial defect in follicle formation at the stage of dictyate arrest, whereas defects in DSB repair (in Dmc1−/− and Msh5−/− mutants) caused more-severe oocyte loss earlier in meiotic prophase, which could be suppressed by eliminating DSB formation.
Recombination defects cause meiocyte loss in the male germ line as well, but the situation is different than that for females. Several mutations that cause fundamentally different defects in meiotic recombination result in spermatocyte apoptosis at what appears to be roughly the same stages in meiosis, described variously as late zygotene to early to mid-pachytene, depending on the means used to stage meiotic progression (reviewed in reference 11). Thus, spermatocytes show an earlier response compared to the effects of the same mutations in oocytes, and this response appears to be qualitatively similar in the presence or absence of persistent DNA damage. However, available data do not rule out the possibility that both damage-dependent and independent responses induce apoptosis at the same developmental stage.
To address these issues, molecular and histological criteria were used to compare spermatocyte development in animals carrying a mutation that eliminates DSB formation (Spo11−/− mutants) to animals carrying mutations that affect the repair of DSBs once they have formed (Dmc1−/− and Msh5−/− mutants). We find that even though apoptosis occurs at the same points in developmental time in all of these mutants, there nevertheless are significant physiological differences between the mutants well before the onset of apoptosis. Epistasis analysis demonstrates that these physiological differences can be attributed at least in part to distinct DNA damage-dependent and independent responses, analogous to the situation in the female germ line. Cytological data implicate a component of the mitotic DNA damage surveillance machinery in responding to defects in the repair of meiotic DSBs. Building on these findings, we also show that Atm−/− spermatocytes show physiological hallmarks of the response to unrepaired DSBs, consistent with a role for ATM in directly promoting the proper repair of DSBs.
MATERIALS AND METHODS
Mice.
Spo11−/−, Dmc1−/−, Msh5−/−, and Atm−/− null mice were previously generated (4, 5, 15, 35). Animals in this study were of C57BL/6 × 129/Sv mixed background. To minimize variability due to strain background, experimental animals were compared to control animals from the same litter or from the same matings. Genotyping was performed by PCR of tail tip DNA, as previously described (13).
Meiotic chromosome preparations and histology.
Testis cell preparations were generated from 2- to 4-month-old mice. Usually, one testis was used for immunohistochemistry while the other was processed for surface spreading according to established techniques (30) or for squashes, in which case testes were frozen in liquid nitrogen and transported on dry ice prior to processing as described previously (32). For histological analyses, testes were fixed in 4% paraformaldehyde or Bouin's solution (Sigma) overnight at 4°C and embedded in paraffin. Staging of spermatogenesis was performed according to the method of Russell (41).
Surface spreads were incubated overnight at room temperature, with the primary antibody diluted in antibody dilution buffer (ADB) (10% goat serum, 3% bovine serum albumin [BSA], 0.05% Triton X-100 in phosphate-buffered saline [PBS]). After one wash in washing buffer 1 (0.4% Kodak Photo-Flo in PBS) and one wash in washing buffer 2 (0.4% Photo-Flo, 0.01% Triton X-100 in PBS) for 5 min each, slides were blocked for 5 min in ADB at room temperature and incubated with the secondary antibody for 1 h at 37°C in the dark. Secondary antibodies were as follows: Alexa 594-labeled goat anti-rat (Molecular Probes, Eugene, OR), 1:500 dilution; tetramethyl rhodamine isothiocyanate-labeled goat anti-mouse (Jackson ImmunoResearch), 1:200; and fluorescein isothiocyanate-labeled goat anti-rabbit (Cedarlane), 1:500. All antibody incubations were performed in a humidified chamber. After 5-min washes with washing buffers 1 and 2, slides were washed for 1 min with washing buffer 3 (0.4% Photo-Flo) and dried 10 min at room temperature in the dark. Coverslips were mounted using ProLong Gold antifade reagent (Molecular Probes) containing 4′,6′-diamidino-2-phenylindole (DAPI) as a counterstain. Images were captured using an Axio2 microscope (Zeiss) connected to a charge-coupled-device (CCD) camera and analyzed using the SlideBook software package (Intelligent Imaging Innovations).
Squash preparations were incubated in PBT (0.15% BSA, 0.1% Tween 20 in PBS) for 60 min before incubation overnight at 37°C with primary antibodies in PBT. Following three 5-min PBS washes, the following pairs of goat secondary antibodies were applied at 1:500 dilutions: anti-rabbit Cy3 (H1t) (Amersham Pharmacia Biotech) and anti-mouse Alexa 488 (XMR) (Molecular Probes) or anti-mouse Cy3 (XMR) (Amersham Pharmacia Biotech) and anti-rabbit Alexa 488 (γH2AX) (Molecular Probes). Both PBT and ADB buffers were made using immunoglobulin G-free protease-free BSA (Jackson ImmunoResearch).
For the histological analysis, 8-μm sections were placed on glass slides, baked overnight at 42°C, and stained by periodic acid-Schiff staining and hematoxylin using routine methods. For immunohistochemical analysis, 8-μm sections were placed on positively charged slides, baked overnight at 56°C, and processed by the Molecular Cytology Core Facility at MSKCC using the automated Discovery staining module (Ventana Medical System). For immunohistochemistry, antigen was unmasked using a cell-conditioning reagent, and slides were incubated for 3 h with the primary antibody. The γH2AX antibody (Upstate Cell Signaling) was used at 2.5 μg/ml, and NBS1 antibody (J. Petrini, Memorial Sloan-Kettering Cancer Center) was used at a 1:500 dilution. Secondary antibodies (ABC kit; Vector Laboratories) were used at 1:200 dilutions and detected with a Ventana Medical System kit. Slides were counterstained with hematoxylin. For NBS1 staining, optimal results were obtained using 4% paraformaldehyde-fixed tissues. The γH2AX and phospho-histone H3 antibodies gave comparable results using either 4% paraformaldehyde or Bouin's fixative; in this report, samples prepared with Bouin's solution are presented. Images were captured using an Axio2 microscope (Zeiss) connected to a CCD camera and analyzed by use of an IPLab v3.55 scientific imaging system (Scanalytics, Inc.).
For XY chromosome fluorescent in situ hybridization (FISH), chromosome spreads were prepared according to the method of Spyropoulos and Moens (45) and stained with anti-SCP3 as described above. After immunostaining, slides were mounted in 50% glycerol in PBS, and the quality of staining was assessed. For subsequent chromosome FISH painting, the coverslip was removed by soaking in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.05% Tween 20, and then slides were washed two times in the same buffer at room temperature. After two further washes in 1× PBS, slides were immersed in 0.05 M MgCl2 in PBS for 5 min at room temperature and then postfixed by immersing for 10 min in 1% formaldehyde and 0.05 M MgCl2 in PBS. Slides were then washed in PBS and dehydrated in an alcohol series.
The hybridization solution containing the X- and Y-specific probes was prepared as follows: 100 ng of fluorescein isothiocyanate-conjugated chromosome-painting X probe (Cambio, United Kingdom) and 100 ng of coumarin-conjugated (ENZO) Y-specific repetitive bacterial artificial chromosome probe Ct7-590p11 (Invitrogen) were mixed with 2 μg of Cot-1 DNA (Invitrogen), denatured for 10 min at 70°C, and incubated for 30 to 45 min at 37°C. To denature the target DNA, slides were immersed in denaturation buffer (70% formamide, 2× SSC, pH 7.5) at 65°C for 3 min, promptly transferred to ice-cold 70% ethanol for 2 min, dehydrated in an alcohol series, and air dried. The hybridization solution was applied to the slides on a slide warmer at 37°C, and a coverslip was immediately applied. Sealed slides were then incubated at 37°C in the dark for 16 h. At the end of the incubation, the coverslip was removed with 50% formamide and 2× SSC, pH 7.5, at 42°C, and slides were washed successively in 2× SSC at 42°C and in distilled water and then air dried. Coverslips were applied, and then slides were examined by epifluorescence microscopy, CCD camera images of nuclei were captured, and the positions were recorded. The coverslips were then removed, and anti-γH2AX staining was performed as described above using a rabbit anti-γH2AX antibody. Slides were examined again by epifluorescence microscopy, and nuclei were rephotographed and images analyzed using the SlideBook program. To evaluate the overlap of X and Y FISH paint with γH2AX bodies, we analyzed only nuclei with obvious accumulations of γH2AX near their peripheries.
Primary antibodies for indirect immunofluorescence.
Sources and dilutions of antibodies were as follows: rat anti-SYCP3, M. A. Handel (Jackson Laboratories) (14), or mouse anti-COR1, P. Cohen (Cornell University) (25), 1:1,000; rabbit anti-γH2AX, W. Bonner (NIH) (39), 1:800; mouse monoclonal anti-γH2AX (Upstate Cell Signaling), 1:500; rabbit anti-NBS1, J. Petrini (MSKCC) (50), 1:300; rabbit anti-histone H1t, P. Moens (York University) (29), 1:200; mouse monoclonal anti-XLR (which recognizes the meiosis-specific family member XMR [17]), H. J. Garchon (INSERM), 1:500. Affinity-purified rabbit anti-TOPBP1 was provided by J. Chen (Mayo Clinic) (52) and was used at a dilution of 1:100. Similar results were obtained with a second rabbit anti-TOPBP1 antiserum generously provided by B. Spyropoulos and P. Moens (33; data not shown).
RESULTS
Fundamentally different recombination defects trigger similar timings of spermatocyte apoptosis. Spermatocytes in Spo11−/−, Dmc1−/−, and Msh5−/− mutants are efficiently eliminated by apoptosis during prophase of meiosis I (5, 12, 15, 35, 40, 53). Earlier findings indicated that cell death in all three mutants is triggered at approximately the same stages in spermatogenesis, as judged by histological criteria (described further below). To rule out subtle differences in timing, we analyzed testis sections from these mutants side by side (Fig. 1).
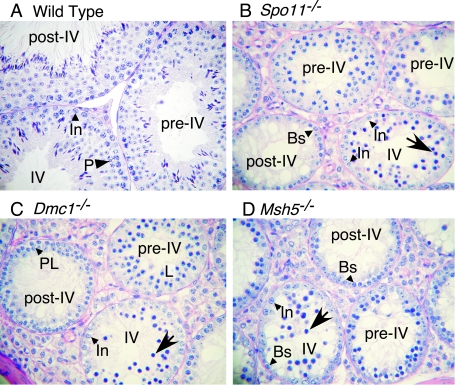
FIG. 1.
Different recombination defects trigger similar timings of spermatocyte apoptosis. (A to D) Periodic acid-Schiff-stained testis sections from wild-type and the indicated mutant mice. Examples of tubules at epithelial stage IV and the stages before (pre-IV) and after (post-IV) stage IV are indicated. Apoptotic cells (arrows) were observed at epithelial stage IV in the mutants, when pachytene spermatocytes (P) are normally present. Earlier-stage spermatocytes present in the mutants are indicated (preleptotene, PL; leptotene, L), as are intermediate (In) and B (Bs) spermatogonia.
Mouse spermatogenesis is highly regular (reviewed in reference 10). Stem cells (spermatogonia) line the periphery of the seminiferous tubule. These cells divide mitotically to replenish the stem-cell population and to give rise to differentiating cells that undergo several rounds of mitotic division before entering meiosis. Spermatocytes proceed through meiotic prophase and the two meiotic divisions to give rise to spermatids, which mature until being released into the lumen of the tubule. Within an adult testis, successive waves of spermatogenesis begin approximately once every 8 to 9 days (31). Specific stages of spermatogenesis have stereotyped lengths, with the entire process taking approximately 34.5 days. Because the initiation of a new cycle occurs before the preceding cycles have finished, the epithelium of each tubule contains a mixture of germ cells at various steps in development, with later cells displaced toward the lumen as new waves of cells enter the pathway. A population of spermatogonia within a small region of a tubule begins a new cycle of germ cell proliferation and differentiation synchronously, but different regions of the tubules are offset from one another with respect to this cycle, so a single histological section will contain germ cells at many different steps of spermatogenesis (some examples are indicated in Fig. 1A).
Because both the period between cycles and the duration of individual developmental steps are so regular, each tubule section can be classified into one of 12 distinct types, referred to as stages I to XII (41). Staging in the wild type is defined according to the constellation of germ cell types present in a tubule section, including late prophase and postmeiotic cells. In most mutants with defects in meiotic recombination, spermatocytes undergo apoptosis prior to the first division (see below) and thus lack cells undergoing postmeiotic steps in development. Nevertheless, it is possible to assign tubule sections from these mutants to stages that are equivalent to the wild-type stages by examining the division cycle of the spermatogonia and by scoring for the presence or absence of early meiotic prophase cells (11, 12, 19). This staging is possible because it is based on premeiotic and early meiotic cell types whose development is not detectably affected by the mutations studied here (e.g., references 5 and 11).
In Spo11−/− males, a number of tubules contained aberrant spermatocytes with condensed nuclei, shown previously by terminal deoxynucleotidyltranserase-mediated dUTP-biotin nick end labeling to be apoptotic (5, 40). These tubules are equivalent to those seen in stage IV in the wild type, as judged by the presence of intermediate-type spermatogonia in late G2 or mitosis and/or of early B spermatogonia (Fig. 1B) (10, 11). The same timing of apoptosis is found for Dmc1−/− mice (Fig. 1C). Side-by-side comparisons demonstrate the same timing for Msh5−/− mutant males as well (Fig. 1D), consistent with previous results (12).
Thus, despite different molecular defects, Spo11−/−, Dmc1−/−, and Msh5−/− spermatocytes undergo apoptosis at the same point with respect to developmental timing. This stage is inferred to be equivalent to mid-pachynema, based on the spermatocyte stage present in stage IV tubules in wild-type animals (41). Because chromosome synapsis defects are found in all three mutants, a simple explanation is that stage IV apoptosis is a response to this common defect. Alternatively, it is possible that distinct cellular defects trigger cell death at the same developmental stages. To address these alternatives, we examined additional molecular markers for meiotic progression.
A pseudo-sex body forms in Spo11−/− mutants.
The sex body is a heterochromatin domain encompassing the XY chromosomes during pachynema (reviewed in reference 20). Sex body chromatin accumulates a number of proteins and protein modifications (see below), and its formation is accompanied by transcriptional inactivation of X- and Y-resident genes. One proposed role is to shield the unsynapsed axes of the XY pair (or persistent DSBs on these axes) from surveillance mechanisms that would otherwise sense these as aberrant. Although the sex body is most prominent during pachynema, early components can be observed at late zygonema (e.g., reference 17).
A prominent marker of sex body formation is the phosphorylation of histone H2AX on Ser-139 to form γH2AX. In somatic cells, γH2AX forms rapidly in the vicinity of DSBs (39). In spermatocytes, γH2AX is generated in response to SPO11-induced DSBs and then disappears as chromosomes synapse and a second wave of γH2AX formation occurs on chromatin surrounding unsynapsed axes (27, 48). In normal males, this latter signal is usually restricted to the chromatin of the sex body. γH2AX is revealed by immunofluorescence on chromosome spreads (Fig. 2A) and as patches of immunoreactive material at the peripheries of pachytene nuclei in testis sections (Fig. 3A).
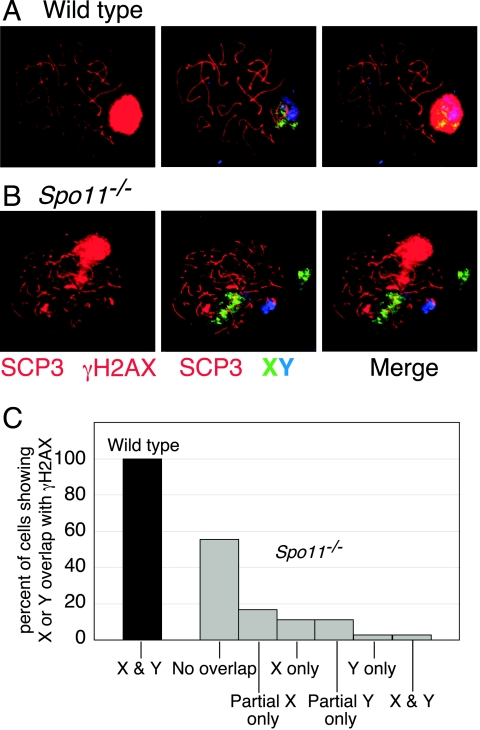
FIG. 2.
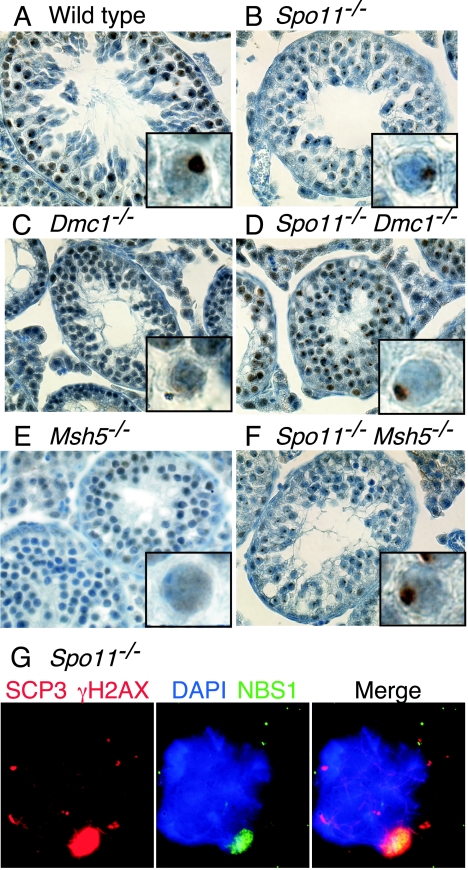
Spo11−/− spermatocytes form pseudo-sex bodies. (A and B) Immunofluorescence and FISH staining of spermatocyte spreads. In wild-type spermatocytes, the X and Y chromosomes colocalize with a discrete domain of γH2AX staining (the sex body). In Spo11−/− spermatocytes the sex chromosomes rarely localize to the domain of γH2AX staining, revealing that this is not a true sex body. (C) Quantification of the frequency of overlap of the sex chromosomes with the γH2AX domain in wild-type and Spo11−/− spermatocytes.
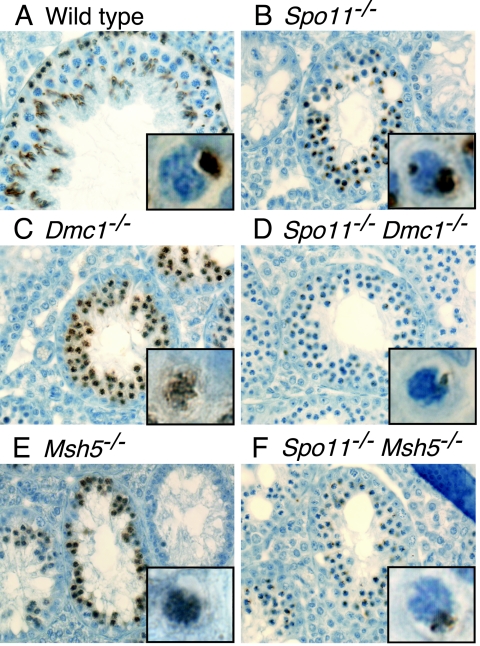
FIG. 3.
Localized accumulation of γH2AX does not occur in recombination mutants that are defective in the repair of SPO11-generated DSBs. Testis sections were stained with anti-γH2AX (brown). Insets show higher-magnification views of individual spermatocytes. As in the wild type (A), the γH2AX signal occurred in discrete domains in Spo11−/− spermatocytes that had progressed sufficiently far in meiotic prophase (B). However, a subset of Spo11−/− spermatocytes contained two localized domains of γH2AX, indicating that sex body formation did not always appear normal in this mutant (inset in panel B). In Dmc1−/− (C) and Msh5−/− (E) spermatocytes, γH2AX was distributed across most of the chromatin. This γH2AX localization defect was suppressed by the introduction of a Spo11 mutation (D and F).
Spo11−/− spermatocytes form localized accumulations of γH2AX that resemble sex bodies (27) (Fig. 2B and 3B). The formation of this γH2AX domain suggested that the X and Y chromosomes may colocalize normally even in the absence of SPO11-dependent recombination. Surprisingly, however, γH2AX accumulation in Spo11−/− spermatocytes did not localize to the X or Y chromosomes. In wild-type pachytene cells, the γH2AX signal of the sex body colocalized completely with the XY pair, as expected (Fig. 2A and C). By contrast, the X and Y chromosomes in Spo11−/− spermatocytes were usually separated from one another and from the γH2AX (Fig. 2B and C). Only 1 of 35 mutant spreads showed substantial overlap of γH2AX with both the X and Y chromosomes (Fig. 2C). This low level of overlap is most likely random. Thus, the formation of a discrete chromatin domain containing γH2AX and other sex body-associated proteins (see below) can be uncoupled from the presence of the X or the Y to form a “pseudo-sex body.” Nevertheless, the timing for pseudo-sex body formation is similar to that for the true sex body.
Absence of a sex body-like structure in mutants with impaired DSB repair.
γH2AX was observed in Dmc1−/− spermatocytes, but in contrast to what was seen in the wild type or in Spo11−/− mutants, the signal was distributed in patches across most of the chromatin and did not form an obvious sex body-like structure (compare Fig. 4A and B with Fig. 4C). Immunostaining of testis sections confirmed this observation (Fig. 3C). It is straightforward to account for the Dmc1−/− spermatocyte pattern if the γH2AX signal reflects the persistence of SPO11-generated DSBs. This signal cannot be attributed to apoptosis, because Spo11−/− spermatocytes became apoptotic at the same stage but did not show this pattern.
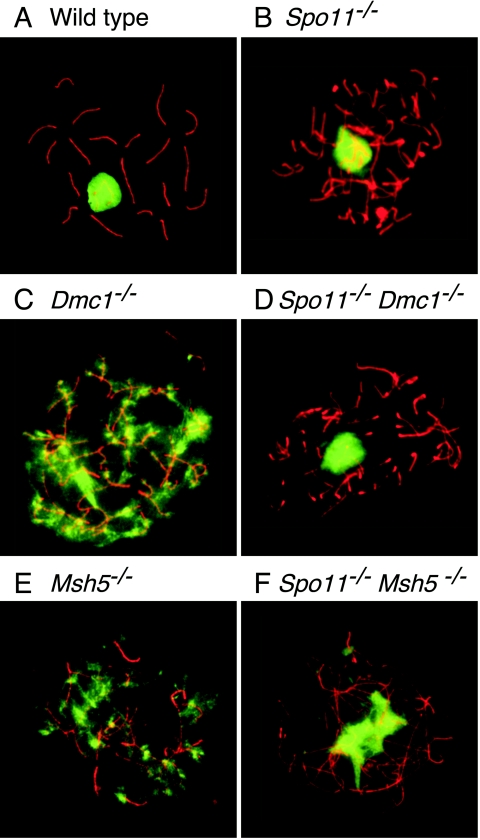
FIG. 4.
Mutants defective in the repair of SPO11-generated DSBs do not form sex body-like domains of γH2AX. Chromosome spreads were stained with anti-SCP3 (red) and anti-γH2AX (green). The γH2AX signal occurred in discrete domains in wild-type (A) and in Spo11−/− (B) spermatocytes but was distributed in patches across most of the chromatin in Dmc1−/− (C) and Msh5−/− (E) spermatocytes. The defect in γH2AX localization in Dmc1−/− and Msh5−/− spermatocytes was suppressed by Spo11 mutation (panels D and F, respectively).
Since it is possible that the abundant, dispersed γH2AX signal in Dmc1−/− spermatocytes might mask a sex body-related signal, we examined the behavior of NBS1 protein, which also localizes to the sex body (Fig. 5A) like its protein partners RAD50 and MRE11 (16). Like γH2AX, NBS1 localized to one or more discrete domains in Spo11−/− spermatocytes (Fig. 5B and G) but not in Dmc1−/− spermatocytes (Fig. 5C).
FIG. 5.
Localized accumulation of NBS1 does not occur in recombination mutants that are defective in the repair of SPO11-generated DSBs. (A to F) Testis sections were stained with anti-NBS1 (brown). As in the wild type (panel A), the NBS1 signal occurred in discrete domains in Spo11−/− spermatocytes that had progressed sufficiently far in meiotic prophase (panel B). In Dmc1−/− (panel C) and Msh5−/− (panel E) spermatocytes, NBS1 did not localize to discrete domains, although the NBS1 localization defects in Dmc1−/− and Msh5−/− spermatocytes were suppressed by the introduction of a Spo11 mutation (panels D and F, respectively). (G) Immunofluorescence staining of a spermatocyte spread with anti-γH2AX and anti-SCP3 in red and anti-NBS1 in green. NBS1 is a component of the sex body (here, the pseudo-sex body), as indicated by its colocalization with γH2AX.
The failure to observe a localized domain of γH2AX or NBS1 suggests that sex body formation is more profoundly defective in Dmc1−/− spermatocytes than in Spo11−/− spermatocytes. Epistasis analysis revealed that Spo11−/− Dmc1−/− spermatocytes were indistinguishable from Spo11−/− single mutants (Fig. 3D, 4D, and 5D). Thus, the failure to form even a pseudo-sex body is due to the persistence of SPO11-dependent DSBs rather than to a role for DMC1 in sex body formation per se.
Previous studies revealed γH2AX patterns in Msh5−/− mutant spermatocytes similar to those of Dmc1−/− spermatocytes, consistent with the persistence of unrepaired or improperly repaired DSBs (27) (Fig. 4E). Moreover, analysis of Msh5−/− females demonstrated that the absence of MSH5 triggers a DNA damage-dependent response in oocytes similar to that in Dmc1−/− mutants (13). We therefore analyzed sex body formation in Msh5−/− spermatocytes. Similar to what is seen with Dmc1−/− spermatocytes, there was no obvious accumulation of γH2AX or NBS1 into discrete domains in Msh5−/− spermatocytes (Fig. 3E, 4E, and 5E). As with Dmc1−/− spermatocytes, the inability of Msh5−/− spermatocytes to form discrete domains of sex-body-related proteins was suppressed by Spo11 mutation (Fig. 3F, 4F, and 5F).
These studies reveal that Spo11 mutation is epistatic to Dmc1 and Msh5 mutations for this marker of meiotic progression and that distinct physiological states result from recombination failure in the presence versus the absence of DSBs.
Deposition of H1t and XMR onto spermatocyte chromatin in recombination mutants.
Meiotic progression was analyzed using additional markers. H1t, a testis-specific isoform of histone H1, replaces somatic H1 on chromatin at mid-pachynema (22). XMR protein is at first faint and dispersed on chromatin at leptonema, becomes more abundant through early zygonema, and then disappears from bulk chromatin and accumulates in the sex body in late zygonema to early pachynema (47) (Fig. 6A through C). The accumulation of XMR in the sex body occurs after the appearance of γH2AX (compare Fig. 6B and C). The deposition of H1t occurs after XMR accumulates in the sex bodies, and H1t persists in postmeiotic cells after the sex bodies have disappeared (Fig. 7A).
FIG. 6.
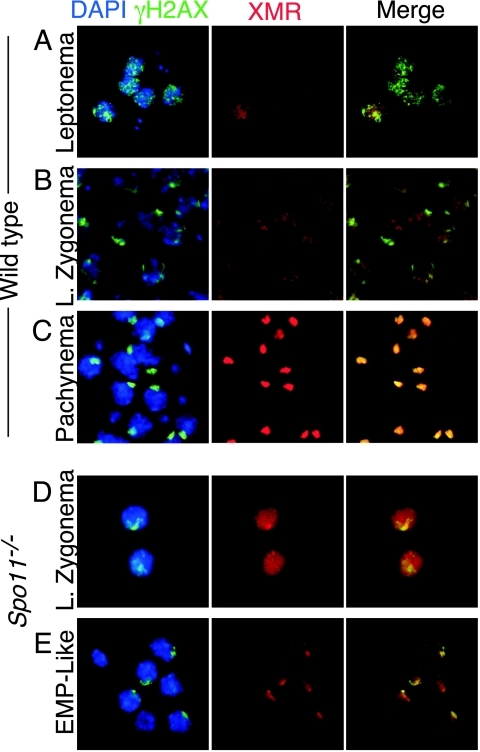
Staining patterns of γH2AX and XMR in wild-type and Spo11−/− spermatocytes in squash preparations of testicular cells. Squash preparations of testicular cells were stained with DAPI (blue), anti-γH2AX (green), and anti-XMR (red). (A to C) Staining patterns in wild-type cells, as previously described (27, 47). At leptonema (panel A), γH2AX is present in focal regions across the chromatin, and XMR staining is very weak. By late (L.) zygonema, γH2AX localization to the sex body becomes apparent, while XMR staining is diffuse (B). By pachynema, both markers colocalize to the sex body (C). (D and E) Staining patterns in Spo11−/− spermatocytes. At late zygonema, γH2AX is localized to a discrete domain, the pseudo-sex body (D), while XMR staining is diffuse. By the early to mid-pachynema (EMP)-like stage, both markers colocalize (E).
FIG. 7.
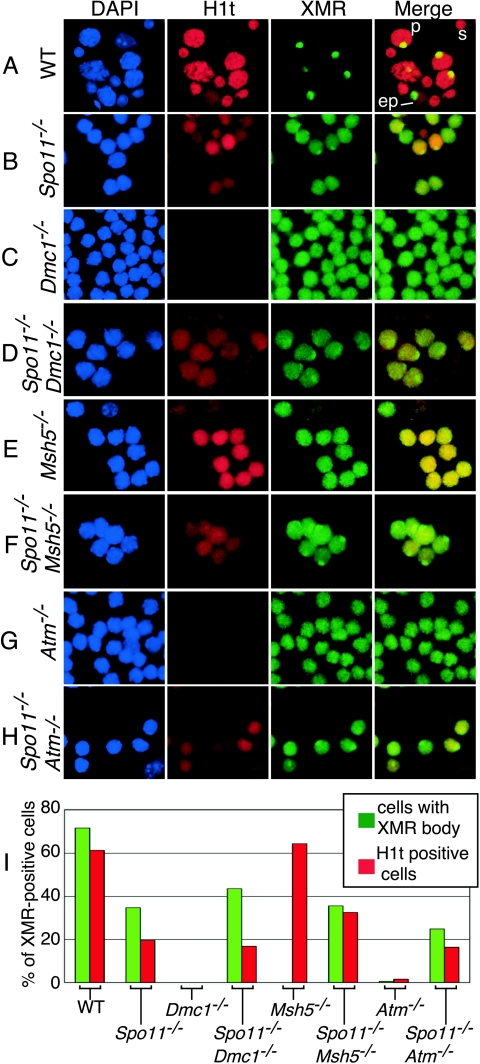
Different recombination mutants show distinct patterns of markers for meiotic progression. (A to H) Squash preparations of testicular cells from mice of the indicated genotypes were stained for histone H1t and XMR. Examples of early pachytene (ep) and mid- to late-pachytene (p) spermatocytes and of a spermatid (s) are shown for the wild type (WT) (A). Some Spo11−/− spermatocytes (B) stain positive for H1t, although generally not as brightly as the wild type, and localize XMR to a discrete domain, although in fewer cells than in the wild type. In Dmc1−/− spermatocytes (C), histone H1t deposition was not detected and XMR was diffuse on chromatin, never accumulating in discrete patches. Spo11−/− Dmc1−/− spermatocytes (D) gave H1t and XMR staining patterns similar to those of Spo11−/− spermatocytes. In Msh5−/− spermatocytes (E), histone Hlt straining was de-tected but XMR was only diffuse. Spo11−/− Msh5−/− spermatocytes (F) gave H1t and XMR staining patterns similar to those of Spo11−/− spermatocytes. Atm−/− spermatocytes behaved similarly to Dmc1−/− spermatocytes, both for the single mutant (G) and when combined with the Spo11−/− mutation (H). (I) XMR-positive cells were counted for those with XMR concentrated in a discrete domain and those that stained positive for histone H1t. The absence of a bar indicates that the frequency of cells in that class was zero (see text).
In Spo11−/− spermatocytes, diffuse XMR was observed on the chromatin, comparable to what is seen in early prophase in the wild type. In addition, a subset of spermatocytes accumulated XMR in the pseudo-sex body (34.6% of XMR-positive cells; n = 393) (Fig. 7B and I and 6D and E). Even in cells containing pseudo-sex bodies, however, XMR staining persisted on bulk chromatin (Fig. 6E and 7B). In these squash preparations, 19.5% (n = 393) of XMR-positive Spo11−/− spermatocytes were positive for histone H1t, although with weaker staining than is seen in mid- to late pachynema in the wild type (Fig. 7B and I). These findings indicate that Spo11−/− spermatocytes reach a stage with properties characteristic of early to mid-pachynema in the wild type.
In Dmc1−/− spermatocytes, XMR was only diffuse on chromatin and never accumulated in a discrete patch (0/476 XMR-positive squashes examined) (Fig. 7C and I). This is consistent with the γH2AX and NBS1 analyses above, which indicated a profound defect in sex body formation. No histone H1t deposition was detectable (Fig. 7C and I). These patterns imply that Dmc1−/− spermatocytes arrest with physiological properties characteristic of an earlier stage than the arrest in Spo11−/− spermatocytes. As noted before, Spo11−/− Dmc1−/− double mutants were indistinguishable from Spo11−/− single mutants (Fig. 7D and I) (n = 407). We conclude that the more severe progression defect in Dmc1−/− spermatocytes is a consequence of the persistence of unrepaired DSBs.
Msh5−/− spermatocytes displayed a distinct phenotype. Similar to Spo11−/− spermatocytes, Msh5−/− spermatocytes were positive for H1t, although the intensity of the H1t staining appeared stronger than that for Spo11−/− spermatocytes (Fig. 7E and I). However, as in Dmc1−/− spermatocytes, XMR staining in Msh5−/− spermatocytes was diffuse and did not accumulate into a discrete domain (Fig. 7E and I) (0/707 XMR-positive cells examined). Consistent with the analyses above, the Msh5−/− defect in the formation of a sex body-like structure was dependent on DSBs, because mutation of Spo11 suppressed this defect (Fig. 7F and I). Thus, Msh5−/− spermatocytes respond to the presence of unrepaired DSBs, yet not in a manner identical to that of Dmc1−/− spermatocytes.
Association of TOPBP1 with meiotic chromosomes with persistent DSBs.
The differences in meiotic progression in Spo11−/− spermatocytes compared to that in Dmc1−/− and Msh5−/− spermatocytes reveal the existence of a previously unknown surveillance mechanism(s) for persistent DNA damage in mammalian male meiosis. In yeast (Saccharomyces cerevisiae), factors that respond to DNA damage in mitotically dividing cells are often required for the arrest of meiotic progression caused by the defective repair of meiotic DSBs (26, 38, 44). To address whether such checkpoint factors respond to persistent DSBs in mammalian meiosis, we examined the behavior of TOPBP1, a BRCT repeat containing protein homologous to S. cerevisiae Dpb11 and Saccharomyces pombe Cut5/Rad4 (52). These yeast proteins have roles in DNA damage checkpoint responses in vegetative cells (1, 42, 49) and in meiosis (33). In human cells, TOPBP1 interacts with other known checkpoint factors, including ATR, and forms foci on chromatin of irradiated cells, consistent with a role in the responses to DNA damage (28, 52). TOPBP1 also forms foci along the axes of spermatocyte chromosomes from leptonema into zygonema (33, 36) (Fig. 8A and B), which colocalize with ATR and are coincident with chromatin regions containing γH2AX (33). The foci are greatly reduced on autosomes as homologous chromosomes synapse (Fig. 8B), but TOPBP1 protein persists on the unsynapsed axes and chromatin of the XY pair from pachynema into diplonema (33) (Fig. 8C and D).
FIG. 8.
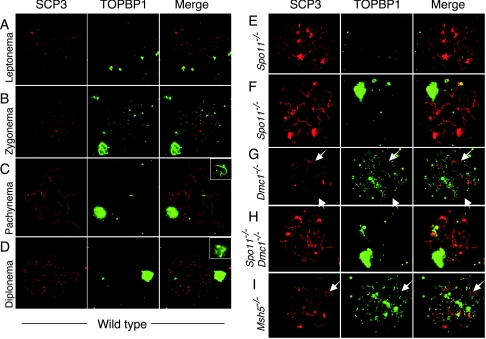
Association of TOPBP1 with meiotic chromosomes in Dmc1−/− and Msh5−/− spermatocytes but not in Spo11−/− spermatocytes. Spermatocyte chromosome spreads were stained with anti-SCP3 (red) and anti-TOPBP1 (green). Merged images are also shown. (A to D) In wild-type cells, TOPBP1 forms foci along the axes of chromosomes in leptonema (A) and zygonema (B), and the foci disappear or are greatly reduced on autosomes as DSBs are repaired and chromosomes synapse (C) in a manner similar that previously observed with a different antibody (33). TOPBP1 protein persists on the unsynapsed axes and chromatin of the XY pair from pachynema into diplonema (B to D). Note that the XY pair in panel B is from an adjacent pachytene nucleus. (E and F) In Spo11−/− spermatocytes, TOPBP1 staining was virtually absent from most of the chromatin, except for the pseudo-sex body when present (F). (G to I) In Dmc1−/− spermatocytes, TOPBP1 accumulated to levels higher than those in the wild type and persisted on chromosome axes and surrounding chromatin without forming obvious pseudo-sex bodies (G). Spo11−/− Dmc1−/− double mutants were indistinguishable from Spo11−/− single mutants (H). Msh5−/− spermatocytes (I) accumulated TOPBP1 in a fashion similar to that of Dmc1−/− spermatocytes. TOPBP1 foci were absent or reduced on many axes undergoing at least limited synaptonemal complex formation (arrows in panels G and I). Matched exposures are shown, except for the insets to panels C and D, which show lower exposures of the TOPBP1 staining in the sex bodies.
TOPBP1 localization was altered in Spo11−/− spermatocytes; in 69% (11/16) of spermatocytes at a stage equivalent to zygonema (i.e., with long axes but little synapsis), TOPBP1 staining was virtually absent (Fig. 8E), demonstrating that TOPBP1 in the wild type responds to SPO11-generated DSBs. In remaining cells of this stage and in the majority (11/15) of later cells with significant amounts of nonhomologous synapsis, TOPBP1 colocalized with γH2AX staining within pseudo-sex bodies (Fig. 8F and data not shown).
By contrast, in Dmc1−/− spermatocytes, TOPBP1 formed abundant, bright foci on chromosome axes in all SCP3-positive spreads examined, with significantly more signal than in the wild type (Fig. 8G). Consistent with other markers, no sex body-like accumulation of TOPBP1 was observed (n = 50). In Msh5−/− spermatocytes as well, TOPBP1 accumulated to higher levels than in the wild type and persisted on chromosome axes and surrounding chromatin without forming obvious pseudo-sex bodies (Fig. 8I). Unlike what was seen for Dmc1−/− spermatocytes, however, rare Msh5−/− spermatocyte spreads showed only faint (5 out of 54 zygotene-like cells examined) or no (1 out of 54) TOPBP1 foci (data not shown). In both Dmc1−/− and Msh5−/− spermatocytes, TOPBP1 foci were absent from many of the axes that had undergone at least limited synaptonemal complex formation (Fig. 8G and I). Spo11−/− Dmc1−/− double mutants were again indistinguishable from Spo11−/− single mutants (Fig. 8H). Thus, TOPBP1 binding to chromosomes in early prophase depends on SPO11-induced DSBs, and TOPBP1 persists on chromosomes when DSBs cannot be repaired.
Atm−/− spermatocytes show hallmarks of the presence of poorly repaired DSBs. Mouse Atm encodes a Ser/Thr kinase that activates cell cycle checkpoints in somatic cells in response to DSBs (43). ATM is also required for normal meiosis, such that Atm−/− spermatocytes have chromosome synapsis defects and undergo apoptosis at stage IV of the seminiferous epithelial cycle (4, 19, 51). In the female germ line, Atm−/− oocytes are eliminated at or prior to follicle formation similar to what is seen in Dmc1−/− mutants, indicating that ATM is required for the proper repair of SPO11-induced DSBs in female meiosis as opposed to simply monitoring defects in DSB repair (13).
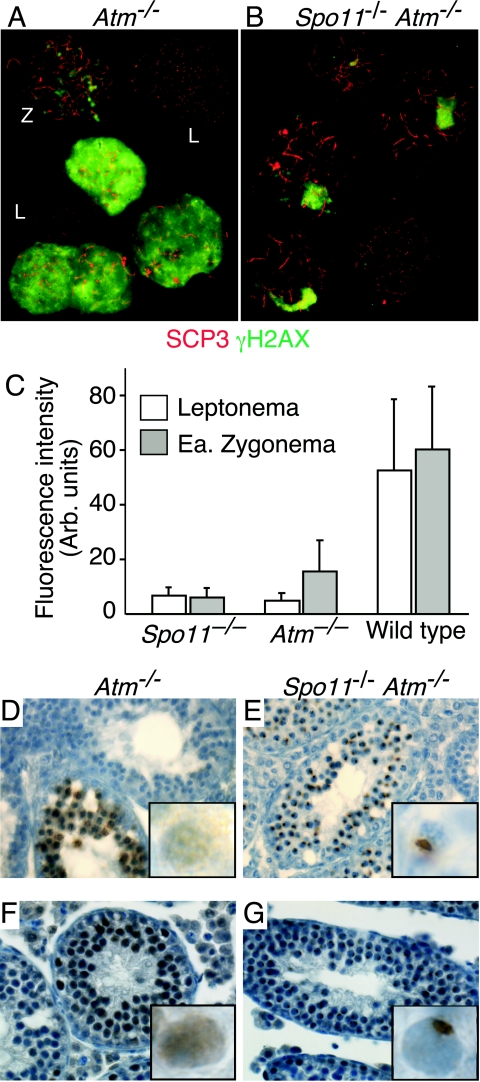
To determine whether ATM provides a similar function in the male germ line, we analyzed sex body-associated proteins and histone H1t in Atm−/− and Spo11−/− Atm−/− males. Leptotene chromosomes from Atm−/− males had little or no γH2AX signal, comparable to what was seen in Spo11−/− males (Fig. 9A and C). Zygotene cells showed a γH2AX signal reduced compared to that of the wild type but slightly above that seen in Spo11−/− cells (Fig. 9A and C). This reduction and/or delay of γH2AX formation indicates that ATM is the principal kinase responsible for H2AX phosphorylation in response to SPO11-induced DSBs, in agreement with other recent observations (18, 48). In addition, a subset (34/75) of Atm−/− spermatocytes was observed with an abnormally high γH2AX signal dispersed across the chromatin (Fig. 9A). These nuclei usually exhibited short, fragmented synaptonemal complexes as well (26/34).
FIG. 9.
Atm−/− spermatocytes show hallmarks of the presence of poorly repaired DSBs. (A and B) Collages of chromosome spreads from Atm−/− and Spo11−/− Atm−/− testes, stained with anti-SCP3 (red) and anti-γH2AX (green). Atm−/− leptotene cells (L) showed little or no γH2AX signal, comparable to Spo11−/− cells. Zygotene cells (Z) had a γH2AX signal that was above that of Spo11−/− cells although reduced compared to that of the wild type. Atm−/− spermatocytes were also observed with abundant γH2AX signal dispersed across the chromatin (bright green staining). Spo11−/− Atm−/− double mutants were indistinguishable from Spo11−/− single mutants. (C) Fluorescence intensity (in arbitrary [Arb.] units) of the γH2AX signal of Atm−/− compared to those of Spo11−/− and wild-type leptotene andearly (Ea.) zygotene spermatocytes. The γH2AX (bright) Atm−/− cells were excluded from this quantification. (D to G) Testis sections of Atm−/− and Spo11−/− Atm−/− mice stained for γH2AX (D and E) or NBS1 (F and G). Insets show higher-magnification views of individual spermatocytes. The defect in sex body formation in Atm−/− cells is apparent in these sections; the Spo11−/− mutation rescues this defect to allow the localization of both γH2AX and NBS1 to pseudo-sex bodies.
Spreads with accumulations of γH2AX similar to sex bodies were rarely observed (1/40 SCP3-positive spreads). This defect was verified in spermatocyte squashes stained for XMR (Fig. 7G and I) and in testis sections stained for γH2AX or NBS1 (Fig. 9D and F, respectively). Histone H1t deposition was nearly as defective as that in Dmc1−/− mutants (Fig. 7G and I). The Atm−/− mutant block to the deposition of histone H1t and the localization of sex body-associated proteins is dependent on the presence of SPO11-induced DSBs, because both processes in Spo11−/− Atm−/− double mutants were indistinguishable from those in Spo11−/− single mutants (Fig. 7H and I and 9B, E, and G). Thus, Atm−/− spermatocytes show characteristics similar to those of Dmc1−/− mutants, indicating that ATM is required for the proper repair of SPO11-induced DSBs in spermatocytes, as in oocytes. Importantly, ATM activity is not required for spermatocyte apoptosis in the absence of SPO11, because the Atm−/− mutation did not ameliorate cell death in Spo11−/− cells (Fig. 9E and G).
DISCUSSION
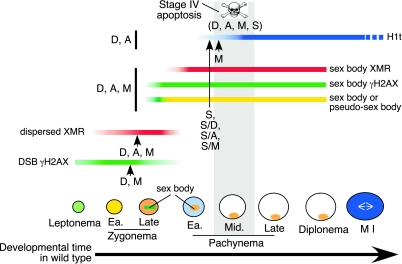
Recombination defects in mammalian meiosis cause arrest and programmed cell death, but it is not always clear whether surveillance mechanisms are responding to the absence of recombination, the improper repair of recombination intermediates, defects in chromosome structure or other processes downstream of recombination, or some combination of these. Mouse oocytes mount distinct DNA damage-dependent and independent responses to recombination defects (13). Unlike oocytes, however, spermatocytes were expected to respond similarly to the presence and absence of DNA damage, given the apparently similar timing of apoptosis in Spo11−/−, Dmc1−/−, and other mutants. To address this apparent sexual dimorphism, we analyzed male meiotic progression in these mutants. Our studies demonstrate that spermatocytes mount distinct responses to different recombination defects, including persistent DSBs (summarized in Fig. 10).
FIG. 10.
Markers of meiotic progression in the wild type and recombination mutants. The stages of spermatogenic development and the patterns for various molecular markers in the wild type are shown. For the mutants analyzed here, the most advanced stage for each marker is indicated with a vertical line or arrow coded as follows: A, the Atm−/− mutant; D, the Dmc1−/− mutant; M, the Msh5−/− mutant; S, the Spo11−/− mutant; S/A, S/D, and S/M, double mutants containing the Spo11−/− mutation along with the Atm−/−, Dmc1−/−, and Msh5−/− mutations, respectively. The gray box indicates the developmental stage when apoptosis occurs. Note the distinct behaviors of molecular markers among the different mutants. Also note the temporal distinction between the onset of physiological defects, as revealed by the molecular markers, and the timing of apoptosis for Spo11−/−, Dmc1−/−, Msh5−/−, and Atm−/− spermatocytes. See the text for further discussion. Ea., early.
Meiotic progression in recombination-defective spermatocytes.
Comparison of Spo11−/− and Dmc1−/− phenotypes is particularly informative because these mutations cause very different molecular defects. Meiotic DSBs do not form in Spo11−/− meiocytes (5, 27, 40), so effects of this mutation must be independent of the presence of unrepaired recombination intermediates. In contrast, in yeast dmc1 mutants (6) and likely in mouse Dmc1−/− mutants as well, DSBs persist as a result of impaired strand invasion. As in Spo11 mutants, Dmc1−/− spermatocytes undergo apoptosis at epithelial stage IV. Nevertheless, analysis of molecular markers reveals that Dmc1−/− spermatocytes are physiologically different from Spo11−/− spermatocytes—DNA damage checkpoint-associated factors (TOPBP1, γH2AX) persist on the chromatin in Dmc1−/− spermatocytes, unlike in the wild type or in Spo11−/− mutants, and the deposition of histone H1t and formation of pseudo-sex bodies occurs in Spo11−/− mutants but not in Dmc1−/− mutants (Fig. 10). We interpret these differences to mean that unrepaired DSBs trigger a regulatory (checkpoint) response in Dmc1−/− spermatocytes, as in oocytes. The epistasis relationship with Spo11−/− spermatocytes supports this conclusion. Importantly, alterations in TOPBP1 and γH2AX behavior in Dmc1−/− cells occur at or before zygonema and thus cannot reflect operation of a “pachytene checkpoint.” It is possible that the absence of sex body formation in Dmc1−/− mutants is a direct consequence of the DSB repair defect rather than a consequence of a regulatory response per se. For example, because NBS1 is involved in responses to unrepaired DSBs (34), it might not be available for sex body formation if it is sequestered elsewhere. While this may explain the lack of sex body formation, it does not, however, satisfactorily account for the behavior of histone H1t, which is not likely to play a role in DSB responses or repair. Thus, our data suggest that meiotic progression is arrested in Dmc1−/− spermatocytes at a stage approximately equivalent to late zygonema in the wild type, significantly earlier than the stage at which apoptosis occurs (Fig. 10).
Although the biochemical role of MSH5 in recombination is not well understood, the persistence of γH2AX and RAD51 foci in Msh5−/− spermatocytes (15, 27), as well as its epistatic relationship with Spo11 in oocytes and spermatocytes (reference 13; this study), indicates that MSH5 is required for proper DSB repair in the mouse, as it is in yeast (e.g., reference 7). Chromosome-associated complexes of TOPBP1 persist and accumulate to higher-than-normal levels in Msh5−/− mutants, as in Dmc1−/− mutants. Notably, however, unlike Dmc1−/− spermatocytes, rare Msh5−/− spermatocytes have faint or no persistent TOPBP1 foci, and histone H1t deposition occurs in Msh5−/− spermatocytes. Thus, the responses in Msh5−/− and Dmc1−/− spermatocytes are overlapping but not identical. This distinction may be related to MSH5 being required for the repair of only a subset of DSBs (7, 24).
Cytological evidence suggests that TOPBP1 functions in meiotic checkpoint responses in the mouse (33). Our findings support this interpretation; chromosome-associated complexes of TOPBP1 are absent in Spo11−/− spermatocytes, whereas they persist and accumulate to higher-than-normal levels in Dmc1−/− and Msh5−/− spermatocytes. Interestingly, TOPBP1 foci were specifically absent from portions of chromosome axes that had undergone synapsis in Dmc1−/− and Msh5−/− mutants. A reasonable interpretation is that the disappearance of TOPBP1 results from the repair of DSBs in these regions. Because synapsis in these mutants occurs between nonhomologous chromosomes, we suggest that this nonhomologous synapsis correlates with the local relaxation of barriers against using the sister chromatid for recombination.
Relationship of recombination progression to sex body formation.
Surprisingly, Spo11−/− spermatocytes form pseudo-sex bodies that rarely include the X or Y chromosomes. Sex body-associated proteins also assemble on unsynapsed portions of autosomes in translocation heterozygotes that otherwise exhibit wild-type meiotic recombination functions (46, 48). These findings argue against the existence of cis-acting sequences that are required to nucleate sex body formation on either the X or Y chromosomes. Instead, it appears that sex body-related structures can form anywhere in the genome but are normally restricted to the XY pair by virtue of these being the only unsynapsed chromosomes. That Spo11−/− spermatocytes show synaptic failure across much of the genome yet assemble pseudo-sex bodies in only a localized region(s) implies, however, that asynapsis is not the sole determinant of these chromatin domains. Persistent SPO11-generated DSBs in unsynapsed regions may normally nucleate the formation of these domains, although they are clearly not a necessary trigger for pseudo-sex body formation.
Even the formation of pseudo-sex bodies was blocked in Dmc1−/− and Msh5−/− spermatocytes, implying that the persistence of global DSBs prevents formation of the sex body. For Dmc1−/− spermatocytes, a DNA damage-dependent regulatory response may arrest cells in a state that is not competent for sex body formation. However, a more direct role for persistent DSBs in preventing sex body formation is suggested by the observation that Msh5−/− spermatocytes progress further than Dmc1−/− spermatocytes, as judged by H1t accumulation, and yet fail to form sex bodies (Fig. 10). Because many sex body-associated proteins are involved in DNA damage responses, persistent DSBs throughout the genome in both Msh5−/− and Dmc1−/− spermatocytes may titrate factors involved in sex body formation.
Atm is required for normal DSB repair in meiosis.
ATM and its relative ATR mediate regulatory responses to DNA damage (43), but these proteins also promote the proper repair of DNA damage (37, 43) and basic chromosomal events in unperturbed cells (8). We show here that Atm-deficient spermatocytes share similarities with Dmc1 mutants (Fig. 10), indicating that a lack of ATM yields a damage-dependent response in males, similar to females (13). These findings indicate that ATM promotes the proper repair of meiotic DSBs. This hypothesis can account for the persistence of putative recombination intermediates in cytological analyses of Atm−/− spermatocytes (2, 4).
In addition to DNA damage, alterations in chromosome structures can also activate ATM in somatic cells (3). It was thus possible that chromosome structure defects in Spo11−/− spermatocytes might trigger a response through the activation of ATM. This possibility is ruled out because ATM activity is not required for spermatocyte loss in the absence of SPO11.
ATM is required for the majority of the γH2AX signal caused by SPO11-induced DSBs in leptotene-zygotene spermatocytes (18, 48; this study). Nonetheless, we find that γH2AX does form at low levels in Atm−/− cells, and in some cells at very high levels, in response to SPO11-generated DSBs. The kinase responsible for this H2AX phosphorylation is presently unknown, but ATR is a likely candidate. γH2AX formation in the (pseudo-) sex body was independent of ATM kinase activity, as determined by epistasis analysis with Spo11−/− spermatocytes. ATR is again implicated as the responsible kinase (46, 48).
Programmed cell death of recombination-defective spermatocytes at epithelial stage IV.
A number of mutants defective for meiotic recombination and/or meiosis-specific chromosome structures undergo apoptosis at approximately the same stage, equivalent to stage IV of the normal seminiferous epithelial cycle (reviewed in references 11 and 21). An important challenge is to understand what triggers cell death. A traditional explanation is that apoptosis reflects the activation of a cell-autonomous regulatory response, often referred to as a pachytene checkpoint (e.g., references 2 and 38). If this is true, then this apoptotic checkpoint is triggered by many different mutations and, importantly, is temporally distinguished and independent from the DSB-dependent response shown here for the Dmc1−/− mutant and other mutants. Either several different checkpoint mechanisms converge on the same apoptosis pathway or apoptosis is caused by a DSB-independent defect that all of these mutants share. Candidates for a common defect could be synapsis or sex body formation. For example, transcriptional repression of the chromatin in the sex body is presumably perturbed in each of the mutants (18a, 48). Thus, it is possible that the failure to silence XY chromosome-resident genes provokes an apoptotic response at pachytene.
As an alternative to this traditional view of a pachytene checkpoint, we propose that the apoptosis of recombination-defective spermatocytes might reflect the operation of a cell-nonautonomous pathway. Spermatogenesis occurs in intimate association with Sertoli cells, which provide intercellular signals that govern the proliferation and differentiation of germ cells (9). We hypothesize that Sertoli cells perform quality control surveillance of recombination-defective spermatocytes and induce apoptosis and that epithelial stage IV represents the developmental time at which this surveillance occurs. Alternatively, Sertoli cells might provide a signal at stage IV that stimulates developmental progression in normal spermatocytes, with the mutants undergoing apoptosis because they are unable to respond properly to this signal. Because extrinsic cues rather than intrinsic ones govern the timing of spermatocyte apoptosis in these models, these scenarios account for why different molecular defects trigger apoptosis at the same stage and why the timing of oocyte loss differs.
Acknowledgments
We thank James Turner (MRC National Institute for Medical Research) for insightful discussions, Margaret Leversha and Lei Zhang (MSKCC, Molecular Cytogenetics Core Facility) for FISH, Katia Manova and Craig Farrell (MSKCC, Molecular Cytology Core Facility) for help with histology, Qingwen Wang (MSKCC) for providing some of the mice used in this study, and Nadine Kolas (Albert Einstein College of Medicine) for initial help with chromosome spreading. We also thank the many investigators who generously provided antibodies and John Schimenti (Cornell), Winfried Edelmann (Albert Einstein College of Medicine), and Raju Kucherlapati (Harvard) for providing mutant mice. M.B. and M.D.G. thank Margherita G. Barchi for her support and members of the Jasin and Keeney labs for comments and suggestions.
This work was supported in part by the Lalor Foundation (M.B. and M.D.G.), an American-Italian Cancer Foundation Fellowship (M.B.), and NIH HD40916 (M.J. and S.K.).
REFERENCES
- 1.Araki, H., S. H. Leem, A. Phongdara, and A. Sugino. 1995. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 92:11791-11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley, T., C. Westphal, A. Plug-de Maggio, and D. G. de Rooij. 2004. The mammalian mid-pachytene checkpoint: meiotic arrest in spermatocytes with a mutation in Atm alone or in combination with a Trp53 (p53) or Cdkn1a (p21/cip1) mutation. Cytogenet. Genome Res. 107:256-262. [DOI] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, C., M. Liyanage, P. B. Moens, M. Tarsounas, K. Nagashima, K. Brown, S. Rottinghaus, S. P. Jackson, D. Tagle, T. Ried, and A. Wynshaw-Boris. 1998. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 125:4007-4017. [DOI] [PubMed] [Google Scholar]
- 5.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 7.Borner, G. V., N. Kleckner, and N. Hunter. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117:29-45. [DOI] [PubMed] [Google Scholar]
- 8.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, C. Y., and D. D. Mruk. 2002. Cell junction dynamics in the testis: Sertoli-germ cell interactions and male contraceptive development. Physiol. Rev. 82:825-874. [DOI] [PubMed] [Google Scholar]
- 10.de Rooij, D. G. 1998. Stem cells in the testis. Int. J. Exp. Pathol. 79:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rooij, D. G., and P. de Boer. 2003. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet. Genome Res. 103:267-276. [DOI] [PubMed] [Google Scholar]
- 12.de Vries, S. S., E. B. Baart, M. Dekker, A. Siezen, D. G. de Rooij, P. de Boer, and H. te Riele. 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Giacomo, M., M. Barchi, F. Baudat, W. Edelmann, S. Keeney, and M. Jasin. 2005. Distinct DNA damage-dependent and independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. USA 102:737-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaker, S., A. Pyle, J. Cobb, and M. A. Handel. 2001. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J. Cell Sci. 114:2953-2965. [DOI] [PubMed] [Google Scholar]
- 15.Edelmann, W., P. E. Cohen, B. Kneitz, N. Winand, M. Lia, J. Heyer, R. Kolodner, J. W. Pollard, and R. Kucherlapati. 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21:123-127. [DOI] [PubMed] [Google Scholar]
- 16.Eijpe, M., H. Offenberg, W. Goedecke, and C. Heyting. 2000. Localisation of RAD50 and MRE11 in spermatocyte nuclei of mouse and rat. Chromosoma 109:123-132. [DOI] [PubMed] [Google Scholar]
- 17.Escalier, D., and H. J. Garchon. 2000. XMR is associated with the asynapsed segments of sex chromosomes in the XY body of mouse primary spermatocytes. Chromosoma 109:259-265. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Capetillo, O., B. Liebe, H. Scherthan, and A. Nussenzweig. 2003. H2AX regulates meiotic telomere clustering. J. Cell Biol. 163:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Fernandez-Capetillo, O., S. K. Mahadevaiah, A. Celeste, P. J. Romanienko, R. D. Camerini-Otero, W. M. Bonner, K. Manova, P. Burgoyne, and A. Nussenzweig. 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4:497-508. [DOI] [PubMed] [Google Scholar]
- 19.Hamer, G., H. B. Kal, C. H. Westphal, T. Ashley, and D. G. de Rooij. 2004. Ataxia telangiectasia mutated expression and activation in the testis. Biol. Reprod. 70:1206-1212. [DOI] [PubMed] [Google Scholar]
- 20.Handel, M. A. 2004. The XY body: a specialized meiotic chromatin domain. Exp. Cell Res. 296:57-63. [DOI] [PubMed] [Google Scholar]
- 21.Hunt, P. A., and T. J. Hassold. 2002. Sex matters in meiosis. Science 296:2181-2183. [DOI] [PubMed] [Google Scholar]
- 22.Inselman, A., S. Eaker, and M. A. Handel. 2003. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet. Genome Res. 103:277-284. [DOI] [PubMed] [Google Scholar]
- 23.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 24.Kolas, N. K., and P. E. Cohen. 2004. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107:216-231. [DOI] [PubMed] [Google Scholar]
- 25.Lipkin, S. M., P. B. Moens, V. Wang, M. Lenzi, D. Shanmugarajah, A. Gilgeous, J. Thomas, J. Cheng, J. W. Touchman, E. D. Green, P. Schwartzberg, F. S. Collins, and P. E. Cohen. 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31:385-390. [DOI] [PubMed] [Google Scholar]
- 26.Lydall, D., Y. Nikolsky, D. K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 27.Mahadevaiah, S. K., J. M. A. Turner, F. Baudat, E. P. Rogakou, P. de Boer, J. Blanco-Rodriguez, M. Jasin, S. Keeney, W. M. Bonner, and P. S. Burgoyne. 2001. Recombinational DNA double strand breaks in mice precede synapsis. Nat. Genet. 27:271-276. [DOI] [PubMed] [Google Scholar]
- 28.Makiniemi, M., T. Hillukkala, J. Tuusa, K. Reini, M. Vaara, D. Huang, H. Pospiech, I. Majuri, T. Westerling, T. P. Makela, and J. E. Syvaoja. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276:30399-30406. [DOI] [PubMed] [Google Scholar]
- 29.Moens, P. B., D. J. Chen, Z. Shen, N. Kolas, M. Tarsounas, H. H. Q. Heng, and B. Spyropoulos. 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106:207-215. [DOI] [PubMed] [Google Scholar]
- 30.Moens, P. B., R. Freire, M. Tarsounas, B. Spyropoulos, and S. P. Jackson. 2000. Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113:663-672. [DOI] [PubMed] [Google Scholar]
- 31.Oakberg, E. F. 1956. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 99:507-516. [DOI] [PubMed] [Google Scholar]
- 32.Page, J., J. A. Suja, J. L. Santos, and J. S. Rufas. 1998. Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. 6:639-642. [DOI] [PubMed] [Google Scholar]
- 33.Perera, D., L. Perez-Hidalgo, P. B. Moens, K. Reini, N. Lakin, J. E. Syvaoja, P. A. San-Segundo, and R. Freire. 2004. TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol. Biol. Cell 15:1568-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrini, J. H., and T. H. Stracker. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458-462. [DOI] [PubMed] [Google Scholar]
- 35.Pittman, D. L., J. Cobb, K. J. Schimenti, L. A. Wilson, D. M. Cooper, E. Brignull, M. A. Handel, and J. C. Schimenti. 1998. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1:697-705. [DOI] [PubMed] [Google Scholar]
- 36.Reini, K., L. Uitto, D. Perera, P. B. Moens, R. Freire, and J. E. Syvaoja. 2004. TopBP1 localises to centrosomes in mitosis and to chromosome cores in meiosis. Chromosoma 112:323-330. [DOI] [PubMed] [Google Scholar]
- 37.Riballo, E., M. Kuhne, N. Rief, A. Doherty, G. C. Smith, M. J. Recio, C. Reis, K. Dahm, A. Fricke, A. Krempler, A. R. Parker, S. P. Jackson, A. Gennery, P. A. Jeggo, and M. Lobrich. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell 16:715-724. [DOI] [PubMed] [Google Scholar]
- 38.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 39.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 40.Romanienko, P. J., and R. D. Camerini-Otero. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6:975-987. [DOI] [PubMed] [Google Scholar]
- 41.Russell, L. D. 1990. Histological and histopathological evaluation of the testis, 1st ed. Cache River Press, Quick Publishing, St. Louis, Mo.
- 42.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 44.Shimada, M., K. Nabeshima, T. Tougan, and H. Nojima. 2002. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 21:2807-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spyropoulos, B., and P. B. Moens. 1994. In situ hybridization of meiotic prophase chromosomes. Methods Mol. Biol. 33:131-139. [DOI] [PubMed] [Google Scholar]
- 46.Turner, J. M., O. Aprelikova, X. Xu, R. Wang, S. Kim, G. V. Chandramouli, J. C. Barrett, P. S. Burgoyne, and C. X. Deng. 2004. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 14:2135-2142. [DOI] [PubMed] [Google Scholar]
- 47.Turner, J. M., S. K. Mahadevaiah, D. J. Elliott, H. J. Garchon, J. R. Pehrson, R. Jaenisch, and P. S. Burgoyne. 2002. Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J. Cell Sci. 115:4097-4105. [DOI] [PubMed] [Google Scholar]
- 48.Turner, J. M., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu, C. X. Deng, and P. S. Burgoyne. 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37:41-47. (First published 5 December 2004; 10.1038/ng1484.) [DOI] [PubMed] [Google Scholar]
- 49.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, B. R., O. K. Mirzoeva, W. F. Morgan, J. Lin, W. Dunnick, and J. H. Petrini. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12:648-653. [DOI] [PubMed] [Google Scholar]
- 51.Xu, Y., T. Ashley, E. E. Brainerd, R. T. Bronson, M. S. Meyn, and D. Baltimore. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411-2422. [DOI] [PubMed] [Google Scholar]
- 52.Yamane, K., X. Wu, and J. Chen. 2002. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22:555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, K., G. Kondoh, Y. Matsuda, T. Habu, Y. Nishimune, and T. Morita. 1998. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1:707-718. [DOI] [PubMed] [Google Scholar]