Abstract
The activation-induced deaminase/apolipoprotein B-editing catalytic subunit 1 (AID/APOBEC) family comprises four groups of proteins. Both AID, a lymphoid-specific DNA deaminase that triggers antibody diversification, and APOBEC2 (function unknown) are found in all vertebrates examined. In contrast, APOBEC1, an RNA-editing enzyme in gastrointestinal cells, and APOBEC3 are restricted to mammals. The function of most APOBEC3s, of which there are seven in human but one in mouse, is unknown, although several human APOBEC3s act as host restriction factors that deaminate human immunodeficiency virus type 1 replication intermediates. A more primitive function of APOBEC3s in protecting against the transposition of endogenous retroelements has, however, been proposed. Here, we focus on mouse APOBEC2 (a muscle-specific protein for which we find no evidence of a deaminating activity on cytidine whether as a free nucleotide or in DNA) and mouse APOBEC3 (a DNA deaminase which we find widely expressed but most abundant in lymphoid tissue). Gene-targeting experiments reveal that both APOBEC2 (despite being an ancestral member of the family with no obvious redundancy in muscle) and APOBEC3 (despite its proposed role in restricting endogenous retrotransposition) are inessential for mouse development, survival, or fertility.
The founder member of the activation-induced deaminase/apolipoprotein B-editing catalytic subunit 1 (AID/APOBEC) family is APOBEC1, the catalytic component of an RNA-editing complex which is restricted to mammals and functions in gastrointestinal tissues to deaminate cytosine to uracil at position 6666 in the RNA encoding apolipoprotein B (26, 41). This deamination leads to the creation of a premature stop codon and to the resulting production of a truncated apolipoprotein B polypeptide. Closely linked to the APOBEC1 gene in both human and mouse is the gene encoding AID (25, 35). The expression pattern and function of AID are quite distinct from those of APOBEC1. AID expression is largely restricted to B lymphocytes (24), where it deaminates cytosines in specific regions of the immunoglobulin locus DNA, acting as a central trigger of antibody gene diversification (27).
There are two further groups of AID/APOBEC proteins; their genes are not linked to the AID/APOBEC1 cluster. APOBEC2 (which has also been designated ARCD1 [1]) is encoded on human chromosome 6. It was first identified as a sequence homologue of APOBEC1 (19) and has been proposed to exhibit deaminating activity on free deoxycytidine (1, 19). However, while APOBEC2 is clearly much more similar to other members of the AID/APOBEC family than it is to any other gene in humans (19), it is nevertheless less related to other members of the AID/APOBEC family than they are to each other (8). It also has a distinct exon/intron structure. APOBEC2 predates APOBEC1 and APOBEC3s, whose homologues have so far been identified only in mammals, while APOBEC2 and AID homologues are found among bony fish as well as other lower vertebrates (8, 50). Expression of APOBEC2 was originally documented as being restricted exclusively to cardiac and skeletal muscle (19), although a lesser level of expression elsewhere was subsequently observed (1). The function of APOBEC2 is not known, although it was originally proposed, by analogy with APOBEC1, to be an RNA-editing enzyme (1, 19). In that case, it would be the only proposed RNA-editing enzyme apart from the adenosine deaminases (36) to be widely expressed among vertebrates. Clearly, it would be interesting to know the effects of APOBEC2 deficiency.
As is the case with APOBEC1, APOBEC3 proteins appear to be restricted to mammals, with mice and rats having single APOBEC3 genes. However, primates harbor substantially expanded loci, with the APOBEC3 cluster in humans containing seven members (8, 15, 42). The first APOBEC3 family member (APOBEC3A, originally designated phorbolin-1) was identified as a protein whose expression was strongly induced on the treatment of human keratinocytes with phorbol ester (21, 34). Analysis of the sequence of human chromosome 22 revealed the existence of a cluster of related sequences, which have been designated APOBEC3A to H (8, 15).
In an independent line of work, APOBEC3G was identified in a subtractive hybridization screen in a nonpermissive human T-cell line for host factors that were responsible for restricting infection by Vif-deficient human immunodeficiency virus type 1 (HIV-1) particles (37). Transfection experiments then revealed that APOBEC3G was packaged into HIV-1 particles and acted to deaminate cytosine to uracil in the first-strand DNA of the HIV replication intermediates (13, 18, 22, 23, 49). Similar antilentiviral activities have subsequently been noted for APOBEC3F and APOBEC3C (3, 17, 20, 45, 48, 51). That APOBEC3F and APOBEC3G can indeed act on HIV-1 during infection of humans is indicated by analysis of the sequences of hypermutated HIV-1 genomes isolated from infected individuals (2, 3, 4, 12, 20, 44). However, the function of several of the human APOBEC3 proteins is unknown. Moreover, the single mouse APOBEC3 has not as of yet been observed to exhibit restricting activity against any mouse retrovirus. Indeed, it has been proposed that APOBEC3 enzymes might provide an ancestral wide defense against the transposition of endogenous retroelements, since both mouse APOBEC3 and human APOBEC3G are able to inhibit the transposition of endogenous mouse retroelements in cotransfection assays in a human cell line (11). Furthermore, the expression of human or mouse APOBEC3 family members in yeast has been shown to be able to inhibit Ty element transposition (10). Clearly, therefore, it would be interesting to know whether the sole APOBEC3 gene in mouse is essential in the prevention of unacceptably high frequencies of transposition of endogenous retroelements.
MATERIALS AND METHODS
Assay of recombinant APOBEC2.
APOBEC2 cDNA was expressed in Escherichia coli either as a His6-tagged protein by use of a pTrc99-based vector (14) or as a fusion with maltose-binding protein (MBP) by use of a pOPTM vector (gift from R. Williams) and purified on Ni-NTA (QIAGEN) or amylose resin (NEB). The abilities of these recombinant proteins (10 μg) as well as of a recombinant APOBEC1 control (30) to deaminate dC in the context of an oligodeoxyribonucleotide were monitored using a set of 5′-biotinylated, 3′-fluorescein-conjugated single-stranded oligonucleotide substrates (SPM317 to 325) that contained dC in a variety of different sequence contexts (2). Successful deamination was then assessed by gel electrophoresis after treatment with uracil-DNA glycosylase/NaOH, as previously described, to cleave the oligodeoxyribonucleotide at sites of dC deamination (2, 30).
Deaminase activity on free dC was monitored following a 10-min incubation of recombinant APOBEC (2 μg) with 3 μCi deoxy[5-3H]cytidine (160 pmol; 18.4 Ci/mmol; Amersham-Pharmacia) in 10 μl deamination buffer prior to the performance of thin-layer chromatography on polyethylene-imine cellulose plates as previously described (2). E. coli cdd dcd strain SO177 (CGSC6127) was used as the cytidine deaminase-/dCTP deaminase-deficient host. 3,4,5,6-Tetrahydrouridine (THU) was obtained from Calbiochem.
Generation and breeding of gene-targeted mice.
The APOBEC2-targeting construct, generated by PCR from 129/Ola mouse DNA, comprises a 1.1-kb neomycin (Neo) resistance cassette (28) inserted as a ClaI fragment between two sections of the APOBEC2 gene. The 5′ APOBEC2 segment extends 2.75 kb from the SpeI site in the first intron through to nucleotide (nt) 282 of exon 2, at which a ClaI site was created by the oligonucleotide used for PCR amplification (5′-GACTAGTTATAGCAAGATAAGCAACCCCTCTGGCAG). The 0.76-kb 3′ APOBEC2 sequence extends from a PCR-generated ClaI site (oligonucleotide 5′-GCCATCGATCCCTGCGTCGCCTGCGCTTGGCCGCCCTTACTCTG) introduced into nucleotide 314 of APOBEC2 exon 2 through to the XbaI site in the second APOBEC2 intron. The targeting construct therefore deletes 32 bp from exon 2 as well as inserting a Neo cassette. A diphtheria toxin A expression cassette (46) was added into the SpeI site of the left arm of the targeting construct. Targeted integration was monitored using PCR-generated probes derived from regions of APOBEC2 flanking either the left (hybridizing a 2,719-bp probe generated by amplification with 5′-GATTACAGACAGCGGCCGCCCCACATCTGACTCTTTA-CTTAGG and 5′-ATAACTA-GTCTCTAGGCTGGGGTGTGGCTTCCTGAGAG to an NdeI/XbaI digest) or right (hybridizing a 3,060-bp probe generated by amplification with 5′-CTTTCTAGATCCCAGTTCCAAAGCCACCACCTC and 5′-AATGAATTCATGACGTCCCGA-CGTATGTTTTGAGTTCTCTG-AGAG to a KpnI/SexAI digest) side of the targeting construct.
For APOBEC3, the targeting construct was generated by PCR from 129/Ola mouse DNA with the 5.6 kb of 5′-flanking APOBEC3 sequence extending from 2.1 kb upstream of the initiator ATG codon through to 148 nt into exon 3 using oligonucleotides (5′-GCGCAGATGCGGCCGCGCACTATGTCCTCCTTTGGATCTG and 5′-CGATCGATCCAGGAACC-TTAGTACCTGC) that introduced terminal NotI and ClaI restriction sites. The 2.9 kb of the 3′-flanking APOBEC3 sequence extending from nucleotide 150 in exon 3 through to nucleotide 2517 of intron 4 was generated by PCR using oligonucleotides (5′-GGATCGATACACACCACAACCTGAGCCTGGAC and 5′-CGGGAATTCGTCGACGCTCTC-AACCTTCCTGCTTACCTGC) that generated terminal ClaI and EcoRI sites. The diphtheria toxin A expression cassette (46) was added into the NotI site in the left arm of the targeting construct. Targeted integration was monitored using PCR-generated probes derived from regions of APOBEC3 flanking either the left (hybridizing a 704-bp probe generated by amplification with 5′-CTATATCACTATAAATCCATTGGTGATTAAGATG and 5′-GCTATCCTTATTTCTTTACAAAGCCAAACTTG to an HincII/AvrII digest) or right (hybridizing a 854-bp probe generated by amplification with 5′-GAGAAGACCCGTCTCTAT-CTAGGCTCTGC and 5′-CATATGAGAGAAATTAAAGCAAAGCTCCTAA to a NdeI/ScaI digest) side of the targeting construct. Polymorphism between 129/Ola and C57BL/6 results in both the EcoNI/AvrII- and HincII/EcoNI-generated fragments from C57BL/6 DNA that hybridize with the left-hand probe being approximately 3.4 kb shorter than those from 129/Ola.
The targeting constructs were transfected by electroporation into E14 (129/Ola) embryonic stem (ES) cells with selection for resistance to G418 (200 μg/ml). Two of the 800 G418-resistant clones transfected with the APOBEC2-targeting construct and 13 of the 500 clones transfected with the APOBEC3-targeting construct harbored targeted integrations into the endogenous loci as judged by Southern blot analysis with both left- and right-hybridizing probes. For both APOBEC2 and APOBEC3, two independent targeted ES cell clones were used to generate chimeric mice by injection into C57BL/6 blastocysts and implantation into pseudopregnant (C57BL/6 × CBA)F1 females. Male chimeric mice were crossed with C57BL/6 females, and the F1 heterozygotes were intercrossed to generate homozygous APOBEC-deficient mice. The animal studies were performed under a project license approved by the Laboratory of Molecular Biology Ethical Review Committee and the United Kingdom Home Office.
Analysis of RNA, protein, and tissues.
Tissues were homogenized in phosphate-buffered saline, and total RNA was extracted using Trizol reagent (Invitrogen). Northern blots of total tissue RNA were hybridized with those that were generated by reverse transcription-PCR (mouse APOBEC3, 5′-GCTGCGTAGCATATGGGACCATTCTGTCTGGGA and 5′-GTCGGTACCTCAAGACATCGGGGGTCCAAGC; mouse APOBEC2, 5′-CCTAATATCCATGGCTCAGAAG-GAAGAGGCCGC and 5′-CATGGTACCCTA-TTCAGGATGTCTGCCAAC; human APOBEC2, 5′-TATAGAATTCCATATGGCCCAGAAGGAAGAGGCTGC and 5′-TATAGAAGGATCCC-TACTTCAGGATGTCTGCCAACTTCTCC).
Western blot assays were carried out using antisera to mouse APOBEC3 and APOBEC2 that had been prepared by immunizing rats and rabbits, respectively, with MBP fusion proteins, and the sera were then depleted of antibodies to MBP by absorption.
Flow cytometric analysis of cell suspensions was performed using fluorescein isothiocyanate-conjugated antibodies to CD4 (L3T4, H129.19), CD3e- (145-2c11), CD45R/B220 (RA3-6B2), Ly-6G, and Ly-6C (Gr-1) (RB6-8C5) and R-phycoerythrin-conjugated antibodies to CD8α (Ly2, 53-6.7), CD45R/B220 (RA3-6B2), and CD11b (Mac-1, M1/70) (all from BD Biosciences Pharmingen) as well as R-phycoerythrin-conjugated goat anti-mouse immunoglobulin M (Santa Cruz Biotechnology Inc.) by use of a BD FACSCalibur analyzer and FCSPress software. Spleen cells were purified on lympholyte M (Cedarlane Labs) density cushions. For histological analyses, tissues were embedded in paraffin, and images of hematoxylin/eosin-stained sections (4 μm) were taken with a Zeiss Axioplan camera system.
RESULTS AND DISCUSSION
Screening recombinant APOBEC2 for deaminase activity.
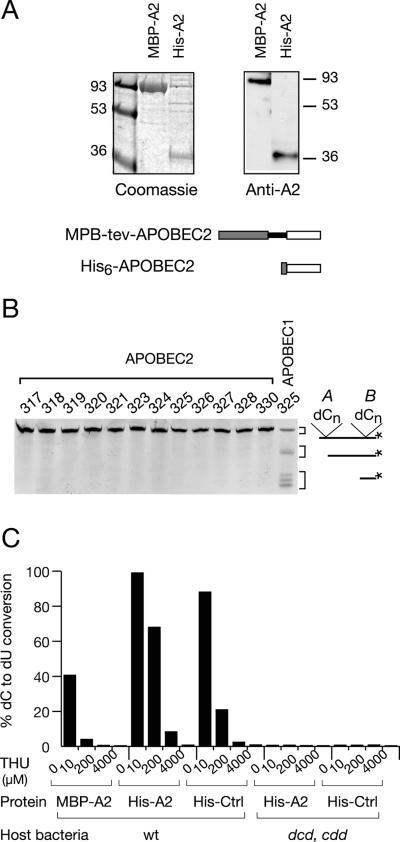
While AID, APOBEC1, and APOBEC3 all have the ability to deaminate cytosine in DNA, as judged by both bacterial mutator and biochemical assays (2, 5, 6, 9, 13, 14, 29, 31, 33, 38, 47), we have previously been unable to observe any activity of APOBEC2 in a mutator assay (14). Biochemical assays with a variety of oligodeoxyribonucleotide substrates also failed to detect any DNA deaminating activating associated with recombinant APOBEC2 (Fig. 1A and B).
FIG. 1.
Testing recombinant APOBEC2 preparations for deaminase activity. (A) His-tagged APOBEC2 and MBP-APOBEC2 fusion proteins were purified from E. coli extracts by affinity chromatography, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, stained with Coomassie blue, and Western blotted with a rabbit anti-APOBEC2 antiserum. Numbers indicate approximate sizes in kilodaltons. (B) No detectable activity of recombinant APOBEC2 on dC in DNA, as judged by the use of a variety of single-stranded 5′-labeled oligodeoxyribonucleotide substrates (317 to 330 as in reference 2) containing two clusters of dC residues (cluster A and cluster B) in different sequence contexts. Following successful deamination (as obtained using the recombinant MBP-APOBEC1 control), the labeled oligonucleotide will yield uracil-containing derivatives which, following uracil-DNA glycosylase/endonuclease treatment and purification on streptavidin-agarose, give rise to shorter, labeled oligonucleotide products. (C) The deoxycytidine deaminase activity present in recombinantAPOBEC2 samples is inhibitable by THU and is undetectable in recombinant APOBEC2 samples prepared from a cdd dcd E. coli host (results for wild-type [wt] E. coli are also shown). Deoxycytidine activity was monitored in recombinant APOBEC2 samples (2 μg) and is expressed as the percentages of the radioactivity in the [(dU)/(dU+dC)] spots following thin-layer chromatography.
However, consistent with previous observations from other groups (1, 19), we find that recombinant APOBEC2 prepared from E. coli (whether as a His-tagged protein or as a fusion with maltose-binding protein) is associated with an activity that deaminates free deoxycytidine (Fig. 1C). We were concerned that this deaminating activity could be due to a low-level contamination of the recombination APOBEC2 by a bacterial deaminase from the E. coli host, since we and others have previously suggested that the contamination of the recombinant proteins by a bacterial deaminase might account for earlier proposals that AID and APOBEC1 can deaminate free deoxycytidine (2, 40). Indeed, we noted that the cytidine-deaminating activity associated with the recombinant APOBEC2 is sensitive to inhibition by THU (Fig. 1C). In this respect, it parallels the E. coli cytidine deaminase (which is highly sensitive to THU [7]) but contrasts with the DNA-deaminating activity of APOBEC1 and APOBEC3G (which are THU resistant [2, 30]). We therefore prepared recombinant APOBEC2 from an E. coli host which is genetically deficient in both cytidine and dCTP deaminases; no cytidine deaminating activity was detected using this recombinant APOBEC2 preparation (Fig. 1C). We conclude that the cytidine deaminating activity that we detected in our other recombinant APOBEC2 preparations is likely to be due to low-level contamination by a host E. coli enzyme; the activity observed in the recombinant APOBEC2 preparations could be accounted for by <0.01% contamination by bacterial cytidine deaminase. Thus, we do not consider that we have any evidence that APOBEC2 is able to deaminate deoxycytidine either as a free nucleotide or in DNA.
APOBEC2-deficient mice.
In order to ascertain whether APOBEC2 was essential for mouse development, gene targeting was used to disrupt the APOBEC2 loci in ES cells, and the targeted ES cell clones were used to generate mouse lines (Fig. 2). Intercrosses of the APOBEC2-targeted heterozygotes gave rise to APOBEC2-targeted homozygotes at the expected Mendelian frequency (Fig. 3A).
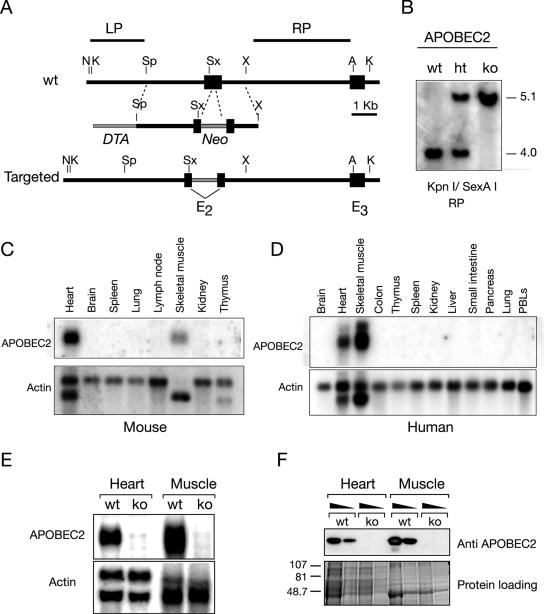
FIG. 2.
Generation of APOBEC2-deficient mice. (A) Targeting construct. The construct contains a Neo selective marker inserted into the second exon of APOBEC2 and includes a flanking diphtheria toxin A-encoding cassette (DTA) to allow selection for homologous integration. A, AatII; K, KpnI; N, NdeI; Sx, SexAI; Sp, SpeI; X, XbaI. (B) Targeted integration of the construct was assessed by Southern blot analysis hybridizing with probe RP to KpnI/SexAI-digested DNA (as shown) as well as with probe LP to NdeI/XbaI digests (not shown). wt, wild type; ht, heterozygous; ko, knockout. Numbers indicate approximate size in kilobases. (C and D) Expression of APOBEC2 RNA in tissues of normal mice and humans as judged by Northern blot analysis. (E) Absence of APOBEC2 RNA in heart and skeletal muscle of homozygous APOBEC2-targeted mice as judged by Northern blot analysis. (F) Absence of APOBEC2 protein in heart and skeletal muscle of homozygous APOBEC2-targeted mice as judged by Western blot analysis of whole-tissue extracts using a rabbit anti-APOBEC2 antiserum. Numbers indicate aproximate sizes in kilodaltons.
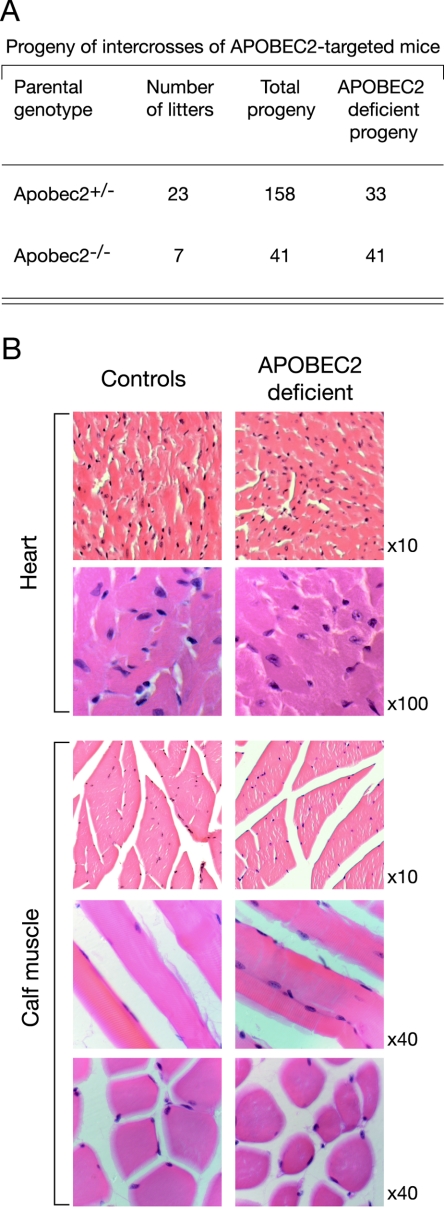
FIG. 3.
Analysis of APOBEC2-deficient mice. (A) Breeding of APOBEC2-targeted mice generates homozygous APOBEC2-deficient mice which are fertile. The data are a compilation of breedings, with the litters deriving from five distinct sets of breedings. (B) Histological analysis of calf and heart muscles reveals no difference between control and APOBEC2-deficient mice. Magnifications are indicated.
Previous studies have revealed that APOBEC2 is either exclusively (19) or preferentially (1) expressed in cardiac and skeletal muscle. In our Northern blot analyses of both human and mouse tissues, we failed to detect APOBEC2, except in heart and skeletal muscle (Fig. 2C and D). Northern blot analysis of heart and skeletal muscle from homozygote APOBEC2-targeted mice revealed that the gene disruption has indeed ablated APOBEC2 expression (Fig. 2E); this conclusion is further supported by Western blot analysis of APOBEC2 protein expression in heart and skeletal muscle using a rabbit anti-APOBEC2 antiserum (Fig. 2F).
We have not detected any major effect of APOBEC2 deficiency on mouse health, fertility, or survival up to 1 year of age; the histological examination of heart and calf muscle also failed to reveal any abnormalities (Fig. 3A and B). This lack of a requirement for APOBEC2 is unlikely to reflect redundancy with regard to any other member of the AID/APOBEC family, since we do not detect transcripts of other family members in muscle (not shown). It does of course remain fully possible that APOBEC2 deficiency may have relatively subtle effects on biochemical or physiological aspects of muscle function or its response to aging or damage, especially given that APOBEC2 has been reported to be up-regulated in the skeletal muscle of old rats (32).
APOBEC3-deficient mice.
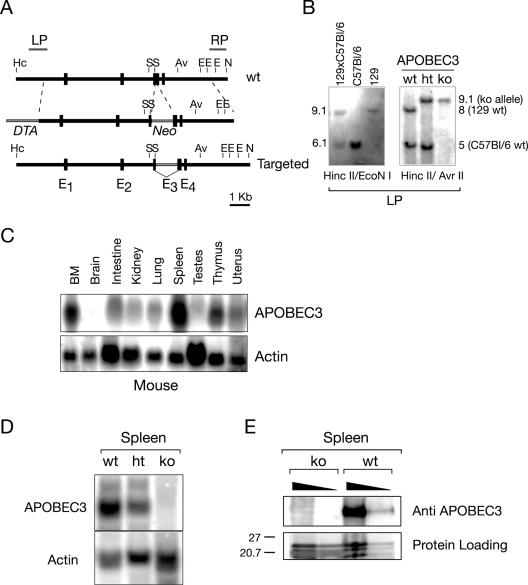
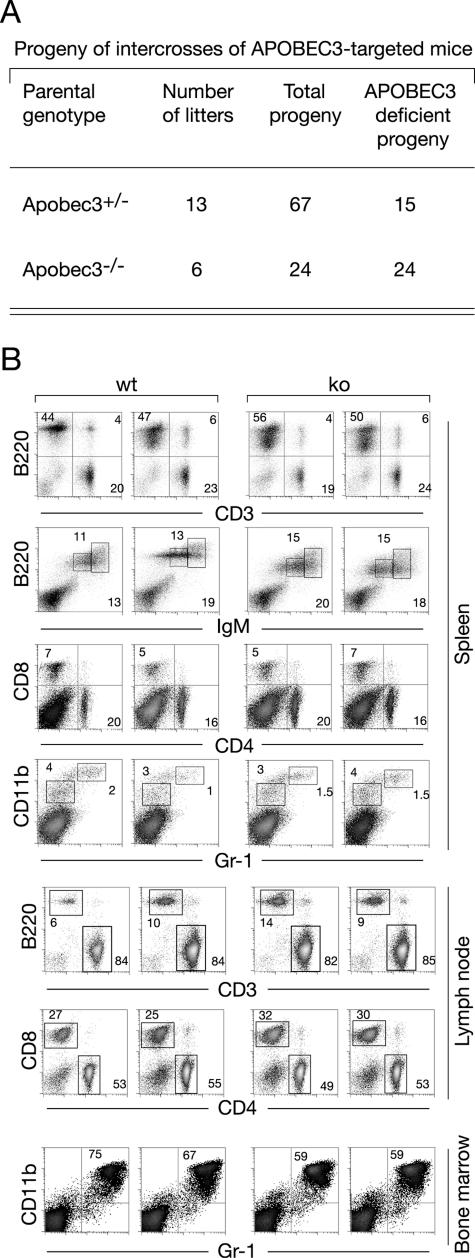
Since mice harbor a single APOBEC3 gene and there is a suggestion that an ancestral function of the APOBEC3 family might be in a providing a defense against endogenous retroelements (10, 11, 43), we were interested in ascertaining whether APOBEC3 deficiency might lead to gross genomic instability or be incompatible with mouse development, survival, or fertility. Gene targeting was used to inactivate the APOBEC3 locus in ES cells; two independently targeted ES cell clones were then used to derive mouse lines. In both cases, it was readily possible to obtain mice homozygous for the targeted gene disruption, as judged by Southern blot analysis (Fig. 4B and 5A). Little is known about the expression pattern of APOBEC3 in mice. We therefore performed a Northern blot analysis of RNA from control animals, which revealed that APOBEC3 is abundantly expressed in spleen as well as in bone marrow (Fig. 4C). A large-scale analysis of the mouse transcriptome (39) is consistent with such a preferential expression of APOBEC3 in mouse lymphoid tissue.
FIG. 4.
Generation of APOBEC3-deficient mice. (A) Targeting construct. The construct contains a Neo selective marker inserted into the third exon of APOBEC3 and includes a flanking diphtheria toxin-encoding cassette (DTA). Av, AvrII; E, EcoNI; Hc, HincII; N, NdeI; S, ScaI. (B) Targeted integration of the construct was assessed by Southern blot analysis hybridizing with probe LP to HincII/AvrII-digested DNA (as shown) as well as with probe RP to NdeI/ScaI digests (not shown). Note [as shown by the Southern blot comparison of the pattern of hybridization of probe LP to HincII/EcoNI-digested DNA from 129/Ola, C57BL/6 and (129/Ola × C57BL/6)F1 mice (129xC57Bl/6) and consistent with sequence databases] that 129/Ola and C57BL/6 mice differ with respect to the sizes of the HincII/EcoNI (and also HincII/AvrII) bands that hybridize with LP, reflecting a 200- to 300-nt deletion in the 129/Ola APOBEC3 first intron, which removes an AvrII and an EcoNI site. wt, wild type; ht, heterozygous; ko, knockout. (C) Expression of APOBEC3 RNA in tissues of normal mice, as judged by Northern blot analysis. (D) Absence of APOBEC3 RNA in spleen of homozygous APOBEC3-targeted mice, as judged by Northern blot analysis. (E) Absence of APOBEC3 polypeptide in splenic extracts of homozygous APOBEC3-targeted mice as judged by Western blot analysis using a rat anti-mouse APOBEC3 antiserum.
FIG. 5.
(A) Breeding of APOBEC3-targeted mice generates homozygous APOBEC3-deficient mice which are fertile. The data are a compilation of breedings established with mice produced from two independent targeted ES cell clones, with the litters deriving from eight distinct sets of breedings. (B) The distribution of splenic lym-phoid subsets is unaffected by APOBEC3 deficiency. The relative proportion of T and B lymphocytes was analyzed by staining with anti-CD3 and anti-CD45R(B220), with T- and B-cell subpopulations further analyzed using antibodies to CD4/CD8 and immunoglobulin M (IgM). Myeloid cells were analyzed by staining for CD11b and Gr-1.
As expected from the nature of the gene disruption, the homozygous gene-targeted mice exhibit a lack of APOBEC3 RNA as well as protein, as judged by Northern and Western blot analysis of spleen (Fig. 4D and E). The mice are nevertheless apparently healthy up to 1 year of age, and we have not observed any increased propensity to tumor development or disease in APOBEC3-deficient mice; both male and female APOBEC3-deficient mice are fertile (Fig. 5A). Although APOBEC3 is most highly expressed in spleen, we have not detected any effects of APOBEC3 deficiency on the abundance of lymphoid subsets (Fig. 5B). While these studies do not exclude the possibility that mouse APOBEC3 might play a role in maintaining genomic integrity, the effect of APOBEC3 deficiency is clearly not so severe that it precludes mouse development, survival, or fertility. Furthermore, it is striking that APOBEC3 is very poorly expressed in testis (Fig. 4C), making it unlikely that if APOBEC3 does indeed play a role in repressing the transposition of endogenous retroelements, this function is fulfilled during the development of the male germ line. With regard to exogenous nucleic acid, we note that while mouse APOBEC3 can act as a restriction element against HIV-1 in human cell line assay systems, it has been found not to act as a restriction factor for a pseudotyped mouse retrovirus (Moloney murine leukemia virus) in similar assay systems (3, 16, 23). It will be interesting in future work to ascertain whether there are any mouse viruses to which APOBEC3-deficient mice do exhibit increased sensitivities.
Acknowledgments
We thank Ted Saunders for help with cell culture; Theresa Langford and staff for animal handling; and Silvo Conticello, Javier Di Noia, and Marc-André Langlois for comments on the manuscript.
The studies were supported in part by grants to M.S.N. from the Association for International Cancer Research and the Leukemia Research Fund.
REFERENCES
- 1.Anant, S., D. Mukhopadhyay, V. Sankaranand, S. Kennedy, J. O. Henderson, and N. O. Davidson. 2001. ARCD-1, an apobec-1-related cytidine deaminase, exerts a dominant negative effect on C to U RNA editing. Am. J. Physiol. Cell Physiol. 281:1904-1916. [DOI] [PubMed] [Google Scholar]
- 2.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585-596. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Borman, A. M., C. Quillent, P. Charneau, K. M. Kean, and F. Clavel. 1995. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G → A hypermutation during negative-strand viral DNA synthesis. Virology 208:601-609. [DOI] [PubMed] [Google Scholar]
- 5.Bransteitter, R., P. Pham, M. D. Scharff, and M. F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F. W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422:726-730. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, R. M., and R. Wolfenden. 1971. Cytidine deaminase from Escherichia coli. Purification, properties and inhibition by the potential transition state analog 3,4,5,6-tetrahydrouridine. J. Biol. Chem. 245:7561-7565. [PubMed] [Google Scholar]
- 8.Conticello, S. G., C. J. Thomas, S. K. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367-377. [DOI] [PubMed] [Google Scholar]
- 9.Dickerson, S. K., E. Market, E. Besmer, and F. N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgibbon, J. E., S. Mazar, and D. T. Dubin. 1993. A new type of G->A hypermutation affecting human immunodeficiency virus. AIDS Res. Hum. Retrovir. 9:833-838. [DOI] [PubMed] [Google Scholar]
- 13.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 14.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10:1247-1253. [DOI] [PubMed] [Google Scholar]
- 15.Jarmuz, A., A. J. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 78:8238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 19.Liao, W., S. H. Hong, B. H. Chan, F. B. Rudolph, S. C. Clark, and L. Chan. 1999. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun. 260:398-404. [DOI] [PubMed] [Google Scholar]
- 20.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 21.Madsen, P., S. Anant, H. H. Rasmussen, P. Gromov, H. Vorum, J. P. Dumanski, N. Tommerup, J. E. Collins, C. L. Wright, I. Dunham, A. J. MacGinnitie, N. O. Davidson, and J. E. Celis. 1999. Psoriasis upregulated phorbolin-1 shares structural but not functional similarity to the mRNA-editing protein apobec-1. J. Investig. Dermatol. 113:162-169. [DOI] [PubMed] [Google Scholar]
- 22.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 23.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 24.Muramatsu, M., V. S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N. O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470-18476. [DOI] [PubMed] [Google Scholar]
- 25.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553-563. [DOI] [PubMed] [Google Scholar]
- 26.Navaratnam, N., J. R. Morrison, S. Bhattacharya, D. Patel, T. Funahashi, F. Giannoni, B. B. Teng, N. O. Davidson, and J. Scott. 1993. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 268:20709-20712. [PubMed] [Google Scholar]
- 27.Neuberger, M. S., R. S. Harris, J. Di Noia, and S. K. Petersen-Mahrt. 2003. Immunity through DNA deamination. Trends Biochem. Sci. 28:305-312. [DOI] [PubMed] [Google Scholar]
- 28.O'Keefe, T., G. T. Williams, S. L. Davies, and M. S. Neuberger. 1996. Hyperresponsive B cells in CD22-deficient mice. Science 274:798-801. [DOI] [PubMed] [Google Scholar]
- 29.Petersen-Mahrt, S. K., R. S. Harris, and M. S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418:99-103. [DOI] [PubMed] [Google Scholar]
- 30.Petersen-Mahrt, S. K., and M. S. Neuberger. 2003. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J. Biol. Chem. 278:19583-19586. [DOI] [PubMed] [Google Scholar]
- 31.Pham, P., R. Bransteitter, J. Petruska, and M. F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103-107. [DOI] [PubMed] [Google Scholar]
- 32.Piec, I., A. Listrat, J. Alliot, C. Chambon, R. G. Taylor, and D. Bechet. 14April2005. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 10.1096/fj.04-3084fje. [DOI] [PubMed]
- 33.Ramiro, A. R., P. Stavropoulos, M. Jankovic, and M. C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452-456. [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen, H. H., and J. E. Celis. 1993. Evidence for an altered protein kinase C (PKC) signaling pathway in psoriasis. J. Investig. Dermatol. 101:560-566. [DOI] [PubMed] [Google Scholar]
- 35.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Genner, I. Tezcan, F. Ersoy, H. Kayserili, A. G. Ugazio, N. Brousse, M. Muramatsu, L. D. Notarangelo, K. Kinoshita, T. Honjo, A. Fischer, and A. Durandy. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102:565-575. [DOI] [PubMed] [Google Scholar]
- 36.Schaub, M., and W. Keller. 2002. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie 84:791-803. [DOI] [PubMed] [Google Scholar]
- 37.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 38.Sohail, A., J. Klapacz, M. Samaranayake, A. Ullah, and A. S. Bhagwat. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su, A. I., M. P. Cooke, K. A. Ching, Y. Hakak, J. R. Walker, T. Wiltshire, A. P. Orth, R. G. Vega, L. M. Sapinoso, A. Moqrich, A. Patapoutian, G. M. Hampton, P. G. Schultz, and J. B. Hogenesch. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99:4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suspene, R. P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng, B., C. F. Burant, and N. O. Davidson. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260:1816-1819. [DOI] [PubMed] [Google Scholar]
- 42.Turelli, P., and D. Trono. 2005. Editing at the crossroad of innate and adaptive immunity. Science 307:1061-1065. [DOI] [PubMed] [Google Scholar]
- 43.Turelli, P., S. Vianin, and D. Trono. 2004. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 279:43371-43373. [DOI] [PubMed] [Google Scholar]
- 44.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanagawa, Y., T. Kobayashi, M. Ohnishi, T. Kobayashi, S. Tamura, T. Tsuzuki, M. Sanbo, T. Yagi, F. Tashiro, and J. Miyazaki. 1999. Enrichment and efficient screening of ES cells containing a targeted mutation: the use of DT-A gene with the polyadenylation signal as a negative selection maker. Transgenic Res. 8:215-221. [DOI] [PubMed] [Google Scholar]
- 47.Yu, K., F. T. Huang, and M. R. Lieber. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279:6496-6500. [DOI] [PubMed] [Google Scholar]
- 48.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, Y., Q. Pan-Hammarstrom, Z. Zhao, and L. Hammarstrom. 2005. Identification of the activation-induced cytidine deaminase gene from zebrafish: an evolutionary analysis. Dev. Comp. Immunol. 29:61-71. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, Y.-H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]