Abstract
Intracellular localization plays an important role in the functional regulation of the cell cycle inhibitor p21. We have previously shown that calmodulin binds to p21 and that calmodulin is essential for the nuclear accumulation of p21. Here, we analyze the mechanism of this regulation. We show that calmodulin inhibits in vitro phosphorylation of p21 by protein kinase C (PKC) and that this inhibition is dependent upon calmodulin binding to p21. Two-dimensional electrophoresis analysis of cells expressing the p21 wild type or p21S153A, a nonphosphorylatable mutant of p21 at position 153, indicates that Ser153 of p21 is a phosphorylable residue in vivo. Furthermore, Western blot analysis using phospho-Ser153-specific antibodies indicates that Ser153 phosphorylation in vivo is induced when PKC is activated and calmodulin is inhibited. The mutation of Ser153 to aspartate, a pseudophosphorylated residue, inhibits the nuclear accumulation of p21. Finally, whereas wild-type p21 translocates to the cytoplasm after PKC activation in the presence of calmodulin inhibitors, p21 carrying a nonphosphorylatable residue at position 153 remains in the nucleus. We propose that calmodulin binding to p21 prevents its phosphorylation by PKC at Ser153 and consequently allows its nuclear localization. When phosphorylated at Ser153, p21 is located at the cytoplasm and disrupts stress fibers.
The cyclin-dependent kinase (cdk) inhibitor p21cip1 is a protein with important roles in cell proliferation, cell cycle checkpoint responses, differentiation, senescence, and apoptosis (16, 19, 40, 44). Although it does not have a catalytic activity, it interacts with a broad range of other proteins, thereby regulating their activities (15). Like other members of the CIP/KIP family, p21 contains conserved cyclin and cdk-binding domains near the amino terminus of the molecule (10, 20, 21, 34, 39). These sequences allow p21 to bind almost all cyclin/cdk complexes. While this binding leads to an inhibition of the activity of the cyclin/cdk2 complex and, to a lesser extent, cyclin/cdc2, its effect on the cyclin D/cdk4 complex is very different (11, 29). p21 allows the assembly of the cyclin D/cdk4 complex and regulates its nuclear localization. This is due to the fact that, on one hand, binding of p21 to cytoplasmic cyclin D/cdk4 provides the complex with a nuclear localization signal (NLS) and, on the other, binding of p21 to nuclear cyclin D/cdk4 blocks exposure of the nuclear export sequences of cyclin D (2, 11). The C-terminal half of p21 is different from that of other CIP/KIP family members. It binds to proliferating cell nuclear antigen (PCNA), inhibiting its role as a processivity factor for DNA polymerase δ in vitro (56). Other cyclin A/E and cdk4-binding domains have been found in the C-terminal region of p21 (4, 47). Furthermore, other proteins, such as calmodulin (CaM) (52), SET (17), GADD45 (25), c-Myc (26), C/EBP (53), CARB (35, 54), and C8 (54) bind to the C terminus, and their binding domains overlap with each other and with the PCNA-binding domain. Moreover, other proteins, such as E2F, procaspase 3, SAPK, and ASK1, also bind to p21 (15). While the functions of all of these interactions are not yet fully understood, some may affect the cell cycle and others may affect apoptosis or cell differentiation, placing p21 as a connecting point between different functional pathways.
The interaction of p21 with different proteins can be regulated by phosphorylation. In fact, phosphorylation of the C-terminal region of p21 regulates its association with other proteins such as PCNA, as well as its stability and cellular localization (60).
A functional regulation of p21 may also be exerted by its cellular localization. Only the nuclear form of p21 is able to inhibit cell cycle progression (41), while cytoplasmic p21 seems to be a positive modulator of cell survival (7, 14). p21 is localized in the cytoplasm during monocytic and neuronal differentiation, where it interacts with the kinases SAP and ASK1 and Rho-kinase (3, 45, 50). It also interacts with procaspase 3 in the mitochondria to inhibit caspase 3 activation and to resist Fas-mediated cell death (48, 49). Furthermore, cytoplasmic p21 prevents the formation of stress fibers in Ras-transformed cells (5, 31). Considering also that the levels of p21 expression and cytoplasmic localization are highly increased in various cancers (6, 58, 59), the analysis of the mechanisms regulating the cellular localization of p21 is important in the fields of cell proliferation and survival.
CaM is a Ca2+-binding protein that acts as a transducer of intracellular Ca2+ signals (13, 37). When bound to Ca2+, CaM is able to bind to other proteins (CaM-binding proteins [CaMBPs]), directly regulating their activities (23, 28, 57). Through the action of these CaMBPs, like CaM-dependent kinases II and IV, calcineurin, hnRNP A2, hnRNP C, and others, CaM regulates a great variety of cellular processes, such as gene expression, protein translation, and protein phosphorylation (1, 27, 36, 46, 55). Consequently, CaM has been implicated in a large number of cellular events, including fertilization, cell division, cell differentiation, and neuronal signaling. Progression through G1 and exit from mitosis are sensitive to changes in the intracellular concentration of CaM. Furthermore, the addition of specific anti-CaM drugs to cell cultures inhibits the reentry of growth-arrested cells into the cell cycle (G0/G1 transition), the progression into and through S phase, and the entry and exit from mitosis (12, 33). We have previously reported that the addition of CaM inhibitors to cultured fibroblasts during G1 inhibits pRb phosphorylation and cdk4 activation (51). Furthermore, we have shown that CaM binds to the C terminus of p21 near the NLS sequence and that the inhibition of CaM inhibits nuclear accumulation of p21, cdk4, and cyclin D1 (41, 51, 52).
Here, we characterize the CaM-binding domain of p21 and assess whether the interaction of CaM with p21 is essential for the nuclear translocation of p21. Our data show that the interaction of CaM with p21 is not in itself essential for the nuclear translocation of p21, but it inhibits the phosphorylation of p21 by protein kinase C (PKC) and, as a consequence, prevents the cytoplasmic localization of p21.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
Expression plasmids for green fluorescent protein (GFP)-p21 and glutathione S-transferase (GST)-p21 fusion proteins were constructed by single PCR as indicated in reference 41 but with reverse oligonucleotides carrying the appropriate mutations. Digestions with NdeI and HindIII allowed cloning into a modified pGEX-KG vector, and digestions with EcoRI and BamHI allowed cloning into pEGFP-C1 vector (Clontech). GFP-p21NLS codes for a p21 with the nuclear localization signal mutated as described previously (41). The plasmid pMT2HAp21 for the N-terminal expression of the hemagglutinin (HA) tag in front of the complete p21 cDNA was a gift from C. Serra (IDIBAPS, Barcelona, Spain). To obtain the Ser153-to-Ala mutant (HA-p21S153A), single PCR was also performed with the appropriate forward and reverse oligonucleotides. The forward oligonucleotide incorporated an EcoRI site and the reverse oligonucleotide incorporated an XhoI site to allow digestion of the PCR product with these two enzymes and subcloning into the same pMT2HA plasmid. PCRs using a forward oligonucleotide with an EcoRI site and a reverse oligonucleotide with an XhoI site were used to subclone the different p21 mutants in the pMT2HA plasmid.
Protein expression, purification, and pulldown assays.
GST-p21-containing plasmids were transformed into Escherichia coli strain BL21(DE3) carrying the pLysS plasmid. Expression and purification were performed by following the manufacturer's instructions (Amersham Biosciences, Inc.) with minor modifications (17). Human recombinant CaM was also expressed in E. coli and purified as previously described (55). Peptides (from the peptide facility of the University of Barcelona) and purified proteins were covalently bound to BrCN-activated Sepharose 4B, as indicated by the manufacturer (Pharmacia-Biotech.). Pulldown assays were performed as follows: 1 to 10 μg of purified protein was incubated for 1 h at 4°C with 20 μl of either CaM-Sepharose (1:1, vol/vol), CT-p21-Sepharose (1:1, vol/vol) (where CT represents the C terminus of p21), or peptide-Sepharose (1:1, vol/vol) in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 1% Triton X-100 in the presence of either 0.1 mM CaCl2 or 1 mM EGTA. After centrifugation, nonbound proteins were collected and bound proteins were washed three times in the same buffer and eluted directly with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis Laemmli loading buffer, electrophoresed, and analyzed by Coomassie blue or silver staining.
Cell culture and transfections.
NIH 3T3 and COS7 cells were grown in Dulbecco's minimum essential medium supplemented with 10% donor calf serum or 5% fetal calf serum, respectively. Transient expression of the different p21 mutants was achieved by transfecting the cells with the appropriate expression vectors and using Effectene (QIAGEN) or Lipofectamine (Invitrogen Life Technology) according to the manufacturer's instructions.
PKC activity assays.
Ten micrograms of the different GST-CT-p21 mutants to be used as substrates was incubated with a mixture containing 25 mM HEPES (pH 7.5), 10 mM MgCl2, 0.5 mM dithiothreitol (DTT), and 25 μM ATP in the presence or absence of CaM (20 μg) and the presence of either 0.1 mM CaCl2 or 1 mM EGTA in a final volume of 30 μl. The reaction was initiated by the addition of 0.25 μl of rat brain PKC catalytic fragment (Biomol) and [γ-32P]ATP (300 Ci/mmol) (Amersham Biosciences). Kinase reactions were carried out for 30 min at 30°C and stopped by the addition of Laemmli sample buffer. Samples were run on 12% SDS-polyacrylamide gels, stained with Coomassie blue to confirm the same loading of GST-p21 in each lane, destained, and dried. Phosphorylation was detected and quantified by phosphorimaging (Bio-Rad).
BIAcore analysis.
The PCNA protein was immobilized on a CM5 sensor chip by the amine coupling method. Purified GST-CTp21WT (91 to 164 amino acids [aa]) or GST-CTp21S153D (91 to 164 aa) proteins were diluted to various concentrations with HBS-EP buffer (Biacore), and each sample was injected separately at a flow rate of 15 μl/min for 3 min. A final regeneration of the matrix was performed with 0.1% (wt/vol) SDS. The interactions among proteins and PCNA were detected and presented as a sensogram by plotting the resonance units against time. The kinetic results were calculated using BIAevaluation software (version 3.0.2.; Amersham Pharmacia Biotech).
Immunocytochemistry and fluorescence microscopy.
For intracellular localization analysis of GFP fusion proteins, transfected cells were grown on coverslips and fixed in 4% paraformaldehyde-phosphate-buffered saline (PBS) (140 mM NaCl, 5 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.2]) for 20 min at room temperature. Coverslips were then washed three times (5 min each) in PBS and mounted on glass slides with Mowiol (Calbiochem). For the detection of HA-p21 fused protein, cells were fixed as described above, permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature, blocked for 20 min with 1% bovine serum albumin (BSA) in PBS, and then incubated for 1 h at 37°C in a humidified atmosphere with a monoclonal anti-HA antibody, clone 12CA5 (no. 1583816; Roche), at a 1:200 dilution. Coverslips were then washed three times (5 min each) in PBS and incubated for 45 min at 37°C with an Alexa594- or Alexa488-conjugated anti-mouse antibody (dilution 1:500; Jackson). Coverslips were washed and mounted as indicated above and analyzed by fluorescence microscopy. For each mutant and condition, the experiment was repeated at least three times, and each time the percentage of cells (from a total of 300) with exclusively nuclear signals was calculated. To detect stress fibers, phalloidin conjugated with Alexa594 (1:500 dilution; Molecular Probes) was added during the incubation with the secondary antibody. Analysis of bromodeoxyuridine (BrdU) incorporation in cells was done as described previously (41).
Cell fractionation.
Cells grown in p60 dishes containing COS7 cells at 80% confluence were scraped in PBS, pelleted (3 min at 2,000 rpm), and washed twice with the same buffer. Cells were resuspended in 300 μl of buffer A (10 mM HEPES [pH 7.4], 1 mM EDTA, 1 mM DTT) containing protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 0.1 U/ml aprotinin, 1 mM orthovanadate, 50 mM NaF), and cell membranes were disrupted by 15 passages through a 23-gauge needle. Disruption of the cell membranes was confirmed under a microscope. Then, 300 μl of buffer B (50 mM HEPES [pH 7.4], 500 mM sucrose, 10 mM Mg2SO4) was added and the lysed cells were centrifuged for 15 min at 2,500 rpm. The supernatant (cytoplasmic fraction) was collected, aliquoted, and stored at −20°C, while the pellet was resuspended in 300 μl of buffer A plus 300 μl of buffer B, rehomogenized by five passages through a 23-gauge needle, and centrifuged for 10 min at 2,000 rpm on a 1.5-ml cushion of 0.88 M sucrose. The pellet was washed once with 500 μl of buffer A plus 500 μl of buffer B and centrifuged for 10 min at 1,700 rpm. The pellet (nuclear fraction) was resuspended with 100 μl of 2% SDS and 67 mM Tris-HCl (pH 6.8) and sonicated twice for 20 s to shear the DNA.
All processes were performed at 4°C. The protein contents of the cytoplasmic and nuclear fractions were measured according to the Lowry method and subjected to Western blot analysis.
Western blot analysis and antibodies.
Proteins were resolved in SDS-polyacrylamide gels (30) and transferred to Immobilon-P membranes for 2 h at 60 V. The sheets were then preincubated for 1 h at room temperature with TBST (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20) containing 5% defatted milk powder or 5% BSA (anti-phospho-Ser153p21 Western blot). Western blots were probed for 1 h at room temperature with anti-p21 rabbit polyclonal (sc-397; 1:200 dilution [Santa Cruz]), anti-phospho-Ser153p21 rabbit polyclonal (2 μg/ml), anti-lamin B goat polyclonal (sc-6217; 1:200 dilution [Santa Cruz]), anti-GAP mouse monoclonal (05-178; 1 μg/ml [Upstate Biotech]), anti-pMARCKS (no. 2741; 1:500 dilution [Cell Signaling]), or anti-HA mouse monoclonal, clone 12CA5 (1583816; 1 μg/ml [Roche]), antibody. After being washed in TBST (three times for 10 min each), sheets were incubated with the appropriate peroxidase-coupled secondary antibodies (1:2,000 dilution; Bio-Rad) for 1 h at room temperature, washed twice in TBST and once in Tris-buffered saline, and visualized by enhanced chemiluminescence reaction (Amersham). Rabbit anti-phosho-Ser153p21-specific antibodies were raised against phosphopeptide MTDFYHS*KRRLIFC (Isogene Life Science) and affinity purified.
Two-dimensional gel electrophoresis. (i) Sample preparation.
Harvested cells (1.5 × 106) were lysed in 200 μl of buffer L, containing 0.3% SDS, 50 mM Tris-HCl (pH 8), and 200 mM dithioerythritol (DTE); samples were then boiled for 5 min and refreshed for 2 min on ice. Homogenates were then incubated with a mixture of DNase I and RNase A (final concentrations of 250 μg/ml each) for 10 min on ice. Next, protein precipitation was performed in an 80% final concentration of cold acetone for 20 min on ice, followed by centrifugation at 14,000 rpm for 15 min at 4°C. After the protein pellet was dried completely, it was resuspended in 200 μl of buffer U, containing 9 M urea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 65 mM DTE, and 0.1% ampholytes (Bio-Lyte 3/10, no. 163-1113; Bio-Rad), and centrifuged at 14,000 rpm for 15 min at 4°C. Protein quantification of the supernatant was carried out using the Bradford assay (8a), and 50 μg of the sample was diluted to a final volume of 125 μl with buffer U.
Phosphatase treatment was performed on the harvested cells with λ phosphatase (400 U/μl; Calbiochem) in 20 μl buffer P (150 mM Tris-HCl [pH 8], 2 mM NaCl, 2 mM EDTA, 10% glycerol, 0.2% Nonidet P-40, 5 mM DTT, 2 mM MnCl2, 100 μg/ml BSA containing protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 0.1 U/ml aprotinin]) for 30 min at 30°C.
(ii) Performance of 2D electrophoresis.
Two-dimensional (2D) first-dimension electrophoresis was performed as isoelectric focusing (IEF) with precast, immobilized pH gradient IPG gel strips (ReadyStrip IPG Strip, 7 cm, pH 3 to 10; no. 163-2000 [Bio-Rad]) by using a PROTEAN IEF system (no. 165-4000; Bio-Rad). Sample application and rehydration of the strips were carried out using the passive method according to the manufacturer's instructions (Bio-Rad). Next, focusing was performed at 12,500 to 20,000 V per hour. IEF gels were equilibrated for 10 min in 1 ml of buffer E (6 M urea, 0.375 M Tris [pH 8.8], 2% SDS, 20% glycerol, and 2% [wt/vol] DTE), and the second-dimension run was carried out as described by Laemmli (30). After electrophoresis, gels were transferred to Immobilon-P transfer membranes (Millipore) and immunoblotted, essentially as described above.
RESULTS
Characterization of the CaM-binding domain of p21.
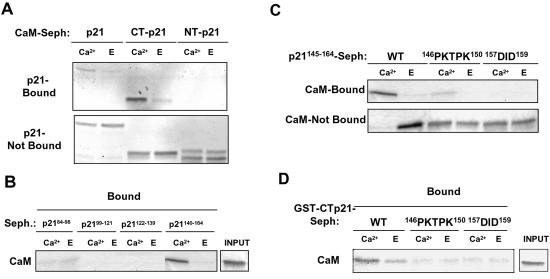
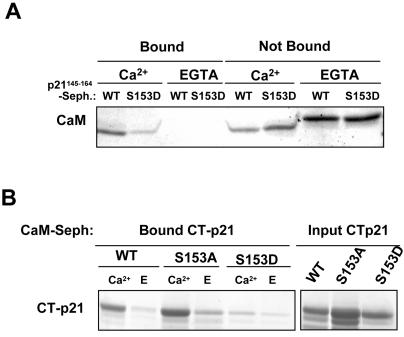
We have previously reported that p21 is able to bind CaM and that this binding was mimicked by a peptide corresponding to the last 20 C-terminal amino acids of p21 (52). To further characterize the CaM-binding domain of p21, GST-NT-p21 (aa 1 to 90) (where NT represents the N terminus of p21) and GST-CT-p21 (aa 91 to 164) were expressed and their binding abilities to CaM-Sepharose were analyzed. As shown in Fig. 1A, only the C-terminal moiety of p21 was able to bind to CaM. This binding occurred specifically in the presence of Ca2+. A number of peptides covering all of the C-terminal half of p21 were subsequently synthesized and coupled to Sepharose. Analysis of CaM binding to these peptides confirmed that the most relevant CaM-binding domain of p21 was in the p21140-164region (Fig. 1B).
FIG. 1.
Analysis of the CaM-binding domain of p21. (A) CaM-Sepharose pulldown analysis. Full-length p21, NT-p21 (1 to 90 aa), and CT-p21 (91 to 164 aa) were expressed as GST fusion proteins, and their binding abilities to CaM-Sepharose were analyzed in the presence of either Ca2+ or EGTA (E). (B) Peptide-Sepharose pulldown analysis. Binding of purified CaM to the different p21 peptides coupled to Sepharose was performed in the presence of either Ca2+ or E. (C) Binding of purified CaM to the p21145-164 peptide containing the indicated mutations and coupled to Sepharose was performed in the presence of either Ca2+ or E. (D) Binding of purified CaM to GST-CT-p21 containing the indicated mutations and coupled to Sepharose was performed in the presence of either Ca2+ or E. All pulldown assays were performed as indicated in Materials and Methods. Total bound and nonbound fractions (A and C) and samples containing the same amount of CaM used for each of the pulldown assays (INPUT) (B and D) are shown. After electrophoresis, proteins were either Coomassie blue (A and B) or silver (C and D) stained. Seph, Sepharose.
One of the most common CaM-binding domains consists of an amphipathic α-helix. Our previous results indicate that the C-terminal region of p21 (aa 145 to 164) can adopt an α-helical conformation (18). In fact, when the p21145-164 peptide was mutated either to restrain α-helix formation (peptide p21145-164with the sequence 146PKTPK150 instead of the wild-type [WT] 146SMTDF150 sequence) or to reduce hydrophobicity (peptide p21145-164 with the sequence 157DID159 instead of 157LIF15), binding to CaM was clearly reduced (Fig. 1C). Moreover, GST-CT-p21 constructs harboring the same mutations were also unable to bind CaM (Fig. 1D). Our data are consistent with the fact that the C-terminal amphipathic α-helix domain of p21 is essential for CaM binding.
CaM inhibits PKC-mediated phosphorylation of p21 in vitro.
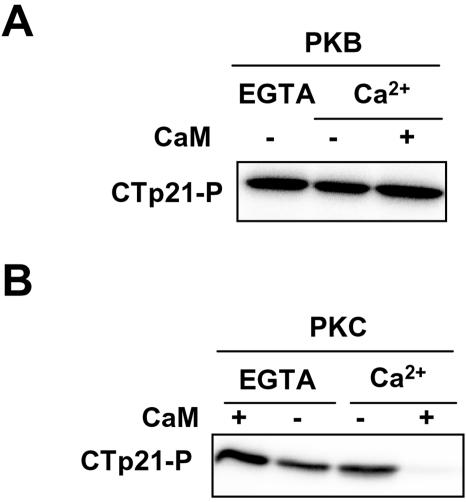
There is increasing evidence that some residues of the C-terminal region of p21 might be phosphorylated by various kinases, including protein kinase B (PKB) and PKC, and that in some cases, this phosphorylation regulates the nuclear localization of the protein (32, 43, 60). Thus, we aimed to analyze whether CaM was interfering with the phosphorylation of GST-CT-p21 by PKB or PKC. Results revealed that both kinases were able to phosphorylate GST-CT-p21 (Fig. 2). Interestingly, phosphorylation of p21 by PKC was inhibited by CaM, whereas p21 phosphorylation by PKB was not. The effect of CaM on p21 phosphorylation by PKC was specific because the inhibition was observed in the presence of Ca2+ but not in the presence of EGTA (Fig. 2B). SET and E7, proteins that also interact with the C-terminal region of p21, did not effect p21 phosphorylation by PKC (data not shown).
FIG. 2.
Effect of CaM on p21 phosphorylation by PKB and PKC. Purified GST-CT-p21 (CTp21-P) was phosphorylated in vitro, as indicated in Materials and Methods, by either PKB (A) or PKC (B) in the presence (+) or absence (−) of EGTA, Ca2+, or CaM, as indicated in the figure.
Pseudophosphorylation of p21 Ser153 increases the cytoplasmic localization of p21.
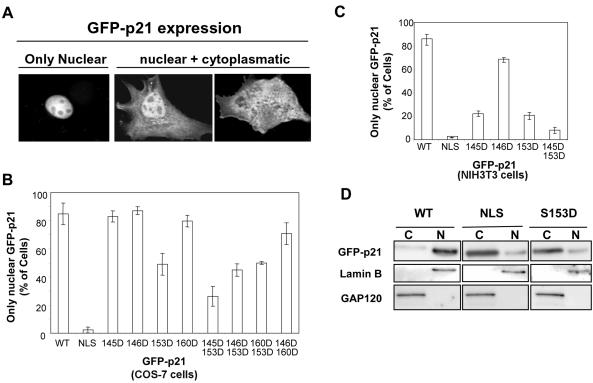
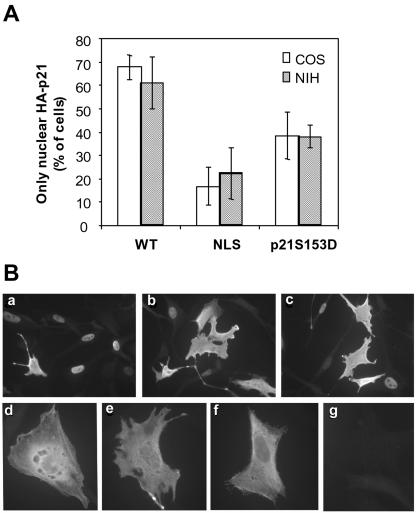
We further aimed to identify putative phosphorylation sites in the CaM-binding domain of p21 that could be involved in the regulation of the intracellular localization of this protein. To this end, p21WT, p21NLS (41), and p21 mutants mimicking phosphorylation of the C-terminal region of p21 were fused to GFP and expressed in COS cells. After transfection, we quantified the percentage of cells showing only nuclear localization (Fig. 3A). As shown in Fig. 3B, GFP-p21S153D showed an increased cytoplasmic localization. In contrast, all of the other single mutants, GFP-p21T145D, GFP-p21S146D, and GFP-p21S160D, did not significantly alter their nuclear localizations. We also analyzed the intracellular distribution of double mutants. Results revealed that the simultaneous mutation of both T145D and S153D increased the cytoplasmic localization of p21 (Fig. 3B). In contrast, the association of either S146D or S160D with S153D led to levels of cytoplasmic p21 similar to those observed in the S153D single mutant. It has previously been described that in HER-2/neu 3T3 transformed fibroblasts, the phosphorylation of T145 by AKT induced a cytoplasmic localization of p21 (60). Transfections were repeated in NIH 3T3 cells, and in agreement with these results, GFP-p21T145D had a major cytoplasmic localization in NIH 3T3 cells (Fig. 3C). Interestingly, the S153D mutation also inhibited in these cells the accumulation of GFP-p21 into the nucleus, and again an additive response was observed with the double T145D S153D mutant. The effect of the S153D mutation on the intracellular localization of p21 was confirmed by cell fractionation, followed by Western blotting of COS cells. As shown in Fig. 3D, the S153D mutant of p21 was observed mainly in the cytoplasm, supporting that phosphorylation of S153 is crucial for the cytoplasmic localization of p21. To confirm the effect of Ser153 phosphorylation on the nuclear localization of p21, the localization of p21 fused to HA was also analyzed. In concordance with results obtained with GFP-fused proteins, the percentage of cells with HA-p21 only in the nucleus was lower when p21 was pseudophosphorylated at Ser153 than in the WT p21 (Fig. 4). Moreover, when expressing HA-p21NLS and HA-p21S153D, cells with almost no nuclear signal could be seen (Fig. 4B). This was not observed in GFP-fused mutants (Fig. 3A), most probably because GFP itself has some affinity for the nucleus.
FIG. 3.
Intracellular localizations of the different mutants of GFP-p21. COS or NIH 3T3 cells were transfected with GFP-p21 containing the indicated mutations, and intracellular localizations of the expressed GFP-p21 proteins were analyzed 24 h later. Fluorescence microscopy was performed to determine the intracellular localizations of the expressed proteins. (A) Examples of confocal images of COS cells with GFP-p21 expressed exclusively in the nucleus (Only Nuclear) or in the nucleus plus different degrees of cytoplasmic intensity (nuclear + cytoplasmic) are shown. COS cells (B) or NIH 3T3 cells (C) with “only nuclear” GFP-p21 expression were counted for each indicated mutant of p21. The data represent the means of at least three different experiments, and the bars represent standard deviations. (D) COS cells were transfected with the indicated mutants of p21, and 24 h later, cells were harvested and nuclear (N) and cytoplasmic (C) fractions were obtained, as indicated in Materials and Methods. Samples corresponding to the same percentage (5% of total fraction) of cytoplasmic fraction and nuclear fraction were electrophoresed, and the amount of GFP-p21 in each fraction was analyzed by Western blotting. GAP120 and lamin B were used as markers of the cytoplasmic and nuclear fractions, respectively.
FIG. 4.
Intracellular localization of HA-p21S153A. COS or NIH 3T3 cells were transfected with HA-p21WT, HA-p21NLS, or HA-p21S153D, and intracellular localizations of the expressed HA-p21 proteins were analyzed 24 h later by immunocytochemistry. (A) Cells with only nuclear p21 expression were counted for each construct. The data represent the means of three different experiments, and the bars represent standard deviations. (B) Representative fluorescent microscopy images of HA-p21WT (a), HA-p21NLS (b), and HA-p21S153D (c) transfections of NIH 3T3 cells, higher magnifications of HA-p21S153D-transfected NIH 3T3 cells with cytoplasmic p21 (d, e, and f), and a nontransfected cell (g) are shown.
In vivo phosphorylation of p21 at Ser153 is regulated by CaM and PKC.
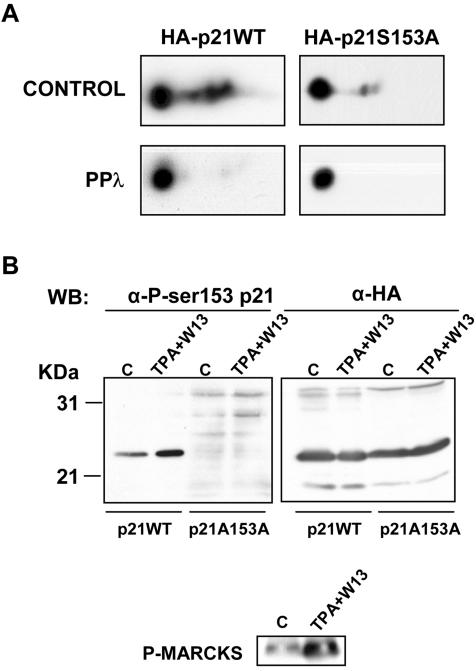
We first analyzed whether the Ser153 residue of p21 was phosphorylated in vivo. COS cells were transfected with HA-p21WT or HA-p21S153A (a nonphosphorylatable mutant of p21 at position 153). Protein extracts were subsequently separated by 2D gel electrophoresis, and the mobility of HA-p21 was analyzed by Western blotting with anti-HA antibodies. As shown in Fig. 5A, HA-p21WT showed a pattern of several spots that, when treated with phosphatase lambda, were reduced to the more basic spot, indicating that HA-p21 was in fact a phosphoprotein in vivo. On the contrary, HA-p21S153A showed a major basic spot, and the pattern was not greatly changed upon phosphatase treatment. Thus, Ser153 is a major phosphorylation site of p21 in COS cells. To analyze whether this phosphorylation was regulated by CaM and PKC, an antibody that specifically recognized the phosphorylated form of p21 at Ser153 was generated. Confirming the results obtained with 2D gel electrophoresis, phospho-Ser153 p21 was detected in COS cells transfected with HA-p21WT, whereas no signal was observed in cells transfected with HA-p21S153A. These results indicate that the antibody specifically recognized phosphorylation of Ser153. When cells were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA) in order to activate PKC plus W13 to inhibit CaM, phosphorylation of p21 at Ser153 increased (Fig. 5B). To confirm the activation of PKC by TPA treatment of COS cells, phosphorylation of MARCKS (myristilated alanine-rich C kinase substrate) was also determined (Fig. 5B).
FIG. 5.
p21 is phosphorylated at Ser153 in vivo. COS cells were transfected with HA-p21WT or HA-p21S153A for 48 h. (A) Control or phosphatase λ (PPλ)-treated cells were processed for 2D gel electrophoresis (see Materials and Methods), and transferred membranes were processed for Western blotting with anti-HA antibodies. (B) Proliferating COS cells not treated (C) or treated with TPA (100 nM) and W13 (1.5 μg/ml) for 45 min (TPA+W13) were electrophoresed and processed for Western blotting (WB) with either anti-phospho-Ser153p21 (α-P-ser153 p21) antibodies or anti-HA (α-HA) antibodies as a control of HA-p21 expression and loading of the gel. Samples were also blotted with anti-phospho-MARCKS antibody (bottom panel).
Phosphorylation of p21 at S153 by PKC induces the cytoplasmic localization of p21.
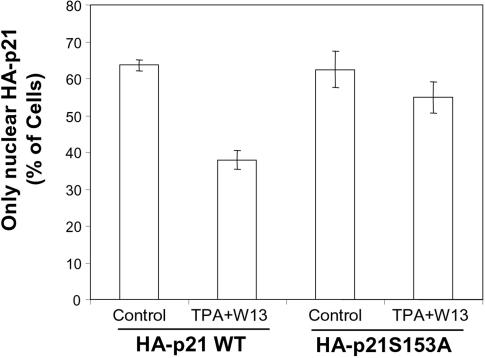
The results described above strongly suggest that phosphorylation of Ser153 of p21 by PKC might be important for the regulation of the intracellular localization of p21. Thus, we aimed to further confirm this hypothesis. To this end, COS cells were transfected with HA-p21WT or HA-p21S153A expression vector and the intracellular distributions of these proteins were analyzed by immunocytochemistry with anti-HA antibodies. As can be seen in Fig. 6, the treatment of cells with TPA plus W13 induced a 40% decrease in the nuclear localization of the HA-p21WT protein. In contrast, this treatment did not significantly affect the intracellular distribution of the HA-p21S153A protein. These results indicate that phosphorylation of the S153 residue of p21 is critical for the cytoplasmic localization of this protein in response to PKC activation plus CaM inhibition. Furthermore, when HA-p21S153A was expressed in NIH 3T3 cells, an increased nuclear localization (85% of the transfected cells) from that of HA-p21WT (62% of transfected cells) was observed, indicating that in proliferating NIH 3T3 cells, phosphorylation of Ser153 of p21 was relevant. In NIH 3T3 cells, localization of HA-p21S153A was also not modified upon treatment with TPA plus W13.
FIG. 6.
Effect of CaM inhibition plus PKC activation on the intracellular location of HA-p21S153A. COS cells grown on coverslips were transfected with expression plasmids for HA-p21WT and HA-p21S153A and, after 24 h, left untreated (Control) or treated for 45 min with TPA (100 nM) plus W13 (1.5 μg/ml) (TPA+W13). Immunolocalization of HA fusion proteins was performed, as indicated in Materials and Methods, by using anti-HA antibodies. Cells with only nuclear expression of HA-p21 were counted. The graph represents the means of three different experiments, and the bars represent standard deviations.
Pseudophosphorylation of p21 at Ser153 inhibits binding of p21 to CaM.
We further studied whether the phosphorylation of p21 at S153 affects the interaction of p21 with CaM. The associations of CaM with p21145-164 (the WT CaM-binding domain) and with the same peptide containing an S153D mutation (to mimic a phosphorylation status) were analyzed (Fig. 7A). We also analyzed the associations of CaM with GST-CTp21WT, GST-CTp21S153D, and GST-CTp21S153A (Fig. 7B). In both experiments, pseudophosphorylation of S153 strongly inhibited the binding of CaM to the CaM-binding domain of p21.
FIG. 7.
Analysis of CaM binding to p21WT, p21S153D, and p21S153A. (A) Synthetic WT p21145-164 or S153D p21145-164 peptides were coupled to Sepharose. Binding of purified CaM to these peptides was analyzed by pulldown assay, as indicated in Materials and Methods, in the presence of either Ca2+ or EGTA. Total bound and total nonbound CaM fractions were electrophoresed and Coomassie blue stained. (B) CT-p21WT, CT-p21S153A, and CT-p21S153D were expressed as GST fusion proteins, and their binding abilities to CaM-Sepharose were analyzed in the presence of either Ca2+ or EGTA (E), as indicated in Materials and Methods. The total bound fraction and a sample containing the same amount of p21 used for each of the pulldown assays (Input) were electrophoresed and Coomassie blue stained. Seph, Sepharose.
Binding of CaM to p21 is not in itself essential to induce nuclear translocation of p21.
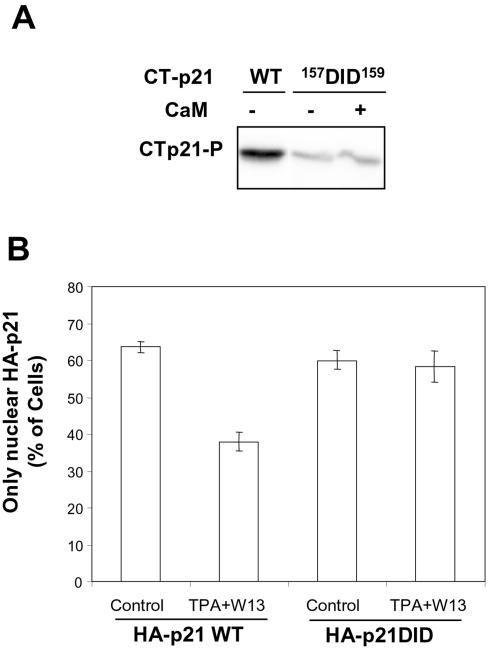
Finally, we wanted to discard the possibility that p21S153D was more cytoplasmic due directly to its incapacity to bind CaM. To this end, we studied the intracellular distribution of a p21 mutant (GST-CTp21157DID159) that did not bind CaM (Fig. 1C) and that is not phosphorylated by PKC (Fig. 8A). COS cells were transfected with HA-p21WT or HA-p21157DID159 expression vector, and the intracellular distributions of the fusion proteins were analyzed by immunocytochemistry with anti-HA antibodies. The proportions of cells with only nuclear protein were high and similar in both cases, indicating that the interaction of p21 with CaM was not in itself essential to induce the nuclear translocation of p21. Furthermore, corroborating the fact that the p21 HA-p21157DID159 protein could not be phosphorylated by PKC, the intracellular distribution of HA-p21157DID159 was not affected by TPA plus W13 treatment (Fig. 8B).
FIG. 8.
p21157DID159 is not phosphorylated by PKC and has a nuclear localization. (A) Purified GST-CTp21WT and GST-CTp21157DID159 were phosphorylated in vitro by PKC in the presence of Ca2+ plus or minus CaM, as indicated in the figure. (B) COS cells grown on coverslips were transfected with expression plasmids for HA-p21WT and HA-p21157DID159 and, after 24 h, left untreated (Control) or treated for 45 min with TPA (100 nM) plus W13 (1.5 μg/ml) (TPA+W13). Immunolocalization of HA fusion proteins was performed, as indicated in Materials and Methods, by using anti-HA antibodies. Cells with only nuclear expression of HA-p21 were counted. The graph represents the means of three different experiments, and the bars represent standard deviations.
Ability of p21 phosphorylated at Ser153 to disrupt stress fibers and to bind to PCNA.
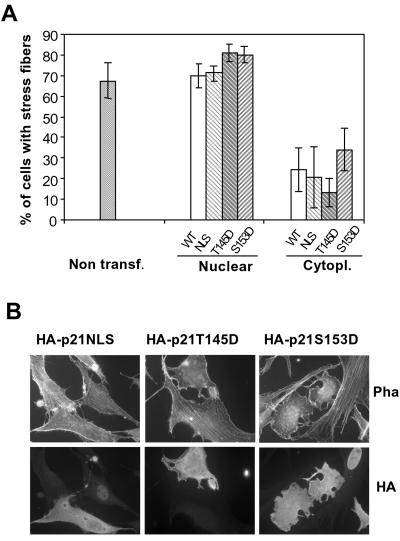
It is known that nuclear and cytoplasmic p21 have very different roles. While DNA synthesis inhibition is achieved mainly by nuclear p21 and is partially due to PCNA inhibition, stress-fiber disruption is an effect of cytoplasmic p21. We analyzed whether cytoplasmic HA-p21S153D or HA-p21S145D inhibited stress-fiber formation with the same efficiency as cytoplasmic HA-p21WT or cytoplasmic HA-p21NLS. To this end, NIH 3T3 cells were transfected with the different constructs, and immunocytochemistry to detect HA was performed together with phalloidin staining. The percentage of cells with stress fibers for each p21 mutant among the cells that presented p21 either in the nucleus or in the cytoplasm was measured. As shown in Fig. 9A and B, when present in the cytoplasm, HA-p21 (WT, NLS, S153D, or T145D) disrupted stress fibers with the same efficiency. On the contrary, in the few cells expressing HA-p21S153D, HA-p21S45D, or HA-p21NLS only in the cell nucleus, stress fibers were not affected, as was the case with HA-p21WT cells. We conclude that in NIH 3T3 cells, phosphorylation of p21 at Ser153 or at Thr145 inhibits the nuclear translocation of p21 and consequently induces disruption of the stress fibers.
FIG. 9.
Effect of HA-p21S153D expression on stress fibers. The indicated HA-p21 constructs were transfected into NIH 3T3 cells, and after 24 h, immunocytochemistry to detect cells expressing HA and phalloidin-Alexa staining (Pha) were performed as indicated in Materials and Methods. (A) Graphic representation of the percentage of cells with stress fibers among the cells that presented p21 either in the nucleus or in the cytoplasm (Cytopl.) for each p21 mutant. The graph represents the means of three different experiments, and the bars represent standard deviations. Non transf., nontransfected. (B) Representative fluorescent microscopy images showing the disruption of stress fibers by cytoplasmic p21 (NLS, T145D, or S153D) and not nuclear p21.
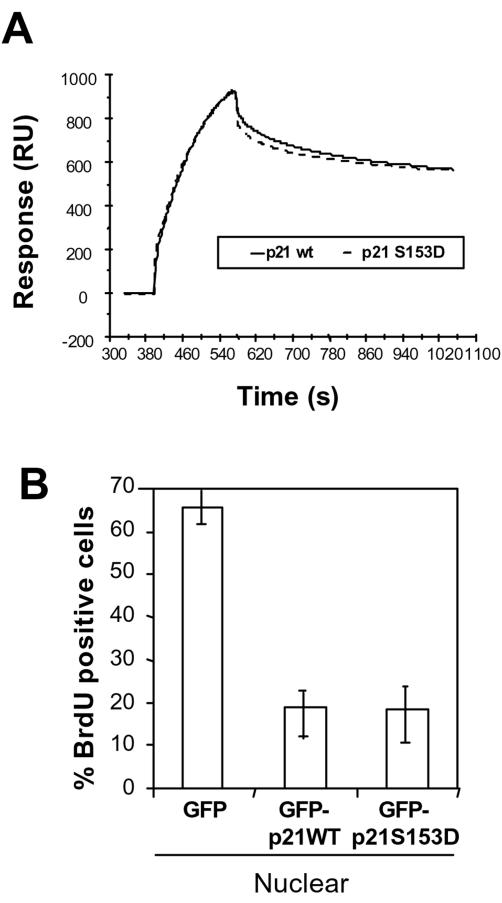
Since Ser153 is in the PCNA-binding domain of p21, we also analyzed whether phosphorylation of this residue could affect binding to PCNA. BIAcore analysis with GST-CTp21WT and GST-CTp21S153D indicated that both proteins had the same affinity for PCNA: 1.3 μM and 1.4 μM, respectively (Fig. 10A). Furthermore, when located in the nucleus, both proteins GFP-p21WT and GFP-p21S153D equally inhibited DNA synthesis measured as BrdU incorporation (Fig. 10B). In conclusion, when p21 phosphorylated at Ser153 can enter into the nucleus, most probably because it is not forming high-molecular-weight complexes, it is able to inhibit DNA synthesis.
FIG. 10.
Effect of p21 Ser153 pseudophosphorylation on PCNA binding. (A) Binding abilities of purified CT-p21WT and CT-p21S153D to PCNA were analyzed by BIAcore as indicated in Materials and Methods. Response curves obtained with 2 μM of CT-p21 are shown. RU, resonance units. (B) COS cells were transfected with GFP-p21WT and GFP-p21S153D and, after 24 h, incubated for 10 h with BrdU. BrdU immunostaining was performed as indicated in Materials and Methods. The percentages of BrdU-positive cells among the cells with nuclear GFP-p21 (WT or S153D) and the nontransfected cells are indicated in the graph. The bars represent standard deviations.
DISCUSSION
There is evidence indicating that the different functions of p21 rely, at least in part, on its intracellular localization. While nuclear p21 inhibits cell proliferation, cytoplasmic p21 does not and, in contrast, is able to increase survival and cell motility. Due to the quite opposite roles of this protein in these two compartments, it is of great interest to know how its translocation is regulated.
We have previously shown that CaM is able to bind to the C terminus of p21 and that, in fibroblasts, the inhibition of CaM prevents its translocation to the nucleus (52). The mechanism by which CaM regulates the intracellular localization of p21 was analyzed here. We have determined that the only CaM-binding domain of p21 is located within the p21145-164 region. The p21 protein has been shown to be phosphorylated at serine and threonine residues included in the CaM-binding domain by a number of different kinases (24, 43, 60). Furthermore, at least one of these phosphorylations was shown to inhibit its nuclear accumulation (60). We analyzed a possible role for CaM in the regulation of p21 phosphorylation. In agreement with published results, p21 was phosphorylated in vitro by PKB and by PKC (24, 43, 60). We have shown here that CaM specifically inhibits phosphorylation by PKC but not by PKB and that it is Ca2+ dependent for this inhibition. The inhibition of protein phosphorylation by CaM binding has already been described for other CaMBPs, like neurogranin, neuromodulin, and MARCKS (9). Furthermore, in all of these cases (and also shown here for p21), once the CaMBP is phosphorylated, its affinity for CaM is significantly reduced. It has been suggested that, as a consequence, CaM and PKC obstruct each other's action because of the embedding of PKC phosphorylation sites in the CaM-binding domain. Interestingly, the PKC isoforms that are able to phosphorylate p21 (PKC η and PKC ζ) are Ca2+ independent (24, 42); consequently, at low Ca2+ concentrations, when CaM does not bind to p21, they would be able to phosphorylate it.
A controversy exists over the effect of the phosphorylation of the p21 C terminus. Zhou et al. (60) showed that p21T145D has a cytoplasmic localization in Her-2/neu-overexpressing cells and that this threonine is phosphorylated by PKB. For other cell types, it has been shown that PKB phosphorylates Thr145 and Ser146 (32) and that, while phosphorylation of Thr145 inhibits binding of p21 to PCNA, phosphorylation of Ser146 increases p21 stability. Finally, Scott et al. (42, 43) demonstrated that phosphorylation of p21 at Ser146 by an atypical PKC also leads to decreased binding of p21 to PCNA but to an increased instability of p21 in HeLa and HCT116 cells. We have mutated all serines and threonines present in the CaM-binding domain to aspartate, to mimic phosphorylation of this residue, and analyzed the intracellular localization of the GFP-p21 proteins. In COS cells, the substitution of a pseudophosphorylated residue for Thr145, Ser146, or Ser160 did not have a significant effect on the nuclear localization of GFP-p21, but when Ser153 was changed to aspartate, a significant decrease in the percentage of cells with p21 located exclusively in the nucleus was observed. These results were confirmed by Western blot analysis after cell fractionation. Interestingly, when double mutants were analyzed, an additive response was observed between the S153D and T145D substitutions. In contrast to COS cells, in NIH 3T3 cells, a T145D mutation reduced the nuclear accumulation of p21. The S153D mutation effect appears to be more general, because it was observed for both COS and NIH 3T3 cells. Most probably, the phosphorylations of Thr145 and Ser153 collaborate in inhibiting nuclear accumulation of p21. Then, if in some cell type, phosphorylation of one of the residues is constitutively high, phosphorylation of the other residue could strongly affect the localization of the protein. The role of Ser153 phosphorylation in reducing the nuclear accumulation of p21 was confirmed by using HA-fused p21.
We focused on the analysis of Ser153 phosphorylation because it is a novel phosphorylation site of p21 and because phosphorylation of Thr145 has been reported to be mediated by AKT and not by PKC. Two-dimensional gel analysis showed that when HA-p21WT is expressed in COS cells, different spots appear, indicating different phosphorylation states of the protein. In contrast, phosphorylation spots were drastically reduced when HA-p21S153A was expressed, indicating that Ser153 is a major phosphorylated-residue and suggesting that its phosphorylation may favor further phosphorylation in other residues. Western blot analysis using anti-phosphoSer153-specific antibody definitively showed that p21 is phosphorylated in vivo at this residue and that this phosphorylation can be increased when PKC is activated in the absence of active CaM. In agreement with our previous results for normal rat kidney cells (52), we have now shown that in COS cells, the inhibition of CaM plus TPA treatment decreases the nuclear localization of the WT p21. Interestingly, this is not the case when residue 153 cannot be phosphorylated (p21S153A). Considering these results together with the fact that S153D is the only single mutation that exerts a significant inhibition of nuclear accumulation of p21 in these cells, we conclude that CaM prevents phosphorylation of p21 by PKC at serine 153 and, consequently, enhances its nuclear accumulation. Inhibition of nuclear accumulation of p21S153D was not due to a lack of CaM binding, since a p21 mutant that did not bind CaM and could not be phosphorylated by PKC translocated into the nucleus with the same efficiency as the WT p21. Furthermore, reduced nuclear localization of p21S153D cannot be due to a reduced affinity to PCNA (that could act as a nuclear anchor), because we have shown that phosphorylation of Ser153 does not change the affinity of p21 for PCNA. Most probably, a negative charge at position 153 of p21 after its phosphorylation by PKC interferes with the binding of importin α to the NLS region. A reduced affinity of importin α to classical nuclear localization sequences after phosphorylation of residues surrounding the region has already been described (22). Considering also the results by Zhou et al. (60), we propose that both kinases, PKC phosphorylating Ser153 and AKT phosphorylating Thr145, could collaborate in inhibiting the nuclear accumulation of p21.
During the G1 phase of a normal cell cycle, most cellular p21 localizes into the cell nucleus, and if CaM is inhibited, cytoplasmic p21 increases (51, 52). This suggests that CaM is at that moment important to the prevention of p21 phosphorylation by the active PKC found in the cell. Interestingly, colocalization of p21 and CaM has been reported under these conditions (52). Cytoplasmic accumulation of p21 in correlation with PKC activation has been reported in monocytic differentiation (3) and in thrombin-induced myofibroblasts (8). It would be interesting to know if phosphorylation at Ser153 occurs and if it also correlates with a decrease in intracellular free Ca2+. Furthermore, while the last version of the manuscript was in preparation, it was reported that the nuclear kinase Mirk/dyrk1B can phosphorylate p21 at Ser153, causing p21 to localize in the cytoplasm during differentiation of C2C12 myoblasts (38). Thus, including these and our results, up to three different kinases have been shown to be involved in the regulation of the intracellular localization of p21. We show here that CaM prevents phosphorylation by PKC but not by AKT. It would be interesting to show whether CaM could also inhibit phosphorylation by Mirk/dyrk1B and whether this kinase and PKC phosphorylate p21 in different intracellular compartments.
Cytoplasmic and nuclear p21 have different functions: while nuclear p21 inhibits DNA synthesis, functioning as a tumor suppressor, cytoplasmic p21 has the oncogenic role of disrupting stress fibers and inhibiting apoptosis (5, 14). We analyzed here whether phosphorylation of p21 at Ser153 affects only the intracellular distribution of p21 or also its functionality in each intracellular compartment. We show that Ser153 phosphorylation does not interfere with the capacity of cytoplasmic p21 to disrupt stress fibers and that, in cells in which p21 phosphorylated at Ser153 is able to enter the nucleus, maybe because it is not complexed to other proteins, it can inhibit DNA synthesis with the same efficiency as p21WT. This correlates with the fact that phosphorylation of Ser153 does not affect PCNA binding.
We propose that CaM prevents phosphorylation of p21 by PKC, favoring the nuclear translocation and tumor suppressor functions of p21. Consequently, an inhibition of CaM function (e.g., decrease in free Ca2+) together with an activation of PKC could favor the oncogenic function of p21.
Acknowledgments
This study was supported by grants SAF2000-52, SAF2001-2901, SAF2004-02159, and GEN 2003-2043-C08-01 from the Ministerio de Educación y Ciencia (Spain). Aina Rodríguez-Vilarrupla and Neus Abella are recipients of predoctoral fellowships from the Ministerio de Educación y Ciencia (Spain).
We thank Anna Bosch (Serveis Científico-Tècnics, Universitat de Barcelona, Campus Medicina, IDIBAPS) for the technical assistance with confocal microscopy.
REFERENCES
- 1.Agell, N., R. Aligue, V. Alemany, A. Castro, M. Jaime, M. J. Pujol, E. Rius, J. Serratosa, M. Taules, and O. Bachs. 1998. New nuclear functions for calmodulin. Cell Calcium 23:115-121. [DOI] [PubMed] [Google Scholar]
- 2.Alt, J. R., A. B. Gladden, and J. A. Diehl. 2002. p21Cip1 promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J. Biol. Chem. 277:8517-8523. [DOI] [PubMed] [Google Scholar]
- 3.Asada, M., T. Yamada, H. Ichijo, D. Delia, K. Miyazono, K. Fukumuro, and S. Mizutani. 1999. Apoptosis inhibitory activity of cytoplasmic p21Cip1/WAF1 in monocytic differentiation. EMBO J. 18:1223-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, K. L., S. Lain, R. Fahraeus, C. Smythe, and D. P. Lane. 1996. Cell-cycle arrest and inhibition of Cdk4 activity by small peptides based on the carboxy-terminal domain of p21WAF1. Curr. Biol. 7:71-80. [DOI] [PubMed] [Google Scholar]
- 5.Besson, A., R. K. Assoian, and J. M. Roberts. 2004. Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors? Nat. Rev. Cancer 4:948-955. [DOI] [PubMed] [Google Scholar]
- 6.Biankin, A. V., J. G. Kench, A. L. Morey, C.-S. Lee, S. A. Biankin, D. R. Head, T. B. Hugh, S. M. Henshall, and R. L. Sutherland. 2001. Overexpression of p21WAF1/CIP1 is an early event in the development of pancreatic intraepithelial neoplasia. Cancer Res. 61:8830-8837. [PubMed] [Google Scholar]
- 7.Blagosklonny, M. V. 2002. Are p27 and p21 cytoplasmic oncoproteins? Cell Cycle 1:391-393. [DOI] [PubMed] [Google Scholar]
- 8.Bogatkevich, G. S., E. Gustilo, J. C. Oates, C. Feghali-Bostwick, R. A. Harley, R. M. Silver, and A. Ludwicka-Bradley. 2005. Distinct PKC isoforms mediate cell survival and DNA synthesis in thrombin-induced myofibroblasts. Am. J. Physiol. Lung Cell. Mol. Physiol. 288:L190-L201. [DOI] [PubMed] [Google Scholar]
- 8a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarthy, B., P. Morley, and J. Whitfield. 1999. Ca2+-calmodulin and protein kinase Cs: a hypothetical synthesis of their conflicting convergences on shared substrate domains. Trends Neurosci. 22:12-16. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., P. K. Jackson, M. W. Kirschner, and A. Dutta. 1995. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature 374:386-388. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, M., P. Olivier, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21cip1 and p27kip1 CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18:1571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, W. Y. 1980. Calmodulin plays a pivotal role in cellular regulation. Science 207:19-27. [DOI] [PubMed] [Google Scholar]
- 13.Chin, D., and A. R. Means. 2000. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10:322-328. [DOI] [PubMed] [Google Scholar]
- 14.Coqueret, O. 2003. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 13:65-70. [DOI] [PubMed] [Google Scholar]
- 15.Dotto, G. P. 2000. p21WAF1/Cip1: more than a break to the cell cycle? Biochim. Biophys. Acta 1471:M43-M56. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 17.Estanyol, J. M., M. Jaumot, O. Casanovas, A. Rodriguez-Vilarrupla, N. Agell, and O. Bachs. 1999. The protein SET regulates the inhibitory effect of p21Cip1 on cyclin E-cyclin-dependent kinase 2 activity. J. Biol. Chem. 274:33161-33165. [DOI] [PubMed] [Google Scholar]
- 18.Esteve, V., N. Canela, A. Rodriguez-Vilarrupla, R. Aligue, N. Agell, I. Mingarro, O. Bachs, and E. Perez-Paya. 2003. The structural plasticity of the C terminus of p21Cip1 is a determinant for target protein recognition. Chembiochem 4:863-869. [DOI] [PubMed] [Google Scholar]
- 19.Gartel, A. L., and A. L. Tyner. 2002. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 1:639-649. [PubMed] [Google Scholar]
- 20.Goubin, F., and B. Ducommun. 1995. Identification of binding domains on the p21Cip1 cyclin-dependent kinase inhibitor. Oncogene 10:2281-2287. [PubMed] [Google Scholar]
- 21.Harper, J. W., G. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 22.Harreman, M. T., T. M. Kline, H. G. Milford, M. B. Harben, A. E. Hodel, and A. H. Corbett. 2004. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J. Biol. Chem. 279:20613-20621. [DOI] [PubMed] [Google Scholar]
- 23.Ikura, M. 1996. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 21:14-17. [PubMed] [Google Scholar]
- 24.Kashiwagi, M., M. Ohba, H. Watanabe, K. Ishino, K. Kasahara, Y. Sanai, Y. Taya, and T. Kuroki. 2000. PKC eta associates with cyclin E/cdk2/p21 complex, phosphorylates p21 and inhibits cdk2 kinase in keratinocytes. Oncogene 19:6334-6341. [DOI] [PubMed] [Google Scholar]
- 25.Kearsey, J. M., P. J. Coates, A. R. Prescott, E. Warbrick, and P. A. Hall. 1995. GADD45 is a nuclear cell cycle regulated protein which interacts with p21Cip1. Oncogene 11:1675-1683. [PubMed] [Google Scholar]
- 26.Kitaura, H., M. Shinshi, Y. Uchikoshi, T. Ono, T. Tsurimoto, H. Yoshikawa, S. M. M. Iguchi-Ariga, and H. Ariga. 2000. Reciprocal regulation via protein-protein interaction between c-Myc and p21cip1/waf1/sdi1 in DNA replication and transcription. J. Biol. Chem. 275:10477-10483. [DOI] [PubMed] [Google Scholar]
- 27.Klee, C. B., H. Ren, and X. Wang. 1998. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273:13367-13370. [DOI] [PubMed] [Google Scholar]
- 28.Klee, C. B., and T. Vanaman. 1982. Calmodulin. Adv. Protein Chem. 35:213-321. [DOI] [PubMed] [Google Scholar]
- 29.La Baer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S., and D. M. Helfman. 2004. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/Cofilin pathway. J. Biol. Chem. 279:1885-1891. [DOI] [PubMed] [Google Scholar]
- 32.Li, Y., D. Dowbenko, and L. A. Lasky. 2002. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J. Biol. Chem. 277:11352-11361. [DOI] [PubMed] [Google Scholar]
- 33.Lu, K. P., and A. R. Means. 1993. Regulation of the cell cycle by calcium and calmodulin. Endocr. Rev. 14:40-58. [DOI] [PubMed] [Google Scholar]
- 34.Luo, Y., J. Hurwitz, and J. Massague. 1995. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature 375:159-161. [DOI] [PubMed] [Google Scholar]
- 35.McShea, A., T. Samuel, J. T. Eppel, D. A. Galloway, and J. O. Funk. 2000. Identification of CIP-1-associated regulator of cyclin B (CARB), a novel p21-binding protein acting in the G2 phase of the cell cycle. J. Biol. Chem. 275:23181-23186. [DOI] [PubMed] [Google Scholar]
- 36.Means, A. R. 2000. Regulatory cascades involving calmodulin-dependent protein kinases. Mol. Endocrinol. 14:4-13. [DOI] [PubMed] [Google Scholar]
- 37.Means, A. R., and J. R. Dedman. 1980. Calmodulin—an intracellular calcium receptor. Nature 285:73-77. [DOI] [PubMed] [Google Scholar]
- 38.Mercer, S. E., D. Z. Ewton, X. Deng, S. Lim, T. R. Mazur, and E. Friedman. Mirk/dyrk1B mediates survival during the differentiation of C2C12 myoblasts. J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]
- 39.Nakanishi, M., R. S. Robetorye, G. R. Adami, O. M. Pereira-Smith, and J. R. Smith. 1995. Identification of the active region of the DNA synthesis inhibitory gene p21Sdi1/CIP1/WAF1. EMBO J. 14:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noda, A., Y. Ning, S. F. Venable, O. M. Pereira-Smith, and J. R. Smith. 1994. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res. 211:90-98. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Vilarrupla, A., C. Diaz, N. Canela, H. P. Rahn, O. Bachs, and N. Agell. 2002. Identification of the nuclear localization signal of p21cip1 and consequences of its mutation on cell proliferation. FEBS Lett. 531:319-323. [DOI] [PubMed] [Google Scholar]
- 42.Scott, M. T., A. Ingram, and K. L. Ball. 2002. PDK1-dependent activation of atypical PKC leads to degradation of the p21 tumour modifier protein. EMBO J. 21:6771-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott, M. T., N. Morrice, and K. L. Ball. 2000. Reversible phosphorylation at the C-terminal regulatory domain of p21Waf1/Cip1 modulates proliferating cell nuclear antigen binding. J. Biol. Chem. 275:11529-11537. [DOI] [PubMed] [Google Scholar]
- 44.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 45.Shim, J., H. Lee, J. Park, H. Kim, and E. J. Choi. 1996. A non-enzymatic p21 protein inhibitor of stress-activated protein kinases. Nature 381:804-806. [DOI] [PubMed] [Google Scholar]
- 46.Soderling, T. R. 1999. The Ca-calmodulin-dependent protein kinase cascade. Trends Biochem. Sci. 24:232-236. [DOI] [PubMed] [Google Scholar]
- 47.Sung, Y. H., J. Shin, and W. Lee. 2001. Solution structure of p21Waf1/Cip1/Sdi1 C-terminal domain bound to Cdk4. J. Biomol. Struct. Dyn. 19:419-427. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki, A., Y. Tsutomi, K. Akahane, T. Araki, and M. Miura. 1998. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 17:931-939. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, A., Y. Tsutomi, N. Yamamoto, T. Shibutani, and K. Akahane. 1999. Mitochondrial regulation of cell death: mitochondria are essential for procaspase 3-p21 complex formation to resist Fas-mediated cell death. Mol. Cell. Biol. 19:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka, H., T. Yamashita, M. Asada, S. Mizutani, H. Yoshikawa, and M. Tohyama. 2002. Cytoplasmic p21Cip1/WAF1 regulates neurite remodeling by inhibiting Rho-kinase activity. J. Cell Biol. 158:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taulés, M., E. Rius, D. Talaya, A. López-Girona, O. Bachs, and N. Agell. 1998. Calmodulin is essential for cyclin-dependent kinase 4 (Cdk4) activity and nuclear accumulation of cyclin D1-Cdk4 during G1. J. Biol. Chem. 273:33279-33286. [DOI] [PubMed] [Google Scholar]
- 52.Taulés, M., A. Rodríguez-Vilarrupla, E. Rius, J. M. Estanyol, O. Casanovas, D. B. Sacks, E. Pérez-Payá, O. Bachs, and N. Agell. 1999. Calmodulin binds to p21Cip1 and is involved in the regulation of its nuclear localization. J. Biol. Chem. 274:24445-24448. [DOI] [PubMed] [Google Scholar]
- 53.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villalonga, P., C. López-Alcalá, M. Bosch, A. Chiloeches, N. Rocamora, J. Gil, R. Marais, C. J. Marshall, O. Bachs, and N. Agell. 2001. Calmodulin binds to K-Ras but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell. Biol. 21:7345-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waga, S., G. J. Hannon, D. Beach, and B. Stillman. 1994. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369:574-578. [DOI] [PubMed] [Google Scholar]
- 57.Weinstein, H., and E. L. Mehler. 1994. Ca2+-binding and structural dynamics in the functions of calmodulin. Annu. Rev. Physiol. 56:213-236. [DOI] [PubMed] [Google Scholar]
- 58.Winters, Z. E., N. C. Hunt, M. J. Bradburn, J. A. Royds, H. Turley, A. L. Harris, and C. J. Norbury. 2001. Subcellular localisation of cyclin B, Cdc2 and p21WAF1/CIP1 in breast cancer: association with prognosis. Eur. J. Cancer 37:2405-2412. [DOI] [PubMed] [Google Scholar]
- 59.Xia, W., J.-S. Chen, X. Zhou, P.-R. Sun, D.-F. Lee, Y. Liao, B. P. Zhou, and M.-C. Hung. 2004. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin. Cancer Res. 10:3815-3824. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, B. P., Y. Liao, W. Xia, B. Spohn, M. H. Lee, and M. C. Hung. 2001. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 3:245-252. [DOI] [PubMed] [Google Scholar]