Abstract
The Mdm2 oncoprotein regulates abundance and activity of the p53 tumor suppressor protein. For efficient degradation of p53, Mdm2 needs to be phosphorylated at several contiguous residues within the central conserved domain. We show that glycogen synthase kinase 3 (GSK-3) phosphorylated the Mdm2 protein in vitro and in vivo in the central domain. Inhibition of GSK-3 rescued p53 from degradation in an Mdm2-dependent manner while its association with Mdm2 was not affected. Likewise, inhibition of GSK-3 did not alter localization of p53 and Mdm2 or the interaction of Mdm2 and MdmX. Ionizing radiation, which leads to p53 accumulation, directed phosphorylation of GSK-3 at serine 9, which preceded and overlapped with the increase in p53 levels. Moreover, expression of a GSK-3 mutant where serine 9 was replaced with an alanine reduced the accumulation of p53 and induction of its target p21WAF-1. We therefore conclude that inhibition of GSK-3 contributes to hypophosphorylation of Mdm2 in response to ionizing rays, and in consequence to p53 stabilization.
The activity of the p53 tumor suppressor protein is mainly controlled by the Mdm2 protein (for a review see reference 36). Mdm2 prevents the interaction of p53 with factors of the basal transcription machinery and mediates p53 degradation by cellular proteasomes (18, 27, 28). Efficient degradation of p53 requires phosphorylation of several contiguous residues in the central domain of the Mdm2 protein (amino acids [aa] 220 to 260 [3]). Under normal growth conditions, this domain is highly phosphorylated (3, 16). Replacement of most of the phosphorylated residues with an alanine but not with an aspartic acid interferes with p53 degradation, indicating that phosphorylation of these sites is obligatory for p53 degradation (3). We recently identified casein kinase I delta (CKIδ) as a kinase that phosphorylates the Mdm2 protein at serine 244 within the central domain (39). In addition, there are two consensus sites for glycogen synthase kinase 3 (GSK-3).
GSK-3 is a kinase that is involved in numerous cellular processes as diverse as glucose metabolism, protein synthesis, cell proliferation, microtubule dynamics, cell motility, and Wnt signaling (for a review see references 14 and 40). At present, at least 47 protein substrates have been reported, ranging from metabolic enzymes through structural proteins to transcription factors. The phosphorylation of GSK-3 substrates is usually directed by the presence of another phosphorylated residue (priming site), optimally located 4 amino acids C-terminal to the site of GSK-3 phosphorylation [S/T-XXX-S(P)/T(P)] (13). Other residues can occasionally substitute for a priming serine or threonine, but this is typically associated with an up-to-100-fold drop in activity (1, 9). GSK-3 exists in two isoforms, GSK-3α and GSK-3β. Both forms are constitutively active and ubiquitously expressed in mammalian tissues (41, 42). Inactivation of GSK-3 is triggered by activation of several signaling pathways (for a review see reference 40) that either prompt phosphorylation of serine 9 (serine 21 for GSK-3α), thus turning the amino-terminal domain of GSK-3 into a pseudosubstrate (8, 12, 32, 33, 34), or disrupt multiprotein complexes that contain GSK-3 and its substrates (10). Interestingly, both modes of GSK-3 inactivation appear to be completely separated, e.g., inhibition of GSK-3 in response to Wnt signaling is independent of phosphorylation of serine 9. Conversely, inactivation of GSK-3 by phosphorylation of serine 9 does not lead to stabilization of β-catenin or activation of LEF-1 (31, 44).
Cellular stress, like ionizing radiation (IR), leads to the activation of the p53 tumor suppressor protein. The protein is rescued from degradation, accumulates to high levels, and activates transcription of its target genes. The mechanism that leads to p53 stabilization is not as yet fully understood. Several reports discuss either disruption of p53/Mdm2 complexes by phosphorylation of p53 or Mdm2 or hypophosphorylation of the central domain of the Mdm2 protein (3, 7, 17, 25). Interestingly, among the sites that are hypophosphorylated after IR in the Mdm2 protein are the two putative GSK-3 consensus sites.
To gain further insight into the regulation of p53 stability under normal growth conditions and in response to IR, we searched for kinases that phosphorylate the central domain of the Mdm2 protein. Here we report that GSK-3 phosphorylated the central domain of the Mdm2 protein at sites among which the presence of phosphorylated residues is obligatory for p53 degradation. Moreover, overexpression of GSK-3 decreased p53 expression levels while inhibition of GSK-3 rescued p53 from degradation. Degradation of p53 was prevented in the presence of GSK-3 inhibitors without changes in subcellular localization of p53 or Mdm2. The analysis of p53 and Mdm2 in several assays did not provide any evidence for an additional activity of GSK-3 that could account for the regulation of p53 degradation, apart from phosphorylation of Mdm2. We therefore conclude that GSK-3 regulates p53 stability via phosphorylation of Mdm2.
Ionizing radiation (which causes hypophosphorylation of Mdm2 at two GSK-3 consensus sites) inactivated GSK-3, and this inactivation preceded and partly overlapped with p53 accumulation. Most importantly, expression of a GSK-3 mutant, where serine 9 has been replaced with an alanine, reduced the accumulation of p53 after ionizing radiation. We therefore conclude that inhibition of GSK-3 contributes to the stabilization of p53 in response to ionizing radiation.
MATERIALS AND METHODS
Plasmids.
The plasmids pcDNA3-mdm2, pcDNA3-p53, and pDWM 659 Myc-Mdm2 and the plasmid for His-tagged ubiquitin have been described previously (3). The Mdm2 mutants pcDNA3-mdm2-Δ200-300, pcDNA3-mdm2-S240A, pcDNA3-mdm2-S254A, pcDNA3-S240A/S254, pGex4T2-mdm2-S240A/S254A, and pGex4T2-mdm2-S244A/S258A and the GSK-3 mutant GSK-3β-S9A were created by site-directed mutagenesis. Flag-tagged GSK-3β and GSK-3β S9A were created by PCR using primers encoding the tag. For pGEX4T2-mdm2(1-206), pGEX4T2-mdm2(226-300), and pGEX4T2-mdm2(296-491), the Mdm2 sequences were amplified by PCR and ligated into the pGEX 4T-2 vector (Amersham). pSuper-GSK-3β and pSuper.neo.gfp-Mdm2 were constructed by ligating the respective oligonucleotides into pSuper and pSuper.neo-gfp (Oligoengine). Primer sequences are available on request. The plasmid pMACS KK.II was purchased from Miltenyi Biotec. pcDNA3-GSK-3β and pcDNA3-GSK-3β-R96A were given to us by Thilo Hagen, and HA-pcDNA3-mdmX was given to us by Martin Scheffner.
Antibodies.
The following antibodies were used in the study: CM-1 (anti-p53, polyclonal; David Lane), DO-1 (anti-p53, monoclonal; Santa Cruz), Ab-2 (anti-p53, monoclonal; Oncogene Science), 4B2 (anti-Mdm2, monoclonal; David Lane), PC10 (anti-proliferating nuclear cell antigen [PCNA], monoclonal, Santa Cruz), anti-GSK-3β (monoclonal; BD Transduction Laboratories), anti-phospho-Ser9-GSK-3β (polyclonal; Cell Signaling), anti-glutathione S-transferase (anti-GST) (polyclonal; Rockland), 9E10 (anti-c-Myc, monoclonal; Oncogene Science), and H-164 (anti-p21, polyclonal; Santa Cruz).
Cell lines and their treatments.
U2OS, H1299, 293T, and Cos-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 100 units/ml penicillin-streptomycin at 37°C and 5% CO2 in a humidified atmosphere. Cells were transiently transfected by calcium phosphate or Jet-PEI (PolyPlus) or with Lipofectamine (Invitrogen) according to the manufacturer's instructions.
For U2OS cells overexpressing Flag-GSK-3β and Flag-GSK-3β/S9A, cells were transfected with pcDNA3-Flag-GSK-3β and pcDNA3-Flag-GSK-3β-S9A, respectively, and selected for 10 days with neomycin (300 μg/ml).
For selection of cells expressing Mdm2 or GSK-3β short interfering RNA (siRNA), the MACSelect transfected cell selection kit (Miltenyi Biotec) was used according to the manufacturer's instructions.
Alsterpaullone (Calbiochem) was used at a final concentration of 10 μM, bisindolylmaleimide IX (Calbiochem) at a final concentration of 2 μM, bisindolylmaleimide I (Calbiochem) at a final concentration of 4 μM, lithium chloride at a final concentration of 50 mM, cycloheximide (Sigma) at a final concentration of 30 μg/ml, puromycin at a final concentration of 1.5 μg/ml, and MG132 at a final concentration of 10 μM.
Cells were irradiated in culture medium with 7.5 grays using a 60Co γ source with a dose rate of 2 grays/min.
Ubiquitylation assay, in vivo radiolabeling, and peptide mapping.
Ubiquitylation assay, in vivo radiolabeling, and peptide mapping were performed as described in reference 3.
Immunofluorescence staining, immunoprecipitation, SDS-PAGE, and Western blotting.
Immunofluorescence staining, immunoprecipitations, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotting were performed as described previously (3). Detection of phosphorylated GSK-3 was performed according to the manufacturer's instructions (Cell Signaling).
Western blot membranes were stripped by incubation at 50°C (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 50 mM dithiothreitol) for 40 min followed by two washes with phosphate-buffered saline.
RNA isolation, Northern blotting, and RT-PCR.
RNA was isolated and blotted according to standard procedures. For radiolabeling the Prime-a-Gene kit (Promega) was used according to the manufacturer's recommendations. Probes comprised the open reading frame of mouse p53 and a 1,200-bp PstI fragment of the rat glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH). Reverse transcription-PCR (RT-PCR) was performed using the Abi7000 system according to the manufacturer's recommendations.
In vitro kinase assays.
GST-Mdm2 proteins were expressed in Escherichia coli and purified according to the instructions of the supplier of the pGEX4T-2 vector (Amersham). Phosphorylation with recombinant kinases was performed according to the manufacturer's recommendations (CKIδ, New England Biolabs; GSK-3β, Upstate Biotechnology) using 50 ng to 1 μg of purified protein. Priming was carried out in 10 μl at 30°C for 15 min, and then CKIδ was inactivated at 72°C for 90 min and the sample was adjusted to GSK-3 phosphorylation conditions and incubated for 20 min at 30°C in the presence of [γ-32P]ATP. After heat denaturation, samples were diluted 1:1 with 2× SDS-PAGE sample buffer and loaded onto an SDS-10% polyacrylamide gel.
RESULTS
GSK-3 phosphorylates the Mdm2 oncoprotein.
The Mdm2 protein is the most important regulator of the p53 protein. In order to target the p53 protein for proteasomal degradation, the central domain of the Mdm2 protein needs to be phosphorylated at several residues (3). The only kinase that is known as yet to phosphorylate this domain of the Mdm2 protein is casein kinase I (39). In order to get a better understanding of p53 regulation, we searched for additional protein kinases that phosphorylate this important domain of the Mdm2 protein.
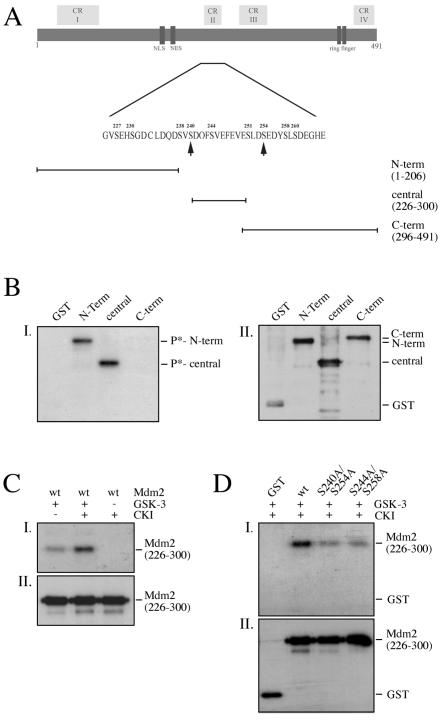
Since serine 240 and serine 254 of mouse Mdm2 present potential phosphorylation sites for GSK-3 (Fig. 1A), we investigated whether the Mdm2 protein may be a substrate for GSK-3. We expressed the N-terminal domain (aa 1 to 206), the central domain (aa 226 to 300), and the C-terminal domain (aa 296 to 491) of the Mdm2 protein as GST fusion proteins, purified the proteins by using the GST tag, and performed kinase assays with recombinant GSK-3 and [γ-32P]ATP. In this experimental setting, GSK-3 phosphorylated the N-terminal and central parts of the Mdm2 protein (Fig. 1B) while neither the C-terminal part of the Mdm2 protein nor the GST tag was phosphorylated.
FIG. 1.
GSK-3 phosphorylates Mdm2 in vitro. (A) Schematic drawing of the murine Mdm2 protein showing the conserved regions (CR), functional domains, and the localization of the deletion mutants. Arrows point to putative GSK-3 phosphorylation sites. NLS, nuclear localization signal; NES, nuclear export signal; N-term, N-terminal domain; central, central domain; C-term, C-terminal domain. (B) One microgram of GST or GST-Mdm2 fusion protein was incubated with 50 ng of recombinant GSK-3β and [γ-32P]ATP, separated on an SDS-12% polyacrylamide gel, and transferred to an Immobilon-P blotting membrane. The membrane was exposed to X-ray film (I) prior to hybridization with a GST antibody to determine the amount of GST and GST fusion proteins (II). (C) Fifty nanograms of GST-Mdm2 (aa 226 to 300) was primed with CKIδ and cold ATP. After 15 min, the kinase was heat inactivated and the reaction was adjusted to conditions for GSK-3β. GSK-3β and [γ-32P]ATP were added, incubated for 20 min, and processed as described in the legend to panel B. D: Fifty nanograms of wild-type GST-Mdm2 (aa 226 to 300) or GST-Mdm2 with the indicated substitutions was primed with CKIδ, heat inactivated, and incubated with recombinant GSK-3β and [γ-32P]ATP. After the kinase assay, the samples were processed as described in the legend to panel B. wt, wild type.
GSK-3 is a kinase that phosphorylates primed substrates with a 100-fold increase in activity. In eucaryotic cells, the Mdm2 protein is heavily phosphorylated at several contiguous residues in the central domain including GSK-3 priming sites (3). We recently identified CKIδ as a kinase that phosphorylates one of the priming sites for GSK-3 (serine 244) in the central domain of the Mdm2 protein (39). Consistent with an increase in activity in the presence of primed substrates, prephosphorylation of Mdm2 by CKIδ significantly enhanced phosphorylation of Mdm2 by GSK-3 (Fig. 1C). Conversely, replacing the priming sites serine 244 and serine 258 with an alanine considerably reduced Mdm2 phosphorylation (Fig. 1D).
In order to determine the sites that are phosphorylated by GSK-3, we replaced the serines in the two GSK-3 consensus sites with an alanine (S240A/S254A). Phosphorylation of mutant Mdm2 was strongly decreased compared to that of wild-type GSK-3, indicating that GSK-3 preferentially phosphorylates the consensus sites (Fig. 1D).
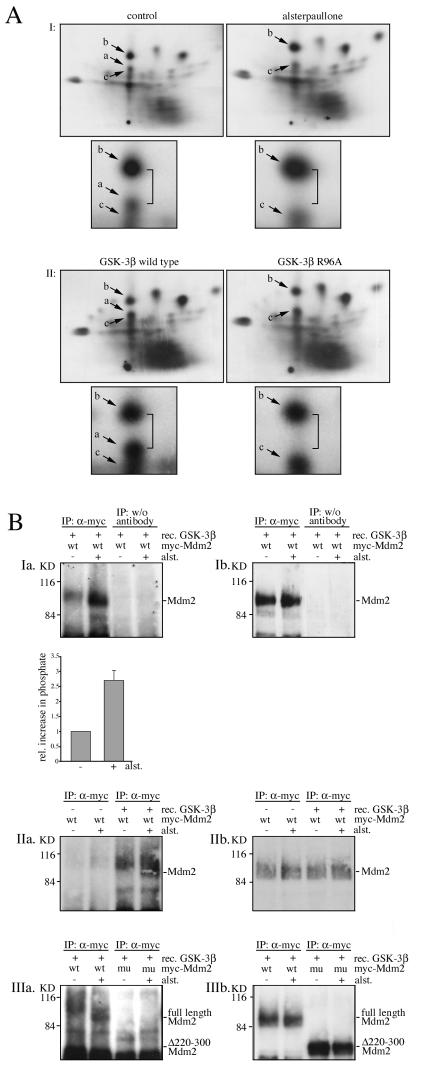
After having established that GSK-3 phosphorylates the Mdm2 protein in vitro, we explored whether GSK-3 phosphorylates the Mdm2 protein also under physiologic conditions. We transfected Cos-7 cells with Myc-tagged Mdm2, labeled the cells with [32P]orthophosphate, and performed two-dimensional peptide analysis. As shown previously (3, 16), the Mdm2 protein was constitutively phosphorylated at a number of sites. Treatment with the GSK-3 inhibitor alsterpaullone changed this pattern of phosphorylation. In particular a single peptide in the center of the map (peptide a, Fig. 2A) became invisible, due to the loss of phosphorylation. The loss of peptide a is particularly apparent because of the increased distance between the two labeled spots which is highlighted by the insets in Fig. 2A. Correspondingly, when we overexpressed wild-type GSK-3 in Cos-7 cells, phosphorylation of the same peptide (peptide a) was increased (Fig. 2A). Conversely, when we transfected the GSK-3 mutant (R96A), which is inactive against primed substrates, instead of wild-type GSK-3 into cells, phosphorylation of peptide a was strongly reduced (Fig. 2A).
FIG. 2.
The Mdm2 protein is phosphorylated in a GSK-3-dependent way in vivo. (A) (I) Cos-7 cells were transfected with Myc-tagged Mdm2, labeled with [32P]orthophosphate in the presence or absence of alsterpaullone, and subjected to two-dimensional phosphopeptide mapping. (II) Cos-7 cells were transfected with Myc-tagged Mdm2 and wild-type GSK-3β or GSK-3β-R96A, incubated for 3 hours with [32P]orthophosphate, and subjected to two-dimensional phosphopeptide mapping. Arrows point to changes in the peptide pattern in re-sponse to the treatment. a, b, and c are individual peptides of the Mdm2 protein. Peptide a is partly overlapping with peptide c. Smaller pictures below the maps show insets with higher magnification of the altered area. (B) Cos-7 cells were transfected with a cDNA expressing Myc-tagged full-length Mdm2 (wt) or a mutant lacking the central domain (mu, Δ220-300). Cells were treated for 4 hours with alsterpaullone (+) or left untreated for control (−). The Myc-tagged Mdm2 proteins were precipitated and incubated with [γ-32P]ATP in the presence or absence of recombinant GSK-3β. The reaction mixture was loaded onto an SDS-10% polyacrylamide gel, transferred onto a blotting membrane, and exposed to an X-ray film (a, kinase assay) prior to hybridization with the 4B2 anti-Mdm2 antibody for expression control (b, Western blot).
To further corroborate the result that GSK-3 phosphorylates the Mdm2 protein in vivo, we employed a second approach. This approach was based on the principle that, if Mdm2 is un- or underphosphorylated in the cell, it should be possible to immunoprecipitate this unphosphorylated (or poorly phosphorylated) Mdm2 protein from cell extracts and phosphorylate it by recombinant GSK-3 in vitro (in the presence of [γ-32P]ATP). On the other hand, if the phosphorylation sites are modified in the cells prior to immunoprecipitation of the Mdm2 (e.g., in the absence of any inhibitor), it should be impossible to incorporate any further phosphate into these sites in vitro. Along this line, we immunoprecipitated the Mdm2 protein from cells and incubated it with recombinant GSK-3 and [γ-32P]ATP. Under these experimental conditions, we found a basal level of phosphorylation, which is probably due to phosphorylation in vitro at sites that are usually not phosphorylated in vivo (Fig. 2B, Ia, IIa, and IIIa). However, and most importantly, phosphorylation of the Mdm2 protein was significantly enhanced when the Mdm2 protein was derived from cells that were treated with the GSK-3 inhibitor alsterpaullone (Fig. 2B, Ia, IIa, and IIIa), indicating that the inhibitor prevented phosphorylation of the Mdm2 protein at sites that are phosphorylated by GSK-3. In contrast, phosphorylation of mutant Mdm2, where the central domain was deleted (Δ200 to 300), was not enhanced by the treatment with alsterpaullone (Fig. 2B). This result is consistent with the premise that GSK-3 phosphorylates the Mdm2 protein in vivo within the central domain.
To further support the concept that GSK-3 phosphorylates Mdm2 directly and not via the regulation of a bridging kinase, we transfected 293T cells with Mdm2, immunoprecipitated GSK-3, and determined the amount of coprecipitating Mdm2 protein. As we show in Fig. 3, Mdm2 and GSK-3 were coprecipitated, indicating that both proteins interact in vivo.
FIG. 3.
GSK-3 coprecipitates with the Mdm2 protein. 293T cells were transfected with Myc-tagged Mdm2 where indicated. GSK-3 was immunoprecipitated, and coprecipitating Mdm2 was monitored by Western blotting using the 9E10 anti-Myc antibody. IgG, immunoglobulin G.
Inhibition of GSK-3 prevents p53 degradation.
Phosphorylation of serines in the central domain of the Mdm2 protein is vital for p53 degradation. In the area that needs to be phosphorylated to allow p53 degradation are the two GSK-3 consensus sites. If GSK-3 phosphorylates the Mdm2 protein in vivo in the central domain, then inhibition of GSK-3 should rescue p53 from degradation.
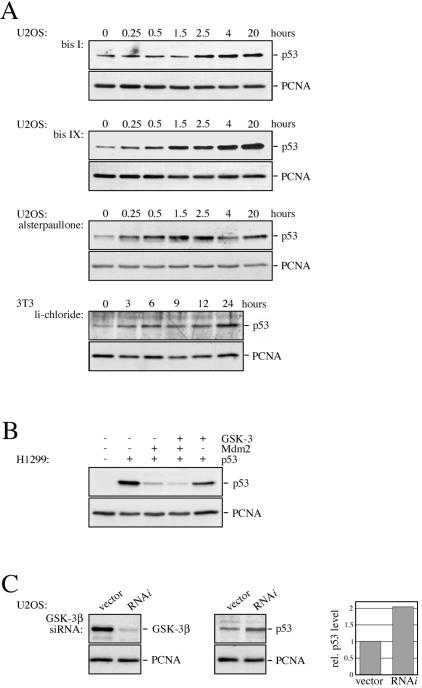
To test this hypothesis, we treated U2OS and mouse 3T3 cells with a range of GSK-3 inhibitors (lithium chloride, alsterpaullone, bisindolylmaleimide I, and bisindolylmaleimide IX) and determined p53 protein levels by Western blotting. Consistent with the notion that GSK-3 phosphorylates the central domain of the Mdm2 protein at sites where phosphorylation is obligatory for p53 degradation, the tumor suppressor protein accumulated within less than 2 hours in the presence of each of the GSK-3 inhibitors (Fig. 4A) while treatment with the drug vehicle alone had no effect (data not shown). Furthermore, overexpression of GSK-3β reduced p53 protein levels (Fig. 4B) while downregulation of GSK-3β by siRNA increased p53 expression (Fig. 4C and 5C).
FIG. 4.
Inhibition of GSK-3 leads to the accumulation of p53. (A) U2OS cells were incubated with bisindolylmaleimide I (bis I), bisindolylmaleimide IX (bis IX), alsterpaullone, or lithium chloride (li-chloride) for the indicated time, and p53 expression levels were monitored by Western blotting. Hybridization with the anti-PCNA antibody PC-10 was performed for loading control. (B) H1299 cells were transfected with pcDNA3-p53, pcDNA3-mdm2, and pcDNA3-GSK-3β, where indicated. Thirty-six hours after transfection, cells were harvested and probed for the presence of p53. (C) U2OS cells were cotransfected with pSuper-GSK-3β expressing GSK-3β RNAi and a puromycin resistance gene. Cells were selected for 3 days with puromycin and probed for the presence of p53 and GSK-3β. p53 and PCNA levels were quantified and plotted. p53 levels of vector-transfected cells were set to 1.
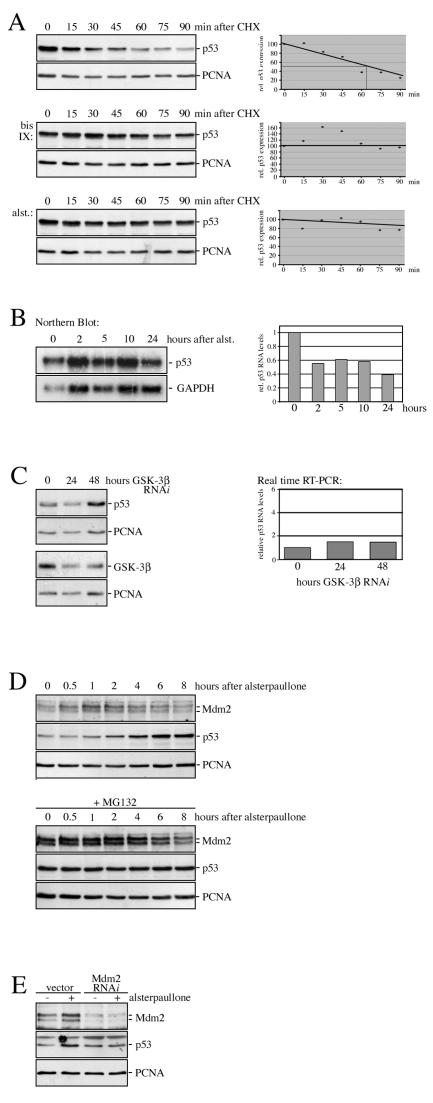
FIG. 5.
Inhibition of GSK-3 prevents p53 degradation. (A) U2OS cells were treated with alsterpaullone (alst.) or bisindolylmaleimide IX (bis IX) for 4 hours or left untreated for control. At the indicated time points, cycloheximide (CHX) was added and p53 protein levels were determined. p53 and PCNA levels were quantified and relative p53 expression was plotted, whereas relative p53 expression levels at thetime of CHX addition were set to 100. (B) U2OS cells were treated with alsterpaullone (alst.) for the indicated time. Poly(A)+ RNA was prepared and probed for the presence of p53 and GAPDH RNA by Northern blotting. p53 and GAPDH RNA levels were quantified, and relative p53 RNA levels were plotted, whereas the relative p53 RNA level of untreated cells was set to 1. (C) U2OS cells were transfected with pMACS KK.II and pSuper-GSK-3β expressing GSK-3β RNAi or pMACS KK.II and vector control (0). GSK-3β RNAi-transfected cells were harvested after 24 and 48 h and sorted by magnetic beads. An aliquot of the cells was analyzed for p53 and GSK-3β expression by Western blotting. From the remaining cells, total RNA was prepared and p53 and β-actin were amplified by real-time RT-PCR. Relative p53 RNA levels were plotted, whereas the relative p53 level of vector-transfected cells was set to 1. (D) U2OS cells were incubated with the proteasome inhibitor MG132 for 4 hours or left untreated for control. Alsterpaullone was added, and the cells were harvested after the indicated time. p53 and Mdm2 protein levels were determined by Western blotting. (E) U2OS cells were transfected with pMACS KK.II and pSuper.gfp/neo encoding Mdm2 siRNA, or pSuper.gfp/neo vector for control. Cells were treated with alsterpaullone for 2 hours, and transfected cells were sorted by using magnetic beads. Mdm2 and p53 protein levels were determined by Western blotting.
Inhibition of GSK-3 rescues p53 from proteasomal degradation.
To further define the molecular level of p53 regulation by GSK-3, we determined the half-life of the p53 protein in the presence and absence of GSK-3 inhibitors. In untreated cells, we observed a half-life of the p53 protein of about 65 min (Fig. 5A). Treatment of cells with alsterpaullone or bisindolylmaleimide IX prolonged the half-life of the p53 protein considerably (Fig. 5A). The accumulation of the p53 protein in response to GSK-3 inhibition is entirely posttranscriptional, since p53 RNA levels were even slightly reduced in the presence of alsterpaullone (Fig. 5B). Similar results were obtained when GSK-3β was inhibited by small interfering RNA (siRNA) (Fig. 5C). Moreover, inhibition of the 26S proteasome by the proteasome inhibitor MG132 prevented the induction of p53 by alsterpaullone, suggesting that a functional proteasome is required for the response (Fig. 5D). Most importantly, stabilization of p53 in response to inhibition of GSK-3 was fully dependent on the presence of the Mdm2 protein. Downregulation of Mdm2 by siRNA blocked p53 accumulation in response to GSK-3 inhibition completely (Fig. 5E). Interestingly, p53 levels did not increase considerably in the absence of the Mdm2 protein.
Inhibition of GSK-3 stabilizes the p53 protein despite its association with and ubiquitylation by Mdm2.
Several independent principles could be responsible for the increase in p53 abundance in response to GSK-3 inhibition. Conceivable mechanisms are (i) phosphorylation of p53 by GSK-3, (ii) alteration of cellular localization of p53 or Mdm2, (iii) dissociation of p53/Mdm2 complexes, (iv) dissociation of Mdm2/MdmX complexes, or (v) interference with p53 ubiquitylation.
We first tested whether GSK-3 may regulate p53 stability by phosphorylating the tumor suppressor protein itself. However, comparison of the two-dimensional peptide maps from alsterpaullone-treated and nontreated control cells showed no alteration in the phosphorylation pattern of the p53 protein (data not shown).
Next, we investigated if mimicking GSK-3 inhibition by replacing the GSK-3 phosphorylation sites in Mdm2 with alanine prevents p53 degradation. As we show in Fig. 6A, II, replacing serine 254 or serine 240 together with serine 254 with an alanine rescued p53 from degradation. In contrast, replacing serine 240 alone with an alanine did not interfere with p53 degradation. However, when serine 240 together with serine 238 was replaced with an alanine, p53 degradation was hampered (3). This result indicated that hypophosphorylation of Mdm2 at the GSK-3 consensus site 254 could trigger the accumulation of p53 in response to GSK-3 inhibition.
FIG. 6.
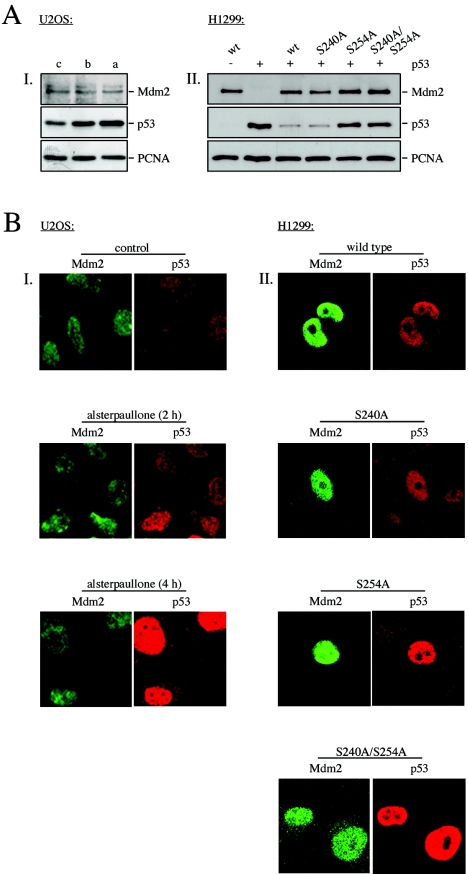
Inhibition of GSK-3 does not alter the cellular localization of p53 or Mdm2. (A) (I) U2OS cells were treated with bisindolylmaleimide IX (b) or alsterpaullone (a) for 4 hours or left untreated for control (c). Mdm2, p53, and PCNA levels were determined by Western blotting. (II) H1299 cells were transfected with pcDNA3-p53 and wild-type pcDNA3-mdm2 or pcDNA3-mdm2 possessing the indicated point mutations. Twenty-four hours after transfection, cells were analyzed for the presence of p53, Mdm2, and PCNA by Western blotting. (B) (I) U2OS cells were treated with alsterpaullone and harvested after 2 and 4 hours. Localization of Mdm2 and p53 was monitored by immunofluorescence staining. The Mdm2 protein is stained in green, and the p53 protein in red. (II) H1299 cells were transfected with pcDNA3-p53 and pcDNA3-mdm2 encoding wild-type Mdm2 or Mdm2 harboring the indicated mutations. Twenty-four hours after transfection, cells were analyzed for the expression of Mdm2 and p53 by immunofluorescence staining. The Mdm2 protein is stained in green, and the p53 protein is in red.
To further explore the mechanism of p53 accumulation in response to GSK-3 inhibition, we investigated the subcellular localization of p53 and Mdm2 in response to inhibition of GSK-3 or in the presence of mutant Mdm2 by immunofluorescence analysis. In nontreated control cells, we observed a rather weak but exclusively nuclear staining for p53 and Mdm2. Two hours after addition of alsterpaullone, nuclear abundance of p53 was significantly enhanced and the levels increased even further within the next 2 hours. At the same time, the signal for the Mdm2 protein remained largely unchanged (Fig. 6A, I, and 6B, I). In summary, both p53 and Mdm2 proteins were localized in the nucleus in the presence and absence of alsterpaullone. No cytoplasmic staining was evident for p53 or Mdm2. Notably, the p53 protein accumulated in the presence of alsterpaullone despite the presence of Mdm2 in the same cellular compartment (Fig. 6B, I). Correspondingly, when we replaced serine 254 or serine 240 and serine 254 with an alanine, the p53 protein also accumulated in the nucleus despite the presence of Mdm2 in the same cellular compartment (Fig. 6A, II, and 6B, II).
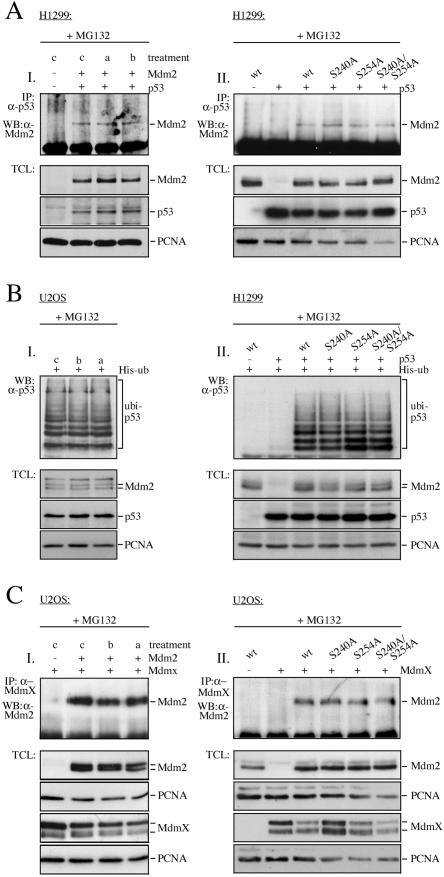
Another possibility that could lead to the accumulation of p53 upon GSK-3 inhibition would be disruption of p53/Mdm2 complexes. We therefore investigated whether p53 and Mdm2 interacted with each other in the presence of GSK-3 inhibitors. We immunoprecipitated the p53 protein from cells that had been treated with the GSK-3 inhibitor alsterpaullone or bisindolylmaleimide IX or left untreated for control and determined the level of associated Mdm2 protein by Western blotting. To obtain similar levels of p53 in all samples despite the stabilization of p53 in the presence of GSK-3 inhibitors, we incubated the cells several hours beforehand with the proteasome inhibitor MG132.
Consistent with earlier reports (3), p53 and Mdm2 coprecipitated in control cells. Interestingly, both proteins also coprecipitated in inhibitor-treated cells. Moreover, similar levels of the Mdm2 protein coprecipitated with p53 in inhibitor-treated and control cells, showing that the activity of GSK-3 did not influence the strengths of the interaction of p53 and Mdm2 (Fig. 7A, I). Correspondingly, the interaction of Mdm2 mutants, where the GSK-3 phosphorylation sites were replaced with an alanine, with p53 was indistinguishable from the interaction of p53 and wild-type Mdm2 (Fig. 7A, II). Likewise, the association of Mdm2 with MdmX, another partner of the Mdm2 protein that is required for p53 degradation (21), was not affected by the presence of alsterpaullone or bisindolylmaleimide IX (Fig. 7C, I) or by replacement of GSK-3 phosphorylation sites with an alanine (Fig. 7C, II).
FIG. 7.
Inhibition of GSK-3 leads to the accumulation of ubiquitylated p53. (A) (I) H1299 cells were transfected with pcDNA3-p53 and pcDNA3-mdm2. Twenty-four hours after transfection, cells were treated with MG132 for 5 hours. Two hours prior to harvest, alsterpaullone (a) or bisindolylmaleimide IX (b) was added or the cells were left untreated for control (c). (II) H1299 cells were transfected with pcDNA3-p53 or with pcDNA3-p53 and wild-type pcDNA3-mdm2 or pcDNA3-mdm2 harboring the indicated mutations. Twenty-four hours after transfection, cells were treated with MG132 for 5 hours. IP: p53 was precipitated and associated Mdm2 protein was determined by Western blotting. TCL: total cellular lysate was probed for the presence of Mdm2, p53, and PCNA. (B) (I) U2OS cells were transfected with a plasmid encoding His-tagged ubiquitin. Twenty-four hours after transfection, cells were treated with MG132 for 5 h. Two hours prior to harvest, alsterpaullone (a) or bisindolylmaleimide IX (b) was added or cells were left untreated for control (c). (II) H1299 cells were transfected with a plasmid encoding His-tagged ubiquitin, pcDNA3-p53, and wild-type pcDNA3-mdm2 or pcDNA3-mdm2 harboring the indicated mutations. Twenty-four hours after transfection, cells were treated with MG132 for 5 hours. Aliquots of the cells wereanalyzed for the presence of Mdm2 and p53 by Western blotting (TCL). The remaining cells were lysed in guanidinium lysis buffer. Ubiquitylated proteins were purified by adsorption to Ni2+-nitrilotriacetic acid agarose and probed by Western blotting for the presence of ubiquitylated p53 (ubi-p53). (C) (I) U2OS cells were transfected with HA-pcDNA3-mdmX and pcDNA3-mdm2 where indicated. Twenty-four hours after transfection, cells were treated with MG132 for 5 hours. Two hours prior to harvest, alsterpaullone (a) or bisindolylmaleimide IX (b) was added or cells were left untreated for control (c). (II) U2OS cells were transfected with HA-pcDNA3-mdmX and wild-type pcDNA3-mdm2 or pcDNA3-mdm2 harboring the indicated mutations. Twenty-four hours after transfection, cells were treated with MG132 for 5 hours. IP: MdmX was immunoprecipitated with an anti-HA antibody, and associated Mdm2 was monitored by Western blotting. TCL: total cellular lysate was probed for the presence of Mdm2 and MdmX by Western blotting.
Last but not least, we determined whether p53 protein ubiquitylation is altered in the presence of GSK-3 inhibitors or by mutant Mdm2. We transfected U2OS cells with His-tagged ubiquitin, purified ubiquitylated proteins by adsorption to Ni2+ agarose, and probed the ubiquitylated proteins for the presence of p53 by Western blotting. To obtain similar levels of the p53 protein, we incubated the cells for 5 h with MG132.
As shown in Fig. 7B, I, similar levels of ubiquitylated p53 protein were retrieved in the presence and absence of GSK-3 inhibitors. Also replacing serines in the GSK-3 consensus site of the Mdm2 protein did not grossly alter p53 ubiquitylation although there may have been a slight decrease in p53 ubiquitylation in the presence of the S254A and S240A/S254A mutant (Fig. 7B, II).
GSK-3 is inactivated after ionizing radiation.
Ionizing radiation leads to hypophosphorylation of the central domain of the Mdm2 protein (3). Within the hypophosphorylated region are the two GSK-3 consensus sites. If Mdm2 is a physiologic target of GSK-3, then we have to conclude that GSK-3 should be inactivated after ionizing radiation.
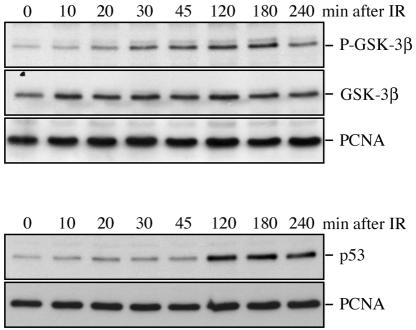
To investigate whether GSK-3 is inactivated in response to ionizing radiation, we used phosphorylation-specific antibodies that are directed against serine 9 of GSK-3β. Phosphorylation of serine 9 inactivates GSK-3β by turning the amino terminus of the kinase into a pseudosubstrate that competes with authentic GSK-3 substrates for the active site (reviewed in reference 14).
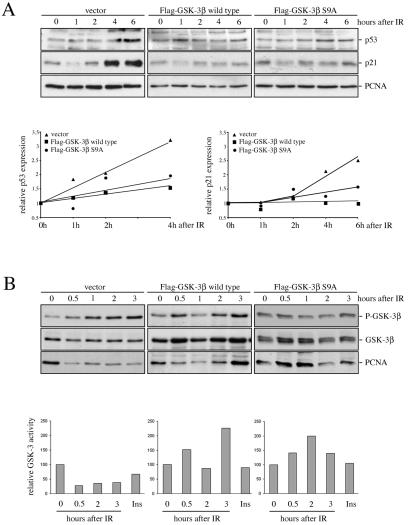
Ionizing radiation led within less than 20 minutes to a strong increase in GSK-3β phosphorylation, and this phosphorylation was retained for more than 3 hours (Fig. 8). Thus, phosphorylation of GSK-3 preceded and overlapped with the increase in p53 abundance, which was detectable within less than 2 hours (Fig. 8). More importantly, overexpression of wild-type GSK-3β or a GSK-3β mutant, where serine 9 was replaced with an alanine and which was, hence, insensitive to ionizing radiation, significantly reduced p53 accumulation after ionizing radiation (Fig. 9A). Induction of the p53 target gene p21WAF1 in response to ionizing radiation was equally reduced upon exogenous expression of wild-type GSK-3β or GSK-3β S9A. Determination of GSK-3β activity against a synthetic primed substrate revealed a strong correlation of its activity with p53 levels. GSK-3 activity was reduced by about 50% in cells transfected with vector while it was not inhibited in cells transfected with wild-type or mutant GSK-3β (Fig. 9B). In fact we instead observed a slight increase in GSK-3 activity in response to ionizing radiation, which was also reflected by reduced phosphorylation of GSK-3β in GSK-3β wild-type- and -S9A-transfected cells compared to vector-transfected cells (Fig. 9B). According to these results we conclude that inactivation of GSK-3 contributes to the stabilization of p53 after ionizing radiation.
FIG. 8.
GSK-3β is inactivated after ionizing radiation. U2OS cells were irradiated with 10 grays, harvested after the indicated time, and analyzed by Western blotting. The membrane was incubated with an antibody against phosphorylated GSK-3β, stripped, and rehybridized with a pan-GSK-3β antibody or with the Ab-2 antibody (anti-p53).
FIG. 9.
Expression of a constitutively active GSK-3β mutant reduces p53 accumulation after ionizing radiation. U2OS cells were transfected with Flag-tagged wild-type GSK-3β or Flag-tagged GSK-3β-S9A or with vector for control, selected for 6 days with neomycin, irradiated with 7.5 grays (IR), and harvested after the indicated time. (A) p53 and p21 expression were determined by Western blotting. Expression levels were quantified, and relative p53 and p21 expression levels were plotted. The graph shows mean values of three (p21) or four (p53) independent experiments. Relative p53 or p21 expression of nonirradiated cells was set to 1. (B) An aliquot of cells was used to determine the amount of phosphorylated GSK-3β and total GSK-3β by Western blotting. From the remaining cells, GSK-3β was precipitated and incubated with phospho-glycogen synthase peptide 2 in the presence of [γ-32P]ATP. An aliquot of the reaction mixture was spotted onto a P81 phosphocellulose membrane and counted in a scintillation counter. Counts obtained from nonirradiated cells were set to 100. For control, cells were treated with insulin (Ins) for 5 min.
DISCUSSION
GSK-3 phosphorylates Mdm2 in vitro and in vivo.
The antiproliferative activity of the p53 protein qualifies this tumor suppressor protein as a target for antitumor therapy. Therefore, ways to activate p53 are under intense investigation. Activation of p53 by small molecules, like kinase inhibitors, is of particular interest since they are effective in their action, easily administered to patients, and cell permeable, at least if carrying appropriate side chains. The possibility of activating p53 by kinase inhibitors arose from the observation that p53 degradation is regulated by phosphorylation of the central domain of the Mdm2 protein (3). In a search for kinases that phosphorylate the central domain of Mdm2, we came across GSK-3. Here we show that GSK-3 phosphorylated Mdm2 in vitro and in vivo.
Several lines of evidence support the model that the Mdm2 protein is a physiologic substrate for GSK-3. Firstly, Mdm2 was dephosphorylated when cells were incubated with inhibitors of GSK-3 (Fig. 2A). Secondly, mimicking dephosphorylation of the area that is phosphorylated by GSK-3 by replacing serines with an alanine affected the function of the protein in a manner that is consistent with the physiological effect of its antagonist (Fig. 4 and 6). Thirdly, ionizing radiation that leads to the inactivation of GSK-3 towards primed substrates (Fig. 12D in reference 35) decreases phosphorylation of Mdm2 at GSK-3 consensus sites (3). Fourthly, the kinetics of Mdm2 hypophosphorylation is similar to the kinetics of GSK-3 inactivation (3) (Fig. 8 and 9B), and last but not least, GSK-3 and Mdm2 interact with each other (Fig. 3).
Phosphorylation of Mdm2 by GSK-3 was not unexpected, as sequences within the central domain of the Mdm2 protein are in complete harmony with the requirements of a GSK-3 target. Two distinct patches in the central domain of the Mdm2 protein match with the GSK-3 consensus motif, and several serines within this domain are usually highly phosphorylated, including serine 244 and serine 258, thus providing primed residues within the GSK-3 consensus motif (3, 16). Several lines of evidence support the hypothesis that Mdm2 is a primed substrate for GSK-3. Firstly, Mdm2 is constitutively highly phosphorylated within the area of the GSK-3 consensus motifs (3). Secondly, CKI, which phosphorylates GSK-3 priming sites on β-catenin (2), has recently been identified as a kinase that phosphorylates Mdm2 at serine 244, one of the GSK-3 priming sites on the Mdm2 protein (39). Thirdly, transfection of the GSK-3 mutant R96A, which does not phosphorylate primed substrates, is incapable of increasing Mdm2 phosphorylation.
Nevertheless, despite all the evidence that Mdm2 is directly phosphorylated by GSK-3 we cannot entirely exclude the possibility that another kinase that is closely regulated by GSK-3 mediates Mdm2 phosphorylation in living cells. Similarly, a protein other than Mdm2 that can be (co-)precipitated with the 9E10 antibody, which is of similar size, and could be phosphorylated in a GSK-3-dependent manner.
A postubiquitylation function of Mdm2 is regulated by GSK-3.
Consistent with the concept of being a physiologic kinase for the central domain of Mdm2, inhibition of GSK-3 rescued p53 from degradation by cellular proteasomes. Most importantly, in response to GSK-3 inhibition, the capacity of Mdm2 to ubiquitylate the p53 protein was not affected and thus ubiquitylated p53 protein accumulated in the absence of active GSK-3.
The Mdm2 protein has different activities that are required for p53 degradation. It binds the p53 protein with its N-terminal domain and conjugates ubiquitin molecules to C-terminal lysines of the p53 protein via its C-terminal ubiquitin ligase function. Once the p53 protein is fully ubiquitylated, a postubiquitylation function of the Mdm2 protein, which is not as yet fully characterized, mediates p53 degradation by cellular proteasomes. This postubiquitylation function of the Mdm2 protein is located in the central region and activated by phosphorylation. Since ubiquitylated p53 protein accumulates in response to GSK-3 inhibition and since the accumulation depends on the Mdm2 protein and functional proteasomes, it is most likely the postubiquitylation function of Mdm2 that is regulated by GSK-3. This idea is moreover supported by preliminary data from our group and the group of Steven Grossman (University of Massachusetts). It is moreover consistent with the observation that GSK-3 phosphorylated the central domain of Mdm2 and that it is inhibition of GSK-3 and not, e.g., activation that leads to p53 accumulation.
In contrast to previous reports (29, 35), we were unable to detect phosphorylation of p53 by GSK-3, although we found the kinase associated with p53. Most likely, the discrepancy is due to different experimental conditions. We moreover consider posttranslational modifications of p53 as being unlikely to trigger stabilization of p53 in response to GSK-3 inhibition for the following reasons. (i) The accumulation of p53 fully depends on the presence of Mdm2 and cannot be mimicked by other p53-specific E3 ligases such as Cop-1, topors, or Pirh2 (11, 20, 30). (ii) Neither the association with Mdm2 nor the ubiquitylation of p53 is affected by GSK-3 inhibition. (iii) E2F-1 and Mdm2, which are also targets for Mdm2-mediated degradation (4, 22), also accumulated in response to GSK-3 inhibition (data not shown).
The fact that GSK-3 inhibition does not interfere with p53 ubiquitylation also rules out other factors, e.g., translocation and sequestration of p53 and Mdm2 in different subcellular compartments, dissociation of p53/Mdm2 complexes, or dissociation of Mdm2 and MdmX, and indicates that GSK-3 inhibition interferes with p53 degradation at a step beyond ubiquitylation.
Apart from in response to GSK-3 inhibition, accumulation of ubiquitylated p53 protein has been observed in the presence of Mdm2 mutants where phosphorylation sites of the central domain were replaced with an alanine (including GSK-3 consensus sites [3]) and in response to ionizing radiation (24), which reduces phosphorylation of the central domain of Mdm2 (3). The precise mechanism by which p53 degradation is prevented in all these cases is unclear. One possibility is that the absence of active GSK-3 retained ubiquitylated p53 in the nucleus, thus interfering with its degradation by cytoplasmic proteasomes. In this context it is of interest that activation of GSK-3 is reported to promote degradation of p53 by enhancing its export into the cytoplasm (29). Nevertheless, several reports show that p53 is also degraded by nuclear proteasomes (19, 43). Alternatively, Mdm2 phosphorylation could regulate the association with hHR23, which is important for p53 degradation (6, 15).
Interestingly, downregulation of Mdm2 by siRNA had only a minor effect on p53 expression, most probably due to the presence of alternative E3 ubiquitin ligases such as Pirh2, topors, and COP1 (11, 20, 30) that most likely mediate p53 degradation in the absence of Mdm2.
Ionizing radiation inactivates GSK-3 towards primed substrates.
Ionizing radiation leads to hypophosphorylation of several individual phosphorylation sites in the central domain of Mdm2, including both GSK-3 consensus sites (3). It is therefore valid to speculate that, if Mdm2 is a true GSK-3 substrate, GSK-3 activity should be inhibited after IR. Consistent with the results of Turenne and Price (35), we found that GSK-3β was rapidly inactivated in response to IR towards primed substrates. Moreover, inactivation of GSK-3 preceded p53 accumulation. Residues of the Mdm2 protein that are hypophosphorylated in response to IR need to be phosphorylated to allow p53 degradation. In consistency with this principle, accumulation of p53 in response to ionizing radiation was reduced when we overexpressed a mutant form of GSK-3β (S9A) that is insensitive to inactivation by IR since it can no longer be phosphorylated at serine 9. However and to our own surprise, when we overexpressed wild-type GSK-3β, we observed the same effect. Notably, the reduction in p53 accumulation in response to IR upon overexpression of wild-type GSK-3 was accompanied by a decrease in IR-mediated phosphorylation of GSK-3. In addition, we were unable to detect a decrease in GSK-3 activity in response to IR or after treatment of cells with insulin (data not shown) in cells that overexpress wild-type GSK-3 or the S9A mutant. The reason for the failure to inhibit GSK-3 in response to IR when the kinase is overexpressed is unclear. It is, though, possible that it may be caused by the fact that GSK-3 is inhibited only partially at the dose that we used in our experiments. Upon overexpression, there might be enough GSK-3 activity left to phosphorylate and thus activate the Mdm2 protein.
While we and Turenne and Price have observed that GSK-3 is inhibited in response to ionizing radiation (35), there are also reports that show that GSK-3 is activated in response to stress (29, 37, 38). The reason for this discrepancy is unclear. It is, however, possible that the inconsistency is caused by the fact that different stimuli lead to p53 stabilization via different mechanisms. Different mechanisms of p53 accumulation in response to cellular stress are reflected, e.g., by different kinetics of p53 accumulation and by differences in Mdm2 expression and posttranslational modifications (3, 4, 5, 23, 25, 26). It would therefore be conceivable that IR inactivates GSK-3 while camptothecin enhances its activity. Another option would be that GSK-3 activity towards primed and nonprimed substrates is differently affected by cellular stress. This idea is supported by our observation that the activity of GSK-3 was reduced towards phospho-glycogen synthase peptide 2 in response to IR while its activity towards the tau protein, which is a nonprimed substrate, was increased (data not shown).
Our data show that GSK-3 regulates p53 abundance by phosphorylation of Mdm2. Inhibition of GSK-3 leads to the accumulation of transcriptionally active p53. Accordingly, preventing Mdm2 phosphorylation by kinase inhibitors is a valid approach to increase p53 activity. In this context it is particularly interesting that GSK-3 is efficiently inhibited by lithium chloride, a substance that is already used in the clinic to treat patients with neurological disorders. Most importantly, despite the numerous activities of GSK-3 in the cell, treatment with lithium chloride does not stimulate strong side effects, which makes this compound and other GSK-3 inhibitors a potentially interesting tool for controlling cancer.
Acknowledgments
We thank Martin Scheffner, Thilo Hagen, and David Lane for plasmids and antibodies and Axel Knebel, Philip Cohen, and Nick A. Morrice for helpful comments. We are very grateful to Andy Nestl for help with the real-time RT-PCR and to Margaret Ashcroft for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), grant Bl 526/1-2.
REFERENCES
- 1.Alt, J. R., J. L. Cleveland, M. Hannink, and J. A. Diehl. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent nuclear transformation. Genes Dev. 14:3102-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amit, S., A. Hatzubai, Y. Birman, J. S. Andersen, E. Ben-Shushan, M. Mann, Y. Ben-Neriah, and I. Alkalay. 2002. Axin-mediated CKI phosphorylation of beta-catenin at Ser45: a molecular switch for the Wnt pathway. Genes Dev. 16:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, C., T. Hay, D. W. Meek, and D. P. Lane. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 22:6170-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, C., A. Sparks, and D. P. Lane. 1999. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 19:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, C., A. Knebel, A. Radler-Pohl, C. Sachsenmaier, P. Herrlich, and H. J. Rahmsdorf. 1994. DNA-damaging agents and growth factors induce changes in the program of expressed gene-products through common routes. Environ. Mol. Mutagen. 24:3-10. [DOI] [PubMed] [Google Scholar]
- 6.Brignone, C., K. E. Bradley, A. F. Kisselev, and S. R. Grossman. 2004. A post-ubiquitination role for Mdm2 and hHR23A in the p53 degradation pathway. Oncogene 23:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 8.Cross, D. A. E., D. R. Alessi, P. Cohen, M. Andjelkovich, and B. A. Hemmings. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785-789. [DOI] [PubMed] [Google Scholar]
- 9.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, V. W., R. H. Chen, and F. McCormick. 2000. Differential regulation of glycogen synthase kinase 3 β by insulin and Wnt signaling. J. Biol. Chem. 275:32475-32481. [DOI] [PubMed] [Google Scholar]
- 11.Dornan, D., I. Wertz, H. Shimizu, D. Arnott, G. D. Frantz, P. Dowd, K. O'Rourke, H. Koeppen, and V. M. Dixit. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86-92. [DOI] [PubMed] [Google Scholar]
- 12.Fang, X., Y. Lu, S. X. Yu, Y. Lu, R. C. Bast, J. R. Woodgett, and G. B. Mills. 2000. Phosphorylation and inactivation of glycogen synthase kinase by protein kinase A. Proc. Natl. Acad. Sci. USA 97:11960-11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiol, C. J., A. M. Mahrenholz, Y. Wang, R. W. Roeske, and P. J. Roach. 1987. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262:14042-14048. [PubMed] [Google Scholar]
- 14.Frame, S., and P. Cohen. 2001. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glockzin, S., F. Ogi, A. Hengstermann, M. Scheffner, and C. Blattner. 2003. Stabilization of polyubiquitinated forms of p53 by the DNA repair protein hHR23. Mol. Cell. Biol. 23:8960-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay, T. J., and D. W. Meek. 2000. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett. 478:183-186. [DOI] [PubMed] [Google Scholar]
- 17.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 18.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein Mdm2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 19.Joseph, T. W., and U. M. Moll. 2003. Analysis of nuclear and cytoplasmic degradation of p53 in cells after stress. Methods Mol. Biol. 234:211-217. [DOI] [PubMed] [Google Scholar]
- 20.Leng, R. P., Y. Lin, W. Ma, H. Wu, B. Lemmers, S. Chung, J. M. Parant, G. Lozano, R. Hakem, and S. Benchimol. 2003. Pirh2, a p53-inducible ubiquitin protein ligase, promotes p53 degradation. Cell 112:779-791. [DOI] [PubMed] [Google Scholar]
- 21.Linares, L. K., A. Hengstermann, A. Ciechanover, S. Muller, and M. Scheffner. 2003. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. USA 100:12009-12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loughran, Ö., and N. B. La Thangue. 2000. Apoptotic and growth-promoting activity of E2F modulated by MDM2. Mol. Cell. Biol. 20:2186-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, X., and D. P. Lane. 1993. Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell 75:765-778. [DOI] [PubMed] [Google Scholar]
- 24.Maki, C. G., and P. M. Howley. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meek, D. W. 1998. Multisite phosphorylation and the integration of stress signals at p53. Cell. Signal. 10:159-166. [DOI] [PubMed] [Google Scholar]
- 27.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 28.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein Mdm2 conceals the activation domain of tumour suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 29.Qu, L., S. Huang, D. Baltzis, A.-M. Rivas-Estilla, O. Pluquet, M. Hatzoglou, C. Koumenis, Y. Taya, A. Yoshimura, and A. E. Koromilas. 2004. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3β. Genes Dev. 18:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajendra, R., D. Malegaonkar, P. Pungaliya, H. Marshall, Z. Rasheed, J. Brownell, L. F. Liu, S. Lutzker, A. Saleem, and E. H. Rubin. 2004. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 279:36440-36444. [DOI] [PubMed] [Google Scholar]
- 31.Scanga, S. E., L. Ruel, R. C. Binari, B. Snow, V. Stambolic, D. Bouchard, M. Peters, B. Calvieri, T. W. Mak, J. R. Woodgett, and A. S. Manoukian. 2000. The conserved PI3′K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 19:3971-3977. [DOI] [PubMed] [Google Scholar]
- 32.Stambolic, V., and J. R. Woodgett. 1994. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem. J. 303:701-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland, C., and P. Cohen. 1994. The α-isoform of glycogen synthase kinase-3 from rabbit skeletal muscle is inactivated by p70 S6 kinase or MAP-kinase-activated protein kinase-1 in vitro. FEBS Lett. 338:37-42. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland, C., I. A. Leighton, and P. Cohen. 1993. Inactivation of glycogen synthase kinase-3 β by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem. J. 296:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turenne, G. A., and B. D. Price. 2001. Glycogen synthase kinase 3 beta phosphorylates serine 33 of p53 and activates p53's transcriptional activity. BMC Cell Biol. 2:1471-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 37.Watcharasit, P., G. N. Bijur, L. Song, J. Zhu, X. Chen, and R. S. Jope. 2003. Glycogen synthase kinase-3β (GSK-3β) binds to and promotes actions of p53. J. Biol Chem. 278:48872-48879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watcharasit, P., B. N. Bijur, J. W. Zmijewski, L. Song, A. Zmijewska, X. Chen, G. V. W. Johnson, and R. S. Jope. 2002. Direct activating interaction between glycogen synthase kinase-3β and p53 after DNA damage. Proc. Natl. Acad. Sci. USA 99:7951-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter, M., D. Milne, S. Dias, R. Kulikov, U. Knippschild, C. Blattner, and D. Meek. 2004. Protein kinase CK1δ phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry 43:16356-16364. [DOI] [PubMed] [Google Scholar]
- 40.Woodgett, J. R. 2001. Judging a protein by more than its name: GSK-3. Sci. STKE 2001:RE12. [DOI] [PubMed] [Google Scholar]
- 41.Woodgett, J. R. 1991. cDNA cloning and properties of glycogen synthase kinase-3. Methods Enzymol. 200:564-577. [DOI] [PubMed] [Google Scholar]
- 42.Woodgett, J. R. 1990. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 9:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xirodimas, D. P., C. W. Stephen, and D. P. Lane. 2001. Co-compartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, H., J. Mao, L. Li, and D. Wu. 1999. Suppression of glycogen synthase kinase activity is not sufficient for leukemia enhancer factor-1 activation. J. Biol. Chem. 274:30419-30423. [DOI] [PubMed] [Google Scholar]