Abstract
Homologous recombination is vital to repair fatal DNA damage during DNA replication. However, very little is known about the substrates or repair pathways for homologous recombination in mammalian cells. Here, we have compared the recombination products produced spontaneously with those produced following induction of DNA double-strand breaks (DSBs) with the I-SceI restriction endonuclease or after stalling or collapsing replication forks following treatment with thymidine or camptothecin, respectively. We show that each lesion produces different spectra of recombinants, suggesting differential use of homologous recombination pathways in repair of these lesions. The spontaneous spectrum most resembled the spectra produced at collapsed replication forks formed when a replication fork runs into camptothecin-stabilized DNA single-strand breaks (SSBs) within the topoisomerase I cleavage complex. We found that camptothecin-induced DSBs and the resulting recombination repair require replication, showing that a collapsed fork is the substrate for camptothecin-induced recombination. An SSB repair-defective cell line, EM9 with an XRCC1 mutation, has an increased number of spontaneous γH2Ax and RAD51 foci, suggesting that endogenous SSBs collapse replication forks, triggering recombination repair. Furthermore, we show that γH2Ax, DSBs, and RAD51 foci are synergistically induced in EM9 cells with camptothecin, suggesting that lack of SSB repair in EM9 causes more collapsed forks and more recombination repair. Furthermore, our results suggest that two-ended DSBs are rare substrates for spontaneous homologous recombination in a mammalian fibroblast cell line. Interestingly, all spectra showed evidence of multiple homologous recombination events in 8 to 16% of clones. However, there was no increase in homologous recombination genomewide in these clones nor were the events dependent on each other; rather, we suggest that a first homologous recombination event frequently triggers a second event at the same locus in mammalian cells.
A number of human diseases show a defect in homologous recombination and a predisposition to cancer or premature aging (26, 50). The involvement of homologous recombination in maintaining the stability of the genome may provide a link with the disease phenotype found in these patients (49). Recombination repair is often referred to as an error-free repair pathway and advantageous to the cell. However, recombination is a double-edged sword and may also result in loss of heterozygocity, leading to inactivation of tumor suppressor genes, or cause gross gene rearrangements that can activate proto-oncogenes (39, 47). Although endogenous lesions can induce these homologous recombination events, we know virtually nothing about the lesions that trigger spontaneous homologous recombination. There are three principal lesions which could be responsible, DNA double-strand breaks (DSBs) and stalled or collapsed replication forks.
DNA double-strand breaks are known to be substrates for homologous recombination repair in bacteria, yeasts, and mammals (see reference 49 for a review). However, it is reported that nonhomologous end joining is the main pathway involved in repair of DSBs in mammalian cells (36) and that homologous recombination-deficient cells are fully competent in repairing γ-ray-induced DSBs (51). In addition a DSB with two ends is rare (18) and is unlikely to be the endogenous recombinogenic lesion, it is therefore more likely that homologous recombination is primarily involved in repair of substrates other than two-ended DSBs.
Previous studies show that homologous recombination is essential for repair of stalled replication forks in bacteria and in mammalian cells (21, 24, 33, 41). In bacteria and yeasts, a stalled replication fork may reverse and form a chicken foot intermediate, which may then serve as a substrate for homologous recombination or trans-lesion synthesis (24, 25, 40). This could be a spontaneous lesion, which could trigger homologous recombination.
A third substrate for homologous recombination is a collapsed replication fork. It has been suggested that in this case, unlike a stalled fork, the replication fork does not reverse and no chicken foot structures are formed. Instead, a collapsed fork is produced when a single-strand break (SSB) is converted into a DSB during replication or when an early intermediate of a stalled replication fork is processed by an endonuclease, e.g., Mus81 (6, 7). This collapsed fork only contains one free DNA end. Free ends of this type are known to trigger homologous recombination-mediated break-induced replication in both mammalian and yeast cells (1, 10), making this lesion another candidate for the lesion triggering spontaneous homologous recombination.
Here, we determined the recombinant products produced spontaneously, following DSB induction with the I-SceI restriction endonuclease (17), after treatment with thymidine to stall replication forks (21) and following treatment with camptothecin, which causes replication forks to collapse (3, 32, 43, 48). We show that DSBs and stalled and collapsed replication forks all produce a different spectrum of recombinants and that the spontaneous spectra most resemble that of a collapsed replication fork. We also show that following camptothecin treatment DNA single-strand breaks are converted into DSBs only in the presence of replication. The lesion formed at this collapsed replication fork triggers a sister chromatid exchange and the recombinant products resemble the spectra of products formed following spontaneous recombination. Furthermore, our results suggest that two-ended DSBs are rare spontaneous substrates for homologous recombination in a mammalian fibroblast cell line. Interestingly, all spectra showed evidence of multiple homologous recombination events (in 8 to 16% of clones) but there was not a genomewide increase in homologous recombination in these clones nor were the events dependent on each other; rather, we suggest that a first homologous recombination event frequently triggers a second event at the same locus in mammalian cells.
MATERIALS AND METHODS
Cell lines.
SPD8 (12) and SPD8 cells containing the SCneo recombination reporter construct, named S8SN.11 cells (38), were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin (60 μg/ml), and streptomycin (100 μg/ml) at 37°C under an atmosphere containing 5% CO2. The medium was supplemented with 6-thioguanine (5 μg/ml) to prevent survival of cells that undergo spontaneous reversion of the hprt gene.
SCneo recombination assay.
A confluent petri dish (100 mm) of S8SN.11 cells was expanded from 103 cells in order to avoid contamination with spontaneous G418-resistant clones. From this plate, 1.5 × 106 S8SN.11 cells were plated 4 h prior to 24-hour treatment with thymidine (10 mM) or camptothecin (100 nM; in 0.2% dimethyl sulfoxide, final concentration). For I-SceI-induced recombination the plates were transiently transfected for 5 h with the pCMV3nls-I-SceI expression vector as described elsewhere (38). Following treatments, cells were replated in normal medium to determine cloning efficiency (two plates with 500 cells/plate) or in selection medium containing 1 mg/ml G418 (three plates with 3 × 105 cells/plate) to identify recombinants.
In fluctuation assays, 103 cells were plated in each well of a six-well plate and grown to near confluence before treatment with thymidine or camptothecin for 24 h. Following treatment the cells were trypsinized and plated onto selection plates (25 cells/mm2) with G418 or G418 and HAsT (50 μM hypoxanthine, 10 μM l-azaserine, 5 μM thymidine). Only one G418-resistant colony was isolated and expanded from the resistant colonies originating from each well. Isolating just one colony from each lineage is important in order to avoid isolating G418-resistant clones originating from the same recombination event.
HPRT recombination assay.
Contamination of spontaneous HAsT-resistant clones is not a problem in the hprt recombination assay since these are killed with medium containing 6-thioguanine prior to the recombination assay. Thus, when starting the hprt recombination assay, all cells are hprtmut, and this creates a low background recombination frequency. We inoculated 1.5 × 106 cells from each S8SN.11 G418-resistant clone without 6-thioguanine overnight. In treatment experiments, the cells were treated for 4 h with 200 nM camptothecin and/or 3 μM aphidicolin. The cells were subsequently washed in PBS and all cells were left for 48 h to recover before being replated in normal medium to determine cloning efficiency (two plates with 500 cells/plate) or in medium containing HAsT to select for recombinants (three plates with 3 × 105 cells/plate). To determine spontaneous recombination events cells were grown for 3 days in nonselective medium prior to replating for cloning and selection. All plates were stained, using methylene blue in methanol (4 g/liter), after 7 days (for cloning efficiency) and after 10 days (for selection).
Characterization of spectrum of recombinants.
About 40 G418-resistant clones were isolated following each treatment and genomic DNA extracted; 10 μg of phenol-chloroform purified DNA from each clone was digested with restriction endonucleases and separated by agarose gel electrophoresis. Southern blotting was carried out as previously described using the S2neo fragment as a probe (38).
Pulsed-field gel electrophoresis.
Prior to treatment 1.5 ×106 cells were inoculated overnight. S8SN.11 were treated for 6 h with or without 200 nM camptothecin and/or 3 μM aphidicolin and AA8 and EM9 cells were treated for 24 h with increasing doses of camptothecin. Cells were then trypsinized and 106 cells were melted into each 0.7% agarose insert. These inserts were incubated in 0.5 M EDTA, 1% N-laurylsarcosyl, proteinase K (1 mg/ml) at 50°C for 48 h and then washed four times in Tris-EDTA buffer prior to loading onto a 0.7% agarose (chromosomal grade) gel. Separation by pulsed-field gel electrophoresis was for 24 h (Bio-Rad; 120° angle, 60 to 240 seconds switch time, 4 V/cm). The gel was subsequently stained with ethidium bromide for analysis.
Cell cycle analysis.
We plated 1.5 ×106 S8SN.11 cells 4 h prior to a 24-hour treatment with thymidine (10 mM) or camptothecin (100 nM; in 0.2% dimethyl sulfoxide final concentration). For I-SceI-induced recombination the plates were transiently transfected with the pCMV3nls-I-SceI expression vector as described elsewhere (38). For the final 30 min of treatment 10 μM bromodeoxyuridine was added to the medium after which cells were fixed in 70% methanol and left overnight at −20°C. After washing in phosphate-buffered saline (PBS) cells were incubated for 30 min in 2 M HCl, washed twice in PBS and once in PBS containing 0.1% bovine serum albumin and 0.2% Tween 20 (PBS-BT); 2 μl anti-bromodeoxyuridine antibody (Dako) was then added directly to the cell pellet and left for 20 min in the dark. Cells were washed twice in PBS-BT and incubated for 20 min in the dark in 50 μl fluorescein isothiocyanate-conjugated rabbit anti-mouse antibody (Dako) diluted 1:10 in PBS-BT. After a final wash in PBS cells were stained with propidium iodide-RNase A solution (50 mg/ml propidium iodide, 100 mg/ml RNase A) for at least 30 min. Samples were analyzed by flow cytometry (Becton-Dickinson FACSort, 488 nm laser).
Immunofluorescence.
Cells were plated onto coverslips 24 h prior to 24-h treatments as indicated. Following treatments the medium was removed and coverslips rinsed once in PBS at 37°C and fixed and stained as described previously (38).
The primary antibodies used in this study were rabbit polyclonal anti γ-H2AX (Trevigen) at a dilution of 1:500 and rabbit polyclonal anti-RAD51 (H-92, Santa Cruz) at a dilution of 1:1,000. The secondary antibody was Alexa 555 goat anti-rabbit F(ab′)2 immunoglobulin G antibody (Molecular Probes) at a concentration of 1:500. Antibodies were diluted in PBS containing 3% bovine serum albumin. DNA was stained with 1 μg/ml To Pro (Molecular Probes).
Images were obtained with a Zeiss LSM 510 inverted confocal microscope using planapochromat 63×/1.4-numerical-aperture oil immersion objective and excitation wavelengths of 546 and 630 nm. Through focus maximum projection images were acquired from optical sections 0.50 μm apart and with a section thickness of 1.0 μm. Images were processed using Adobe PhotoShop (Abacus Inc).
The frequencies of cells containing γ-H2Ax or RAD51 foci were determined in at least three separate experiments. At least 300 nuclei were counted on each slide. Nuclei containing more than five γ-H2Ax foci or 10 RAD51 foci were classified as positive.
RESULTS
Homologous recombination occurs following induction of DSBs and stalling or collapsing of replication forks.
The SCneo vector was integrated as a single copy into the genome of the SPD8 Chinese hamster cell line, to produce the cell line S8SN.11 (Fig. 1) (16, 38). We then investigated homologous recombination events occurring between the two nonfunctional neoR genes in the SCneo construct. Following homologous recombination a functional neoR gene may be formed either via short-tract gene conversion, long-tract gene conversion, or a sister chromatid exchange. Short-tract gene conversion results in a 4-kb product, while a sister chromatid exchange and long-tract gene conversion result in a 7.3-kb product (Fig. 1B) (16).
FIG. 1.
Homologous recombination is induced by an I-SceI-induced DSB, thymidine, and camptothecin. (A) Structure of the SCneo recombination substrate containing two nonfunctional copies of the neoR gene. (B) Upon homologous recombination, a functional neoR gene can be produced by short tract gene conversion, which results in a 4.0-kb product or a sister chromatid exchange/long-tract gene conversion, which restores a neoR gene within a 7.3-kb product (16). (C) Homologous recombination frequencies (as measured by formation of G418-resistant clones) in S8SN.11 cells following transient transfection with pCMV3xnlsI-SceI or treatment with thymidine (10 mM) or camptothecin (100 nM) for 24-hour). The mean recombination frequency and standard deviation of at least three experiments are shown. Two and three stars designate statistical significance versus the control in the t test: P < 0.01 and P < 0.001, respectively.
In the S8SN.11 cell line the spontaneous frequency of homologous recombination, as measured by reversion to a functional neoR gene, was 1.8 × 10−6 (Fig. 1C). Upon transient transfection of S8SN.11 cells with the pCMV3xnlsI-SceI vector, which overexpresses the rare-cutting I-SceI restriction endonuclease, a DSB is specifically introduced in the S2neo repeat (Fig. 1B). The frequency of homologous recombination was 1,000-fold higher in pCMV3xnlsI-Sce-transfected cells compared with the control, confirming earlier reports that a restriction endonuclease-induced DSB triggers homologous recombination (17). Homologous recombination was also induced in S8SN.11 cells following a 24-hour thymidine treatment (Fig. 1C). Thymidine treatment depletes only the dCTP pool and results in stalled replication forks but not detectable DSBs (21). In this case, homologous recombination was only induced eightfold; this low level is probably due to the genomewide effect of thymidine compared to the targeted DSB induced with I-SceI.
A 24-hour treatment with the topoisomerase I inhibitor camptothecin induced homologous recombination 20-fold (Fig. 1C). Topoisomerase I relaxes supercoiled DNA during transcription or replication by introducing a transient SSB, which is subsequently religated. Camptothecin stabilizes these topoisomerase I cleavage complexes, leaving SSBs unligated. It has been shown that these SSBs are converted into DSBs during replication and produce collapsed replication forks (3, 32, 43, 48), which are presumably repaired by homologous recombination (1).
To test if homologous recombination induced by camptothecin is the consequence of SSBs collapsing replication forks, we determined if replication is required for camptothecin-induced DSBs or homologous recombination. We cotreated cells with camptothecin and the DNA polymerase inhibitor aphidicolin (15). We found a reduced number of camptothecin-induced DNA fragments released in pulsed-field gel electrophoresis when cotreating cells with aphidicolin (Fig. 2A), in agreement with previously published data (53). We also found DNA fragments released following a 6-hour aphidicolin treatment alone, in agreement with the notion that DSBs form at stalled replication forks (21, 31). However, additional treatment with camptothecin did not increase the amount of DNA fragments released in pulsed-field gel electrophoresis, indicating that camptothecin-induced DSBs require replication (Fig. 2A) and most likely result from the collision between the camptothecin-stabilized cleavable complexes and DNA polymerase.
FIG. 2.
DSBs and homologous recombination induced by camptothecin depend on replication. (A) Pulsed-field gel electrophoresis showing DNA fragmentation following a 6-hour treatment with camptothecin (200 nM) and/or aphidicolin (3 μM). (B) Homologous recombination measured as the frequency of reversion to HAsT-resistant colonies in S8SN.11 cells following a 6-hour treatment with camptothecin (200 nM) and/or aphidicolin (3 μM). The average (column) and standard deviation (error bars) of three experiments are depicted.
To investigate if camptothecin-induced homologous recombination is triggered in response to camptothecin-induced DSBs we measured homologous recombination between the repeats in a 5-kb tandem duplication mutation in the hprt gene located on the single X chromosome giving a nonfunctional protein and resistance to 6-thioguanine (12). Reversion by homologous recombination to a functional hprt gene can be selected for using HAsT. We treated SPD8 cells with camptothecin and/or aphidicolin and found that camptothecin potently induces homologous recombination (Fig. 2B), in line with earlier results (1). Also, we found that aphidicolin treatment alone triggers homologous recombination, confirming previous reports that it triggers homologous recombination for repair of stalled replication forks (2, 31). Furthermore, we report that treatment with aphidicolin abolishes the camptothecin-induced homologous recombination (Fig. 2B). These data together support the hypothesis that SSBs induced by camptothecin do indeed collapse replication forks forming DSBs which are substrates for homologous recombination.
Recombination products formed depend on the recombination substrate.
The three different treatments (i.e., I-SceI, thymidine, and camptothecin) represent three different ways of inducing homologous recombination (11). We isolated about 40 individual G418-resistant colonies following each treatment (and a no-treatment control) in order to investigate which homologous recombination pathways are triggered. A fluctuation assay was used to avoid isolation of colonies originating from the same recombination event. Also, only one colony was isolated from each lineage. The presence of a 7.3-kb product (from a sister chromatid exchange/long-tract gene conversion) or a 4-kb product (from short-tract gene conversion) was determined by Southern blotting (Fig. 3A). The spectra of recombinants were significantly different (P = 6 × 10−10 in χ2 test) suggesting that the three lesions trigger differential use of homologous recombination pathways (Fig. 3B).
FIG. 3.
(A) Southern blot identifying the presence of the 4.0-kb product or 7.3-kb product in isolated G418-resistant clones. (B) Relative frequency of each recombinant product formed in approximately 40 clones isolated following either control, I-SceI, thymidine (10 mM), or camptothecin (100 nM) treatments. Numbers above the bars indicate the number of clones found in each category. (C) Number of recombination events triggered following control, I-SceI, thymidine (10 mM), or camptothecin (100 nM) treatments.
It is possible that the difference in recombination products found could be a reflection of different homologous recombination pathways being active at different phases of the cell cycle. This could be the case, since we see that thymidine-treated cells are arrested in the early S phase and that camptothecin arrests cells in the late S/G2 phase of the cell cycle; while I-SceI or control cells do not arrest (Fig. 4). However, we have shown previously that similar I-SceI-induced spectra are produced when cells are synchronized in the G1, S, and M phases of the cell cycle (34), and that they all resemble the spectrum produced in unsynchronized cells, so the differential spectra observed following different forms of damage are unlikely to be related to cell cycle effects of the agents used.
FIG. 4.
Cell cycle analysis of S8SN.11 cells following treatment with I-SceI or after a 24-hour treatment with thymidine (10 mM) or camptothecin (100 nM). A 30-minute bromodeoxyuridine (BrdU) treatment indicates replicating cells. DNA content is measured through propidium iodine staining.
I-SceI-induced homologous recombination products are different from those that result from spontaneous recombination.
We confirmed that an I-SceI-induced DSB produces mainly short-tract gene conversion products (Fig. 3B) (16, 35). This is consistent with current models for homologous recombination triggered by DSBs, resulting in gene conversion (see Fig. 8A). In contrast, we demonstrate that spontaneous homologous recombination involves mainly a sister chromatid exchange/long-tract gene conversion (Fig. 3B). This is statistically significant different from DSB-induced recombinants in χ2 test (P = 2.3 × 10−8) and suggests that a DSB, producing two ends, is a rare substrate for homologous recombination in mammalian fibroblast cell lines.
FIG. 8.
Pathways for homologous recombination in mammalian cells. (A) Homologous recombination repair of a two-end DSB with synthesis-dependent strand annealing results in gene conversion with no crossover (11, 29). (B) A stalled replication fork may reverse and form a half-chicken foot structure with one protruding end. This structure is efficiently cleaved by the Mus81/Eme1 complex (6, 7) and results in a collapsed fork, which is repaired with break-induced replication (see below). Full reversal of the replication fork leads to a chicken foot structure that has one free 3′ end that is able to initiate homologous recombination to restore replication. Resolution of double Holliday junctions with topoisomerase III/BLM leads to gene conversion with no crossover (54). (C) A persisting SSB causes a collapsed replication fork which contains a one-ended DSB; this is repaired by break-induced replication (10). This collapsed replication fork is likely to be a common spontaneous substrate for homologous recombination and therefore for causing a sister chromatid exchange. For a more detailed description of the above pathways see reference 11. Dotted lines represent newly synthesized strands.
Spectrum of recombinant products formed following replication fork stalling does not resemble the spontaneous or DSB-induced spectra.
Homologous recombination has been shown to have an important role in the repair of stalled replication forks in mammalian cells (21, 33). Chinese hamster cells deficient in homologous recombination show delayed progress through the cell cycle (8, 9, 45) and hypersensitivity to agents that stall replication forks (21), suggesting that homologous recombination might also have an important role in repairing spontaneously formed stalled forks.
Here, we report that 46% of thymidine-induced homologous recombination products were formed by short-tract gene conversion and 37% by long-tract gene conversion or a sister chromatid exchange (Fig. 3B), and this spectrum of recombinants differs from both the spontaneous (P = 0.014 in χ2 test) and the I-SceI-induced spectra (P = 0.010 in χ2 test). This implies that homologous recombination induced at stalled replication forks, at least in the recombination reporter system used here, results in differential use of multiple pathways.
Spontaneous homologous recombination primarily repairs collapsed replication forks produced by spontaneous SSBs.
The camptothecin-induced spectrum of recombinant products consist mainly of a sister chromatid exchange or long-tract gene conversion (Fig. 3B), implying that different homologous recombination pathways may be employed at these lesions. This is could be predicted from the break-induced replication model for recombination (11) (Fig. 8C).
The spectrum of recombinants induced by camptothecin is very similar to the spontaneous spectrum (P = 0.515 in χ2 test). Thus, it seems probable that a collapsed replication fork, produced when an SSB is converted to a DSB at a replication fork, is a common lesion triggering homologous recombination.
Earlier work showing that SSB-repair-defective cells show a hyperrecombination phenotype supports the notion that SSBs are substrates for spontaneous homologous recombination (37, 46, 52). To test this further, we investigated if the defect in SSB repair found in the XRCC1-deficient EM9 cells results in an increased amount of spontaneously formed lesions. Using pulsed-field gel electrophoresis we detected an increase in spontaneously formed DSBs in untreated EM9 cells compared to wild-type AA8 cells (Fig. 5). Furthermore, we found an increased level of DNA fragments released from camptothecin treatments in EM9 cells (Fig. 5). In comparison, there is no increase in DNA fragments from camptothecin treatments in recombination-deficient irs1SF cells or end joining-deficient V3-3 cells (1).
FIG. 5.
SSB repair-deficient EM9 cells (XRCC1 mutated) accumulate more DSBs following camptothecin treatment. Pulsed-field gel electrophoresis of XRCC1-deficient (EM9) and wild-type (AA8) cells following a 24-hour treatment with increasing doses of camptothecin.
The level of DNA damage and in particular DSBs may also be quantified through visualization of γH2Ax focus formation (Fig. 6A) (27). We found that a higher number of EM9 cells contained γH2Ax foci compared to AA8 wild-type cells (statistically significant in t test, P < 0.05; Fig. 6C), as previously shown (5), presumably as a result of the defect in SSB repair. Furthermore, a treatment with camptothecin triggered a massive γH2Ax focus response in 74% of EM9 cells (Fig. 6A) and the number of EM9 cells carrying γH2Ax foci was higher than when adding the control EM9 with the camptothecin-induced γH2Ax foci in AA8 cells, suggesting the effect is synergistic. An increase in both DSBs and γH2Ax foci in EM9 cells suggests that stabilized as well as spontaneous SSBs collapse more replication forks in EM9 cells (Fig. 5 and 6).
FIG. 6.
Elevated levels of spontaneous and camptothecin-induced γH2Ax foci and RAD51 foci in SSB repair-deficient EM9 cells. (A) Visualization of γH2Ax foci (green) and DNA (blue) in control and treated AA8 and EM9 cells. (B) Visualization of RAD51 foci (red) and DNA (blue) in control and treated AA8 and EM9 cells. (C) Percentage of cells with >5 γH2Ax foci. (D) Percentage of cells with >10 RAD51 foci. The average (column) and standard deviation (error bars) of at least three experiments are depicted.
We also wanted to investigate whether the increased level of collapsed forks correlated with an increase in homologous recombination. We visualized RAD51 foci formation in wild-type AA8 and XRCC1-deficient EM9 cells (Fig. 6B) and found an increased amount of spontaneously formed RAD51 foci (Fig. 6D), in agreement with previous results (5) and consistent with a hyperrecombination phenotype in EM9 cells (46). In addition, we found that following camptothecin-treatment almost all of the EM9 cells contain RAD51 foci. The effect of camptothecin was synergistic on the level of RAD51 foci, i.e., we found more RAD51 foci than would be expected from adding the effect of camptothecin in wild-type AA8 cells to the background level of RAD51 foci in EM9 cells (Fig. 6D). This supports our hypothesis that collapsed replication forks resulting from spontaneous or stabilized SSBs trigger homologous recombination.
Multiple homologous recombination events in mammalian cells.
Between 8 and 16% of the clones isolated showed the presence of both the 4-kb and 7.3-kb fragments (Fig. 3B). These could result from one complex recombination event or following two subsequent recombination events triggered by either one or several DNA lesions. To address this, we subcloned four double mutants and isolated 8 to 10 colonies from each; from these we extracted DNA and performed Southern blotting. In agreement with previous results (16), we found that the double recombinant clones were a mixed population with either a functional 4-kb or 7.3-kb product (Fig. 7A). Thus, the two different functional neoR genes were present on different DNA molecules and were separated following mitosis. If one complex recombination event had occurred the two functional neoR genes would have occasionally ended up on the same DNA molecule. As this was not the case in any of the clones tested, we assume that in our double recombinants two subsequent homologous recombination events have taken place.
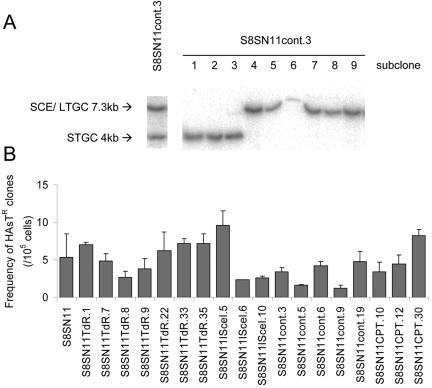
FIG. 7.
Clones carrying both the 7.3-kb and 4-kb recombination products are mosaic and have normal recombination levels. (A) Southern blot of DNA isolated from the G418-resistant S8SN11contr.3 cell line and subclones thereof. (B) Homologous recombination frequency in the hprt gene of the G418-resistant S8SN.11 clones, carrying both the 7.3-kb and 4-kb recombination products. The average (column) and standard deviation (error bars) of at least three experiments are depicted.
During I-SceI-induced recombination, it is easy to envisage two consecutive recombination events occurring where the I-SceI restriction endonuclease cuts at the restriction sites present in both copies of the sister chromatids (16). Surprisingly, we found that double recombinants also occurred in the spontaneous and thymidine- and camptothecin-induced spectra of recombinants; the frequency of double recombinants was 2 × 10−7, 1.2 × 10−6 and 2.8 × 10−6, respectively (Fig. 3C). This cannot be explained by induction of two specific DSBs, as in the case of I-SceI. Instead, it suggests that the first recombination event triggered a second one at the same locus. Alternatively, it could reflect a general increase in homologous recombination in these specific clones, perhaps as a result of a mutation in a gene suppressing homologous recombination.
To investigate the global level of homologous recombination in the double recombinant clones we measured recombination at an independent locus. In addition to the SCneo construct, the S8SN.11 cell line carries the 5-kb duplication mutation in the hprt gene that can be used to assay for homologous recombination (12). We found that at the hprt loci none of the double recombinant clones had any difference in recombination frequency compared with the parental cell line (Fig. 7B), showing that the multiple homologous recombination events in the double recombinant clones cannot explained by globally high homologous recombination levels.
Homologous recombination events in SCneo and the hprt gene are unrelated.
As an alternative explanation, the double recombinants could be the result of a transient increase in genome-wide homologous recombination. Here, we again utilize the fact that the S8SN.11 cells carry two substrates for homologous recombination. A transient increase in homologous recombination would affect both recombination loci and give an increased recombination rate at both sites. Thus, we determined the recombination rate of clones resistant to both HAsT and G418. In fluctuation assays, when seeding 250 million cells with both selective agents on >1,000 selection plates, we found no clone resistant to both G418 and HAsT, i.e., the spontaneous double recombination rate is lower than 9.8 × 10−11 (Table 1).
TABLE 1.
Average recombination rate and standard error in the hprt gene and SCneo constructs as determined by fluctuation assays in S8SN.11 cells
| Treatment | Recombination rate
|
Expected recombination rate, SCneo and hprt | Observed recombination rate, SCneo and hprt | |
|---|---|---|---|---|
| hprt | SCneo | |||
| None | 5.5 ± 0.3 × 10−6 | 2.5 ± 0.8 × 10−6 | 1.4 × 10−11 | < 9.8 × 10−11 |
| Thymidine (10 mM) | 1.6 ± 0.4 × 10−5 | 7.2 ± 2.5 × 10−6 | 1.1 × 10−10 | < 9.6 × 10−11 |
If homologous recombination events at different loci occur as independent events the expected frequency of double recombinants would be 1.4 × 10−11. Thus, we would need to plate 10 times more cells to determine if multiple spontaneous homologous recombination events occur independently. To circumvent this obstacle, we exploited the fact that thymidine induces homologous recombination in both the hprt gene and the SCneo substrate (21). Following a 24-hour treatment with 10 mM thymidine, we seeded cells on about 1,000 selection plates in the presence of both HAsT and G418. We found no clone resistant to both agents (rate < 9.6 × 10−11). In this case, the expected frequency of double recombinants would be 1.1 × 10−10. Thus, a homologous recombination event induced at a stalled replication fork is independent of another homologous recombination event occurring at a distant locus.
DISCUSSION
Here, we confirm earlier studies showing that in mammalian cells homologous recombination is involved in the repair of DSBs (17) and stalled (21, 33) and collapsed (1) replication forks.
In agreement with previous reports we found that an I-SceI-induced DSB mainly induces short-tract gene conversion (16). This is consistent with models for two-end repair (44), especially the synthesis-dependent strand-annealing model (Fig. 8A) (11, 29) which explains why we see suppressed crossover products in mitotic homologous recombination in mammalian cells (16, 30). Comparison of the recombinant products formed after treatment with I-SceI with those formed following spontaneous homologous recombination showed that while I-SceI-induced homologous recombination involves mainly short-tract gene conversion, spontaneous homologous recombination involves mainly a sister chromatid exchange/long-tract gene conversion (statistically significant difference in χ2 test, P = 2.3 × 10−8). A DSB, producing two ends, is therefore unlikely to be a common lesion triggering spontaneous homologous recombination. This is not surprising given that γ-ray-induced two-ended DSBs in homologous recombination-deficient mammalian cells are repaired as quickly as in wild-type cells (50) and that less than half of two-ended DSBs are repaired by homologous recombination, even in the presence of a homologous template (16, 36).
A significant portion of the mammalian genome contains repeated sequences that may be used for repair of two-ended DSBs. However, unlike in our reporter construct they contain single nucleotide differences; this reduces the efficiency of homologous recombination dramatically (20). Thus, the 1,000-fold induction of homologous recombination following introduction of a site-specific DSB with pCMV3xnlsI-SceI reflects an artificial situation and may not be the way two-ended DSBs are repaired normally. The artificial nature of our substrates may also influence the spectra we find.
The thymidine-induced homologous recombination products resulted from both short-tract gene conversion and a sister chromatid exchange/long-tract gene conversion and the spectrum of recombinants differs from both the spontaneous (P = 0.014 in χ2 test) and the I-SceI-induced spectra (P = 0.010 in χ2 test). This indicates that homologous recombination at a stalled replication fork results in differential use of homologous recombination pathways to restore replication. However, it is interesting that here thymidine only induced homologous recombination eightfold, while previously homologous recombination has been shown to be highly important in repairing stalled replication forks (21). This contradiction in results may be because homologous recombination in the recombination reporter construct involves finding homology 3 to 4 kb away from the recombination site. It has previously been reported that recombination tracts at stalled replication forks are short (22). Therefore, the recombination reporter is likely to pick up only a few homologous recombination events produced at stalled replication forks, and this may cause a bias in the spectrum produced. In conclusion, although we did not find that the spontaneous spectrum of recombinants overlap with the one induced at stalled replication forks, stalled replication forks may still be common substrates for homologous recombination.
A mechanism of homologous recombination at a stalled replication fork has been suggested for bacteria and yeasts. It involves fork regression, possibly caused by positive torsional strain in the DNA (28) or by enzymatic action (24, 25) (Fig. 8B). An early replication intermediate, a half-chicken foot, may be produced when the first strand initially reverses. This is likely to be a substrate for the human Mus81-Eme1 endonuclease complex (6, 7). If this early structure is cleaved it will produce a one-ended DSB identical to that produced at a collapsed replication fork, which is likely to be repaired by break-induced replication (Fig. 8C). On the other hand, if the fork fully regresses, a full chicken foot structure will form, subsequent strand invasion and resolution of the double Holliday junction formed could then result in gene conversion; this may involve the BLM-topoisomerase III complex and produce a noncrossover product (54) (Fig. 8B). This model explains how differential homologous recombination pathways may be used at stalled replication forks and clarifies why we see both gene conversion and a sister chromatid exchange/long-tract gene conversion following thymidine-mediated stalling of replication forks.
We present that both camptothecin-induced DSBs and homologous recombination depend on replication fork elongation. This shows that growing replication forks may collapse when they run into SSBs and that these one-ended DSBs trigger homologous recombination. This is in agreement with previous results showing that camptothecin-induced homologous recombination is associated with the S phase (1). The spectrum of recombinants induced by camptothecin contained mainly a sister chromatid exchange/long-tract gene conversion events. This fits well with the break-induced replication model for replication restart that results in a sister chromatid exchange (Fig. 8C). In addition, the camptothecin-induced spectrum was very similar to the spontaneous spectrum (P = 0.515 in χ2 test). Thus, it seems probable that a collapsed replication fork, produced when an SSB is converted to a DSB at a replication fork, is a common lesion triggering spontaneous homologous recombination.
In contrast to DSBs, SSBs are frequent endogenous DNA lesions in cells (104/cell/day) (18). SSBs can be induced directly by free radicals or more commonly as a consequence of repair of apurinic sites caused by depurination or repair of deaminated cytosine or other damaged bases (18). Some of these SSBs are likely to collapse replication forks and produce DSBs with only one free end, similar to those induced by camptothecin.
Further support that an SSB is a common spontaneous substrate that can induce homologous recombination comes from cells deficient in SSB repair. These cells e.g., XRCC1- or PARP-1-deficient cells, have endogenous levels of homologous recombination and a sister chromatid exchange 10-fold higher than that seen in wild-type cells (37, 46, 52). Here, we show that XRCC1-deficient EM9 cells have a high level of spontaneous DSBs and γH2Ax foci, likely representing an elevated level of spontaneously collapsed replication forks. Furthermore, we report an elevated level of spontaneous RAD51 foci, indicating that the collapsed replication forks trigger homologous recombination, resulting in the higher sister chromatid exchange levels found in EM9 cells. In addition, we find that camptothecin synergistically triggers DSBs, γH2Ax foci, and homologous recombination, indicating that an increase in unrepaired SSBs is the lesion triggering collapsed forks and homologous recombination in these cells.
If the spontaneous lesion triggering the elevated level of damage and RAD51 foci in EM9 cells and the camptothecin-produced lesions were different, the effect on homologous recombination and DSB formation following camptothecin treatment in EM9 cells would be additive. Since the effect is synergistic, we suggest that the lack of SSB repair in EM9 causes the elevated homologous recombination response. We explain the synergistic effect by an increased amount of unrepaired camptothecin-induced SSBs in EM9 SSB repair-deficient cells than in AA8 wild-type cells. Increased amounts of camptothecin-induced SSBs will result in more collapsed forks in EM9 cells, resulting in a synergistic effect on both DSBs and recombination.
Further reports from the literature support the notion that the level of SSBs is critical to regulate spontaneous homologous recombination. For instance, several Saccharomyces cerevisiae genes involved in oxidative stress response, e.g., tsa1, skn7, and yap1, were found, in a genomewide screen, to strongly suppress gross chromosomal rearrangements (14), further suggesting that in regard to spontaneous recombination SSBs are a highly relevant lesion.
Although sister chromatid exchanges are known to be the result of homologous recombination (42), it is unclear whether they represent the majority of spontaneous homologous recombination events. To address this issue we calculated the expected recombination rate in the SCneo construct and the hprt gene (12). There are about six sister chromatid exchanges per cell generation in the cells used here (23). If these were responsible for spontaneous homologous recombination, the expected recombination rate on the 1-kb SCneo and 5-kb hprt substrates would be 10−6 and 5 × 10−6, respectively (Table 2). The actual spontaneous recombination rates in the SCneo and hprt substrates were 2.5 × 10−6 and 5.5 × 10−6, respectively. Thus, it is likely that observed sister chromatid exchanges represent most of the spontaneous homologous recombination events that we detect in our assays.
TABLE 2.
Average observed and expected recombination rates if all spontaneous homologous recombination events are sister chromatid exchanges in the hprt gene and SCneo construct in S8SN.11 cellsa
| Locus | Recombination rate | Target size (kb) | Expected recombination rate (target size × 6/6 × 109) |
|---|---|---|---|
| SCneo | 2.5 ± 0.8 × 10−6 | 1 | 1 × 10−6 |
| hprt | 5.5 ± 0.3 × 10−6 | 5 | 5 × 10−6 |
The average rate in the full genome is six sister chromatid exchanges/cell. Data are from reference 23.
Other reports show that nonreciprocal exchange or gene conversion executes half or more than half of spontaneous intrachromosomal recombination events (4, 19). One reason for this discrepancy with our results could be the length of recombination tracts used to assay recombination. The SCneo substrate used here has a long stretch of homology between the 5′ deletion in the 3′ neo repeat and the I-SceI site in 3′ S2neo and this allows homology searching and facilitates the exchange events. A shorter homology overlap, as used in other reports, may make the recombination event prone to gene conversion (19). In addition, we cannot distinguish between a sister chromatid exchange and long-tract gene conversion in the SCneo system used here. Thus, a significant proportion of the 7.3-kb products we detected may in fact have resulted from long-tract gene conversion. Lastly, the fact that we used exponentially growing cells with a short cell cycle grown in a highly oxidized environment may also have made our system biased for replication fork collapse and thus for a sister chromatid exchange, and therefore, there might be a reduced contribution of SSB-induced homologous recombination in animal models (13).
We found that all the spectra of recombinants had a portion of clones carrying both the 7.3-kb and 4-kb products. We subcloned the double recombinant clones and found that they were mosaic, i.e., carrying either a 4-kb or a 7.3-kb product (Fig. 7A). This is likely to be the result of multiple homologous recombination events rather than of one complex recombination event, since in the latter case the two functional copies would occasionally end up on the same DNA molecule, a scenario that we did not observe. In the case of I-SceI-induced recombination, double recombinants have been reported previously and probably result when the I-SceI restriction endonuclease cuts both copies of the sister chromatids, which subsequently are repaired using different pathways (16). However, thymidine and camptothecin do not induce recombination specifically in the SCneo substrate and are therefore unlikely to induce damage on both sister chromatids. Instead, we suggest three possible ways through which multiple recombination products could have occurred: (i) that the first recombination event triggered a second event at the same locus, (ii) that the first recombination event triggered a genome-wide increase in homologous recombination, or (iii) that a subset of cells have higher recombination levels, possibly due to mutations in genes suppressing homologous recombination.
We isolated 18 clones containing both long-tract and short-tract gene conversions and found that their recombination levels are no different from that of the control. Thus, these clones had no overall higher level of homologous recombination, and we can exclude possibility iii. We also found that homologous recombination events occurring at two different loci occur independently of each other, showing that a first recombination event does not trigger other recombination events genome-wide (ii). Thus, we suggest that the multiple recombination events occur because a first recombination event triggers a second at the same locus in mammalian cells. The results we find here may again be biased due to the fact that we are limited to detect recombination events occurring between repeated sequences that has full homology. These regions are not common in the genome.
It is interesting that the frequency of double recombinants following thymidine and camptothecin treatments was 6- and 24-fold higher than control, respectively (Fig. 3C). This suggests that double recombination events are also formed following these treatments. The fold increase in double recombination events is similar to the overall fold induction of homologous recombination with thymidine (eightfold) and camptothecin (20-fold). Thus, it appears that the formation of double recombinants at the same locus is dependent on the first recombination event regardless of how this is induced. We speculate that a second recombination event might be triggered at the same locus by unresolved or misresolved recombination intermediates and/or by the fact that proteins involved in homologous recombination are abundant at the locus (in RAD51 foci). In addition, open chromatin structures might favor a second homologous recombination event at the same loci.
In conclusion, we show that the substrate for spontaneous homologous recombination is likely to be endogenous SSBs that collapse replication forks. Furthermore, DSBs with two ends, which are rare, are unlikely to be the usual substrates for homologous recombination in mammalian cells. Several pathways for homologous recombination are used in repair of stalled replication forks. Our results further suggest that sister chromatid exchanges visualized in cells are likely products following spontaneous recombination, using a break-induced replication mechanism for reactivating collapsed replication forks. Finally, our data suggest that, while recombination events in mammalian cells are unlikely to affect recombination events at a distant locus, a first recombination event is likely to trigger a second recombination event at the same locus.
Acknowledgments
We thank Mark Meuth, Angela Cox, and a number of colleagues within the recombination field for discussions.
The Biological & Biotechnological Sciences Research Council, the Swedish Cancer Society, the Swedish Research Council, and Yorkshire Cancer Research supported this work financially. N.S.G. has a scholarship from the Kerman University of Medical Science, Iran.
REFERENCES
- 1.Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235-1245. [DOI] [PubMed] [Google Scholar]
- 2.Arnaudeau, C., E. Tenorio Miranda, D. Jenssen, and T. Helleday. 2000. Inhibition of DNA synthesis is a potent mechanism by which cytostatic drugs induce homologous recombination in mammalian cells. Mutat. Res. DNA Repair 461:221-228. [DOI] [PubMed] [Google Scholar]
- 3.Avemann, K., R. Knippers, T. Koller, and J. M. Sogo. 1988. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol. Cell. Biol. 8:3026-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag, R. J., and R. M. Liskay. 1991. Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse cells. Mol. Cell. Biol. 11:4839-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, H. E., N. Schultz, H. D. Thomas, K. M. Parker, D. Flower, E. Lopez, S. Kyle, M. Meuth, N. J. Curtin, and T. Helleday. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose)polymerase. Nature 434:913-917. [DOI] [PubMed] [Google Scholar]
- 6.Ciccia, A., A. Constantinou, and S. C. West. 2003. Identification and characterization of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 278:25172-25178. [DOI] [PubMed]
- 7.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 8.Fuller, L. F., and R. B. Painter. 1988. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res. 193:109-121. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, C. S., P. J. Simpson, C. R. Wilson, and J. Thacker. 2000. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2:757-761. [DOI] [PubMed] [Google Scholar]
- 10.Haber, J. E. 2000. Lucky breaks: analysis of recombination in Saccharomyces. Mutat. Res. 451:53-69. [DOI] [PubMed] [Google Scholar]
- 11.Helleday, T. 2003. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 532:103-115. [DOI] [PubMed] [Google Scholar]
- 12.Helleday, T., C. Arnaudeau, and D. Jenssen. 1998. A partial hprt gene duplication generated by non-homologous recombination in V79 Chinese hamster cells is eliminated by homologous recombination. J. Mol. Biol. 279:687-694. [DOI] [PubMed] [Google Scholar]
- 13.Hendricks, C. A., K. H. Almeida, M. S. Stitt, V. S. Jonnalagadda, R. E. Rugo, G. F. Kerrison, and B. P. Engelward. 2003. Spontaneous mitotic homologous recombination at an enhanced yellow fluorescent protein (EYFP) cDNA direct repeat in transgenic mice. Proc. Natl. Acad. Sci. USA 100:6325-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, M. E., A. G. Rio, A. Nicolas, and R. D. Kolodner. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100:11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikegami, S., T. Taguchi, M. Ohashi, M. Oguro, H. Nagano, and Y. Mano. 1978. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 275:458-460. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 19.Liskay, R. M., and J. L. Stachelek. 1983. Evidence for intrachromosomal gene conversion in cultured mouse cells. Cell 35:157-165. [DOI] [PubMed] [Google Scholar]
- 20.Lukacsovich, T., and A. S. Waldman. 1999. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics 151:1559-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundin, C., K. Erixon, C. Arnaudeau, N. Schultz, D. Jenssen, M. Meuth, and T. Helleday. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 22:5869-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundin, C., N. Schultz, C. Arnaudeau, A. Mohindra, L. T. Hansen, and T. Helleday. 2003. RAD51 is involved in repair of damage associated with DNA replication in mammalian cells. J. Mol. Biol. 328:521-535. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka, A., C. Lundin, F. Johansson, M. Sahlin, K. Fukuhara, B. M. Sjoberg, D. Jenssen, and A. Onfelt. 2004. Correlation of sister chromatid exchange formation through homologous recombination with ribonucleotide reductase inhibition. Mutat. Res. 547:101-107. [DOI] [PubMed] [Google Scholar]
- 24.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 25.Michel, B., G. Grompone, M. J. Flores, and V. Bidnenko. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA 101:12783-12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohaghegh, P., and I. D. Hickson. 2002. Premature aging in RecQ helicase-deficient human syndromes. Int. J. Biochem. Cell Biol. 34:1496-1501. [DOI] [PubMed] [Google Scholar]
- 27.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 28.Postow, L., C. Ullsperger, R. W. Keller, C. Bustamante, A. V. Vologodskii, and N. R. Cozzarelli. 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 276:2790-2796. [DOI] [PubMed] [Google Scholar]
- 29.Prado, F., F. Cortes-Ledesma, P. Huertas, and A. Aguilera. 2003. Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 42:185-198. [DOI] [PubMed] [Google Scholar]
- 30.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12:3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothkamm, K., I. Kruger, L. H. Thompson, and M. Lobrich. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23:5706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan, A. J., S. Squires, H. L. Strutt, and R. T. Johnson. 1991. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res. 19:3295-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saintigny, Y., F. Delacote, G. Vares, F. Petitot, S. Lambert, D. Averbeck, and B. S. Lopez. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20:3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh-Gohari, N., and T. Helleday. 2004. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 32:3683-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saleh-Gohari, N., and T. Helleday. 2004. Strand invasion involving short tract gene conversion is specifically suppressed in BRCA2-deficient hamster cells. Oncogene 23:9136-9141. [DOI] [PubMed] [Google Scholar]
- 36.Sargent, R. G., M. A. Brenneman, and J. H. Wilson. 1997. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 17:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber, V., D. Hunting, C. Trucco, B. Gowans, D. Grunwald, G. De Murcia, and J. M. De Murcia. 1995. A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc. Natl. Acad. Sci. USA 92:4753-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultz, N., E. Lopez, N. Saleh-Gohari, and T. Helleday. 2003. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 31:4959-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sengstag, C. 1994. The role of mitotic recombination in carcinogenesis. Crit. Rev. Toxicol. 24:323-353. [DOI] [PubMed] [Google Scholar]
- 40.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 41.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strumberg, D., A. A. Pilon, M. Smith, R. Hickey, L. Malkas, and Y. Pommier. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20:3977-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 45.Tebbs, R. S., Y. Zhao, J. D. Tucker, J. B. Scheerer, M. J. Siciliano, M. Hwang, N. Liu, R. J. Legerski, and L. H. Thompson. 1995. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl. Acad. Sci. USA 92:6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, L. H., K. W. Brookman, L. E. Dillehay, A. V. Carrano, J. A. Mazrimas, C. L. Mooney, and J. L. Minkler. 1982. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res. 95:427-440. [DOI] [PubMed] [Google Scholar]
- 47.Tischfield, J. A. 1997. Loss of heterozygosity or: how I learned to stop worrying and love mitotic recombination. Am. J. Hum. Genet. 61:995-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsao, Y. P., A. Russo, G. Nyamuswa, R. Silber, and L. F. Liu. 1993. Interaction between replication forks and topoisomerase I-DNA cleavable complexes: studies in a cell-free SV40 DNA replication system. Cancer Res. 53:5908-5914. [PubMed] [Google Scholar]
- 49.van Gent, D. C., J. H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 50.Venkitaraman, A. R. 2002. Cancersusceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 51.Wang, H., Z. C. Zeng, T. A. Bui, E. Sonoda, M. Takata, S. Takeda, and G. Iliakis. 2001. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene 20:2212-2224. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z. Q., L. Stingl, C. Morrison, M. Jantsch, M. Los, K. Schulze-Osthoff, and E. F. Wagner. 1997. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11:2347-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, J., M. B. Yin, G. Hapke, K. Toth, and Y. M. Rustum. 2002. Induction of biphasic DNA double strand breaks and activation of multiple repair protein complexes by DNA topoisomerase I drug 7-ethyl-10-hydroxy-camptothecin. Mol. Pharmacol. 61:742-748. [DOI] [PubMed] [Google Scholar]
- 54.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]