Abstract
Histone chaperones are a group of proteins that aid in the dynamic chromatin organization during different cellular processes. Here, we report that the human histone chaperone nucleophosmin interacts with the core histones H3, H2B, and H4 but that this histone interaction is not sufficient to confer the chaperone activity. Significantly, nucleophosmin enhances the acetylation-dependent chromatin transcription and it becomes acetylated both in vitro and in vivo. Acetylation of nucleophosmin and the core histones was found to be essential for the enhancement of chromatin transcription. The acetylated NPM1 not only shows an increased affinity toward acetylated histones but also shows enhanced histone transfer ability. Presumably, nucleophosmin disrupts the nucleosomal structure in an acetylation-dependent manner, resulting in the transcriptional activation. These results establish nucleophosmin (NPM1) as a human histone chaperone that becomes acetylated, resulting in the enhancement of chromatin transcription.
The eukaryotic genome is packaged into a complex and compact nucleoprotein structure, the chromatin, composed of repeating subunits called nucleosomes. Each nucleosome is composed of 146 bp of DNA wrapped around a proteinaceous scaffold, which is an octamer of four core histones (H3, H4, H2A, and H2B) (41, 64). The hallmark of chromatin is its dynamic nature, which is necessary for the regulation of nuclear processes that require access to the genetic information. Several strategies exist to alter the dynamic properties of chromatin, including both enzymatic and nonenzymatic methods. Posttranslational modification of the histones and other chromatin-associated proteins is one such strategy (27, 66). Interestingly, different modifications of the core histone tails (notably, acetylation, methylation, phosphorylation, etc.) set up an environment to facilitate chromatin dynamics in conjunction with other remodeling systems (46, 51, 62). The role of reversible acetylation in the regulation of chromatin transcription has long been documented (3). The switching of the histone subunit composition with conditional histone variants is another strategy (reference 5 and references therein). Active chromatin regions have been found to contain the H3.3 histone variant, which is enriched in modifications associated with transcriptional activity (43, 65).
Prominent among the nonenzymatic machineries that are involved in the alteration of chromatin structure, replication-dependent and -independent chaperones play a significant role (40). The function of histone chaperones in chromatin replication is well established, while its involvement in other nuclear processes is not well documented (40, 60). Histone chaperones are known to be involved in chromatin reorganization in a replication-independent manner (45). Human histone chaperone nucleophosmin, a nucleolar protein that shuttles between the nucleolus and nucleoplasm, shares a 50% homology to the N terminus of the Xenopus nucleoplasmin protein (10, 14) and possesses a unique C-terminal domain with a low homology to known human proteins. Xenopus nucleoplasmin has been shown to enhance transcription factor binding to mononucleosomes (15). Though thought to play a role in ribosome assembly, nucleophosmin has recently been shown to interact with the tumor suppressor proteins p53, Rb, and ARF (7, 16, 36, 37). Its role in stabilizing p53 is well documented (16, 37). The function of ARF is also regulated by nucleophosmin (7). Nucleophosmin binds to several cellular and viral proteins that include the human immunodeficiency virus Rev and Tat proteins (20), hepatitis virus δ antigen (24), and the transcription factor YY1 (25). It is significantly abundant in tumor and growing cells compared to normal cells (7, 14). Thus, presumably, it may also play a crucial role in transcriptional regulation in vivo.
Transcription from a chromatin template converts the promoter region from a nuclease-resistant site to a nuclease-sensitive one (19). This suggests that transcriptional activity results from a deficiency of histones in the promoter region. Recent reports have demonstrated a genome-wide loss of nucleosomes in the regulatory region (39), suggesting that nucleosome depletion could be a key step in transcriptional activation. Several factors have been implicated in this process, though very few have been validated. Histone hyperacetylation has been implicated in this process (52), which could lead to a partial loss of nucleosomes. The RNA polymerase II elongation complex (FACT) is involved in the rearrangement of histones (53). ATP-dependent remodeling factors SWI/SNF and RSC complexes have been demonstrated to facilitate the removal of the H2A-H2B dimer (11). Histone chaperones have also been suggested to play a role in the removal of histones during transcriptional activation. Recent evidence suggests that the yeast antisilencing factor Asf1 assists in the removal of histones, which consequently enhances the transcriptional activity of PHO5 and PHO8 (1, 2).
Acetylation of histones has been associated with active genes. Further impetus to this observation came from the fact that several transcriptional coactivators were found to possess histone acetyltransferase (HAT) activity (56). Members of the p300/CBP family and the GNAT family of HATs have been shown to acetylate nonhistone proteins, which earn them the new title of factor acetyltransferases (56, 66). Numerous transcription factors are the targets of the factor acetyltransferase activity, generally resulting in an increase in their DNA-binding ability upon acetylation (reference 4 and references therein). Acetylation of nonhistone chromatin proteins has a wide variety of functional consequences based on the nature and location of these proteins in the cell. For example, acetylation of the HMGN1 and -N2 proteins by PCAF reduces its affinity for the nucleosomes (23). Acetylation of the HMGB1 protein by CBP has been suggested to increase its affinity toward distorted DNA and promote DNA bending (50).
In this work, we report the involvement of human nucleophosmin in the regulation of acetylation-dependent chromatin transcription. Interestingly, nucleophosmin becomes acetylated by p300, which is essential for the activation of transcription, in conjunction with histone acetylation. The histone chaperone activity of nucleophosmin has been implicated in this function. We have demonstrated that acetylation enhances the chaperone activity with respect to core histones, while it promotes nucleosomal disruption in conjunction with acetylated core histones. This suggests that transcriptional activation in the presence of nucleophosmin occurs through the disassembly of nucleosomes. Thus, this report establishes nucleophosmin as a novel human histone chaperone involved in the activation of chromatin transcription in an acetylation-dependent manner.
MATERIALS AND METHODS
Cloning of NPM1.
NPM1 was subcloned between the NcoI and XhoI sites of the pET28b vector by PCR-based subcloning from a cDNA clone. The various deletion mutants were cloned between the NcoI and XhoI sites of the pET28b vector by the PCR-based subcloning technique. The sense and antisense mammalian expression clones were cloned into the HindIII and XhoI sites of pcDNA 3.1(+) through PCR-based subcloning.
Purification of human core histones and recombinant proteins.
Human core histones were purified from the HeLa nuclear pellet as described earlier (35). Acetylated core histones were purified from HeLa cells treated with 5 mM sodium butyrate and 125 nM trichostatin A (TSA) as described above for the core histones. The FLAG epitope-tagged PCAF was purified from the recombinant baculovirus-infected insect cell line Sf21 by immunoaffinity purification using M2 agarose (Sigma) (35). Full-length p300 was also purified from the recombinant baculovirus-infected Sf21 cells as a His6-tagged protein (34). The His6-tagged nucleosome assembly protein 1 (NAP1) used for the in vitro chromatin assembly was purified from Escherichia coli cells as reported previously (35), and the FLAG-tagged chimeric activator Gal4-VP16 was expressed in E. coli and purified by immunoaffinity purification with M2 agarose (34). The individual Xenopus histones were expressed and purified from E. coli cells as described earlier (42). The His6-tagged Drosophila topoisomerase core catalytic domain was purified as described above.
Ni-NTA pulldown assays.
One microgram of the His6-tagged protein (full-length NPM1, its deletion mutants, or acetylated NPM1) was mixed with indicated amounts of the core histones, acetylated core histones, or the individual histones in the interaction buffer (20 mM Tris-HCl [pH 7.9], 20% glycerol, 0.2 mM EDTA [pH 8.0], 0.1% Nonidet P-40, 2 mM phenylmethylsulfonyl fluoride, 300 mM KCl, 30 mM imidazole) along with the Ni-nitrilotriacetic acid (NTA) His Bind resin (Novagen). The mixture was incubated for 2 h at 4°C on a rotary shaker. After the beads were extensively washed in the interaction buffer, the proteins were extracted from the beads into the sodium dodecyl sulfate (SDS) sample buffer, separated on an SDS-12% polyacrylamide gel electrophoresis (PAGE) gel, and either visualized by Coomassie brilliant blue (CBB) staining or blotted and probed with appropriate antibodies.
Histone transfer assay.
The nucleosome assembly activity of NPM1 and its deletion mutants was measured as described earlier (45) with a few modifications (see Fig. 3A). Briefly, 200 ng of closed circular G5ML (34) plasmid was relaxed by preincubation with the Drosophila TopoI core catalytic domain in the assembly buffer (34) at 37°C for 40 min. Indicated amounts of HeLa core histones or acetylated histones and full-length NPM1, the deletion mutants, or the acetylated NPM1 were preincubated in the assembly buffer at 37°C for 40 min. The two reactions were mixed and incubated further at 37°C for 20 min. The reaction was stopped by incubating at 37°C for 30 min in an equal volume of stop buffer (100 mM NaCl, 20 mM EDTA [pH 8.0], 1% SDS, 0.2 mg/ml proteinase K, 0.25 mg/ml glycogen). Following this reaction, the plasmids were extracted using phenol-chloroform and precipitated with ethanol. The plasmids were separated on a 1% agarose gel and visualized by staining with ethidium bromide. For the dose-dependent assays with acetylated NPM1, the postrelaxation incubation was reduced to 10 min to perceive the difference in the histone transfer compared to the unacetylated NPM1.
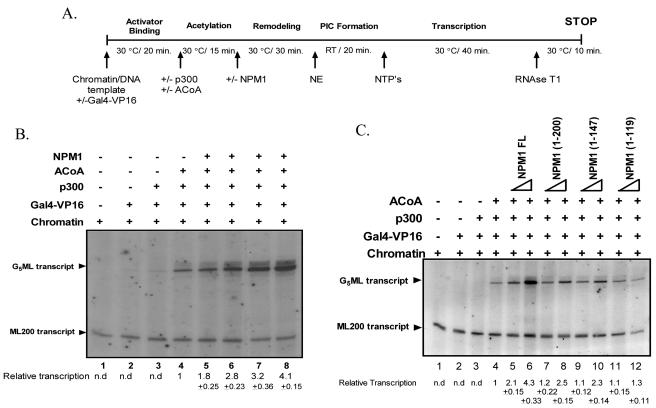
FIG. 3.
Nucleophosmin stimulates acetylation-dependent chromatin transcription. (A) Schematic representation of the in vitro transcription protocol. RT, room temperature; PIC, preinitiation complex. (B) Freshly assembled chromatin (28 ng) was subjected to the protocol described for panel A. NPM1 stimulates chromatin transcription in a dose-dependent manner. Lane 1, without activator; lanes 2 through 8, with Gal4-VP16 (50 ng); lanes 3 through 8, with p300 (25 ng); lanes 4 through 8, with acetyl-CoA (ACoA; 1.5 μM); lanes 5 through 8, 1, 10, 30, and 50 pmol of full-length NPM1, respectively. (C) Freshly assembled chromatin (28 ng) was subjected to the protocol described for panel A. Lane 1, without activator; lanes 2 through 12, with Gal4-VP16 (50 ng); lanes 3 through 12, with p300 (25 ng); lanes 4 through 12, with acetyl-CoA (1.5 μM). Lanes 5, 7, 9, and 11, have 5 pmol and lanes 6, 8, 10, and 12 have 30 pmol of full-length (FL) NPM1 or the respective deletion mutants added as indicated. The relative transcription per lane (in severalfold activation over the acetylation-dependent transcription lane [lane 4]) was determined by phosphorimage analysis (Fuji) and presented under each lane along with the standard deviation. n.d., not determined.
In vitro chromatin assembly.
A chromatin template for in vitro transcription experiments was assembled and characterized as described earlier (34).
In vitro transcription assay.
Transcription assays were essentially carried out as described elsewhere (34) with minor modifications. The scheme of transcription is enumerated in Fig. 3A. Briefly, 28 ng of DNA or an equivalent amount of chromatin template was incubated with 30 ng of activator (Gal4-VP16) in a buffer containing 4 mM HEPES (pH 7.8), 20 mM KCl, 2 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 10 mM sodium butyrate, 0.1 mg/ml bovine serum albumin, and 2% glycerol (35). Twenty nanograms of p300 and 1.5 μM acetyl coenzyme A (acetyl-CoA) was added and the reaction incubated at 30°C for 15 min. Indicated amounts of NPM1 or its deletion mutants were added to the reaction, which was incubated at 30°C for 30 min. HeLa nuclear extract (NE; 5 μl, which contains ∼8 mg/ml protein) was added to initiate the preinitiation complex formation. Transcription reaction was started by the addition of NTP mix (12 mM concentrations of ATP and CTP, 0.5 mM UTP, 2 mM 3′ O-methyl GTP) and [α-32P]UTP after the preinitiation complex formation. The incubation was continued for 40 min at 30°C. Transcription was terminated by the addition of 250 μl of stop buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl, 1% SDS, 0.025 ng/μl tRNA). The 32P-radiolabeled transcript was extracted with phenol-chloroform, ethanol precipitated, and dried and the pellet dissolved in loading dye (8 M urea, 0.005% bromphenol blue, and xylene cyanol in 1× Tris-borate-EDTA [TBE]) and analyzed on a urea-5% polyacrylamide gel. Gels were then dried and subjected to autoradiography at −70°C. Quantification of transcription was done by using the Fuji BAS system and analyzed by using ImageQuant software. Quantitation of DNA and chromatin transcription data represent three independent experiments. In the case of interrupted transcription experiments, 10 nM lysyl-CoA, a p300 HAT-specific inhibitor, was added immediately after the acetylation reaction and the reaction was incubated for 15 min at 30°C. The rest of the reaction was carried out as described above.
In vitro acetylation assay and mass acetylation.
In vitro acetylation assays were performed as described elsewhere (33). Indicated amounts of proteins (see figure legends) were incubated at 30°C for 30 min in a 30-μl final reaction volume consisting of 50 mM Tris-HCl (pH 8.0), 10% (vol/vol) glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM EDTA (pH 8.0), 10 mM sodium butyrate, and 0.5 μl of 3.3 Ci/mmol [3H]acetyl-CoA. The reaction mixture was then blotted onto P-81 filter paper. Radioactive counts were recorded on a Wallac 1409 liquid scintillation counter. To visualize the radiolabeled acetylated protein, the reaction products were precipitated with 25% trichloroacetic acid, washed with ice-cold acetone, and resolved electrophoretically on an SDS-15% polyacrylamide gel and subjected to fluorography using a solution containing 22.5% 2,5-diphenyloxazole in Me2SO (9). Gels were dried, and autoradiography was performed at −70°C for 1 to 2 days. For the mass acetylation reaction, 12 μg of NPM1 and 100 ng full-length p300 were incubated in the presence of 50 μM acetyl-CoA at 37°C for 1.5 h. To achieve efficient acetylation, p300 and acetyl-CoA were added at every 30-min interval.
MALDI analysis of acetylated nucleophosmin.
Mass-acetylated protein was separated on an SDS-12% PAGE gel, the band excised out, and in-gel protease digestion carried out in ammonium bicarbonate buffer as described elsewhere (54). The different proteases used were trypsin, Asp-N, and Glu-C (Sigma). The resultant peptides were extracted in 50% acetonitrile-5% formic acid and the peptide masses analyzed on a Bruker Daltonics matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) machine.
Luciferase assay.
293T cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle medium with 10% fetal bovine serum. The mammalian expression constructs of p53, Gal4-VP16, and NPM1 sense and antisense constructs used in this study were placed under a cytomegalovirus (CMV) promoter. The p53-responsive reporter construct pG13luc contains 13 consensus p53-binding sites in tandem followed by a polyomavirus promoter, which drives the luciferase gene (28). The Gal4-VP16-responsive reporter construct pG10luc contains 10 Gal4-binding sites in tandem followed by the luciferase gene. These reporter constructs were used to probe into the effect of NPM1 overexpression on the p53- and Gal4-VP16-mediated gene expression in vivo. The CMV-driven β-galactosidase (pCMV-βgal) construct was used as an internal control. Prior to transfection, cells were seeded at 0.6 × 106 cells per 60-mm-diameter dish and transfected with the different plasmid constructs by using Lipofectamine 2000 Plus (Invitrogen) according to the manufacturer's protocol. An empty vector was used to keep the total amount of DNA constant in each transfection. Luciferase and β-galactosidase activities were measured 48 h after the transfection by luciferase assay and β-galactosidase assay systems according to the procedure provided by the manufacturer (Promega). The transient transfection assay was performed three times independently to monitor average relative luciferase activity.
In vivo acetylation status of nucleophosmin.
293T cells were treated with 5 mM sodium butyrate, 100 nM trichostatin A, and 1 mCi/ml [3H]sodium acetate for a period of 24 h, following which the cell lysate was prepared in radioimmunoprecipitation assay buffer and nucleophosmin was immunoprecipitated with the monoclonal antibody against NPM1 (Sigma). The immune complex was separated on an SDS-12% PAGE gel and subjected to fluorography as described above. Gels were dried, and autoradiography was performed at −70°C for 1 to 2 days. In a separate experiment, the pulldown complexes were separated on an SDS-12% PAGE gel, blotted onto a nitrocellulose membrane, and probed with antibody against acetylated lysine (Calbiochem).
Acetylation of NPM1 under transcription conditions.
In vitro transcription reactions were carried out on a chromatin template in the presence of [3H]acetyl-CoA as described in Fig. 5A and terminated following the remodeling step. The samples were precipitated using trichloroacetic acid to a final concentration of 25% and separated on an SDS-12% PAGE gel. The gel was then subjected to fluorography as described above. Gels were dried, and autoradiography was performed at −70°C for 2 to 4 days.
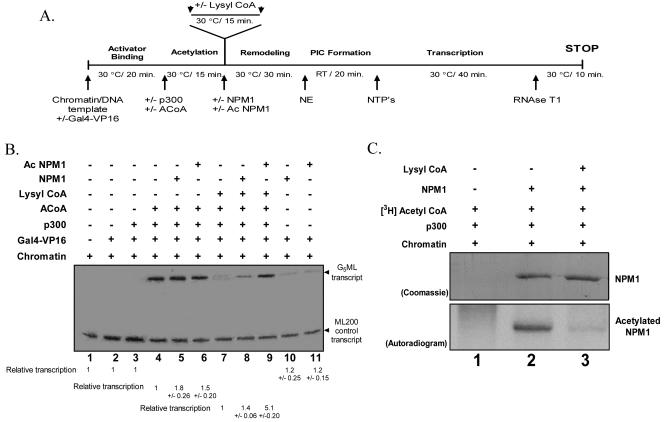
FIG. 5.
Acetylation of nucleophosmin is critical for transcriptional activation from a chromatin template. (A) Schematic representation of in vitro transcription protocol modified to include the interrupted acetylation step (addition of the p300-specific inhibitor lysyl-CoA). RT, room temperature; PIC, preiniation complex. (B) Freshly assembled chromatin (28 ng) was subjected to the protocol described for panel A without or with Gal4-VP16, full-length p300, or 1.5 μM acetyl-CoA (ACoA) and without or with the p300-specific HAT inhibitor lysyl-CoA, along with 30 pmol (each) of full-length NPM1 or acetylated NPM1 as indicated. The relative transcription per lane (in severalfold activation over the respective control lanes marked with the number 1) was determined by phosphorimage analysis (Fuji) and presented under each lane along with the standard deviation, calculated from three independent experiments. (C) Acetylation status of NPM1 under the transcription conditions. Chromatin was subjected to the transcription protocol enumerated for panel A in the presence of [3H]acetyl-CoA, terminating the reaction after the remodeling step with (lane 2) or without (lane 3) lysyl-CoA, in the presence (lanes 2 and 3) or absence (lane 1) of NPM1. The top panel shows the Coomassie blue-stained gel, while the bottom panel shows the autoradiogram.
RESULTS
In vitro binding preference of nucleophosmin to core histones.
Histone chaperones are a class of histone-interacting proteins that aid in a wide variety of processes that involve chromatin (40). This class of proteins has been postulated to possess the ability to alter local chromatin structure and facilitate nuclear processes that include transcription. In a classical report, it was shown that a Xenopus histone chaperone nucleoplasmin enhances transcription factor binding to mononucleosomes by displacing the nucleosomal histones (15). Human nucleophosmin/B23 (NPM1) shows 50% similarity with the N-terminal region of Xenopus nucleoplasmin (14, 18), which has been demonstrated to interact with the histone pair H2A-H2B (6). We investigated the histone interaction pattern for nucleophosmin. We cloned, expressed, and purified full-length His6-tagged nucleophosmin from E. coli. The authenticity of the recombinant protein was confirmed using the NPM1 monoclonal antibody (see the supplemental material) and mass spectrometric analysis.
We used the Ni-NTA pulldown assay to map the interaction profile of nucleophosmin with the core histones. Highly purified human core histones from HeLa cells (see the supplemental material) were incubated along with the recombinant His6-tagged nucleophosmin and the Ni-NTA beads in the interaction buffer containing 300 mM salt (see Materials and Methods). Full-length nucleophosmin could pull down all four core histones, while the beads alone did not pull down any protein (Fig. 1B, top panel, lane 3 versus lane 2). We also carried out immunoblots on the pulldown product with antibodies against the core histones, which validated the earlier statement (Fig. 1B, Western blot, lane 3 versus lane 2). Most histone chaperones have been documented to possess a preferential affinity toward either the H3-H4 tetramer or the H2A-H2B dimer (40, 61). Our data suggest that nucleophosmin interacts either with all the core histones or at least with one partner each in the H3-H4 tetramer or H2A-H2B dimer. We repeated the interaction of nucleophosmin with recombinant individual histones from Xenopus (42) and probed the blot with antibodies raised against H3, H2A, and H2B. Interestingly, nucleophosmin strongly interacted with H3, H4, and H2B, while it did not interact with H2A (Fig. 1C, lanes 1). None of the individual histones were pulled down by using Ni-NTA beads alone (Fig. 1C, lanes 5). These results indicate that H2A in HeLa core histones is pulled down by nucleophosmin as a dimer, along with H2B.
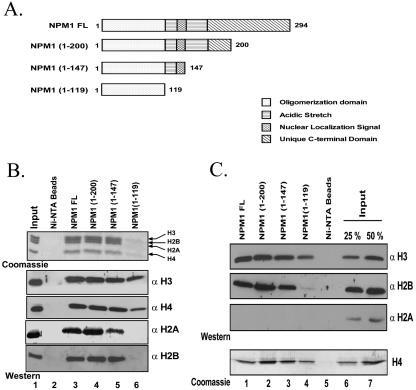
FIG. 1.
Nucleophosmin interacts with the core histones preferentially. (A) Schematic representation of the C-terminal deletion mutants of nucleophosmin with the key to domain organization. (B and C) Interaction of the His6-tagged, full-length (FL) nucleophosmin and its deletion mutants (1 μg) with HeLa core histones (5 μg) (B) or individual recombinant histones (C) was investigated using a Ni-NTA pulldown assay. The beads were loaded on an SDS-12% PAGE gel and visualized by CBB staining. The same was blotted on a polyvinylidene difluoride membrane and probed with antibodies against different histones as indicated.
Nucleophosmin has a modular structure, with a hydrophobic N terminus, important for oligomerization; two acidic stretches separated by the bipartite nuclear localization signal; and a unique C-terminal domain (Fig. 1A). We wanted to map the precise region of NPM1 that interacts with the histones. We subcloned different His6-tagged C-terminal truncations of NPM1 (Fig. 1A) that sequentially eliminated the unique C-terminal domain (NPM1 1-200), the second acidic stretch and nuclear localization signal (NPM1 1-147), and finally, the first acidic stretch (NPM1 1-119), leaving the oligomerization domain intact. These deletion mutants were expressed and purified from E. coli (see the supplemental material). We then used the Ni-NTA pulldown assay to test the histone-interacting ability of the deletion mutants. The mutants NPM1 1-200 and 1-147 could interact with both core histones (Fig. 1B, lanes 4 and 5 versus lanes 3) and the individual recombinant histones (Fig. 1C, lanes 2 and 3 versus lanes 1) to the same extent as the full-length protein. However, the N-terminal oligomerization domain (NPM1 1-119) interacted very feebly with the H3-H4 tetramer (Fig. 1B, lanes 6 versus lanes 3), while in the case of the individual histones, the binding of histone H2B was completely abolished (Fig. 1C, second panel, lane 4 versus lanes 1 through 3). Histones H3 and H4 could still interact, albeit with less efficiency (Fig. 1C, lanes 4 versus lanes 1 through 3). These results are in contrast to those in an earlier report wherein nucleophosmin was suggested to possess a preference for histone H3 (48). In our report, we have shown that human nucleophosmin interacts with both the histone tetramer and the dimer, despite a lack of interaction with histone H2A. Most histone chaperones do not require the acidic stretch for the histone interaction (20, 58). Taken together, these data show that nucleophosmin can interact with the H3-H4 tetramer through the N terminus and the H2B-H2A dimer through the first acidic stretch.
Histone transfer activity of nucleophosmin.
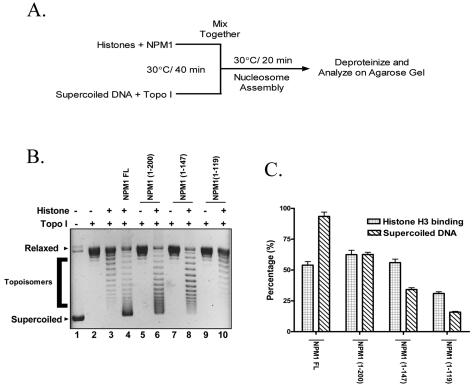
The ability of nucleophosmin to assemble nucleosomes on a plasmid was tested as a measure of its function as a histone chaperone. The plasmid supercoiling assay (Fig. 2A) measures the assembly activity on the basis of the introduction of negative supercoils on a relaxed template (47). The full-length nucleophosmin could introduce negative supercoils on the plasmid DNA in the presence of TopoI and core histones (Fig. 2B, lane 4). In order to map the chaperone domain and to correlate it with the histone interaction ability of nucleophosmin, we tested the different deletion mutants for their assembly activity. Interestingly, we found that the chaperone activity of nucleophosmin dropped by 40% upon the deletion of 94 C-terminal amino acids (NPM1 1-200) (Fig. 2B, lane 6 versus lane 4) and by 70% upon the deletion of 147 C-terminal amino acids (NPM1 1-147), compared to the full-length nucleophosmin (Fig. 2B, lane 8 versus lane 4). Interestingly, the N-terminal oligomerization domain did not show any histone transfer activity (Fig. 2B, lane 10 versus lane 4). These two activities of nucleophosmin were compared by quantifying the percentage of histone H3 bound as well as the percentage of supercoiled DNA formed in each case. A gradual loss of chaperone activity was noted upon the sequential deletion of the C-terminal domain, while the ability to interact with the core histones remained intact for all mutants except the N-terminal oligomerization domain (Fig. 2C). This suggested that the histone-binding ability of nucleophosmin is functionally separate from the chaperone activity. Interestingly, the histone-binding region is at the N terminus while the chaperone activity is at the C terminus, suggesting a distinct modular nature of the nucleophosmin protein. These results indicate that the histone interaction ability of human nucleophosmin is necessary but not sufficient for its histone transfer ability.
FIG. 2.
The histone chaperone activity of nucleophosmin is independent of its histone interaction. (A) Schematic depiction of the histone transfer assay to test the histone chaperone function of NPM1. (B) Histone transfer assay was carried out with 500 ng of histones and 250 ng of either full-length (FL) NPM1 or the deletion mutants. Lane 1, supercoiled DNA; lane 2, DNA relaxed by TopoI; lane 3, relaxed DNA with core histones; lane 4, relaxed DNA with core histones and full-length NPM1. Relaxed DNA without (lane 5) or with (lane 6) core histones and NPM1 1-200, relaxed DNA without (lane 7) or with (lane 8) core histones and NPM1 1-147, and relaxed DNA without (lane 9) or with (lane 10) core histones and NPM1 1-119 are shown. (C) Comparative analysis of the amount of supercoiled DNA and the histone H3-binding ability of NPM1 FL or each of the deletion mutants. The supercoiled DNA formed was quantified using Image Gauge software (Fuji) and expressed as a percentage of the total supercoiled DNA used. The percentage of histone H3 binding was calculated similarly.
Nucleophosmin stimulates acetylation-dependent chromatin transcription.
Based on its ability to interact with and transfer the histones, we investigated the role of nucleophosmin in transcriptional regulation. For this purpose, the in vitro transcription assay based on a nucleosomal array template was used with necessary modifications. The chromatin template was assembled on a previously relaxed pG5ML array plasmid (34) by incubating it with HeLa core histones and recombinant mouse NAP1. The assembly of chromatin was confirmed using the supercoiling and partial MNase digestion assays.
Transcription assays followed the protocol represented in Fig. 3A. The different steps include activator binding, histone acetylation by p300 and acetyl-CoA, incubation with nucleophosmin, preinitiation complex assembly, and the final transcription step. HeLa NE was used as a source of the general transcription factors and cofactors. Nucleophosmin is an abundant protein in the nucleus of HeLa cells. In order to understand the exact role of nucleophosmin in transcription, we immunodepleted nucleophosmin from NE (see the supplemental material). The nucleophosmin-depleted NE was found to be equally efficient as undepleted NE, as characterized by activator-dependent transcription from a DNA template (see the supplemental material). Interestingly, the exogenous addition of nucleophosmin did not affect the transcription from the DNA template (see the supplemental material).
Addition of nucleophosmin to the chromatin transcription reaction in the absence of acetylation (without the addition of p300 and acetyl-CoA), up to 50 pmol, could not activate transcription (see the supplemental material). In agreement with previously published results (34), a 10-fold transcriptional activation was observed upon the addition of p300 and acetyl-CoA (see the supplemental material), confirming the transcriptional competence of the system. However, the ectopic addition of nucleophosmin in a dose-dependent manner up to 50 pmol into the acetylation-dependent chromatin transcription system showed a further ∼4-fold increase over the acetylated chromatin lane (Fig. 3B, lane 8 versus lane 4). We also tested the ability of the different deletion mutants for activating chromatin transcription. Two different concentrations of each deletion mutant (5 and 50 pmol) were tested out. NPM1 1-200 and 1-147 showed a marginal activation at 50 pmol (Fig. 3C, lanes 8 and 10 versus lane 6), while the NPM1 1-119 mutant showed no enhancement in chromatin transcription (Fig. 3C, lanes 11 and 12 versus lane 4). These results hint at a correlation between transcriptional activation and the histone transfer activity of nucleophosmin and not a dependence on its histone-interacting ability alone. Most significantly, nucleophosmin could enhance the transcription from the chromatin template only when p300 and acetyl-CoA were present.
Nucleophosmin becomes acetylated both in vitro and in vivo.
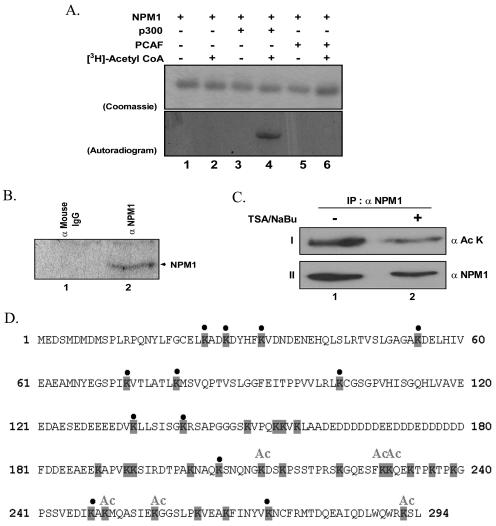
The absolute dependence of nucleophosmin on the acetylation step for the transcriptional activation tempted us to investigate whether NPM1 itself becomes acetylated by p300. We carried out an in vitro acetylation assay (see Materials and Methods) using p300 and PCAF as histone acetyltransferases and [3H]acetyl-CoA. The activities of both acetyltransferases were normalized using histones as substrate (data not shown). The normalized acetyltransferases were then assayed for their ability to acetylate nucleophosmin. Interestingly, it was found that nucleophosmin becomes acetylated specifically by p300 and not by PCAF (Fig. 4A, lanes 4 versus lanes 6). p300 has a specificity to acetylate histones H3 and H4 in vitro and all core histones in vivo, apart from a range of nonhistone substrates which include the transcriptional coactivator PC4 (33), p53 (22), different high-mobility group proteins (12, 23), and human immunodeficiency virus Tat (29), among others. Notably, this is the first report on the acetylation of a histone chaperone.
FIG. 4.
Nucleophosmin becomes acetylated both in vitro and in vivo. (A) In vitro gel HAT assay carried out with NPM1 as a substrate without (lanes 1 and 2) or with (lanes 3 and 4) the activity-normalized p300. The acetylation reactions were also tested with PCAF (lanes 5 and 6) as the histone acetyltransferases. Lanes 2, 4, and 6 show the acetylation reaction in the presence and lanes 1, 3, and 5 in the absence of [3H]acetyl-CoA. The gel was then subjected to CBB staining and fluorography and exposed to an X-ray film. Top panel shows the CBB-stained gel, while the bottom panel shows the autoradiogram. (B) In vivo acetylation of nucleophosmin. Nucleophosmin was immunoprecipitated with the monoclonal antibody (lane 2) from 293T cells treated with deacetylase inhibitors (500 ng/ml TSA and 5 mM sodium butyrate) as well as [3H]sodium acetate. The immunoprecipitate was analyzed on an SDS-12% PAGE gel and processed as described for panel A. Mouse preimmune serum was used as an immunoprecipitation control (lane 1). IgG, immunoglobulin G. (C) In vivo acetylation of nucleophosmin under normal growth conditions. Nucleophosmin was immunoprecipitated (IP) with the monoclonal antibody from 293T cells treated with deacetylase inhibitors (500 ng/ml TSA and 5 mM sodium butyrate). The immunoprecipitate was analyzed on an SDS-12% PAGE gel, blotted, and probed with antibody against acetylated lysine (panel I). The lysates used in the pulldown assay were analyzed on an SDS-12% PAGE gel, blotted, and probed with the antibody against NPM1 (panel II). (D) Protein sequence of nucleophosmin showing the acetylation sites. The lysine residues are shaded. The lysines that are acetylated are marked “Ac,” while the ones that are not acetylated are marked with a bullet. Lysines that could not be assigned as either of the above have been left unmarked.
We went on to map the exact acetylation sites on nucleophosmin by subjecting the in vitro-acetylated protein to MALDI-TOF analysis following its protease digestion. Interestingly, most acetylation sites were found to lie in the C-terminal domain of nucleophosmin. Though five acetylation sites could be assigned confirmedly, there are a few more sites that are yet to be annotated (Fig. 4D and Table 1).
TABLE 1.
Acetylation sites on nucleophosmin
| Mass (Da)
|
Sequence
|
Sequence | Protease | |||
|---|---|---|---|---|---|---|
| Observed | Unmodified | Δ | Start | End | ||
| 1,276.7755 | 1,234.4529 | 42.3226 | 246 | 256 | DIKAKMQSIE | Glu-C |
| 1,274.4924 | 1,232.4270 | 42.0654 | 286 | 294 | DLWQWRKSL | Asp-N |
| 851.2379 | 766.8719 | 84.3660 | 227 | 232 | SFKKQE | Glu-C |
| 848.9597 | 806.4076 | 42.5521 | 251 | 257 | MQASIEK | Trypsin |
| 689.3119 | 647.3107 | 42.0012 | 207 | 212 | SNQNGK | Trypsin |
To ensure the acetylation of nucleophosmin in vivo, it was immunoprecipitated from 293T cells that were previously treated with histone deacetylase inhibitors and [3H]sodium acetate. The immunoprecipitated protein was separated by SDS-PAGE and the label detected by fluorography (Fig. 4B, lane 2). As a control for the nonspecific pulldown, we used anti-mouse immunoglobulin G with the treated lysate (Fig. 4B, lane 1). These results showed that indeed nucleophosmin becomes acetylated in vivo. In order to confirm that NPM1 was acetylated under normal growth conditions, unlike under growth-restrictive conditions in the presence of TSA, we repeated the experiment in the presence and absence of TSA-NaBu. Interestingly, we found that the NPM1 was acetylated under normal and growth-restrictive conditions (Fig. 4C, top panel). As reported earlier, we also noted that there was a drop in the levels of the NPM1 protein itself upon treatment with TSA (Fig. 4C, bottom panel) (13). Taken together, these results confirm that a substantial population of NPM1 is present in the acetylated form under normal growth conditions.
Acetylation of NPM1 is essential for the stimulation of chromatin transcription.
Nucleophosmin stimulates acetylation-dependent chromatin transcription and itself becomes acetylated. Based on these two observations, we wanted to dissect out the requirement of nucleophosmin acetylation on its ability to activate chromatin transcription. In order to address this question, the scheme of transcription was modified as depicted in Fig. 5A. In order to differentiate between the effect of acetylated and unacetylated nucleophosmin on chromatin transcription, the acetylation step was interrupted with the help of the p300-specific HAT inhibitor lysyl-CoA (38). This ensures that the exogenously added unacetylated nucleophosmin, added after the acetylation step, does not become acetylated by the p300 HAT. The transcription reaction was carried out without the addition of lysyl-CoA as a control. Since the reaction was subjected to a longer time point, without the addition of lysyl-CoA, the difference in transcriptional activation dropped ∼1.8- to 2-fold (Fig. 5B, lanes 5 and 6 versus lane 4) compared to 2.5-fold in the earlier experiment (Fig. 3B).
Interrupted acetylation impairs chromatin transcription, producing ∼10-fold reduction compared to the uninterrupted reaction (Fig. 5B, lane 7 versus lane 4). Exogenous addition of nucleophosmin to this reaction produced a modest increase in the transcript levels (Fig. 5B, lane 8 versus lane 7). Interestingly, the addition of preacetylated nucleophosmin to the interrupted acetylation reaction produced a ∼5-fold increase in the transcript levels (Fig. 5B, lane 9 versus lane 8). These data suggest that the acetylation of nucleophosmin is an essential requirement in the activation of transcription from the chromatin template.
In order to further clarify whether the acetylation of nucleophosmin alone is sufficient for transcriptional activation, we carried out an acetylation-independent transcription experiment with the exogenous addition of unacetylated and acetylated nucleophosmin. The absence of the characteristic activation in either case (Fig. 5B, lanes 10 and 11 versus lane 2) argues for the fact that acetylation of the chromatin template is a necessary prerequisite for the transcriptional enhancement brought about by acetylated nucleophosmin.
In order to ensure that the addition of lysyl-CoA indeed inhibits the HAT activity of p300, we carried out the transcription reaction in the presence of [3H]acetyl-CoA and analyzed the reaction for the acetylation status of NPM1. The data clearly indicate that NPM1 acetylation is substantially reduced in the presence of lysyl-CoA (Fig. 5C, lanes 3 versus lanes 2). These results clearly establish the fact that acetylation of NPM1 is essential for the enhancement of acetylation-dependent chromatin transcription.
p300-mediated acetylation of nucleophosmin and histones leads to chromatin disassembly.
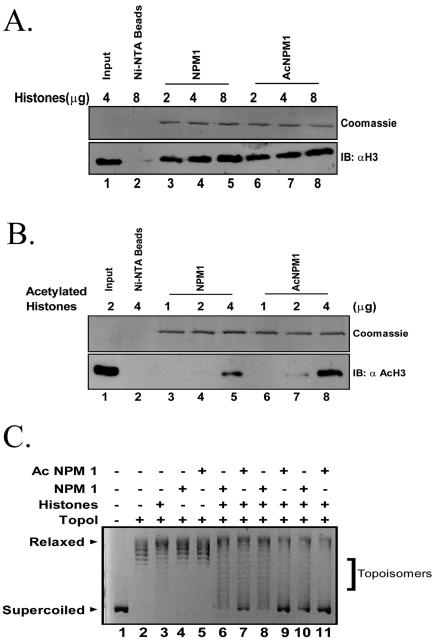
We have shown that the nucleophosmin-mediated activation of chromatin transcription also requires the acetylation of the histone chaperone. In order to investigate the functional attributes conferred on nucleophosmin upon acetylation by p300, the histone interaction profile of the modified protein was probed into. Since transcriptional activation from a chromatin template requires the acetylation of histones, the interaction of both acetylated and unacetylated nucleophosmins with increasing amounts of human hyperacetylated histones (see Materials and Methods) was also investigated using the Ni-NTA pulldown method. Though unacetylated or acetylated nucleophosmin did not show any preference for unacetylated histones (Fig. 6A, lanes 6, 7, and 8 versus lanes 3, 4, and 5, respectively), acetylated nucleophosmin showed a marked preference for acetylated histones (Fig. 6B, lanes 8 versus lanes 5). Quantification of these data using ImageQuant software (Fuji Films) revealed a threefold increase in the pulldown of acetylated histones by acetylated nucleophosmin. These results indicate an increased affinity for acetylated histones upon the acetylation of nucleophosmin. This also raises the possibility that acetylated nucleophosmin may facilitate the removal of acetylated histones from the chromatin.
FIG. 6.
Acetylation of nucleophosmin and core histones potentiates their interaction and nucleosome disruption. (A) Effect of acetylation on NPM1 and histone interaction. Indicated amounts of normal (untreated) core histones purified from the HeLa nuclear pellet were tested for their interaction with 1 μg of either NPM1 (lanes 3, 4, and 5) or acetylated NPM1 (lanes 6, 7, and 8) using the Ni-NTA pulldown assay. Ni-NTA beads alone, without NPM1 or acetylated NPM1, were used as a negative control (lane 2). Four micrograms of the sample was loaded as input (lane 1). The top panel is the Coomassie blue-stained gel showing the amounts of the pulled-down proteins, while the bottom panel shows the Western immunoblot (IB) with antihistone H3. (B) Effect of acetylation on NPM1 and acetylated histone interaction. Indicated amounts of hyperacetylated core histones purified from the HeLa nuclear pellet were tested for their interaction with 500 ng of either NPM1 (lanes 3, 4, and 5) or acetylated NPM1 (lanes 6, 7, and 8) using the Ni-NTA pulldown assay. Ni-NTA beads alone, without NPM1 or acetylated NPM1, were used as a negative control (lane 2). Two micrograms of the sample was loaded as input (lane 1). The top panel is the Coomassie blue-stained gel showing the amounts of the pulled-down proteins, while the bottom panel shows the Western blot with antibodies against acetylated histone H3. (C) Acetylated NPM1 transfers histones more efficiently than NPM1. Histone transfer was tested with 500 ng of core histones and increasing concentrations of either NPM1 (lanes 6, 8, and 10) or the acetylated NPM1 (lanes 7, 9, and 11). Lane 1, supercoiled DNA; lane 2, DNA relaxed by TopoI; lanes 3 through 11, relaxed DNA along with core histones (lane 3), NPM1 (lane 4), or acetylated NPM1 alone (lane 5) and 0.5, 1, or 5 pmol (lanes 6 and 7, 8 and 9, and 10 and 11, respectively) of either NPM1 and acetylated NPM1, respectively.
We then checked the chaperone activity of unacetylated and acetylated nucleophosmin with HeLa core histones and HeLa hyperacetylated histones. The unacetylated nucleophosmin showed no alteration in its ability to transfer acetylated histones onto the DNA template compared to the unacetylated histones (data not shown). Surprisingly, acetylated nucleophosmin showed an increased chaperone activity with unacetylated core histones in a dose-dependent manner (Fig. 6C, lanes 6 through 11) while it demonstrated a reduced ability to transfer acetylated histones, as supported by the prevalence of intermediate forms (data not shown). These data suggest that the transcriptional activation of a chromatin template by nucleophosmin requires the acetylation of both the histone and the histone chaperone, presumably leading to the depletion of the acetylated histones.
Nucleophosmin acts as a transcriptional enhancer in vivo.
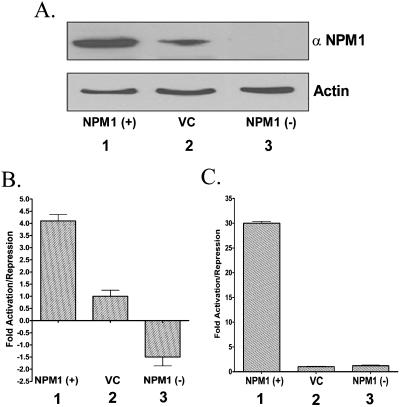
We have established the fact that nucleophosmin can indeed activate HAT-dependent chromatin transcription by the disruption of the nucleosomes in an in vitro system. To validate these results in the cellular environment, we proceeded to clone nucleophosmin into a mammalian expression vector [pcDNA 3.1(+)]. Nucleophosmin was cloned in both the sense and antisense directions to achieve the overexpression and silencing of nucleophosmin expression, respectively. These mammalian expression clones were transfected into the 293T cells and checked for their ability to alter the expression of nucleophosmin. The sense clone NPM1 (+) could overexpress up to four times more (Fig. 7A, lanes 1 versus lanes 2), while the antisense clone NPM1 (−) abolished the expression of nucleophosmin by ∼80% (Fig. 7A, lanes 3 versus lanes 2) as revealed by Western blot analysis on the lysates of transfected cells.
FIG. 7.
Nucleophosmin induces p53-mediated transcription in vivo. (A) The lysates used in the luciferase assays [NPM1 (+) (lane 1), vector-transfected control (VC) (lane 2), NPM1 (−) (lane 3)] were separated on an SDS-12% PAGE gel and immunoblotted with anti-NPM1 or anti-actin as indicated. (B and C) 293T cells were transiently transfected either with 1 μg of pG13luc (B) or pG10luc (C), 250 ng of pIRES puro-p53 (B) or Gal4-VP16 (C), or 1 μg of pCMV-βgal and with 2 μg of either pcDNA3.1, pcDNA3.1 NPM1 (+), or pcDNA3.1 NPM1 (−). The p53-driven luciferase activity was measured and normalized with the beta-galactosidase activity. The severalfold increase or decrease in the luciferase activity over the VC (bar 2) in the case of NPM1 (+) (bar 1) and NPM1 (−) (bar 3) has been presented as a bar diagram.
We used the p53-responsive synthetic reporter gene pG13luc, which contains 13 p53-binding sites upstream of the luciferase gene construct (28), to test the effect of nucleophosmin coexpression. The p53-responsive pG13luc vector was cotransfected along with the mammalian expression vector for p53 and either one of the nucleophosmin sense, antisense, or parental vector clones. The β-galactosidase gene under the CMV promoter was cotransfected for internal control. The normalized reporter gene expression was plotted as the increase or decrease over the parental vector control. Concomitant with our chromatin transcription data, we found that overexpression of nucleophosmin produces a fourfold increase in reporter gene expression (Fig. 7B, bar 1 versus bar 2), while silencing the nucleophosmin expression reduces the transactivation ∼1.5-fold (Fig. 7B, bar 3 versus bar 2). Interestingly, it also suggests that the histone chaperone nucleophosmin could be an important determinant for the p53-mediated gene expression.
NPM1 and p53 are known to interact with each other (16, 37), resulting in a functional modulation of p53. In order to test the global effect of NPM1 overexpression, we repeated the reporter assay using a different transactivator, Gal4-VP16, and a Gal4-responsive luciferase cassette. Interestingly, cotransfection of Gal4-VP16 and NPM1 (+) resulted in a 30-fold increase in the reporter gene expression (Fig. 7C, bar 1 versus bar 2), while knockdown of the NPM1 gene expression did not produce any change in the reporter gene expression (Fig. 7C, bar 3 versus bar 2). These results clearly demonstrate that the histone chaperone nucleophosmin induces transcription globally in vivo, probably through the disruption of the chromatin template.
DISCUSSION
The regulation of chromatin transcription is a complex phenomenon that involves the sequential remodeling of chromatin followed by the recruitment of the general transcription machinery to a targeted promoter (17, 35, 59). The critical event in this sequence is the remodeling step that results in a greater accessibility of chromatin, thereby promoting the formation of the preinitiation complex. A large number of factors that remodel chromatin have been defined. The common target for all these factors is the histones, which are either posttranslationally modified (acetylation, phosphorylation, methylation, etc.) (27, 32, 51) or rearranged or disrupted by a host of energy-dependent remodeling factors (30, 62). In this report, we introduce a member of the human histone chaperone family, nucleophosmin, as a novel chromatin-remodeling factor which is involved in the activation of chromatin transcription. Furthermore, we also report that this histone chaperone is functionally regulated by its acetylation, a posttranslational modification first reported in the case of histone chaperones.
Nucleophosmin, like other histone chaperones, is capable of interacting with the core histones. It interacts with both the H3-H4 tetramer and the H2A-H2B dimer concomitantly, resulting in the pulldown of the histone octamer. This is possible due to the fact that these two subspecies of the octamer bind to separate domains of nucleophosmin, as demonstrated in this report. A similar model has been proposed for Xenopus nucleoplasmin (6). Another interesting feature of the interaction map is that nucleophosmin does not interact with histone H2A, while it interacts with H3, H4, and H2B (Fig. 1C). It is on the basis of its interaction with H2B that nucleophosmin pulls down H2A from core histones (Fig. 1B). Domainwise mapping of the histone-interacting region reveals that the N-terminal hydrophobic oligomerization domain is responsible for binding to the H3-H4 tetramer, while the first acidic stretch is necessary for binding to H2B. This is similar to what has been observed in the case of the Saccharomyces cerevisiae Asf1 protein, wherein the acidic stretch has been shown to interact with H2A-H2B (58). But unlike nucleophosmin, this property was observed only when the N-terminal region of Asf1 was deleted. Most histone chaperones show a preference for either the H3-H4 tetramer (CAF1) (61) or the H2A-H2B dimer (NAP1) (references 21, 26, and 49 and references therein), but we could not detect any preference for either subspecies for nucleophosmin.
The C-terminal domain of nucleophosmin is unique and possesses the histone chaperone activity. It is interesting to note that the two related activities of histone interaction and histone transfer ability (histone chaperone) are located at opposing ends of the protein. Indeed, C-terminal truncations of nucleophosmin abrogate the chaperone activity while the histone interaction remains intact. It has been reported earlier that the C terminus of nucleophosmin possesses the nucleic acid-binding site (63). This observation leads to the speculation that nucleic acid binding could facilitate histone assembly. It is interesting to note that the existence of a nucleic acid-binding domain is not documented for other histone chaperones.
We have shown in this report that the human histone chaperone nucleophosmin can activate transcription from a chromatin template. Exogenous addition of nucleophosmin does not affect DNA transcription, suggesting that nucleophosmin targets the chromatin and not the general transcription factors for its action. This study employed a highly purified nonenzymatic chromatin assembly system in conjunction with an array template (34). With this system, we avoided the usage of other chromatin-remodeling complexes (ACF complex, etc.) to achieve the physiological spacing of the chromatin template. The 5S rDNA nucleosome-positioning sequence flanking the transcription cassette imposed a constraint for nucleosome spacing on the cassette, allowing for the assembly of around 3 or 4 nucleosomes. This system responds to the sequential addition of Gal4-VP16, p300, and acetyl-CoA to result in transcriptional activation. An interesting feature is the near absence of transcriptional activity in the absence of acetylation. The exogenous addition of nucleophosmin into this system produced a 2.5-fold increase in the transcript levels. The addition of C-terminal deletions of nucleophosmin did not show the characteristic enhancement of transcription (Fig. 3C), suggesting the importance of the chaperone activity.
Interestingly, we noticed a dependence on acetylation for the enhancement of chromatin transcription by nucleophosmin. Further probing into this phenomenon revealed that nucleophosmin is acetylated by p300, which is necessary for transcriptional activation. Several nonhistone proteins, particularly transcription factors, become acetylated. While acetylation generally produces an enhancement in the DNA-binding ability for some factors (e.g., p53 [22]), it may also decrease the affinity for DNA in some cases (YY1 [67] and HMG I/Y [57]). We have shown in this report that the acetylation of nucleophosmin results in enhanced affinity toward acetylated histones while demonstrating an increased chaperone activity with unacetylated histones compared to the unacetylated nucleophosmin. This is the first report wherein the acetylation of a histone chaperone enhances its affinity toward acetylated histones.
Recent reports have demonstrated that transcription from the chromatin template proceeds through a depletion of the histones on the promoter region (8, 11, 31). A genome-wide scan of nucleosome occupancy on the active regulatory regions in S. cerevisiae revealed a depletion of nucleosomes (39), though the factor(s) responsible for this depletion is still speculative. The yeast histone chaperone Asf1 has been demonstrated to deplete nucleosomes from the active chromatin region (1, 2). Interestingly, Asf1 interacts with the Bramha complex, an ATP-dependent chromatin-remodeling system involved in the process of transcriptional activation (44). In a previous report, NAP1 was also shown to facilitate the removal of the H2A-H2B dimer from the nucleosome (26). We have shown transcription activation by nucleophosmin in vivo by the activation of the p53-dependent reporter expression assay. Previously published data suggest that NPM1 enhances the p53 stability, which could be one of the causal effects on the observed enhancement in the transactivation by NPM1. We have established the fact that NPM1 is an activator of chromatin transcription. The fact that the increased stability of p53 or the inhibition of phosphorylation in the presence of NPM1 could cause the transcriptional activation as represented in Fig. 7 cannot be discounted, but the drop in the transactivation below the basal levels (i.e., in the presence of p53 alone) when cotransfected with the antisense construct of NPM1 cannot be explained by the same argument. If increased p53 stability were the only reason, transactivation in the presence of the NPM1 antisense construct would be the same as that of p53 alone. We therefore hypothesize that NPM1 may act as an important coactivator in p53-mediated transcriptional activation. In light of our data and the above presumption, the drop in the transactivation by p53 in the absence of NPM1 could be attributed to the lack of necessary histone chaperone function. However, in order to establish the global role of NPM1 to induce transcriptional activation, we have also used a different activator-driven reporter construct (Gal4-VP16 with pG10luc) (Fig. 7C). In agreement with our hypothesis, the results show a robust increase in the transactivation potential in the presence of NPM1. The cotransfection with the NPM1 antisense construct did not alter the reporter gene expression. Presumably, this occurs due to the presence of yet-unidentified factors that could replace the necessity of NPM1 for Gal4-VP16. Taken together, these data suggest that NPM1 indeed has a global role to play in the activation of transcription from a chromatin template in vivo.
Involvement of a histone chaperone in the activation of chromatin transcription, and which is regulated by acetylation, provides insight into the possible mechanism of action. It was demonstrated earlier that the recruitment of p300 by the activator brings about the promoter-proximal acetylation of the histones (34, 55, 59). In the event of the recruitment and subsequent acetylation of the histone chaperone nucleophosmin, histone removal over the promoter region may be facilitated due to the increased affinity among the modified proteins (nucleophosmin and histones), resulting in enhanced transcription. The necessity of histone chaperone activity in this process points to the fact that nucleophosmin does not act merely as a histone sink but may be actively involved in the process of histone depletion. These observations regarding the acetylation-dependent transcriptional activation by a histone chaperone experimentally establish histone chaperones as unique chromatin-remodeling machinery, which may play a significant role in the removal of nucleosomal histones in a replication-independent manner. By employing histone chaperones, cellular machineries may help in the replacement of the active chromatin-specific histone variant (H3.3 [43]) and/or remove methylated histones to regulate chromatin transcription.
Supplementary Material
Acknowledgments
This work was financially supported by CSIR, India, and JNCASR.
We thank J. Kadonaga, Y. Nakatani, and P. A. Cole for invaluable reagents and Dipankar Chaterjee for consent to use the MALDI-TOF instrument. V.S. is a Senior Research Fellow of CSIR.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 3.Allfrey, V. G., R. Faulkner, and A. E. Mirsky. 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51:786-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. [DOI] [PubMed] [Google Scholar]
- 5.Angelov, D., A. Verdel, W. An, V. Bondarenko, F. Hans, C. M. Doyen, V. M. Studitsky, A. Hamiche, R. G. Roeder, P. Bouvet, and S. Dimitrov. 2004. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 23:3815-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnan, C., N. Saperas, C. Prieto, M. Chiva, and J. Ausio. 2003. Interaction of nucleoplasmin with core histones. J. Biol. Chem. 278:31319-31324. [DOI] [PubMed] [Google Scholar]
- 7.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2003. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 9.Bonner, W. M., and R. A. Laskey. 1974. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur. J. Biochem. 46:83-88. [DOI] [PubMed] [Google Scholar]
- 10.Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379-390. [DOI] [PubMed] [Google Scholar]
- 11.Bruno, M., A. Flaus, C. Stockdale, C. Rencurel, H. Ferreira, and T. O. Hughes. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell 12:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catez, F., J. H. Lim, R. Hock, Y. V. Postnikov, and M. Bustin. 2003. HMGN dynamics and chromatin function. Biochem. Cell Biol. 81:113-122. [DOI] [PubMed] [Google Scholar]
- 13.Cecconi, D., A. Scarpa, M. Donadelli, M. Palmieri, M. Hamdan, H. Astner, and P. G. Righetti. 2003. Proteomic profiling of pancreatic ductal carcinoma cell lines treated with trichostatin-A. Electrophoresis 24:1871-1878. [DOI] [PubMed] [Google Scholar]
- 14.Chan, W.-Y., Q.-R. Liu, J. Borjigin, H. Busch, O. M. Rennert, L. A. Tease, and P.-K. Chan. 1989. Characterization of the c-DNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry 28:1033-1039. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., B. Li, and J. L. Workman. 1994. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 13:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo, E., J. C. Marine, D. Danovi, B. Falini, and P. G. Pelicci. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4:529-533. [DOI] [PubMed] [Google Scholar]
- 17.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 18.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8:841-853. [DOI] [PubMed] [Google Scholar]
- 19.Elgin, S. C. 1981. DNAase I-hypersensitive sites of chromatin. Cell 27:413-415. [DOI] [PubMed] [Google Scholar]
- 20.Fankhauser, C., E. Izaurralde, Y. Adachi, P. Wingfield, and U. K. Laemmli. 1991. Specific complex of human immunodeficiency virus type 1 Rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol. Cell. Biol. 11:2567-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii-Nakata, T., Y. Ishimi, A. Okuda, and A. Kikuchi. 1992. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem. 267:20980-20986. [PubMed] [Google Scholar]
- 22.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 23.Herrera, J. E., K. Sakaguchi, M. Bergel, L. Trieschmann, Y. Nakatani, and M. Bustin. 1999. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol. Cell. Biol. 19:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, W. H., B. Y. Yung, W. J. Syu, and Y. H. Lee. 2001. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 276:25166-25175. [DOI] [PubMed] [Google Scholar]
- 25.Inouye, C. J., and E. Seto. 1994. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nucleolar phosphoprotein B23. J. Biol. Chem. 269:6506-6510. [PubMed] [Google Scholar]
- 26.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300 mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 27.Jaskelloff, M., and C. L. Peterson. 2003. Chromatin and transcription: histones continue to make their marks. Nat. Cell Biol. 5:395-399. [DOI] [PubMed] [Google Scholar]
- 28.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B. Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827-830. [DOI] [PubMed] [Google Scholar]
- 29.Kiernan, R. E., C. Vanhulle, L. Schiltz, E. Adam, H. Xiao, F. Maudoux, C. Calomme, A. Burny, Y. Nakatani, K. T. Jeang, M. Benkirane, and C. Van Lint. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 18:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 31.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides, T. 1999. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9:40-48. [DOI] [PubMed] [Google Scholar]
- 33.Kumar, B. R., V. Swaminathan, S. Banerjee, and T. K. Kundu. 2001. p300-mediated acetylation of human transcriptional coactivator PC4 is inhibited by phosphorylation. J. Biol. Chem. 276:16804-16809. [DOI] [PubMed] [Google Scholar]
- 34.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 35.Kundu, T. K., Z. Wang, and R. G. Roeder. 1999. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 19:1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo, M. L., W. den Besten, D. Bertwistle, M. F. Roussel, and C. J. Sherr. 2004. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 18:1862-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurki, S., K. Peltonen, L. Latonen, M. T. Kiviharju, P. M. Ojala, D. Meek, and M. Laiho. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cells 5:465-475. [DOI] [PubMed] [Google Scholar]
- 38.Lau, D. O., A. D. Courtney, A. Vassilev, L. A. Marzilli, R. J. Cotter, Y. Nakatani, and P. A. Cole. 2000. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. J. Biol. Chem. 275:21953-21959. [DOI] [PubMed] [Google Scholar]
- 39.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depeletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 40.Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677:3-11. [DOI] [PubMed] [Google Scholar]
- 41.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 42.Luger, K., T. J. Rechsteiner, A. J. Flaus, M. M. Waye, and T. J. Richmond. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301-311. [DOI] [PubMed] [Google Scholar]
- 43.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munakata, T., N. Adachi, N. Yokoyama, T. Kuzuhara, and M. Horikoshi. 2000. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5:221-233. [DOI] [PubMed] [Google Scholar]
- 46.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 47.Nelson, T., T. S. Hsieh, and D. Brutlag. 1979. Extracts of Drosophila embryos mediate chromatin assembly in vitro. Proc. Natl. Acad. Sci. USA 76:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okuwaki, M., K. Matsumoto, M. Tsujimoto, and K. Nagata. 2001. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 506:272-276. [DOI] [PubMed] [Google Scholar]
- 49.Park, Y. J., J. V. Chodaparambil, Y. Bao, S. J. McBryant, and K. Luger. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280:1817-1825. [DOI] [PubMed] [Google Scholar]
- 50.Pasheva, E., M. Sarov, K. Bidjekov, I. Ugrinova, B. Sarg, H. Lindner, and I. G. Pashev. 2004. In vitro acetylation of HMGB-1 and -2 proteins by CBP: the role of the acidic tail. Biochemistry 43:2935-2940. [DOI] [PubMed] [Google Scholar]
- 51.Peterson, C. L., and M. C. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 52.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 53.Sims, R. J., S. S. Mandal, and D. Reinberg. 2004. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 16:263-271. [DOI] [PubMed] [Google Scholar]
- 54.Steen, H., A. Pandey, J. S. Anderson, and M. Mann. 2002. Analysis of tyrosine phosphorylation sites in signaling molecules by a phosphotyrosine-specific immonium ion scanning method. Science STKE 2002:P16. [DOI] [PubMed] [Google Scholar]
- 55.Steger, D. J., A. Eberharter, S. John, P. A. Grant, and J. L. Workman. 1998. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl. Acad. Sci. USA 95:12924-12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thanos, D., and T. Maniatis. 1992. The high mobility group protein HMG I (Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell 71:777-789. [DOI] [PubMed] [Google Scholar]
- 58.Umehara, T., T. Chimura, N. Ichikawa, and M. Horikoshi. 2002. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is function both in vivo and in vitro. Genes Cells 7:59-73. [DOI] [PubMed] [Google Scholar]
- 59.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 60.Verreault, A. 2000. De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 14:1430-1438. [PubMed] [Google Scholar]
- 61.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 62.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, D., A. Baumann, A. Szebeni, and M. O. Olson. 1994. The nucleic acid binding activity of nucleolar protein B23.1 resides in its carboxyl-terminal end. J. Biol. Chem. 269:30994-30998. [PubMed] [Google Scholar]
- 64.Wolffe, A. 1998. Chromatin: structure and function, 3rd ed. Academic Press, San Diego, Calif.
- 65.Workman, J. L., and S. M. Abmayr. 2004. Histone H3 variants and modifications on transcribed genes. Proc. Natl. Acad. Sci. USA 101:1429-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, X. J. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32:959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao, Y.-L., W.-M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.