Abstract
The ELL family of proteins function in vitro as elongation factors for RNA polymerase II. Deletion studies have defined domains in mammalian ELL required for transcription elongation activity and RNA polymerase binding in vitro, for transformation of cultured cells when overexpressed, and for leukemogenesis and cell proliferation as part of a leukemic fusion protein. The goal of this study was to identify domains required for chromosome targeting and viability in the unique Drosophila ELL (dELL) protein. Here, we show that an N-terminal domain of dELL is necessary and sufficient for targeting to transcriptionally active puff sites in chromatin, supporting a role for this domain in recruiting dELL to elongating RNA polymerase II. We demonstrate that a central domain of dELL is required for rapid mobilization of ELL during the heat shock response, suggesting a regulatory function for this domain. Unexpectedly, transgenic dELL in which the N-terminal chromosome binding domain is deleted can complement the recessive lethality of mutations in ELL, suggesting that Drosophila ELL has an essential activity in development distinct from its role as an RNA polymerase II elongation factor.
The human ELL gene was originally identified as a translocation partner with the MLL (mixed-lineage leukemia) gene in patients with acute leukemia (26). Subsequent work showed that the ELL family of elongation factors stimulates polymerase II (Pol II) elongation in vitro by increasing the Vmax of Pol II (17, 22, 24). The mechanistic role of ELL in the pathogenesis of leukemia mediated by the MLL-ELL fusion protein remains elusive.
Several domains within mammalian ELL have been identified. The Pol II binding domain and the domain required for the in vitro elongation activity of ELL map to the amino-terminal 373 amino acids (23). A domain that inhibits the initiation of transcription maps within the first 60 amino acids. A lysine-rich domain spans amino acids 447 to 465, and this domain is required for the ability of ELL to transform Rat1 fibroblasts (8). A region homologous to the ZO-1 binding domain of occludin, an accessory protein of tight junctions, maps to amino acids 521 to 616. A C-terminal domain containing this occludin homology is necessary and sufficient to prolong the proliferation of primary mouse myeloid precursors in culture and for leukemogenesis in a mouse model (2, 15). This conserved domain also mediates transcriptional activation in a transfection assay (2). In cells expressing the MLL-ELL fusion protein, p53 is sequestered together with the fusion protein in discrete nuclear foci, an association that depends on the ELL C-terminal occludin homology domain (amino acids 341 to 621) (27). The MLL-ELL fusion protein inhibits the transcription activation activity of p53 in transfected cells, and this inhibition also depends on the same ELL C-terminal domain (27). Thus, functional inactivation of p53 by the MLL-ELL fusion protein could account for the role of MLL-ELL in leukemia. It also suggests that ELL could link DNA damage sensing to transcription.
Recently, we described the unique ELL protein in Drosophila melanogaster, dELL (5). Like its mammalian counterparts, dELL stimulates Pol II elongation in vitro. dELL colocalizes with Pol II at transcriptionally active loci on Drosophila polytene chromosomes and interacts with Pol II in fly extracts. Further, dELL is rapidly mobilized to heat shock loci with phosphorylated Pol II upon heat shock induction. These observations are consistent with a role for dELL as a Pol II elongation factor in vivo.
The gene encoding dELL is essential in Drosophila, and mutations in this gene result in embryonic lethality accompanied by extensive segmentation defects (3). Additionally, dELL mutations appear to preferentially enhance hypomorphic mutations of large genes (3). Whether the essential requirement for dELL in development is related to its role as a Pol II elongation factor is unknown.
To test the functional requirements of conserved domains of the dELL protein, we have taken two approaches. In the first, we molecularly characterized mutant alleles of Su(Tpl), the gene encoding dELL, and compared the effects of different alleles on Ras pathway regulation. In the second approach, we performed in vivo structure-function studies using transgenic flies. Here, we show that the N-terminal domain of dELL is both necessary and sufficient for recruitment to chromatin. We identify an internal region of dELL (amino acids 190 to 759) that is required for efficient mobilization of dELL from developmental genes to heat shock loci after heat shock. Surprisingly, we find that the N-terminal domain is dispensable in order to complement the recessive lethality of dELL mutations. These observations suggest that the essential function of dELL for development may be separable from its role as a Pol II elongation factor.
MATERIALS AND METHODS
Drosophila stocks.
SY3-1BK652, SY3-1DV1050 and SY3-1CS653 were obtained from I. Rebay (Massachusetts Institute of Technology); l (3)76BDs3, SR3-4AXS-485, SR3-4AXS-457, and SR3-4AXS-574 were obtained from J. Kennison (National Institute of Child Health and Human Development). Transgenic flies expressing a six-His epitope-tagged wild-type dELL have been previously described (3). Transgenic flies expressing six-His-tagged dELL deletions in a y1w67c23 background were generated by P-element-mediated germ line transformation. All stocks were maintained on cornmeal-molasses medium.
Sequence analysis of Su(Tpl) alleles.
Genomic DNA was prepared from each heterozygous mutant stock, and the entire dELL open reading frame was amplified as previously described (3). PCR products were purified by a High Pure PCR product purification kit (Roche) and sequenced by a commercial sequencing service (Retrogen).
Plasmid construction.
For recombinant expression, dELL mutant cDNAs were cloned from the full-length cDNA by PCR. Using nested restriction sites (BglII at the 5′ end and NcoI at the 3′ end), the resulting products were inserted into the pRSETb expression vector (Invitrogen), downstream of the in-frame N-terminal six-His tag. dELL(Δ190-759) was generated using two-step PCR. dELL(1-262) in pRSETb was generated during cloning of full-length dELL by introduction of a frameshift resulting in a nonsense mutation.
For transgene constructs, epitope-tagged dELL(Δ1-189), dELL(Δ190-759), and dELL(Δ760-1059) were excised from pRSETb as an NdeI-EcoRI restriction fragment, and both ends were blunted by the Klenow fragment. This restriction fragment was then ligated to SmaI-digested BGΔ26S vector (gift of D. Dorsett, Saint Louis University School of Medicine), which contained the promoter and first intron of the Drosophila Chip gene upstream of the insertion site. Orientation of the insert was confirmed by sequencing, and the resulting construct was digested with EcoRI and NotI to release the promoter, cDNA, and accompanying epitope tags. This fragment was then ligated to the NotI-EcoRI fragment of the pCaSpeRN vector (provided by D. Dorsett). These constructs were then used to generate transgenic flies. The dELL(1-262) cDNA and accompanying epitope tags were excised from pRSETb as an NdeI-EcoRI restriction fragment, with the NdeI end blunted by the Klenow fragment. This restriction fragment was then ligated to the EcoRI-SalI fragment of the HIC-L vector (13), in which the SalI end was also blunted by the Klenow fragment. The resulting construct places the dELL cDNA and accompanying epitope tags downstream of the Hsp70 promoter. Subsequently, the entire cDNA, tags, and promoter sequence were excised from HIC-L as an NotI restriction fragment and ligated into an NotI-restricted pYC1.8 vector (4). Transgenic v36F ry506 larvae carrying this construct were used for polytene chromosome staining after a 30-min heat shock and 2-h recovery at room temperature.
Polytene chromosome staining.
Salivary glands were fixed for 30 s in 2% formaldehyde and then in 45% acetic acid-2% formaldehyde for 3 min as previously described (14) before storage in 67% glycerol-33% phosphate-buffered saline at −20°C. For optimal staining, anti-six-His polyclonal serum (Santa Cruz) (1:300 dilution), dELL polyclonal serum (1:300 dilution), or phospho-Pol II antibody (H5; Covance) (1:500 dilution) was incubated with polytene chromosomes overnight. Appropriate secondary antibodies (Jackson Laboratory) were used at a 1:1,000 dilution, except in the experiment shown in Fig. 2D, where the fluorescein isothiocyanate-conjugated donkey anti-rabbit (Sigma) antibody was used at a dilution of 1:200. Fluorescence detection (except for Fig. 2D) was by epifluorescence using an Olympus BX60 fluorescence microscope with an NB barrier filter for Cy2 detection, and a WG barrier filter for Cy3. Images were recorded with a SPOT charge-coupled-device camera (Diagnostic Instruments, Inc.) using PAX-it imaging software (Midwest Information Systems, Inc.). For Fig. 2D, fluorescence detection was done using a Nikon Microphot microscope equipped with a digital Qimaging Microimager II camera and Northern Eclipse software.
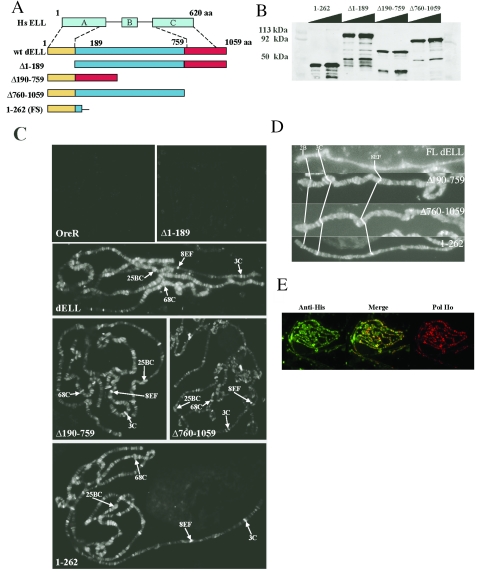
FIG. 2.
The N-terminal domain of dELL is necessary and sufficient for localization to chromatin in vivo. (A) Schematic of dELL wild-type and deletion transgenes used in this study. Yellow indicates the conserved N-terminal domain, and red indicates the C-terminal domain. At top is a schematic of the mammalian ELL protein (Hs ELL), with blocks depicting the characterized domains in the protein sequence as follows: A, N-terminal Pol II binding and elongation activation domain; B, lysine-rich region; C, occludin homology domain. (B) Recombinant dELL mutants were expressed in E. coli and analyzed by Western blotting with anti-dELL polyclonal serum to confirm predicted molecular sizes: dELL(1-262), 42 kDa; dELL(Δ1-189), 95 kDa; dELL(Δ190-759), 56 kDa; and dELL(Δ760-1059), 80 kDa. (C) Immunolocalization pattern for dELL mutants on polytene chromosomes. dELL(Δ1-189) fails to localize to chromosomes in vivo (compare to nontransgenic OreR), while dELL(1-262) exhibits a localization similar to full-length dELL and both dELL(Δ760-1059) and dELL(Δ190-759). Landmark sites on the X (3C and 8EF), second(25BC), and third (68C) chromosomes are indicated with arrows to facilitate comparisons. (D) Distribution of dELL deletion mutant proteins is identical to full-length dELL on the X chromosome. Relative positions of the weakly staining landmark puff at 2B, the strongly staining bands at 3C, and 8EF are indicated in X chromosomes from each transgenic line showing chromosome targeting of dELL. (E) dELL(1-262) colocalizes with phosphorylated Pol II on polytene chromosomes. Chromosomes from transgenic flies expressing dELL(1-262) were fixed and stained with anti-six-His and anti-Pol IIo (H5; Covance). Images were merged to visualize extent of colocalization.
Whole-mount embryo staining.
Embryos were dechorionated and fixed in 4% formaldehyde and stained with anti-six-His polyclonal (Santa Cruz) or phospho-Pol II-specific monoclonal antibody (H5; Covance). Appropriate secondary antibodies [Cy2 for dELL(Δ1-189) and Cy3 for Pol II; Jackson Laboratory] were used at a 1:1,000 dilution, and images were recorded with a SPOT charge-coupled-device camera (Diagnostic Instruments, Inc.) using PAX-it imaging software (Midwest Information Systems, Inc.).
RESULTS
In an effort to identify functional domains of dELL, we have taken two approaches. In the first, we have molecularly characterized mutations in the dELL gene that were identified in several independent genetic screens. In the second, we have engineered specific deletion mutations affecting domains homologous to mammalian domains that were previously associated with biochemical or oncogenic properties in the mammalian ELL protein.
Mutations in Su(Tpl) alleles implicate C-terminal sequences in the essential function of dELL.
Several different mutational screens have yielded mutations in Su(Tpl), the locus that encodes dELL. These include screens for suppressors of the triplolethality of Tpl (20), recessive lethal mutations in the 76D region (11; J. Kennison, personal communication), and mutations affecting the Ras signaling pathway during eye development (10, 16, 18, 19). We previously reported sequence analysis of six alleles that were recovered in these screens (3).
For this study, we obtained and sequenced seven additional mutant alleles of Su(Tpl). All of these alleles suppress the triplolethal phenotype of Tpl (data not shown). The position and nature of each mutational lesion are shown in Fig. 1A. Each mutation results in a truncation of the dELL protein that leaves intact most or all of the domain that shares homology with the Pol II elongation domain of mammalian ELL.
FIG. 1.
(A) Sites of mutations in Su(Tpl) alleles. Mutational lesions are indicated in red above the wild-type amino acid sequence of dELL at the site of the mutation. For frameshift alleles, the resulting out-of-frame peptide is also shown. (B) Effects of Su(Tpl) alleles on sev-Ras1v12 (top) and GMR-yanACT (bottom). The dELL polypeptide is represented by a horizontal bar. Filled intervals represent conserved homology to the N-terminal ELL elongation domain and the C-terminal ZO-1 occludin binding domain. Alleles with an enhancer effect are shown in red, alleles with a suppressor effect are shown in blue, and alleles with no apparent effect are shown in black.
Previous studies using mammalian ELL demonstrated that an N-terminal domain lying between amino acids 50 and 200 is necessary for enhancing RNA Pol II elongation activity in vitro (23). To test whether any of the C-terminally truncated dELL mutants still retain activity, we crossed each allele, together with alleles characterized in our previous study (3), to stocks carrying transgenes that either express a constitutively active Ras1 in the eye (sev-Ras1v12) (10) or overexpress the Ras pathway inhibitor aop (yan) in the eye (GMR-yanACT) (19). For 6 of the 12 alleles tested, a reciprocal effect was observed: alleles that act as dominant suppressors of sev-Ras1v12 act as dominant enhancers of GMR-yanACT and vice versa (Fig. 1B). If we assume that the Su(Tpl)s2 allele, which truncates dELL at 51 amino acids, is a null mutation, then null alleles of dELL are enhancers of the ras1 hyperactivation phenotype and suppressors of the reduced eye phenotype caused by the GMR-yanACT transgene. Thus, the Su(Tpl)XS457 and Su(Tpl)547 alleles behave as nulls in these assays. On the other hand, the Su(Tpl)CS653, Su(Tpl)10, Su(Tpl)XS485, and Su(Tpl)S-192 alleles behave oppositely, suggesting that these are not null alleles, at least with respect to the Ras pathway in the eye.
Nevertheless, all dELL alleles that we have characterized to date are recessive lethal. In some cases, this may be due to mutations that result in an unstable protein product, which would not be informative concerning domain function. The genetic behavior of the Su(Tpl)CS653, Su(Tpl)10, Su(Tpl)XS485, and Su(Tpl)S-192 alleles suggests that they express a functional gene product, indicating that the mutational lesion affects an essential property of the protein. The structures of the dELL mutations we have analyzed suggest the hypothesis that sequences near the C terminus of dELL confer the essential properties observed for this protein. Consistent with this hypothesis, the Su(Tpl)S-192 allele was previously shown to have two amino acid substitutions at two highly conserved residues within the C-terminal domain (3).
An N-terminal domain of dELL is necessary and sufficient for localization to chromatin in vivo.
To test whether a specific domain of dELL is required for chromosome targeting in polytene chromosomes, we generated transgenic flies expressing six-His-tagged engineered deletion mutants of dELL that correspond, based on sequence homology, to the characterized domains of mammalian ELL (Fig. 2A). An N-terminal deletion removed the first 189 amino acids of dELL, which contain homology to the ELL Pol II inhibition domain and most of the sequence previously shown to be necessary for Pol II elongation in vitro (23). A C-terminal deletion removed the last 300 amino acids of dELL, which contain homology to the ELL sequences implicated in leukemogenesis (2, 15), to the minimal p53 binding sequence of ELL (27), and to the ZO-1 binding domain of occludin. An internal deletion removed amino acids 190 to 759 that lie between the N- and C-terminal domains described above and that contain homology to the domain required for the transformation of Rat1 fibroblasts by ELL (8). An independently isolated deletion that expresses only the N-terminal 262 amino acids of dELL was also tested; this peptide contains homology to the ELL Pol II inhibition domain and all of the sequence previously shown to be necessary for Pol II elongation in vitro (23).
Mutant proteins were expressed as recombinant proteins in Escherichia coli and analyzed by Western blotting with polyclonal antibodies to wild-type dELL. As shown in Fig. 2B, each of the recombinant proteins is of the expected size. These cDNAs were used to generate transgenic lines expressing truncated dELL proteins as six-His-tagged proteins under control of the Chip gene promoter, a strategy that we used previously to generate transgenic lines expressing full-length dELL as an epitope-tagged protein (3). Polytene chromosomes were prepared from the salivary glands of transgenic third instar larvae expressing either full-length dELL, a deletion of the conserved N-terminal sequences [dELL(Δ1-189)], a deletion of the sequences between the conserved N- and C-terminal domains [dELL(Δ190-759)], or a deletion of the conserved C-terminal sequences [dELL(Δ760-1059)]. Transgenic proteins were localized using anti-six-His serum. As shown in Fig. 2C, deletion of the N-terminal domain (amino acids 1 to 189) completely abolishes binding of dELL to chromatin in vivo. In contrast, deletion of either the C-terminal (amino acids 760 to 1059) or the internal (amino acids 190 to 759) domain has no detectable effect on the localization or distribution of these proteins on chromosomes. Detailed comparison of staining on the X chromosome demonstrates identical localization of full-length dELL and dELL mutant proteins (Fig. 2D). Furthermore, a transgenic protein including only an N-terminal domain (amino acids 1 to 262) is also recruited to dELL binding sites in chromatin (Fig. 2C, lower right panel), and is colocalized with full-length dELL on the X chromosome (Fig. 2D, bottom panel). In addition, the transgenic protein consisting of amino acids 1 to 262 colocalizes with serine 2 hyperphosphorylated RNA Pol II (Pol IIo) on chromosomes, similar to wild-type dELL (Fig. 2E).
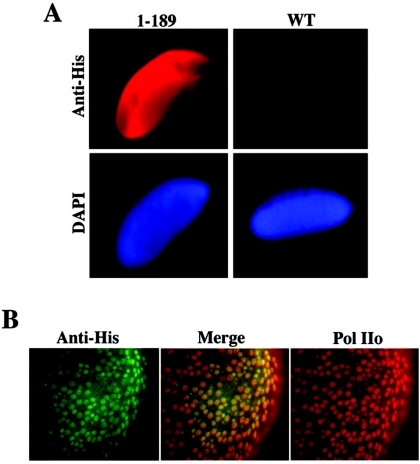
To rule out the possibility that the lack of chromosome targeting of the dELL(Δ1-189) protein is due to lack of expression of the transgene, we performed whole-mount embryo staining with embryos from that transgenic line. As shown in Fig. 3A, this protein is clearly expressed in embryos carrying the transgene, evidenced by anti-His staining. Further, the dELLΔ1-189 mutant colocalizes with phosphorylated RNA Pol II in the nuclei of transgenic embryos (Fig. 3B), demonstrating that removal of the N-terminal domain of dELL does not prevent expression or nuclear localization but only impairs chromosome binding. Taken together, these results identify an N-terminal domain that is both necessary and sufficient to target dELL to active chromatin in vivo, confirming the in vitro studies indicating that this domain of mammalian ELL was required for interaction and increasing the catalytic rate of transcription elongation by RNA Pol II.
FIG. 3.
dELL(Δ1-189) is expressed and localized to the nucleus in Drosophila embryos. (A) Transgenic embryos expressing dELL(Δ1-189) or nontransgenic Oregon R embryos were fixed and stained with anti-six-His and DAPI (4′,6′-diamidino-2-phenylindole). Staining was visualized by fluorescent microscopy. (B) To confirm nuclear localization, embryos were fixed and immunostained with anti-six-His serum and anti-Pol IIo serum (H14; Covance). Merging of these images confirms that the dELL(Δ1-189) is a nuclear protein, since it colocalizes extensively with Pol IIo.
An internal domain of dELL is required for dissociation from non-heat shock loci upon heat shock.
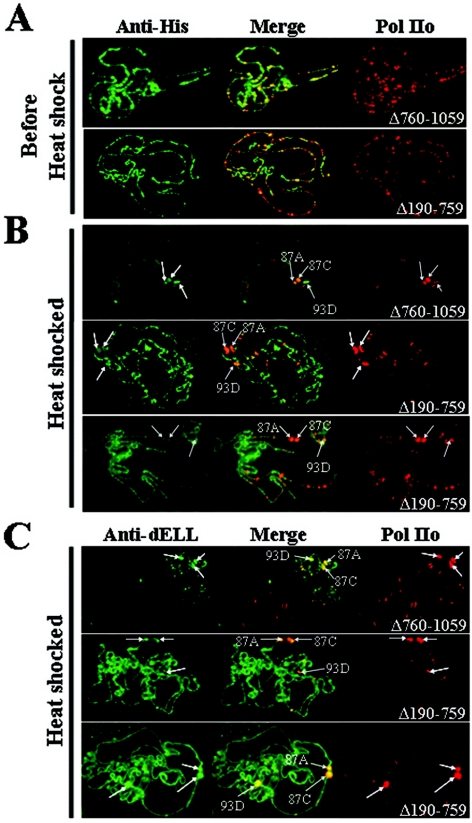
We previously showed that endogenous dELL and transgenic dELL each extensively colocalize on polytene chromosomes with RNA Pol II phosphorylated at serine 5 of the C-terminal domain heptad repeats (5). Figure 4A demonstrates a similar pattern of colocalization for both dELL(Δ190-759) and dELL(Δ760-1059), using antibody specific for phosphorylated serine 2 of the Pol II C-terminal domain, thought to be the elongating form of Pol II (12). Thus, both mutant proteins appear to retain their targeting to sites where actively elongating Pol II is present.
FIG. 4.
dELL(Δ190-759) is retained at developmental loci upon heat shock. (A) Both dELL(Δ760-1059) and dELL(Δ190-759) colocalize with phosphorylated Pol II under non-heat shock conditions. Chromosomes from transgenic flies expressing dELL(Δ760-1059) and dELL(Δ190-759) were fixed and stained with anti-six-His (left side) and anti-Pol IIo (H5; Covance) (right side). Images were merged to determine the extent of colocalization. (B) dELL(Δ190-759) is retained at developmental loci after heat shock. After a 20-min heat shock, chromosomes were preparedand immunostained as above. Unlike dELL(Δ760-1059), dELL(Δ190-759) retains its non-heat shock distribution after heat shock. Major heat shock puff sites are labeled with arrows. (C) Endogenous dELL is still recruited to heat shock puffs in transgenic flies expressing the dELL(Δ190-759) mutant. Chromosomes were prepared as described for panel B, except that anti-dELL polyclonal serum was used rather than anti-six-His serum. In these images, both the widely distributed pattern of localization for dELL(Δ190-759) and the heat shock puff staining observed for wild-type dELL are shown. Major heat shock puff sites are labeled with arrows.
After a brief heat shock, endogenous dELL disappears from most chromosomal sites and is rapidly recruited to the sites of heat shock genes (5). As expected, similar mobilization occurs when wild-type dELL is expressed from a transgene (data not shown). The dELL(Δ760-1059) mutant, which is missing the C terminus, shows an identical redistribution after heat shock (Fig. 4B, upper panel), colocalizing with phosphorylated Pol II. As an additional control, we verified that the transgenic protein follows the endogenous protein by immunostaining with polyclonal antibody to dELL, which detects both proteins (Fig. 4C, upper panel). Thus, the C-terminal occludin homology domain of dELL does not appear to contribute to the redistribution of dELL during heat shock.
Surprisingly, while dELL(Δ190-759) colocalizes with phosphorylated Pol II prior to heat shock (Fig. 4A, lower panels), it remains widely distributed after heat shock induction (Fig. 4B, lower panels), in contrast to both full-length dELL and dELL(Δ760-1059). Phosphorylated Pol II is recruited to heat shock puffs, indicating that the defect seen with the transgenic protein is not due to a failure of Pol II to mobilize to heat shock loci. Furthermore, when these transgenic chromosomes are stained with anti-dELL antibody, a combination of the expected pattern for wild-type dELL (robust mobilization to heat shock puffs) and the pattern observed for dELL(Δ190-759) is seen (Fig. 4C, lower panels). In this case, not only are the chromosomes widely decorated [presumably by the transgenic protein, dELL(Δ190-759)], but the heat shock puff sites are more intensely stained, indicating the presence of endogenous dELL at these loci. Taken together, these data imply that the region of amino acids 190 to 759 of dELL contains a domain that is required for mobilization with Pol II upon heat shock. It also suggests that dELL can bind to chromosomes through interactions independent of Pol II.
The N-terminal domain of dELL is dispensable for genetic complementation of lethal dELL mutations.
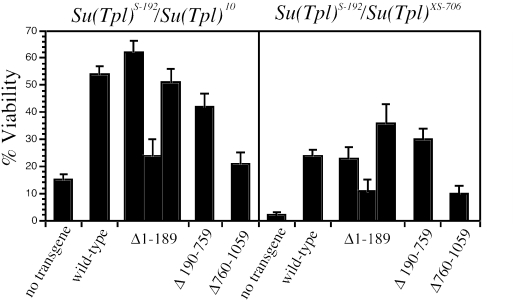
dELL is encoded by the essential gene Suppressor of Triplolethal [Su(Tpl)] in Drosophila, and mutations in dELL are recessive lethal during the late stages of embryogenesis (3). A transgene expressing dELL under the Chip promoter can partially complement this lethality (3). To test the contribution of each of the three functional domains of dELL to the complementation of lethal dELL mutations, we set up crosses to generate semilethal combinations of Su(Tpl) alleles in the presence or absence of dELL deletion mutant transgenes. By choosing allelic combinations of Su(Tpl) that yield some escapers even without a transgene, we could detect weak complementation that might be missed with fully penetrant lethal alleles. Moreover, this design could also detect dominant-negative activity of any deletions.
Surprisingly, deletion of the N-terminal elongation domain homology has little or no effect on the ability of transgenic dELL to complement dELL mutations using two independent transgenic lines in two different heteroallelic combinations of Su(Tpl) alleles (Fig. 5). Similar complementation is observed when amino acids 190 to 759 are deleted, suggesting that the sequences lying between the N- and C-terminal domains are also dispensable for viability. In contrast, the efficiency of complementation by dELL(Δ760-1059) is significantly lower, comparable to crosses lacking any transgene (Fig. 5). Note that the level of dELL(Δ760-1059) present on polytene chromosomes is comparable to the levels observed for full-length transgenic dELL and dELL(Δ190-759), both of which exhibit significant complementation in both heteroallelic combinations of Su(Tpl) used. While it is possible that the efficiency of transgene rescue is quantitatively affected by variation in the levels of expression among the individual transgenes, our data suggest that the N-terminal elongation homology and central domains of dELL are dispensable for the essential function(s) of dELL in development.
FIG. 5.
Transgene rescue of Su(Tpl) lethality. Rescue of flies carrying two different heteroallelic combinations of Su(Tpl) mutants was measured using dELL wild-type and mutant transgenes. Percent viability is computed as the percentage of expected Mendelian survival. Results for three independent transgene inserts are shown for the dELL(Δ1-189) construct. For each genotype, the number of flies scored ranged from 199 to 1,559. The upper 95% confidence interval is indicated above each bar.
DISCUSSION
The goal of this study was to identify functional domains in the ELL family of Pol II elongation factors. Structure-function analysis of mammalian ELL identified an N-terminal domain that is important for Pol II binding and for stimulating Pol II elongation in vitro (23). Consistent with these observations, we find that a homologous N-terminal domain is necessary and sufficient for the recruitment of dELL to sites of elongating Pol II on polytene chromosomes. These are the first studies to implicate the conserved N-terminal domain of an ELL family protein in Pol II transcription in vivo, lending strong support to the in vitro data defining ELL family proteins as Pol II elongation factors. In light of a previous report that purified mammalian ELL interacts with purified Pol II (23), the simplest model to explain our observations is that dELL binds directly to Pol II via its conserved N-terminal domain.
An internal domain regulates dELL localization after heat shock.
In Drosophila, heat shock results in a dramatic reduction in developmental gene transcription, accompanied by intense transcriptional activation of heat shock genes and the recruitment of Pol II and a number of Pol II elongation factors, to heat shock puff sites in polytene chromosomes (1, 5, 6, 9, 14, 21). Endogenous dELL quickly disappears from most chromosomal sites after heat shock, and it is rapidly recruited together with phosphorylated Pol II to major heat shock puffs (5). This mobilization also occurs with the dELL(Δ760-1059) mutant but, surprisingly, not with the dELL(Δ190-759) mutant, which remains associated with developmental loci after Pol II has largely been lost from these sites. This suggests that while the interaction between dELL and Pol II is dependent upon the N-terminal domain, it is also regulated by an internal domain of dELL. One explanation for this result may be that the internal domain of dELL regulates binding of dELL to transcription factors bound at a variety of developmental genes, and this interaction is disrupted upon heat shock. Thus, the dELL(Δ190-759) protein, lacking this regulatory domain, remains bound to developmental loci after heat shock even as Pol II is lost from these loci.
The C-terminal domain of dELL is essential in normal development.
The C-terminal occludin homology domain of mammalian ELL (corresponding to amino acids 839 to 928 in dELL) has been implicated in a number of interactions with potential physiological significance. It is the most highly conserved region of the dELL primary sequence, and it is necessary and sufficient for oncogenic activity in the context of the MLL-ELL fusion protein (2, 15). It has also been implicated in binding the tumor suppressor p53 (25, 27) and in the regulation of cell growth and proliferation (7). In this study, we show that this domain is dispensable for chromosome binding and for recruitment of dELL to heat shock loci. Nevertheless, deletion of this domain results in proteins with little or no ability to complement the recessive lethality of hypomorphic Su(Tpl) alleles. While the exact mechanism of the essential requirement for the ELL C-terminal domain remains unknown, it may be significant that deletions of—and point mutations in—the C-terminal domain were uncovered in four separate screens as dominant suppressors of Ras pathway activation. Perhaps this domain provides a mechanistic link between Ras signaling and transcription.
In light of the requirement for the N-terminal domain for dELL chromosome binding, it was unanticipated that an N-terminal domain deletion could complement the recessive lethality of Su(Tpl) mutations. One obvious interpretation of this result is that the recessive lethality of Su(Tpl) mutations is not due to defects in transcription elongation for one or more essential genes but that the essential requirement for dELL in development lies in a distinct dELL-dependent pathway. dELL is one of several elongation factor homologues in Drosophila, and it is possible that the role of dELL in transcription elongation is functionally redundant to these other factors.
Another possibility is that N-terminally truncated dELL functionally complements the Su(Tpl)S-192 mutant protein, which is present in both complementation assays and which has an intact N-terminal domain but carries missense mutations in the C-terminal domain. This mechanism would suggest that multiple dELL molecules participate in functional protein complexes. Nevertheless, such complexes are presumably extrachromosomal, since the N-terminally truncated dELL does not bind chromosomes even in the presence of wild-type dELL. Future studies aimed at defining the biochemistry of ELL-Pol II complexes should help sort out the mechanism for this genetic interaction.
Acknowledgments
We thank I. Rebay and J. Kennison for fly stocks, D. Michener for help with DNA sequencing sample preparation, and J. Kennison and D. Dorsett for helpful advice and encouragement.
This work was supported by NSF grant MCB 0131414 (J.C.E.), ACS grant RP69921801 (A.S.), NIH grant 1R01CA089455 (A.S.), and a Mallinckrodt Foundation Award to A.S. A.S. is a Scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiMartino, J. F., T. Miller, P. M. Ayton, T. Landewe, J. L. Hess, M. L. Cleary, and A. Shilatifard. 2000. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood 96:3887-3893. [PubMed] [Google Scholar]
- 3.Eissenberg, J. C., J. Ma, M. A. Gerber, A. Christensen, J. A. Kennison, and A. Shilatifard. 2002. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc. Natl. Acad. Sci. USA 99:9894-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fridell, Y. W., and L. L. Searles. 1991. Vermilion as a small selectable marker gene for Drosophila transformation. Nucleic Acids Res. 19:5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber, M., J. Ma, K. Dean, J. C. Eissenberg, and A. Shilatifard. 2001. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J. 20:6104-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber, M., K. Tenny, J. W. Conaway, R. Conaway, J. Eissenberg, and A. Shilatifard. 2005. Regulation of heat shock gene expression by RNA polymerase II elongation factor, elongin A. J. Biol. Chem. 280:4017-4020. [DOI] [PubMed] [Google Scholar]
- 7.Johnstone, R. W., M. Gerber, T. Landewe, A. Tollefson, W. S. Wold, and A. Shilatifard. 2001. Functional analysis of the leukemia protein ELL: evidence for a role in the regulation of cell growth and survival. Mol. Cell. Biol. 21:1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanda, Y., K. Mitani, M. Kurokawa, T. Yamagata, Y. Yazaki, and H. Hirai. 1998. Overexpression of the MEN/ELL protein, an RNA polymerase II elongation factor, results in transformation of Rat1 cells with dependence on the lysine-rich region. J. Biol. Chem. 273:5248-5252. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan, C. D., J. R. Morris, C.-T. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karim, F. D., H. C. Chang, M. Therrien, D. A. Wassarman, T. Laverty, and G. M. Rubin. 1996. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143:315-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. U. R. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- 12.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus, K. W., Y. H. Lee, J. T. Lis, and M. F. Wolfner. 1988. Sex-specific control of Drosophila melanogaster yolk protein 1 gene expression is limited to transcription. Mol. Cell. Biol. 8:4756-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792-803. [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, R. T., C. Lavau, C. Du, F. Simone, P. E. Polak, S. Kawamata, and M. J. Thirman. 2001. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol. Cell. Biol. 21:5678-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxiner, A., T. P. Hecker, Q. N. Phan, and D. A. Wassarman. 1998. A screen for mutations that prevent lethality caused by expression of activated sevenless and Ras1 in the Drosophila embryo. Dev. Genetics. 23:347-361. [DOI] [PubMed] [Google Scholar]
- 17.Miller, T., K. Williams, R. W. Johnstone, and A. Shilatifard. 2000. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J. Biol. Chem. 275:32052-32056. [DOI] [PubMed] [Google Scholar]
- 18.Neufeld, T. P., A. H. Tang, and G. M. Rubin. 1998. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebay, I., F. Chen, F. Hsiao, P. A. Kolodziej, B. H. Kuang, T. Laverty, C. Suh, M. Voas, A. Williams, and G. M. Rubin. 2000. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154:695-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roehrdanz, R. L., and J. C. Lucchesi. 1980. Mutational events in the triplo- and haplo-lethal region (83DE) of the Drosophila melanogaster genome. Genetics 95:355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. T. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301:1094-1096. [DOI] [PubMed] [Google Scholar]
- 22.Shilatifard, A., D. R. Duan, D. Haque, C. Florence, W. H. Schubach, J. W. Conaway, and R. C. Conaway. 1997. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc. Natl. Acad. Sci. USA 94:3639-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shilatifard, A., D. Haque, R. C. Conaway, and J. W. Conaway. 1997. Structure and function of RNA polymerase II elongation factor ELL. Identification of two overlapping ELL functional domains that govern its interaction with polymerase and the ternary elongation complex. J. Biol. Chem. 272:22355-22363. [DOI] [PubMed] [Google Scholar]
- 24.Shilatifard, A., W. S. Lane, K. W. Jackson, R. C. Conaway, and J. W. Conaway. 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271:1873-1876. [DOI] [PubMed] [Google Scholar]
- 25.Shinobu, N., T. Maeda, T. Aso, T. Ito, T. Kondo, K. Koike, and M. Hatakeyama. 1999. Physical interaction and functional antagonism between the RNA polymerase II elongation factor ELL and p53. J. Biol. Chem. 274:17003-17010. [DOI] [PubMed] [Google Scholar]
- 26.Thirman, M. J., D. A. Levitan, H. Kobayashi, M. C. Simon, and J. D. Rowley. 1994. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 91:12110-12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiederschain, D., H. Kawai, J. Gu, A. Shilatifard, and Z.-M. Yuan. 2003. Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL. Mol. Cell. Biol. 23:4230-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]