Abstract
Human immunodeficiency virus type 1 (HIV-1) is able to establish a latent infection during which the integrated provirus remains transcriptionally silent. In response to specific stimuli, the HIV-1 long terminal repeat (LTR) is highly activated, enhancing both transcriptional initiation and elongation. Here, we have identified a specific binding sequence of the nuclear NF-κB-repressing factor (NRF) within the HIV-1 LTR. The aim of this work was to define the role of NRF in regulating the LTR. Our data show that the endogenous NRF is required for transcriptional activation of the HIV-1 LTR in stimulated cells. In unstimulated cells, however, NRF inhibits HIV-1 LTR activity at the level of transcription elongation. Binding of NRF to the LTR in unstimulated cells prevents recruitment of elongation factor DRB sensitivity-inducing factor and formation of processive elongation complexes by hyperphosphorylated RNA polymerase II. Our data suggest that NRF interrupts the regulatory coupling of LTR binding factors and transcription elongation events. This inhibitory mechanism might contribute to transcriptional quiescence of integrated HIV-1 provirus.
In addition to its lytic pathway, human immunodeficiency virus type 1 (HIV-1) is able to enter a latent state in which the transcription of integrated provirus genome is repressed (21). In cells harboring inactive provirus, HIV-1 transcription is exclusively regulated by host cell transcription factors that bind to the HIV-1 long terminal repeat (LTR) (see Fig. 1A, below). The dimeric transcription factor NF-κB plays a central role in the proviral transcription (17). In resting T cells and most established cell lines, regulation of NF-κB activity occurs at several levels, including nuclear shuttling and modulation of its transcriptional activity in the nucleus (8). NF-κB is mainly retained within the cytoplasm by IκB proteins but shuttles permanently in and out of the nucleus (8, 9, 13). In response to a variety of stimuli, including phorbol myristate acetate (PMA), IκB is rapidly degraded and releases NF-κB to translocate into the nucleus (16).
FIG. 1.
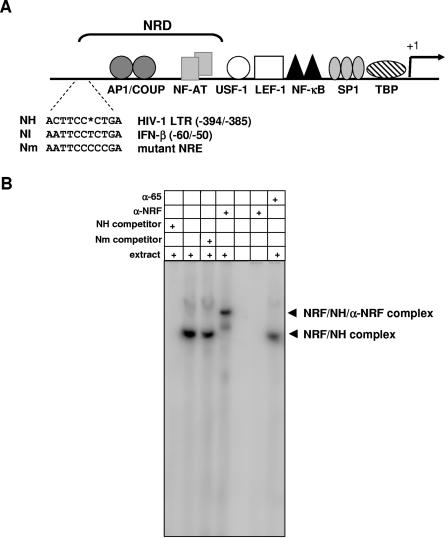
NRF binds to the negative regulatory domain in the HIV-1 LTR. (A) HIV-LTR and proximal binding sites for several transcription factors are illustrated at the top. The NRF binding sequences in the IFN-β promoter (NI) and HIV-1 LTR (NH) as well as a functional mutant of the NRF binding site (Nm) are shown for comparison. Numbers indicate positions of nucleotides with respect to the transcriptional start site of the HIV-1 LTR and IFN-β promoter, respectively (+1). The gap (asterisk) was introduced to obtain maximal homology. (B) Three femtomoles of radiolabeled NH oligonucleotide was incubated with 1 μg nuclear extract from HeLa cells where indicated. A 100-fold concentration of cold competitor or 1 μg of antibody against NRF (α-NRF) or p65 (α-p65) was added to the binding reaction mixtures as indicated. Protein-DNA complexes were analyzed by EMSA. NRF-specific complexes are indicated by arrowheads. The fronts of unbound oligonucleotides were run out of the gel to separate the upper complexes.
Although it was commonly assumed that NF-κB stimulates only transcriptional initiation, it was also found to be implicated in transcriptional elongation (28). Recently, it was shown that NF-κB regulates activity of an immediate-early NF-κB target gene, A20, via elongation factor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF) (1). DSIF has been implicated in controlling the elongation activity of RNA polymerase II (Pol II) both positively and negatively (27, 29). RNA Pol II elongation activity is regulated by a series of phosphorylation events involving the carboxy-terminal domain (CTD) of RNA Pol II (25). During initiation, hypophosphorylated Pol II is able to form a complex with general initiation factors. Following initiation, however, the CTD of RNA Pol II becomes highly phosphorylated at Ser 2 and 5, permitting interaction with several transcription elongation factors (4, 10, 27, 29).
NF-κB-repressing factor (NRF) is a constitutively expressed nuclear transcription factor that binds to beta interferon (IFN-β), interleukin-8 (IL-8), and inducible nitric oxide synthase (iNOS) promoters and represses the basal transcription of these genes (5, 18, 20). Exceptionally in IL-8 transcription, NRF plays an additional role and has been found to be required for IL-1-stimulated IL-8 gene expression (20). Although NRF has been implicated in transcription regulation of several genes, little is known about its molecular mechanism.
Detailed analysis of NRF cDNA revealed that the encoded protein contains at least two functional domains (18). The N-terminal domain spanning amino acids (aa) 1 to 296 inhibits the transcription activity of NF-κB in reporter experiments. In vitro, NF-κB proteins bind to the N-terminal domain of NRF by a direct protein-protein interaction (18). Amino acids 296 to 388 constitute the DNA binding domain of NRF, which is sufficient for binding to IFN-β, IL-8, and iNOS promoters. The transcription activity of the C-terminal domain spanning amino acids 389 to 690 is not yet known. However, this domain is not required for either NRF binding to DNA or its interaction with NF-κB proteins.
By sequence comparison, we identified a potential NRF binding site in the HIV-1 LTR, termed NH. Considering the important role of NF-κB in HIV-1 transcription, we hypothesized that NRF might be involved in LTR regulation. Our data demonstrate that NRF binds to the NH sequence element in the HIV-1 LTR and inhibits basal transcription activity at the level of elongation in unstimulated cells. By binding to the LTR, NRF prevents recruitment of the elongation factor DSIF and, simultaneously, the formation of processive elongation complexes by hyperphosphorylated Pol II.
MATERIALS AND METHODS
Plasmid constructions.
Chloramphenicol acetyltransferase (CAT) reporter plasmid p0 contains a minimal TATA box as described earlier (18). Double-stranded oligonucleotides were inserted to the SphI site of p0 to create pκB, pκB-NI, pκB-NH, and pκB-Nm as follows: NF-κB (sense), 5′-CCATGGGGGACTTTCCGCTGGGGACTTTCCATG-3′; NI (sense), 5′-CCATGGCAATTCCTCTGACATG-3′; NH (sense), 5′-CCATGGCACTTCCCTGACATG-3′; Nm (sense), 5′-CCATGGCAATTCCCCCGACATG-3′.
Expression plasmids p50, p52, p65, and pMBC (empty expression vector) were described earlier (18). Internal control plasmids encoding CAT (pBHECAT), firefly luciferase (pSV2LUC), and Renilla luciferase (pSVRLUC or phRG-B) were described earlier (18). The HIV-1 LTR (isolate ARV-2/SF2, NCBI K02007, 8931/9737) from −1 to −806 with respect to the transcription start site was inserted into pBHELUC containing the luciferase coding region to create pHIVLTR-LUC (18). pM-HIVLTR-LUC was created by mutating the NH site using site-directed mutagenesis (Stratagene) and the following double-stranded oligonucleotides: M1 (sense), 5′-GGTTTGACAGCAAACTAGCAATTCCCCCGATGGCCCGAGAGCTGCATCC-3′; M2 (antisense), 5′-CCATGCAGCTCTCGGGCCATCGGGGGAATTGCTAGTTTGCTGTCAAACC-3′.
The HIV-1 LTR was introduced into the proviral clone pNL4-3.Luc.R-E-, in which the N-terminal 34 amino acids of the nef gene were replaced by the luciferase gene. The env reading frame was disrupted by introducing a frameshift mutation (3).
The BamHI/BsmI and BamHI/XbaI fragments of pHIVLTR-LUC were inserted into pBluescript vector (Stratagene) in the antisense orientation to the T7 promoter to create in vitro transcription plasmids pBLantiLUC102 and pBLantiLUC304 for the synthesis of luciferase antisense probes.
Transferrin receptor cDNA fragment from +61 to +452 was inserted into a pcDNA3 (Invitrogen) vector in the antisense orientation to the T7 promoter to create in vitro transcription plasmid pT7AS-TFR for the synthesis of luciferase antisense probes.
Cell lines and DNA transfection.
HeLa-tTA cells expressing tetracycline-sensitive transactivator protein (18) as well as HeLa and 293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum (FCS). HeLa cells were transfected by calcium phosphate precipitation. Indicated amounts of reporter plasmid and 1 μg of effector plasmids were transfected per 1.5 × 105 cells. CAT and luciferase reporter activities were detected, and the resulting relative light units were calculated as described earlier (18).
TBC and TBC-NRFantisense cell lines were described earlier by transfection of HeLa-tTA cells and maintained in Dulbecco's modified Eagle's medium with 10% FCS, 4.2 μg/ml puromycin, and 500 μg/ml G418 (5, 18, 20).
Jurkat T cells were maintained in RPMI 1640 medium plus 10% FCS and transfected with Nucleofector technology according to the manufacturer's instructions (Amaxa Biosystems). Briefly, for each transfection 107 Jurkat cells were harvested and resuspended in Nucleofector solution with 1 μg HIV-LTR firefly luciferase reporter plasmid and 0.5 μg phRG-B (Promega), which expresses Renilla luciferase, to normalize transfection efficiency. The cell-DNA mixtures were nucleofected, and cells were immediately transferred into 5 ml RPMI 1640 medium plus 10% FCS for 2 h at 37°C. Luciferase reporter activities were detected after stimulation as described earlier (18).
For stimulation, cells were treated with 10 ng/ml PMA in combination with 500 ng/ml ionomycin for the indicated times or, alternatively, with 25 μg/ml poly(rI-rC) (double-stranded RNA [dsRNA]) together with 100 μg/ml DEAE-dextran where indicated.
Western blotting.
Western blot assays were performed as described earlier (18). Endogenous NRF protein was detected using rabbit polyclonal antibody directed against aa 25 to 45 of the NRF protein sequence. Antibodies against p65 (sc8008) and phosphorylated PKR (Thr 446; sc16565) were obtained from Santa Cruz Biotechnology, Inc.
Electrophoretic mobility shift assay (EMSA) and competition assays.
Gel shift analysis was carried out according to the protocol of Fried and Crothers (6). One microgram of HeLa nuclear extracts was incubated with 3 fmol (20,000 cpm) of labeled double-stranded oligonucleotide in the presence of 0.01 U of poly(dI-dC) in 10 mM HEPES, pH 8.0, 5 mM MgCl2, 50 mM KCl, 0.025% bromphenol blue, 0.025% xylene cyanol, and 10% Ficoll at room temperature for 10 min. The indicated amounts of cold competitor, rabbit antibodies against NRF, or p65 were added to the reaction mixtures and incubated for a further 15 min. Samples were analyzed on 8% native polyacrylamide gels that were run at 70 V for at least 8 h. After drying, the gels were exposed overnight to an autoradiographic film.
Pseudotyped HIV-1 virus.
The HIV-1 proviral reporter construct pNL4-3.Luc.R-E- (3) was used for the single-round infection assays. pNL4-3.Luc.R-E- is an env-defective proviral construct which expresses the luciferase reporter gene instead of the nef gene. A construct missing the 3′-LTR U3 sequences (nucleotides 8793 to 9498 in the published HIV-1 NL4-3 sequence), named NL43(e-n-L+)Δ3LTR, served as a negative control. For the generation of pseudotyped viral particles, 293T cells were cotransfected with 6 μg of env-defective proviral NL4-3 luciferase constructs and 3 μg of an expression plasmid encoding the env protein of the vesicular stomatitis virus (VSV). After 24 h viral stocks were aliquoted and frozen at −80°C. The p24 antigen concentration was determined using an HIV enzyme-linked immunosorbent assay provided by the NIH AIDS Research and Reference Reagent Program.
HIV-1 single-round infection assay.
A total of 105 stable transfected HeLa-tTA cells bearing empty vector pTBC or pTBC-NRFantisense were maintained in the presence or absence of 2 μg/ml tetracycline. After 16 h cells were transduced with an aliquot of reporter virus stock solution containing 500 ng of viral p24 antigen for 40 h. Cells were harvested and subjected to a luciferase assay.
S1 protection assay.
HeLa cells were transfected with pHIVLTR-LUC or pM-HIVLTR-LUC. After 48 h, cells were stimulated with PMA or left untreated as described above. Total RNA was extracted from cells using TRIzol reagent (BD Biosciences) and then treated with 10 U of RNase-free DNase (Roche). RNA was then extracted with phenol-chloroform-isoamyl alcohol (Roth) and precipitated in ethanol and 0.6 M LiCl. Antisense probes were prepared using an in vitro T7 transcription system (Promega) and the following templates: for the synthesis of initiation probe, pBLantiLUC102 was linearized with BamHI enzyme; for the synthesis of elongation probe, pBLantiLUC302 was linearized with Bsu36I enzyme; for the synthesis of transferrin receptor probe, pT7AS-TFR was linearized with BsmI enzyme. Each protection was performed on equal amounts of RNA (20 μg) and equal counts per minute of G50 purified in vitro-transcribed antisense RNA as described earlier (19). The specific activity of initiation probe was approximately 10−6 cpm/copy, and that of elongation probe was 0.5 × 10−5 cpm/copy. Protected fragments were analyzed by electrophoresis on 6% polyacrylamide gels containing 6 M urea. The quantitation of autoradiographic fragments was conducted using the PhosphorImager software program.
ChIP assay.
Cells were cross-linked in vivo with 1% formaldehyde for 10 min at 37°C. Cross-linking reactions were stopped by adding 125 mM glycine, and cells were washed in phosphate-buffered saline and then incubated in radioimmunoprecipitation assay lysis buffer (40 mM Tris-HCl [pH 7.05], 120 mM NaPPi, 200 mM NaCl, 1% Triton, 8 mM Na3VO3, 2 mM NaF, 80 mM β-glycerolphosphate, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 10 μM pepstatin) on ice for 10 min. After sonication and centrifugation, 5 μl of the soluble extract was analyzed as input control. DNA fragments bound to various proteins were immunoprecipitated using 1 μg of the following antibodies: anti-DSIF (α-DSIF; sc-13840) was obtained from Santa Cruz Biotechnology, Inc.; α-p65 (AB1606) was obtained from Chemicon. Antibodies against human Pol II (MMS-128P) and phosphorylated RNA Pol II Ser 2 (MMS-129R) and Ser 5 (MM-134R) were obtained from Eurogentec. For immunoprecipitation of the endogenous NRF, 2 μg of each polyclonal antibody directed against aa 256 to 272 and 272 to 288 of the NRF protein sequence was added. Specific LTR and transcribed sequences in the immunoprecipitates were detected by PCR using the following primers: −450, 5′AGTCAGACCTCAGGTACC-3′; +20, 5′CTCTAGAGGATAGAATGG-3′; +110, 5′GGCTATGAAGAGATACGCCCTGG-3′; +300, 5′CCGATAATAACGCGCCCAACACC-3′.
RESULTS
NRF binds to the negative regulatory domain in the HIV-1 LTR.
Sequence comparison of HIV-1 LTR with NRF binding sites in IFN-β (NI) and iNOS promoters revealed a potential NRF binding element in HIV-1 LTR (NH) (Fig. 1A). NH is located within a negative regulatory domain which was previously shown to repress the transcriptional activity of the HIV-1 LTR (15).
To examine whether NRF specifically binds to the HIV-1 LTR, we initially performed an EMSA and competition experiments using NH double-stranded oligonucleotide and nuclear extracts from HeLa cells. As shown in Fig. 1B, NH forms a single complex with endogenous NRF. The formation of NH complex is significantly impaired by addition of specific cold competitor NH. Addition of the previously described mutant NRF binding sequence Nm has no effect on formation of NRF-NH complex (18). To confirm the identity of NRF in the detected NH complex, we added α-NRF antibody. Figure 1B shows that α-NRF antibody forms a supershift complex. Without HeLa nuclear extract, α-NRF antibody does not bind to the NH probe, showing the specificity of the supershift signal. As an additional control, NF-κB α-p65 antibody failed to form a supershift complex.
We next utilized a reporter assay to demonstrate that NRF is able to inhibit the enhancing activity of the NF-κB binding site of HIV-1 LTR by binding to an adjacent NH element. HeLa cells where transiently transfected with reporter plasmids containing NF-κB and NRF binding sites as schematically presented in Fig. 2A. Insertion of HIV-1 NF-κB binding sites in pκB led to a sevenfold activation of the CAT reporter compared to control p0 reporter as demonstrated in Fig. 2B. Additional insertion of the mutant element, Nm, into pκB (pκB-Nm) revealed an identical CAT activity compared to pκB. In contrast, the activity of the NF-κB binding site was significantly reduced by the additional insertion of NH or NI. This indicates the ability of endogenous NRF to specifically bind to NI and NH and thereby repress NF-κB activity.
FIG. 2.
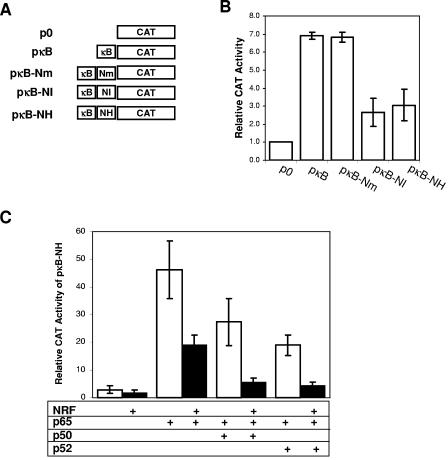
NRF binding site in the HIV-1 LTR inhibits the transcriptional activity of NF-κB. (A) Schematic diagrams of reporter plasmids used for monitoring the NRF and NF-κB interaction. The plasmids contained a minimal synthetic TATA box to direct CAT reporter gene transcription. NF- κB binding sites of the HIV-1 LTR (κB), NRF binding sites of HIV-1 LTR (NH) or IFN-β promoter (NI), and mutated NRF binding element (Nm) were inserted upstream of the TATA box in the indicated orientation. (B) HeLa cells were transfected with 5 μg of the indicated CAT reporter plasmids and 1 μg of a firefly luciferase control plasmid. After 48 h CAT and luciferase reporter activities were determined. CAT activities were normalized to luciferase activities. CAT activity in cells transfected with empty vector p0 was set to 1 in each experiment. The mean activity, expressed as relative CAT activity, is shown ± the standard error of the mean (SEM) from three independent experiments. (C) HeLa cells were cotransfected with 1 μg of reporter plasmid pκB-NH and either 1 μg of an empty control plasmid pMBC (white bars) or NRF-expressing plasmid (black bars). In addition, 1 μg of either p65-expressing vector alone or a combination of expression vectors encoding p50 and p65 (0.5 μg each) or p52 and p65 (0.5 μg each) was cotransfected. After 48 h CAT and luciferase reporter activities were determined. CAT activities were normalized to luciferase activities. CAT activity in cells cotransfected with pκB-NH was set to 1 for each single experiment. The mean activity, expressed as relative CAT activity, is shown ± the SEM from three independent experiments.
To examine the inhibitory action of NRF on different NF-κB homo- and heterodimers, we simultaneously overexpressed both NRF and NF-κB proteins. Figure 2C shows that p65 homodimers and p52-p65 or p50-p65 heterodimers are able to stimulate pκB-NH reporter. These activities are significantly reduced by overexpression of NRF protein (Fig. 2C). In comparison to p65 homodimers, the activity of p52-p65 or p50-p65 heterodimers appears to be more sensitive to NRF. In summary, the data in Fig. 2 confirm the inhibitory interaction of NF-κB and NRF transcription factors.
Endogenous NRF inhibits HIV-1 LTR transcriptional activity.
The experiments described above focused on the isolated NF-κB binding site of the HIV-1 LTR. To explore the role of endogenous NRF in the complete HIV-1 LTR, we used an approved NRF antisense expression system (Tet-Off). Two tetracycline-sensitive expression vectors, empty vector pTBC and pTBC-NRFantisense vector containing NRF antisense sequences, were stably transfected into HeLa-tTA cells to obtain two stable cell lines designated TBC and TBC-NRFantisense. TBC cells served as control, whereas TBC-NRFantisense cells express NRF antisense RNA by withdrawal of tetracycline (20). Western blot analysis using α-NRF antibody showed that the level of endogenous NRF protein is reduced by the induction of NRF antisense RNA (TBC-NRFantisense) (Fig. 3A, −tetracycline). TBC cells fail to reduce the endogenous NRF protein (TBC, + and − tetracycline).
FIG. 3.
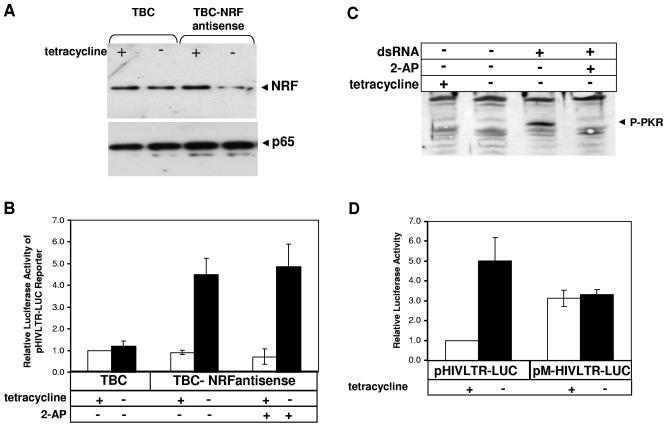
Endogeous NRF inhibits HIV-1 LTR transcriptional activity. HeLa-tTA cells were stably transfected with tetracycline-regulated plasmids containing none (pTBC) or a 300-bp fragment of the NRF cDNA in antisense orientation (pTBC-NRFantisense). Two stable cell lines were obtained and were designated TBC and TBC-NRFantisense. (A) TBC and TBC-NRFantisense cells were cultured in the presence (+) or absence (−) of 2 μg/ml tetracycline. After 30 h, endogenous NRF expression was determined by Western blotting using α-NRF antibody. The NRF-specific protein band is indicated. As control, p65 expression was determined using α-p65 antibody. (B) TBC and TBC-NRFantisense cells were cultured in the presence (white bars) or absence (black bars) of 2 μg/ml tetracycline. After 24 h cells were cotransfected with 1 μg of HIV-1 luciferase reporter plasmid (pHIVLTR-LUC) and 1 μg of CAT-expressing control plasmid. Where indicated, cells were incubated with 10 mM 2-AP. After 48 h cells were harvested and luciferase and CAT activities were determined. In each experiment, luciferase activities were normalized to CAT activities. Luciferase activity of TBC cells in the presence of 2 μg/ml tetracycline was set to 1 in each single experiment. The mean value of the relative luciferase activity is shown ± the standard error of the mean (SEM) from four independent experiments. (C) TBC-NRFantisense cells were incubated with tetracycline where indicated. After 24 h cells were incubated with 10 mM 2-AP, and 4 h later cells were stimulated with synthetic dsRNA where indicated. After 2 h PKR phosphorylation was monitored by direct Western blotting using α-P-PKR antibody. Phosphorylated PKR signal is indicated by an arrowhead. (D) TBC-NRFantisense cells were cultured in the presence (white bars) or absence (black bars) of 2 μg/ml tetracycline. After 24 h cells were cotransfected with 1 μg of HIV-1 luciferase reporter plasmids (pHIVLTR-LUC or pM-HIVLTR-LUC) and 1 μg of CAT-expressing control plasmid. After 48 h cells were harvested and luciferase and CAT activities were determined. Luciferase activities were normalized to CAT expression levels. In each single experiment, luciferase activity of TBC-NRFantisense cells transfected with pHIVLTR-LUC in the presence of 2 μg/ml tetracycline was set to 1. The mean value of the relative luciferase activity is shown ± the SEM from three independent experiments.
Both cell lines, TBC and TBC-NRFantisense, were cultivated in the presence or absence of tetracycline to reduce the expression of endogenous NRF before cells were transfected with pHIVLTR-LUC. In TBC cells, HIV-1 LTR activity is not significantly affected by omitting tetracycline from the medium (Fig. 3B, TBC, −tetracycline). In TBC-NRFantisense cells, in contrast, reduction of endogenous NRF increases the transcriptional activity of the HIV-1 LTR. This suggests an inhibitory role of NRF in the HIV-1 LTR. Since the HIV-1 LTR could be unspecifically activated by dsRNA in TBC-NRFantisense cells, we added 2-aminopurine (2-AP) as a control. 2-AP is a potent inhibitor of dsRNA-dependent kinase (PKR) (12). As shown in Fig. 3B, addition of 2-AP shows no effects on activation of HIV-1 LTR in TBC-NRFantisense cells. This excludes an unspecific activation by dsRNA. In parallel, we examined by Western blotting firstly that PKR is not phosphorylated in TBC-NRFantisense cells and secondly that the 2-AP concentration used in the transfection experiment in Fig. 3B is sufficient to block PKR phosphorylation (Fig. 3C).
In order to demonstrate that the inhibitory role of NRF is due to its binding to the LTR, we mutated the NRF binding sequence in the HIV-1 LTR. TBC-NRFantisense cells were transfected with either pHIVLTR-LUC or pMHIVLTR-LUC lacking the NRF binding site (Fig. 3D). The inhibitory function of NRF was confirmed by mutation of the NRF binding site leading to LTR activation (+tetracycline). Furthermore, downregulation of NRF in TBC-NRFantisense cells (−tetracycline) increases the activity of wild-type LTR but shows no effect on mutant LTR. In summary, the results in Fig. 3 demonstrate that endogenous NRF directly represses the transcriptional activity of HIV-1 LTR by binding to the NH sequence.
Endogenous NRF is required for activation of HIV-1.
Transcriptional activity of HIV-1 LTR can be spontaneously enhanced by viral infection, resulting in activation of cellular regulatory proteins and expression of HIV-1-encoded regulatory proteins, like transactivator protein of HIV-1 (Tat) (7, 23). Both cooperatively stimulate the transcriptional activity of the HIV-1 LTR (26). To further implicate a role for NRF in stimulation of HIV-1 transcription, we utilized a VSV envelope-pseudotyped HIV-1 virus which enabled us to infect HeLa TBC and TBC-NRFantisense cells (schema in Fig. 4A). The pseudotyped HIV-1 virus particles were produced in 293T cells by the simultaneous transfection of a VSV envelope-expressing vector and a mutant HIV-1 construct (pNL4-3.Luc.R-E-). This construct contains a frameshift mutation in the env coding region and a luciferase reporter gene (3). Thus, the generated virus particles are able to transduce mutant HIV-1 genome into TBC and TBC-NRFantisense cells. Since these viral particles are completely incompetent to produce new infective virus, this approach allows monitoring of a single-round infection (22). Therefore, in infected cells, luciferase activity corresponds to the entire transcriptional activity of the HIV-1 LTR in the presence of Tat protein (22).
FIG. 4.
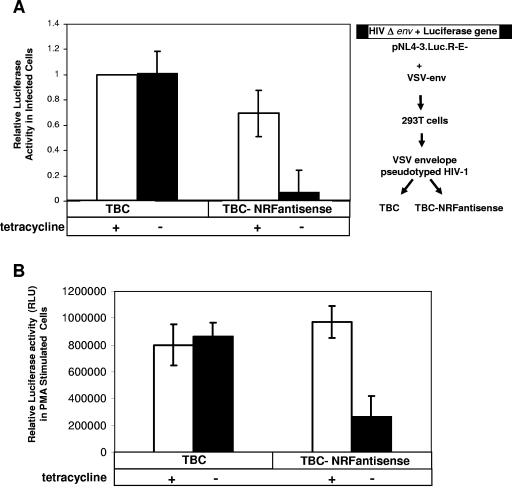
Endogenous NRF is required for activation of HIV-1. (A) On the right side, a schematic description shows the generation of pseudotyped HIV-1 particles in 293T cells (see Materials and Methods for details). TBC and TBC-NRFantisense cell lines (as outlined in the legend of Fig. 3) were cultured in the presence (white bars) or absence (black bars) of 2 μg/ml tetracycline. After 16 h cells were infected with pseudotyped HIV-1 virus (500 ng of viral p24 antigen) containing the luciferase reporter gene. Cells were harvested after 40 h, and luciferase activity was determined. Luciferase activity of TBC cells in the presence of 2 μg/ml tetracycline was set to 1 in each single experiment. The mean value of relative luciferase activity is shown ± the SEM from four independent experiments. (B) TBC and TBC-NRFantisense cells were cultured in the presence (white bars) or absence (black bars) of 2 μg/ml tetracycline. After 24 h cells were cotransfected with 1 μg of HIV-1 luciferase reporter plasmid (pHIVLTR-LUC) and 1 μg of CAT-expressing control plasmid. After 24 h cells were stimulated with 10 ng/ml PMA. Eighteen hours later cells were harvested and luciferase and CAT activities were measured. Luciferase activities were normalized to CAT expression. The mean value of normalized luciferase activity (RLU) of PMA-stimulated transfected cells is shown ± the standard error of the mean from four independent experiments.
TBC and TBC-NRFantisense cells were infected with the pseudotyped HIV-1 virus. As shown in Fig. 4A, TBC cells comprised an equal level of luciferase activity in the presence or absence of tetracycline. TBC-NRFantisense cells showed a small reduction of luciferase activity in the presence of NRF (presence of tetracycline) compared to TBC cells. By reduction of NRF (in TBC-NRFantisense cells), in contrast, luciferase activity was significantly reduced. This indicates that NRF is required for full activation of the HIV-1 LTR upon HIV-1 infection.
As outlined above, cellular pathways also contribute to HIV-1 LTR activation (24, 26). Protein kinase C (PKC) is one of the key signaling intermediates in stimulation of HIV-1 transcription. PMA is a well-characterized activator of classical and novel PKC family members (14). We next investigated the role of endogenous NRF in activation of the HIV-1 LTR after PMA stimulation (Fig. 4B). TBC and TBC-NRFantisense cells were cultivated in the presence or absence of tetracycline. Twenty-four hours later cells were transfected with pHIVLTR-LUC and then stimulated with PMA. PMA stimulation almost led to an equal activation of HIV-1 LTR in TBC cells in the presence or absence of tetracycline. Comparable luciferase activity was detected in TBC-NRFantisense cells in the presence of NRF. In contrast, PMA-mediated activation of the HIV-1 LTR was significantly reduced by reduction of endogenous NRF (TBC-NRFantisense).
The results in Fig. 4A and B demonstrate that NRF is required for activation of the HIV-1 LTR in stimulated cells, in contrast to its inhibitory role in nonstimulated cells, as shown in Fig. 3.
NRF regulates transcription initiation and elongation from HIV-1 LTR.
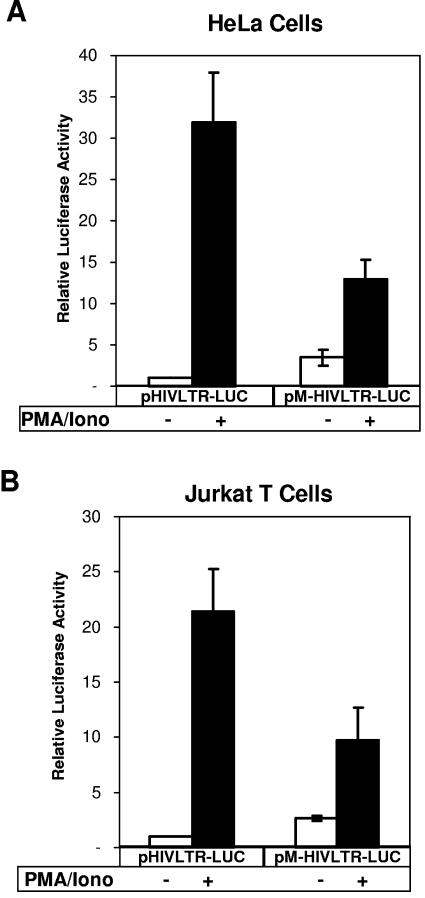
We next utilized wild-type and mutant HIV-1 LTR reporter to prove that NRF binding to LTR is a prerequisite for its effects on transcriptional activity of HIV-1 LTR. In Fig. 5A and B, we compared the wild-type HIV-1 LTR activity (pHIVLTR-LUC) to the mutant HIV-1 LTR lacking a functional NRF binding site (pM-HIVLTR-LUC). The experiments were carried out in HeLa cells (Fig. 5A) and, additionally, in Jurkat T lymphoma cells (Fig. 5B). In comparison with the wild-type LTR, mutation of the NRF binding site leads to basal activation of HIV-1 LTR in nonstimulated HeLa and Jurkat T cells. Following PMA stimulation, the activity of HIV-1 LTR is impaired by blocking NRF binding. These data confirm both the inhibitory function of NRF in nonstimulated cells as well as its activating function after stimulation.
FIG. 5.
NRF binding to LTR is a prerequisite for the effects on transcriptional activity of the HIV-1 LTR. HeLa cells (A) and Jurkat T cells (B) were cotransfected with 10 μg of either wild-type HIV-1 LTR reporter containing an intact NRF binding site (pHIVLTR-LUC) or a mutant HIV-1 LTR reporter lacking the NRF binding site (pM-HIVLTR-LUC) and 2.5 μg of a Renilla luciferase expression plasmid. DNA amounts were kept constant by adding empty expression vector pMBC in each transfection experiment. After 18 h cells were left untreated (white bars) or were stimulated with 10 ng/ml PMA in combination with 500 ng/ml ionomycin (black bars). Cells were lysed 24 h after transfection, and firefly and Renilla luciferase reporter activities were determined. Firefly luciferase activities were normalized to Renilla luciferase activities. Normalized luciferase activity of untreated cells transfected with wild-type LTR reporter (pHIVLTR-LUC) was set to 1 in each single experiment. The mean values of luciferase activities compared to the wild-type LTR reporter activity in untreated cells are shown ± the standard error of the mean from three independent experiments.
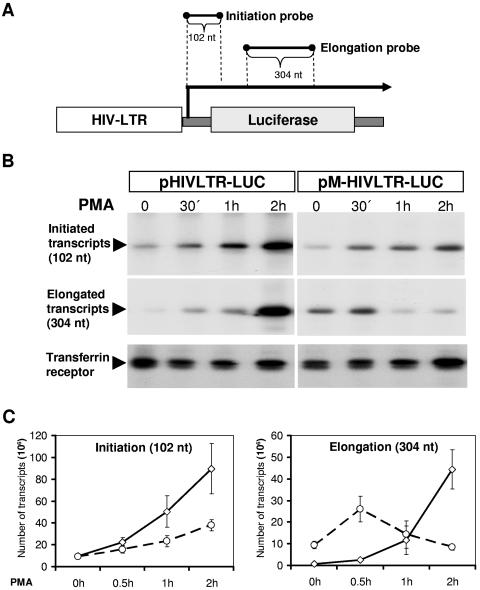
In contrast to NRF antisense-expressing cells, the experimental design in Fig. 5 allowed us to instantly monitor the transcriptional events in the presence or absence of NRF binding to LTR upon PMA stimulation. These reporters were also used for the experiments shown in Fig. 6. In transiently transfected HeLa cells, we compared the level of transcription initiation and elongation mediated by the wild-type LTR (pHIVLTR-LUC) to the mutant LTR (pM-HIVLTR-LUC). We used two different probes able to separately monitor either initiation or elongation of luciferase transcripts by S1 protection analysis. As illustrated in Fig. 6A, hybridization to the proximal probe allows the detection of all initiated transcripts by a 102-nucleotide-long protected fragment. Hybridization to the distal probe detects only those elongated transcripts which have been extended. These transcripts are detected by a protected 304-nucleotide-long fragment. Noteworthy, the distal probe is three times larger than the proximal probe and thereby causes stronger radioactive signals by incorporation of radiolabeled nucleotides (Fig. 6B). To simplify, the number of detected specific transcripts was calculated from three independent experiments and is summarized in Fig. 6C. An additional antisense probe, transferrin receptor, was utilized to control experimental variations. As expected, PMA treatment increases the time-dependent transcription initiation rate from wild-type HIV-1 LTR (Fig. 6C). In comparison, mutant LTR exerts a striking reduction in transcription initiation rate (Fig. 6C). This indicates that NRF improves the transcription initiation rate from the HIV-1 LTR upon PMA stimulation.
FIG. 6.
NRF regulates transcription initiation and elongation from the HIV-1 LTR. (A) The pHIVLTR-LUC plasmid and probes corresponding to initiated or elongated transcripts are illustrated. Lengths of protected regions are indicated. (B) HeLa cells were transfected either with pHIVLTR-LUC or with pM-HIVLTR-LUC as indicated. A Renilla luciferase expression plasmid (phRG-B; Promega) was cotransfected to estimate the transcriptional efficiency in each experiment. After 48 h cells were stimulated with 10 ng/ml PMA for the indicated times or left untreated. Then, cells were harvested and total RNA was purified. Twenty-micrograms aliquots of RNAs were hybridized to antisense initiation or elongation probes of transfected constructs. As control, 10 μg of total RNA was hybridized to transferrin receptor coding sequences. Hybridization products were subjected to the S1 mapping analysis and were analyzed by electrophoresis through 6% denaturing polyacrylamide gel electrophoresis. The autoradiography of protected transcripts is shown. (C) Before electrophoresis the total amounts of protected fragments were measured (in counts per minute). The number of transcripts per 20 μg of total RNA is shown ± the standard error of the mean from three independent experiments.
However, transcription elongation appears to be differently regulated by NRF. In nonstimulated cells, elongation efficiency from wild-type LTR is marginal while the transcription elongation rate from mutant LTR is strikingly higher. This indicates that NRF causes a decline in the transcription elongation rate in nonstimulated cells. Upon PMA stimulation, the number of transcripts was significantly enhanced by wild-type LTR, as expected, whereas the number of elongated transcripts was only temporarily increased by mutant LTR. During longer PMA treatment, the transcription elongation rate returned to the level observed in nonstimulated cells (Fig. 6C, right). We suppose that this is partially, but not completely, caused by the reduction of the transcription initiation rate at the mutant LTR (Fig. 6C, left).
NRF inhibits the recruitment of DSIF to the HIV-1 LTR.
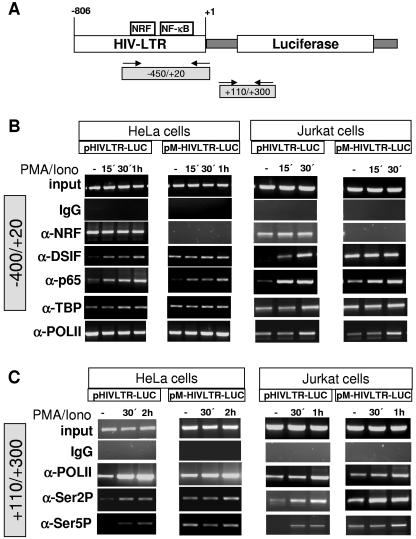
Recently, it was shown that NF-κB can regulate transcription elongation via the elongation factor DSIF (1). Considering the inhibitory effect of NRF on transcription elongation, we investigated recruitment of the elongation factor DSIF to wild-type (pHIVLTR-LUC) and mutant (pM-HIVLTR-LUC) LTRs. In this case we performed parallel experiments in HeLa and Jurkat T cells, presuming a general cell type-independent mechanism. Data were gained by chromatin immunoprecipitation (ChIP) analysis. Cells were transfected with the respective reporter plasmids and were left untreated or stimulated. Transcription factors associated with the LTR were detected using specific primers which amplify the region between −450 and +20 with respect to the transcription start site (Fig. 7A). As shown in Fig. 7B, constant amounts of cross-linked chromatin (input) were added for each PCR. Immunoprecipitation with rabbit immunoglobulin G served as a negative control showing no signal. Vice versa, α-TBP and α-Pol II antibodies reveal a continuous positive signal in ChIP experiments. Most importantly, ChIP using α-NRF antibody detects a significant binding of endogenous NRF to wild-type LTR. Noteworthy, the level of NRF binding to the LTR is not changed upon stimulation. In contrast, mutating the NRF binding site completely abolishes binding of NRF to the LTR. To explore a possible involvement of DSIF in LTR regulation as outlined above, we next compared DSIF recruitment to wild-type and mutant LTR. Prior to stimulation, a very small amount of DSIF binds to wild-type LTR, whereas lack of NRF binding strikingly improves DSIF signal. However, there is no difference of DSIF binding between wild-type and mutant LTR upon stimulation.
FIG. 7.
Recruitment of transcription factors to the HIV-1 LTR. HIV-LTR and proximal binding sites for several transcription factors are illustrated at the top. A ChIP assay was performed using soluble chromatin extract from control or PMA- and ionomycin-stimulated (PMA/Iono) HeLa and Jurkat T cells transfected with wild-type HIV-1 LTR containing intact NRF binding sites (pHIVLTR-LUC) or mutant HIV-1 LTR missing NRF binding sites (pM-HIVLTR-LUC), as indicated. In order to internally control the experiments for variations in transfection efficiency, a Renilla luciferase expression plasmid (phRG-B; Promega) was cotransfected. Cells were harvested, and an aliquot was subjected to a Renilla luciferase assay (data not shown). The extracts were precipitated using the indicated antibodies or left untreated as an internal quality control (input). The precipitated DNAs were used for PCR with primers spanning the core and LTR-proximal region as indicated at the top. Results are representative of four independent ChIP experiments.
Using α-p65 antibody, we noticed that the impact of NRF on DSIF recruitment does not depend on p65 binding to LTR, since p65 binding is almost identical in wild-type and mutant HIV-1 LTR. This is consistent with our previous data, showing that NRF exclusively interferes with transcriptional activity but not with DNA binding activity of p65 (18, 19).
Considering the fact that hyperphosphorylation of the CTD of RNA Pol II correlates with the formation of elongated transcripts, we analyzed distribution of phosphorylated isoforms of Pol II. We observed very low amounts phosphorylated RNA Pol II associated with the LTR (data not shown). Therefore, we next analyzed distribution of Pol II and its phosphorylated isoforms, Ser 2 and Ser 5, within downstream coding sequences (+110/+300) (Fig. 7A). Significant amounts of Pol II were found associated within the downstream region prior to and after PMA stimulation in HeLa and Jurkat T cells (Fig. 7C). The loss of NRF binding to the LTR had no significant effects on Pol II association. However, analysis of phosphorylated forms of Pol II showed that loss of NRF binding elevated the ratio of associated Ser 2- and Ser 5-phosphorylated Pol II in unstimulated cells. Upon stimulation, NRF binding does not influence the ratio of phosphorylated forms of Pol II.
The data in Fig. 7 demonstrate that NRF by its ability to bind to the LTR in unstimulated cells prevents recruitment of DSIF and contributions of hyperphosphorylated isoforms of Pol II.
DISCUSSION
Initiation and elongation of transcription are considered distinctive regulatory events accomplished by different sets of regulatory factors. There is little known about DNA binding transcription factors regulating initiation just as elongation events. In this study, we show for the first time an unusual feature for the transcription factor NRF, regulating both transcriptional initiation and elongation at HIV-1 LTR. In fact, the HIV-1 LTR was previously implicated in both initiation and elongation of proviral transcription in infected cells (7, 23, 26, 28, 30).
The NRF binding site in the HIV-1 LTR was identified by EMSA and reporter experiments. By binding to this site, NRF exerts direct effects on transcriptional events at the HIV-1 LTR. As discussed below, NRF displays a different role in unstimulated and stimulated cells. The effects were consistently confirmed by two different approaches. Firstly, cellular NRF protein expression was reduced by NRF antisense expression; secondly, NRF binding to LTR was eliminated by mutation of the NRF binding site.
In unstimulated cells, NRF, NF-κB, and several regulatory transcription factors are associated with the HIV-1 LTR as demonstrated in ChIP experiments (Fig. 7). Additionally, S1 experiments have shown that this basal transcription complex is capable of initiation, but not of processive elongation (Fig. 6). In this state, the elongation process is apparently inhibited by NRF, since elimination of NRF binding to the LTR provokes the processive elongation of initiated transcripts (Fig. 6). This raises the question of which components of LTR-associated transcription factors mediate transcription elongation in the absence of NRF in these experiments. West et al. demonstrated that overexpressed NF-κB dimers, primarily p65, are able to stimulate transcription elongation in unstimulated cells (28). Therefore, we suggest that in the absence of NRF the basal transcription elongation is at least partially mediated by NF-κB proteins binding downstream of the NRF binding site. We tested this possibility by deletion of the NF-κB site in the HIV-1 LTR. This led to a dramatic reduction of basal transcription initiation; thus, it was most difficult to detect processive elongation of reporter transcripts (data not shown).
NF-κB has been shown to regulate transcription elongation at the A20 promoter via DSIF (1). Interestingly, those authors have identified a negative regulatory element (ELIE) upstream of the NF-κB sites in the A20 promoter that inhibits transcription elongation by the Pol II-DSIF complex (1). Our results indicate that transcription elongation in the absence of NRF is accompanied by the recruitment of elongation factor DSIF (Fig. 6 and 7). We compared the ELIE sequence with NRF binding sites, but we found no significant homology. Thus, NRF is unlikely to be involved in A20 elongation inhibition.
The molecular basis for the NF-κB-mediated recruitment of DSIF is not known, but one possibility is that NF-κB recruits DSIF to the LTR by a direct protein-protein interaction. We tested this possibility by coimmunoprecipitation, but p65 failed to directly bind to DSIF (data not shown). A second possibility is that NF-κB recruits a specific isoform of the Pol II complex which is already engaged with DSIF in unstimulated cells (27, 29). In this case, NRF could affect the ability of NF-κB to recruit the Pol II-DSIF complex to the HIV-1 LTR.
The association of different phosphorylated forms of Pol II is known to correlate with processive transcription elongation (11). The Pol II CTD is composed of multiple repeats of Tyr-Ser-Pro-Thr-Ser-Pro-Ser, which are phosphorylated during transcription (11). Ser 5 phosphorylation was primarily detected at promoter regions, whereas Ser 2 phosphorylation was observed at promoter and downstream coding sequences (2). It was therefore important to determine whether the enhanced processivity we observed in the absence of NRF was a direct consequence of CTD phosphorylation. In ChIP experiments, we observed marginal levels of both Ser 2 and Ser 5 phosphorylation associated with the LTR in nonstimulated cells (data not shown). In downstream coding regions, however, Ser 2 and Ser 5 phosphorylation was markedly detected, although at equally higher levels in the absence of NRF. The appearance of Ser 2 and Ser 5 phosphorylation is consistent with the processivity of elongation in S1 analysis. We therefore propose that NRF binding to the LTR prevents processivity of elongation by phosphorylated Pol II at downstream coding sequences.
The molecular mechanism of NRF action in stimulation of the HIV-1 LTR remains elusive. In PMA-stimulated Jurkat and HeLa cells as well as in pseudotyped HIV-1-infected HeLa cells, the HIV-1 LTR is highly activated by NF-κB and a number of other transcription factors. Accordingly, we found a significant increase in the recruitment of NF-κB to the LTR accompanied by activation of transcription initiation and elongation from wild-type HIV-1 LTR. S1 experiments showed that activation of both initiation and elongation of transcription was impaired, but not completely abolished, by mutating the NRF binding site in the HIV-1 LTR (Fig. 6). As efficiency of transcription elongation directly depends on initiation events, it is difficult to conclude a direct activating role of NRF in processive elongation following specific stimulation.
As examined in ChIP experiments, NRF exerts no effects on recruitment of Pol II, TBP, or NF-κB to the HIV-1 LTR upon stimulation. The question arising from these data is how does NRF coactivate the transcriptional initiation downstream? One possibility is that NRF modifies (directly or indirectly) components of transcriptional machinery, rendering them more active. Alternatively, NRF itself can be modified, thereby positively affecting activity of the Pol II complex. However, we previously tested this possibility using reporter plasmids containing a minimal promoter (5). NRF showed no significant effect on activity of a Pol II-dependent minimal promoter upon stimulation. Thus, we presume that NRF is unlikely to play a true direct activating role.
In summary, the role of NRF in HIV-1 transcription regulation can be considered on two different levels, a basal state in which proviral transcription is inhibited and an activated state in which viral gene transcription is highly activated. The transcriptional active state occurs either immediately after viral entry, which itself stimulates different cellular signaling pathways, or a long time after proviral integration, in which proviral transcription is induced in response to specific stimuli. In the activated state, NRF contributes to the enhancement of transcription initiation and elongation. In the state of proviral transcription latency in which the cellular signaling pathways are not active, NRF inhibits the basal transcription elongation at the HIV-1 LTR. The inhibition of transcription elongation by NRF may guarantee either the specificity of HIV-1 transcription activation by certain stimuli or that randomly initiated transcript will not form a processive elongation complex. Whether these postulated mechanisms can initiate or maintain the proviral transcription latency is yet to be determined. Nonetheless, this study reveals a unique mechanism of transcription repression by NRF which affects the regulatory link between transcription initiation and elongation events.
Acknowledgments
We are grateful to Marta Szamel (Institute for Pharmacology, Medical School Hannover, Germany) and M. G. Gruber (Promega, Madison, Wis.) for helpful discussions. We thank Rabea Gräpel for critical reading of the manuscript.
This work was supported by grants from Deutsche Forschungsgemeinschaft, DFG 457, DFG 45/5-1, and SFB 566.
REFERENCES
- 1.Ainbinder, E., L. Amir-Zilberstein, Y. Yamaguchi, H. Handa, and R. Dikstein. 2004. Elongation inhibition by DRB sensitivity-inducing factor is regulated by the A20 promoter via a novel negative element and NF-κB. Mol. Cell. Biol. 24:2444-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 23:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 4.Dahmus, M. E. 1996. Phosphorylation of mammalian RNA polymerase II. Methods Enzymol. 273:185-193. [DOI] [PubMed] [Google Scholar]
- 5.Feng, X., Z. Guo, M. Nourbakhsh, H. Hauser, R. Ganster, L. Shao, and D. A. Geller. 2002. Identification of a negative response element in the human inducible nitric oxide synthase (hiNOS) promoter: the role of NF-κB repressing factor (NRF) in basal repression of the hiNOS gene. Proc. Natl. Acad. Sci. USA 99:14212-14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fried, M., and D. M. Crothers. 1981. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9:6505-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 9.Karin, M. 1998. The NF-kappa B activation pathway: its regulation and role in inflammation and cell survival. Cancer J. Sci. Am. 4(Suppl. 1):S92-S99. [PubMed] [Google Scholar]
- 10.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 11.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, J. H., E. J. Park, O. S. Kim, H. Y. Kim, E. H. Joe, and I. Jou. 2005. Double-stranded RNA-activated protein kinase is required for the LPS-induced activation of STAT1 inflammatory signaling in rat brain glial cells. Glia 50:66-79. [DOI] [PubMed] [Google Scholar]
- 13.Li, N., and M. Karin. 2000. Signaling pathways leading to nuclear factor-kappa B activation. Methods Enzymol. 319:273-279. [DOI] [PubMed] [Google Scholar]
- 14.Liu, W. S., and C. A. Heckman. 1998. The sevenfold way of PKC regulation. Cell Signal 10:529-542. [DOI] [PubMed] [Google Scholar]
- 15.Lu, Y. C., N. Touzjian, M. Stenzel, T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1990. Identification of cis-acting repressive sequences within the negative regulatory element of human immunodeficiency virus type 1. J. Virol. 64:5226-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.May, M. J., and S. Ghosh. 1997. Rel/NF-kappa B and I kappa B proteins: an overview. Semin. Cancer Biol. 8:63-73. [DOI] [PubMed] [Google Scholar]
- 17.Muthumani, K., A. Y. Choo, D. S. Hwang, M. A. Chattergoon, N. N. Dayes, D. Zhang, M. D. Lee, U. Duvvuri, and D. B. Weiner. 2003. Mechanism of HIV-1 viral protein R-induced apoptosis. Biochem. Biophys. Res. Commun. 304:583-592. [DOI] [PubMed] [Google Scholar]
- 18.Nourbakhsh, M., and H. Hauser. 1999. Constitutive silencing of IFN-beta promoter is mediated by NRF (NF-κB-repressing factor), a nuclear inhibitor of NF-κB. EMBO J. 18:6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nourbakhsh, M., K. Hoffmann, and H. Hauser. 1993. Interferon-beta promoters contain a DNA element that acts as a position-independent silencer on the NF-kappa B site. EMBO J. 12:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nourbakhsh, M., S. Kalble, A. Dorrie, H. Hauser, K. Resch, and M. Kracht. 2001. The NF-kappa B repressing factor is involved in basal repression and interleukin (IL)-1-induced activation of IL-8 transcription by binding to a conserved NF-kappa B-flanking sequence element. J. Biol. Chem. 276:4501-4508. [DOI] [PubMed] [Google Scholar]
- 21.Persaud, D., Y. Zhou, J. M. Siliciano, and R. F. Siliciano. 2003. Latency in human immunodeficiency virus type 1 infection: no easy answers. J. Virol. 77:1659-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75:4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pumfery, A., L. Deng, A. Maddukuri, C. de la Fuente, H. Li, J. D. Wade, P. Lambert, A. Kumar, and F. Kashanchi. 2003. Chromatin remodeling and modification during HIV-1 Tat-activated transcription. Curr. HIV Res. 1:343-362. [DOI] [PubMed] [Google Scholar]
- 24.Rabbi, M. F., L. al Harthi, M. Saifuddin, and K. A. Roebuck. 1998. The cAMP-dependent protein kinase A and protein kinase C-beta pathways synergistically interact to activate HIV-1 transcription in latently infected cells of monocyte/macrophage lineage. Virology 245:257-269. [DOI] [PubMed] [Google Scholar]
- 25.Riedl, T., and J. M. Egly. 2000. Phosphorylation in transcription: the CTD and more. Gene Expr. 9:3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roebuck, K. A., and M. Saifuddin. 1999. Regulation of HIV-1 transcription. Gene Expr. 8:67-84. [PMC free article] [PubMed] [Google Scholar]
- 27.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West, M. J., A. D. Lowe, and J. Karn. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-κB p65 stimulates transcriptional elongation. J. Virol. 75:8524-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, M., L. Deng, V. Lacoste, H. U. Park, A. Pumfery, F. Kashanchi, J. N. Brady, and A. Kumar. 2004. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J. Virol. 78:13522-13533. [DOI] [PMC free article] [PubMed] [Google Scholar]