Abstract
Rhopalosiphum padi virus (RhPV) is an insect virus of the Dicistroviridae family. Recently, the 579-nucleotide-long 5′ untranslated region (UTR) of RhPV has been shown to contain an internal ribosome entry site (IRES) that functions efficiently in mammalian, plant, and insect in vitro translation systems. Here, the mechanism of action of the RhPV IRES has been characterized by reconstitution of mammalian 48S initiation complexes on the IRES from purified components combined with the toeprint assay. There is an absolute requirement for the initiation factors eIF2 and eIF3 and the scanning factor eIF1 to form 48S complexes on the IRES. In addition, eIF1A, eIF4F (or the C-terminal fragment of eIF4G), and eIF4A strongly stimulated the assembly of this complex, whereas eIF4B had no effect. Although the eIF4-dependent pathway is dominant in the RhPV IRES-directed cell-free translation, omission of either eIF4G or eIF4A or both still allowed the assembly of 48S complexes from purified components with ∼23% of maximum efficiency. Deletions of up to 100 nucleotides throughout the 5′-UTR sequence produced at most a marginal effect on the IRES activity, suggesting the absence of specific binding sites for initiation factors. Only deletion of the U-rich unstructured 380-nucleotide region proximal to the initiation codon resulted in a complete loss of the IRES activity. We suggest that the single-stranded nature of the RhPV IRES accounts for its strong but less selective potential to bind key mRNA recruiting components of the translation initiation apparatus from diverse origins.
Translation initiation on most cellular mRNAs occurs in a 5′ cap-dependent manner. However, the number of reports of internal ribosome entry site (IRES) elements that direct translation initiation in a 5′-end-independent manner on some eukaryotic and viral mRNAs is constantly growing (44). First identified in picornavirus RNAs, they have subsequently been found in genomic RNAs of hepatitis C virus, pestiviruses, insect viruses of the Dicistroviridae family, and in the 5′ untranslated regions (UTRs) of many cellular mRNAs (11, 44).
The mechanism of action of only a few of them has been studied in detail (9, 11, 24, 33, 48). Such studies have revealed a strikingly different structural organization of these IRES elements (even between those from the same virus family). This is also manifested in their different requirements for the canonical translation initiation factors or auxiliary mRNA-binding proteins (2). For instance, the IRES elements from cardio- and aphthoviruses and presumably from enteroviruses (family Picornaviridae) require most of the canonical initiation factors for their activity, except for the cap-binding factor eIF4E. To initiate translation initiation at the AUG codon proximal to the foot-and-mouth disease virus and encephalomyocarditis virus IRES elements, the scanning initiation factor eIF1 is also dispensable (27, 34).
In addition to the canonical initiation factors, picornaviral IRES elements require some auxiliary mRNA-binding proteins: La (La autoantigen), polypyrimidine tract binding protein, poly(C) binding protein, UNR (Upstream of N-Ras), etc. (2). However, even picornaviral IRES elements with similar secondary structures have been shown to be distinct in their requirement for a set of additional noncanonical factors (e.g., cardio- versus aphthoviruses) (34). Unlike picornaviruses, the IRES elements from hepatitis C virus and pestivirus RNAs show an absolute requirement for only two canonical initiation factors, eIF2 and eIF3 (30), though the activity of the hepatitis C virus IRES has been reported to be substantially stimulated by La and polypyrimidine tract binding protein (1, 4, 10). Finally, the intercistronic IRESs from some insect Dicistroviridae RNAs (e.g., cricket paralysis virus) need neither translation initiation factors nor the initiator tRNA and are able to bind directly to 80S ribosomes (13, 39, 46). This peculiarity of the intercistronic region in the RNAs from members of the family Dicistroviridae is accounted for by its unique structure. The intercistronic region, presumably mimicking the initiator tRNA, fills the P-site of the 80S ribosome, thereby directing the first aminoacyl-tRNA to the ribosomal A site followed by its pseudotranslocation to the P site, where the formation of the first peptide bond is initiated (13, 46).
These examples clearly demonstrate that there is more than one way for ribopolynucleotides to acquire a structure that would allow the ribosome to bind to it and direct translation initiation in a 5′-end-independent manner. However, there is one common feature about those IRESs studied to date which function in mammalian cells and use mammalian canonical initiation factors and specific mRNA-binding proteins. All of these IRES elements contain highly specific binding sites for some canonical initiation factors (eIF4G and eIF3) or auxiliary mRNA-binding proteins [polypyrimidine tract binding protein, poly(C) binding protein, La, UNR, ITAF45, etc.] (2, 15, 16, 22, 30, 42). Binding sites for polypyrimidine tract binding protein, UNR, and poly(C) binding protein have also been identified in the IRES elements of some cellular mRNAs (24, 33). This accounts, at least in part, for why they are functional only in mammalian cells (7) or even specific tissues (2) and translation systems derived from them.
Recently, the 579-nucleotide-long 5′-UTR of Rhopalosiphum padi virus (RhPV) mRNA has been shown to contain an IRES that functions efficiently in mammalian, plant, and insect in vitro translation systems (38, 47). This suggests that the RhPV IRES may employ a simplified mode of internal ribosome entry. RhPV is an insect virus from the Dicistroviridae family that also includes Plautia stali intestinal virus and cricket paralysis virus. The RNA genomes of these viruses are uncapped and encode two polyproteins in separate open reading frames separated by the intercistronic region (25).
Whereas the structure and function of the intercistronic region as exemplified by the cricket paralysis virus RNA has been well characterized (see above), much less is known about the mode of translation initiation which is employed by the 5′-UTRs of these viral RNAs. Unlike the intercistronic region, the 5′-UTR of RhPV RNA initiates translation at an AUG codon using the initiator tRNA. Strikingly, some large deletions in both the 3′-half and the 5′-half of the RhPV 5′-UTR do not abrogate the RhPV IRES activity, though the exact position of the IRES within the RhPV 5′-UTR has not been determined (47). These intriguing features of the RhPV 5′-UTR prompted us to undertake studies on the mode of function of its IRES.
Here, this mechanism has been characterized by the reconstitution of mammalian 48S initiation complexes from purified components combined with the toeprint assay (5, 27) using both the intact RhPV 5′-UTR and its various deletion derivatives. On the basis of these data and those from probing of the secondary structure of the RhPV 5′-UTR, we suggest that the properties of the IRES are determined by a nonspecific binding of the mRNA recruiting initiation factors within the long unstructured region (380 nucleotides) proximal to the initiation codon. This step is presumably followed by a scanning process which is assisted by the initiation factor eIF1. The single-stranded nature of the RhPV 5′ IRES is therefore proposed to account for its “cross-kingdom” activity in translation initiation.
MATERIALS AND METHODS
Plasmid constructs.
Plasmid pLuc was constructed by inserting a BamHI-XhoI luc sequence-containing fragment from pGEM-Luc (Promega) into the similarly digested pSP72 plasmid (Promega). To obtain pRhPV-Luc, a BamHI fragment from pGEM-CAT/RhPVΔ1/LUC (47) was inserted into BamHI-digested pLuc. To eliminate any possible effect of extraneous vector sequence during mRNA secondary-structure analysis (see below), the remainder of the pSP72 polylinker between BsaI and BamHI in pRhPV-Luc was excised.
A stable stem-loop structure was introduced at the 5′ end of the RhPV IRES-containing mRNA by ligation of the self-annealed and then blunt-ended 5′-AGCTT(CGCGGATCCGCG)5A-3′ oligonucleotide into the SmaI site of pRhPV-Luc, resulting in plasmid pSRhPV-Luc. Deletion mutants of the RhPV 5′-UTR, termed Rd1 to Rd11 (see Fig. 5A), were derived from pSRhPV-Luc by reverse PCR and subsequent PCR-product self-ligation using Pfu Turbo DNA polymerase (Stratagene) and appropriate pairs of oligodeoxynucleotides. Therefore, all these deletion derivatives of the RhPV 5′-UTR contained the stem-loop structure at the 5′ end of the corresponding RNA transcripts.
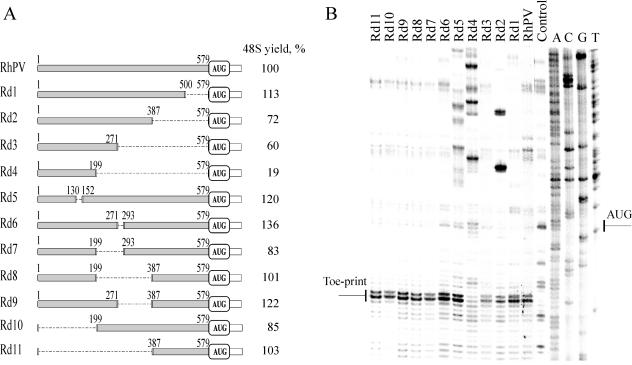
FIG. 5.
Deletion analysis of the RhPV IRES. (A) A schematic representation of the RhPV 5′-UTR deletion derivatives fused to firefly luciferase coding region (for details see Materials and Methods). Although not shown in the scheme, note that all the constructs bear a stable stem-loop structure on their 5′ end. Figures show the terminal nucleotides in the RhPV 5′-UTR that are still present in the constructs. Shaded and open bars represent the RhPV 5′-UTR and luc sequences, respectively. The yield of 48S complexes was estimated from the data of toeprint assays (shown in the right-hand column) and represents the average of three independent experiments. (B). Toeprint analysis of the 48S complex assembly on the RhPV Luc mRNA carrying various deletions within its 5′-UTR. 48S complexes were formed in the presence of eIF1, -1A, -2, -3, and -4A, p100, Met-tRNAiMet and 40S ribosomal subunits. The control lane shows the absence of a toeprint when eIF2 and Met-tRNAiMet were omitted. Other lanes are marked with names of the constructs.
Expression plasmids pET28(His6-eIF4G613-1560) and pET(His6-eIF4A), which were used to produce the recombinant C-terminal fragment (termed p100) of eIF4G and eIF4A, were described previously (27). Similarly, plasmids pQE(His6-eIF1), pET(His6-eIF1A), and pTRM-1, which direct the expression of the recombinant initiation factors eIF1 and eIF1A and the mammalian initiator tRNA, respectively, were also described previously (29, 31, 35). Plasmid pET21(MetRSase-His6) was used to prepare the recombinant Escherichia coli methionyl-tRNA synthetase (41) and was kindly provided by P. Sergiev. The pET(His6-R362Q) expression vector which encodes the eIF4A R362→Q mutant was produced from pET(His6-eIF4A) by reverse PCR using oligonucleotides 5′-AAGGTGGACGGTTTGGCCGTAAAGG-3′ and 5′-GACCGATTCTGTGGATATAGTTTTCC-3′ as the forward and reverse primers, respectively (the introduced mutation is underlined). All molecular cloning manipulations followed standard protocols and the resulting constructs were confirmed by sequencing.
In vitro transcription and translation.
Plasmid pRhPV-Luc and all its derivatives were linearized prior to transcription by digestion with Bpu14I or XbaI for toe-print analysis or in vitro translation, respectively. For sucrose gradient analysis, the RNA was synthesized in the presence of [α-32P]UTP as described previously (27). In vitro translation in rabbit reticulocyte lysates was performed in the presence of [35S]methionine (Amersham Biosciences) essentially as recommended by the manufacturer (Promega). Briefly, 4 μl of rabbit reticulocyte lysate was incubated with 0.2 μl of [35S]methionine (>1,000 Ci/mmol), 0.1 μl of amino acid mixture, and 20 μg/ml of mRNA in a total volume of 7 μl.
RNA secondary-structure analysis.
Chemical and enzymatic RNA secondary-structure probing was carried out as described previously (15). Cleaved or modified RNA was analyzed by primer extension. For this, a set of 32P-labeled oligonucleotides complementary to nucleotides 480 to 499, 366 to 386, 251 to 270, or 130 to 147 of the RhPV mRNA, or nucleotides 3 to 21 of the firefly luciferase coding sequence were used, as appropriate.
Purification of 40S ribosomal subunits, initiation factors, and Met-tRNAiMet.
The 40S ribosomal subunits, eIF3, eIF4B, and eIF4F were purified from cytoplasmic extracts of Krebs-2 and HeLa cells as indicated previously (6), eIF2 was prepared from rabbit reticulocyte lysate according to the protocol described previously (23). Recombinant eIF1 (29), eIF1A (35), eIF4A (27), eIF4A(R362Q) (26, 30), eIF4G613-1560 (27), and MetRSase (41) were purified as described, with the only exception that Mono Q-chromatography was used to further purify eIF4A and eIF4A(R362Q) after Ni-nitrilotriacetic acid column purification. Apparently homogeneous proteins eluted at 250 mM KCl.
In vitro transcribed nonmodified tRNAiMet (31) was aminoacylated using recombinant MetRSase according to the procedure described previously (27) for the total aminoacyl tRNA synthetase pool from E. coli cells.
Assembly and analysis of 48S translation initiation complexes.
For toe-printing analysis ribosomal 48S complexes were assembled in a reaction volume of 20 μl as described earlier (5). The reaction mixture contained 40S subunits (2.5 pmol), an mRNA (0.5 pmol), eIF1 (0.5 μg), eIF1A (0.5 μg), eIF2 (2 μg), eIF3 (3 μg), eIF4A (0.5 μg), eIF4B (0.5 μg) where indicated, eIF4F (0.5 μg) or p100 (27), a C-terminal fragment of eIF4G lacking the eIF4E binding site (0.5 μg), Met-tRNAiMet (5 pmol), ATP (1 mM), and GTP (0.4 mM). Primer extension was performed with the oligonucleotide 5′-TGCAGTTGCTCTCCAGCG-3′ that is complementary to nucleotides 62 to 80 of the firefly luciferase coding sequence. Analysis of the resulting cDNA was performed using denaturing 6% polyacrylamide gel electrophoresis (PAGE) as described before (5, 27). Radioactive bands were visualized with a PhosphorImager (Molecular Dynamics). The yield of 48S complexes was determined as the ratio of the total radioactivity in the toeprint bands to the total radioactivity in the corresponding lanes below and including the toeprint bands using Image Quant 5.0 software.
For sucrose density gradient analysis, performed as described before (5), 48S complexes were assembled in 50-μl volumes, maintaining the concentration of each component as for the toe-printing assays. After 5 min of incubation at 30°C, the reactions were quenched by adjusting the Mg2+ concentration to 30 mM. Then the reaction mixture was layered onto a 5 to 20% sucrose density gradient, and centrifuged in a SW41 rotor (Beckman) at 33,000 rpm for 4.5 h at 4°C.
To assess mRNA integrity during in vitro tests, RNA from the 48S or free RNA peaks of the sucrose gradients was purified by phenol deproteinization, precipitated with ethanol and analyzed by 8% PAGE. Radioactive bands were visualized with a Phosphorimager (Molecular Dynamics).
RESULTS
RhPV IRES displays limited requirements for the canonical translation initiation factors.
We investigated the translation initiation factor requirements for RhPV IRES activity, as determined by the assembly of 48S translation initiation complexes from totally purified translational components (5, 27). This approach has proven to be very fruitful in the elucidation of the function of several viral IRES elements (for review see 11). Briefly, it is based on the primer extension inhibition of reverse transcription from an oligodeoxynucleotide, which is hybridized downstream of the initiation codon of an mRNA. The arrest of reverse transcription always occurs at the same positions, 16 to 18 nucleotides downstream of the A in the AUG initiation codon and only if the initiator tRNA has already formed the codon-anticodon complex with the initiation triplet.
Figure 1 shows the data from toeprinting of the complexes assembled in the complete system (containing all known canonical translation initiation factors needed to form the 48S complex with standard cap-dependent mRNAs (27, 29) or with the omission of one or a group of these factors. The yield of 48S complexes was assessed as described in Materials and Methods and is shown in the upper panel of Fig. 1. The entire 5′-UTR of RhPV RNA linked to the coding region for the N-terminal portion of the luciferase reporter was used throughout these assays, as it was shown earlier that this sequence does not significantly alter the IRES activity compared to the native coding region (47). It should be noted that all RNAs used in these experiments contained a stable stem-loop structure at their 5′ ends as an additional block to the 5′ end-dependent translation initiation. As expected, the presence or absence of such a stem-loop structure in the initial RhPVLuc RNA had no effect on its translational efficiency in rabbit reticulocyte lysates (data not shown).
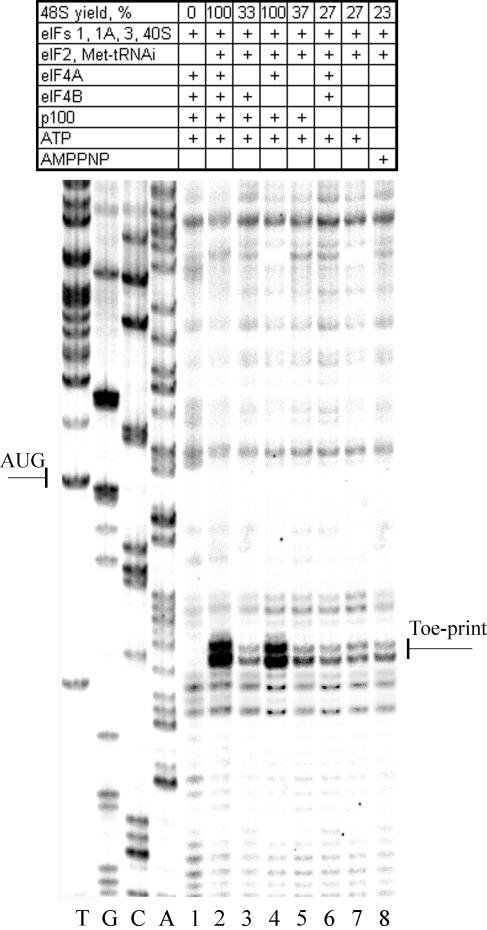
FIG. 1.
Factor requirements for the 48S complex formation on the RhPV-Luc mRNA. Lane 1 corresponds to the control experiment in which eIF2 and Met-tRNAiMet were omitted. The yield of the 48S complexes was estimated as described in Materials and Methods and is presented in the upper line of the table. The values represent the average from three independent assembly experiments. A dideoxynucleotide sequence generated with the same primer was run in parallel (shown on the left of the gel). The AUG initiation codon and the toeprint bands are marked.
The assembly of 48S complexes occurred with a similar efficiency on the RhPV 5′-UTR irrespective of whether native eIF4F plus eIF4A or the recombinant C-terminal fragment of eIF4G (p100) plus eIF4A was used in these tests. This is an expected result since the RhPV RNA is not capped and hence should not require eIF4E or poly(A) binding protein to form 48S initiation complexes. Strikingly, however, the omission of eIF4G or eIF4A or all factors of group 4 did not completely abrogate the formation of the 48S complex on the RhPV IRES (Fig. 1). Some formation of the complex (23% of the maximum yield) was even observed in the presence of a nonhydrolysable analog of ATP, AMPPNP, when the initiation factors of group 4 and ATP were also excluded from the reconstitution system. Under such conditions, the 48S complex formation with standard 5′-end-dependent mRNAs (whether capped or uncapped) does not occur (29) (data not shown). This suggests that the 40S ribosomal subunit is able, albeit with a relatively low efficiency, to accommodate the RhPV IRES and locate its initiation codon without the assistance of the group 4 factors and in the absence of ATP hydrolysis. Remarkably, the omission of eIF4B from the reconstitution system had no effect on the yield of the 48S complex, suggesting the absence of any significant secondary structure in the region of the RhPV 5′-UTR between its 40S ribosome “landing pad” and the initiation codon (6).
Initiation factor eIF1 is essential for the 40S ribosomal subunit to locate the initiation codon of the RhPV RNA.
The unusual requirements of the RhPV IRES for the canonical translation initiation factors are clearly distinct from those which have been found previously for the standard cap-dependent mRNAs or certain viral IRESs studied by the same approach. For instance, cap-dependent mRNAs require the whole set of initiation factors (6, 29). The encephalomyocarditis virus and foot-and-mouth disease virus IRESs show an absolute requirement for the initiation factors eIF4F and eIF4A (or p100 plus eIF4A) and are stimulated to a significant extent by eIF4B (22, 27, 34). However, translation initiation on these IRESs does not involve scanning and hence the scanning factor eIF1 is not required (27, 29). eIF1 is not needed for the hepatitis C virus and related IRESs, either (30).
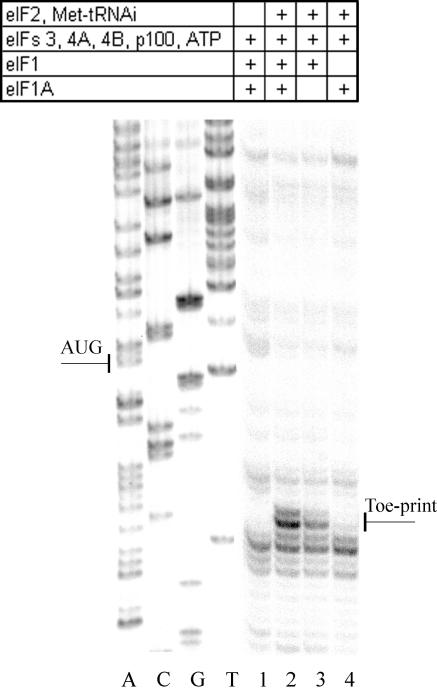
The question arises whether the scanning process is an obligatory step to allow the 40S ribosomal subunit to reach the initiation codon after the primary binding of the subunit to a sequence within the RhPV 5′-UTR. Figure 2 clearly demonstrates that the scanning initiation factor eIF1 is indispensable for the 40S subunit to locate the AUG codon. On the other hand, omission of the other small initiation factor, eIF1A, resulted in a partial, albeit rather significant, inhibition of 48S complex formation. Such an effect of eIF1A omission is in good agreement with its suggested primary role of stimulating the formation and stability of the 43S preinitiation complex (32).
FIG. 2.
Effect of the initiation factors eIF1 and eIF1A on the reconstitution of 48S initiation complexes on the RhPV-Luc mRNA as revealed by toeprint analysis. Lane 1 corresponds to the control in which eIF2 and Met-tRNAiMet were omitted. A dideoxynucleotide sequence generated with the same primer was run in parallel (shown on the left of the gel). The AUG initiation codon and the toeprint bands are marked.
48S translation initiation complexes are formed on the full-length 5′-UTR of RhPV RNA.
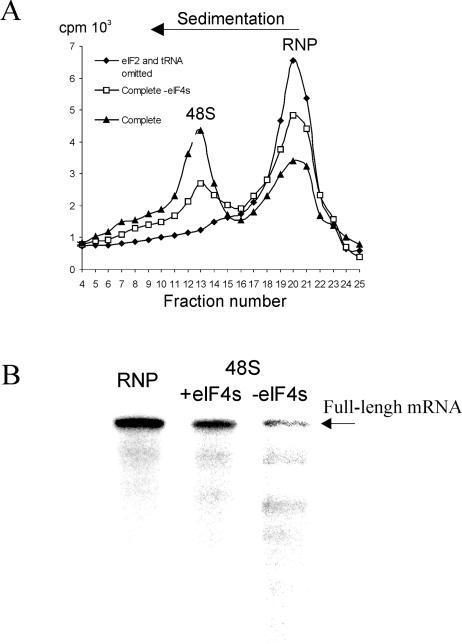
It is possible that the 48S complexes seen in the toeprint bands in Figs. 1 and 2 could have been assembled on shorter RNA fragments which contaminated the initial RNA preparation or were generated from the full-length RNA during the incubation with factors. To exclude this possibility (which was not addressed in similar studies with other IRESes), the transcript containing the RhPV IRES sequence was uniformly labeled with 32P, added to the 48S complex reconstitution mixture, and then the complex was separated by sucrose gradient sedimentation from unbound mRNP (Fig. 3A). The RNA extracted from both peaks was analyzed separately on a denaturing gel. As is evident from Fig. 3B, the 48S complex incorporated the initial full-length transcript added to the reconstitution system whether the complex was assembled with the complete set of initiation factors minus eIF4B or with just eIF1, eIF1A, eIF2, and eIF3. It is not surprising that some degradation of the RNA occurred during sucrose gradient centrifugation but the presence of any full-length RNA offers compelling evidence that the 48S complex does form on the transcript both in the presence and the absence of eIF4A and eIF4G. As expected, in the latter case the yield of the complex is significantly lower.
FIG. 3.
Analysis of the RhPV-Luc mRNA integrity during the 48S complex assembly. (A) Sucrose gradient sedimentation of 48S initiation complexes formed on the RhPV-Luc mRNA with different sets of translation initiation factors. 48S complexes were assembled on the uniformly 32P-labeled mRNA. eIF4B was not present for the assembly of these complexes. Shaded diamonds show a control profile when eIF2 and Met-tRNAiMet were omitted. Shaded triangles and open squares demonstrate the 48S assembly with or without eIF4A and eIF4G, respectively. The peaks corresponding to the 48S complex and free mRNPs are indicated. The direction of sedimentation is indicated. (B). An autoradiograph of the gel after electrophoresis of RNAs isolated from the 48S and mRNP gradient fractions. The position of the full-length transcript is shown by an arrow.
Effect of the dominant negative mutant of eIF4A (R362Q) on protein synthesis directed by the RhPV IRES.
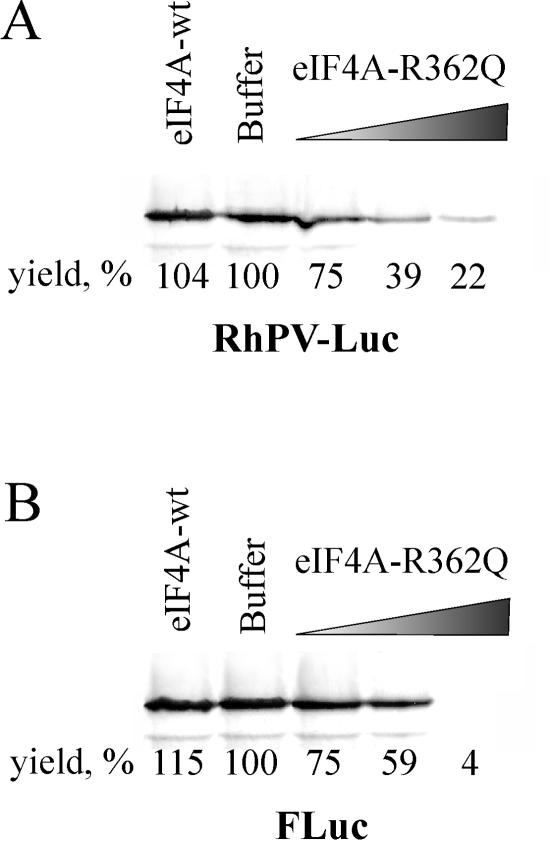
As shown in the previous section, the 48S complex on the RhPV IRES may be reconstituted in the purified system in the absence of eIF4G and eIF4A. However, the strong stimulation of its formation by these initiation factors suggests that under more natural conditions, the RhPV IRES activity should be inhibited by dominant negative mutants of eIF4A, which are able to “freeze” translation initiation complexes in an inactive form, thereby sequestering mRNA (26). Nevertheless, one may expect that in the case of the RhPV IRES some of the initiation complexes would escape this inhibition.
To address this possibility, the effect of the dominant negative mutant eIF4A (R362Q) (26) on the translation of the RhPV IRES-containing mRNA in rabbit reticulocyte lysates was studied and compared with that for a control mRNA, produced from the pFluc plasmid, which could only use a 5′-end-dependent mode of translation initiation. As shown in Fig. 4, the translation of the RhPV IRES-containing mRNA was less sensitive to addition of large amounts of the mutant eIF4A than the control transcript, which was apparently completely inhibited at the highest R362Q concentration, suggesting that some translation initiation could still occur on the RhPV IRES without participation of eIF4G and eIF4A. Addition of wild-type eIF4A had no significant effect on translation efficiency (Fig. 4).
FIG. 4.
Dominant-negative effect of the eIF4A (R362Q) mutant protein on a 5′-dependent translation and translation mediated by the RhPV IRES. Rabbit reticulocyte lysate (7 μl) was incubated with buffer (lane 2), with wild-type eIF4A (0.5 μg) (lane 1), or the eIF4A mutant (0.1, 0.2, and 0.5 μg) (lanes 3 to 5) for 5 min at 30°C and then incubated for 60 min at 30°C with (A) RhPV-Luc or (B) firefly luciferase mRNAs (0.5 μg) as described in Materials and Methods. Translation products were separated by sodium dodecyl sulfate-15% PAGE and analyzed by autoradiography. The yield of translational products represents the average of five independent experiments.
It should be noted that these data may not be directly correlated with those obtained for the assembly of 48S complexes from purified components (Fig. 1). The mechanism of inhibition of translation by dominant negative eIF4A mutants is poorly understood and it is not known why the extent of inhibition depends on the particular mRNA used in these assays (26). The possibility exists that the R362Q mutant of eIF4A may put out of play not only eIF4G plus eIF4A but also other translation initiation components. Furthermore, it may prevent recycling of the translation initiation process which could further complicate such a correlation.
RhPV IRES does not contain specific binding sites for translation initiation components.
In our previous studies (38, 47), the RhPV IRES has not been fully mapped within the 5′-UTR of RhPV RNA, though the effect of some deletions within this 5′-UTR has been tested using the translation of dicistronic RNAs (CAT-RhPV 5′-UTR-Luc) in rabbit reticulocyte lysates, wheat germ extract, and insect cell lysates. Strikingly, deletions of 100 and 200 nucleotides from the 3′ end and 100 nucleotides from the 5′ end of the RhPV 5′-UTR did not abrogate IRES activity (47).
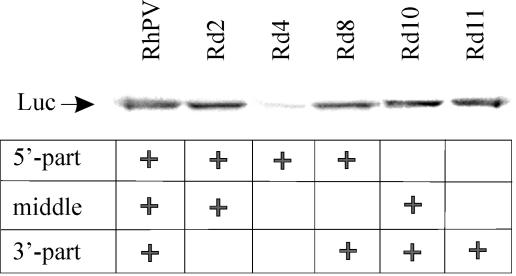
To map the IRES more precisely, additional deletion derivatives have now been prepared (Fig. 5A). To exclude the possibility that some mutant RNAs, especially those with short 5′-UTRs, may use a 5′-end-dependent rather than an IRES-directed mode of translation initiation, a highly stable helix was always included at the 5′ termini of these RNAs. The mutant RNAs were tested for their ability to support the assembly of specific 48S complexes in the toeprint assay (Fig. 5B). From these results one can see that progressive deletions from the 3′ end of the RhPV 5′-UTR only marginally affected RhPV 5′ IRES activity. It should be noted that these mutant RNAs were not only devoid of long sequences from within the 5′-UTR of RhPV RNA, they also differed in sequences proximal to the initiation codon, except for the immediate nucleotide context of the initiation codon.) Shorter deletions (e.g., constructs Rd5 or Rd6) had no adverse effect on the efficiency of 48S assembly. A dramatic drop in RhPV IRES activity was only observed (see Fig. 5B) when the sequence between nucleotides 199 to 579 was completely removed (Rd4).
These data are in good agreement with those obtained by the translation in rabbit reticulocyte lysates of mRNAs carrying large deletions of the RhPV IRES in various parts of its sequence. As seen from Fig. 6, only the deletion of the sequence between positions 199 and 579 had a dramatic effect on the translation efficiency. These results substantially extend and complement our previous data on the deletion analysis of the RhPV IRES (47) and suggest the absence of specific binding sites for translation initiation components, e.g., eIF3, eIF4G, or the 40S ribosomal subunit (see Discussion).
FIG. 6.
Translation in rabbit reticulocyte lysates of mRNAs carrying large deletions in various parts of the RhPV 5′-UTR. The + sign denotes the presence of a particular part of the RhPV 5′-UTR sequence in the RNA transcripts which each encoded the Luc protein. Samples were analyzed by sodium dodecyl sulfate-PAGE and autoradiography. The Luc product is indicated.
40S ribosomal subunit and eIF3 form a stable complex with the RhPV IRES.
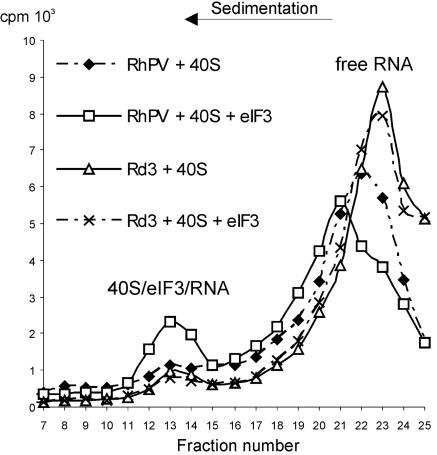
The ability of the RhPV IRES to form 48S complexes in the absence of group 4 factors raised the question as to which other components of the translation initiation apparatus might play the essential role in recruiting this IRES onto the 40S ribosomal subunit. Among them, the best candidates appeared to be the initiation factor eIF3 (whose RNA-binding properties are well documented) and the 40S ribosomal subunit itself. Therefore, the ability of the RhPV IRES to form complexes with eIF3 and the 40S ribosomal subunit was analyzed by sucrose gradient sedimentation. As seen in Fig. 7, the complete RhPV IRES formed a stable complex with the 40S subunit, eIF3 being indispensable for such an interaction. In contrast, the arbitrarily chosen deletion mutant of the RhPV IRES (Rd3) with a strongly reduced IRES activity (see Fig. 5B) did not bind efficiently to the 40S ribosomal subunit. Although no such complexes were observed in the absence of eIF3, it is quite probable that eIF3, and the mRNA-binding channel of the 40S subunit, both contribute to the recruitment of the RhPV IRES onto the 40S ribosomal subunit.
FIG. 7.
Binding of the RhPV IRES to the 40S ribosomal subunit and eIF3. The 32P-labeled RhPV-Luc and Rd3 RNAs were incubated with the 40S alone or with the 40S and eIF3 and the incubation mixtures were analyzed by 5 to 20% sucrose gradient centrifugation. Shaded diamonds show the RhPV plus the 40S, blank squares show the RhPV plus the 40S and eIF3, open triangles and crosses correspond to Rd3 plus 40S and Rd3 plus 40S and eIF3, respectively. The positions of unbound mRNA and the ternary 40S/eIF3/mRNA complex peaks are indicated.
RhPV IRES activity is determined by the single-stranded nature of the 380-nucleotide sequence proximal to the AUG start codon.
The data presented in the previous section indicated that the 40S ribosomal subunit enters onto the RhPV IRES within the sequence 199 to 579 and that various parts of this region can serve as a “landing pad” for the 40S subunit. The question arises as to what structural elements of this region determine such unprecedented features of the RhPV IRES compared with other viral IRES elements studied to date? One of the reasons underlying these features may be the low G+C content (34%) of the 5′-UTR of RhPV. However, the G+C content of the 5′ part of the RhPV IRES (which has a very low IRES activity) and that of its 3′ part (with a high IRES activity) do not differ significantly.
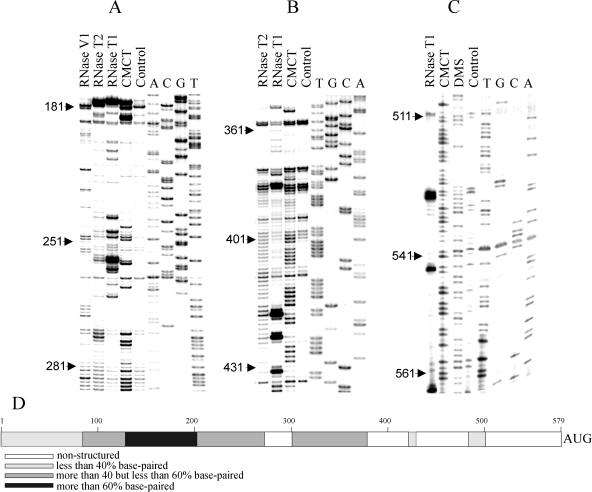
To get some idea of the secondary structure of the 5′-UTR of RhPV RNA, a chemical and enzymatic probing of its nucleotide base-pairing was carried out. Dimethylsulfate and carbodiimide were used for probing the base-paired A and U residues, respectively. For enzymatic probing, a nonspecific nuclease T2 and the guanyl-specific RNase T1 were used to determine unpaired nucleotide bases, whereas the nuclease V1 was used to locate base-paired regions of the RhPV 5′-UTR.
Examples of gels with the carbodiimide, dimethyl sulfate, T1, and T2 probing are presented in Fig. 8A to C. From this analysis combined with the computer-predicted folding of the RhPV RNA 5′-UTR, its secondary structure was generated (to be published elsewhere). Based on this structure, the percent of base-paired nucleotides for different segments of the 5′-UTR of RhPV RNA was estimated (see Fig. 8D). It was found that the 5′-end-proximal part of this 5′-UTR contains well-defined stem-loop structures (this section is presumably involved in transcription of the RhPV RNA), the middle part revealed a rather loose secondary structure, whereas the 3′ segment proximal to the initiation codon was in a single-stranded conformation. Of the 200 nucleotides of this segment, 74% were accessible to nucleases and chemicals. Moreover, no cleavage sites for the V1 nuclease were detected in this region (data not shown). Thus, the key part of the RhPV 5′-UTR that determines its unusual IRES properties possesses a single-stranded conformation.
FIG. 8.
Chemical and enzymatic probings of the secondary structure of the 3′-terminal part of the RhPV-Luc 5′-UTR. (A). The RhPV RNA central part structure probing with RNases T1, T2, and V1, and carbodiimide (CMCT). (B) Chemical (carbodiimide) and enzymatic (RNases T1 and T2) RNA structure probing of the RhPV-Luc 5′-UTR region between nucleotide positions 350 and 430. (C) Chemical (carbodiimide and dimethyl sulfate) and enzymatic (RNase T1) RNA structure probing of the AUG-proximal part of the RhPV-Luc mRNA. In all gels the control lane shows primer extension using nonmodified mRNA as a template. The accessibility of C residues was not tested in the work. (D) A diagram showing the degree of nucleotide base-pairing in different sections of the 5′-UTR of RhPV RNA.
DISCUSSION
We have recently shown that the 5′-UTR of RhPV RNA contains an IRES that functions efficiently in mammalian, plant, and insect in vitro translation systems (38, 47). These unprecedented and intriguing features of the RhPV IRES have prompted us to undertake studies on its structure and function. In this paper, we have presented structural and functional data that shed light on the molecular mechanisms underlying these “cross-kingdom” properties of the RhPV IRES. The most striking feature of this IRES is that it does not lose its translation initiation activity as a result of deletions, both small and large, in any region of its nucleotide sequence. In full agreement with these data, we have also shown that the 3′ sequences can function at least as well as the whole IRES within a dicistronic mRNA context in rabbit reticulocyte lysates and wheat germ extract (Groppelli et al., unpublished observations). This obviously contrasts with other viral IRESs studied to date, where small deletions or even point mutations are able to completely abrogate IRES activity, due to destruction of highly specific binding sites of initiation factors, auxiliary mRNA-binding proteins, or the 40S ribosomal subunit itself (3, 8, 16, 22, 30, 37, 42).
In the case of the RhPV IRES, the specific binding sites for translational components characteristic of other viral IRESs are replaced by a long single-stranded nucleotide sequence, various parts of which can presumably serve as a “landing pad” for the translation initiation apparatus. As evident from the results presented in this study, the longer this single-stranded region, the better. We speculate that this tandem array of potential landing pads may increase the probability and the rate of binding of the 40S ribosomal subunit and/or mRNA-recruiting translation initiation factors (e.g., eIF4G and eIF3) to the IRES. The minimum size of an individual landing pad has not been determined in this work. However, it should be large enough to efficiently accommodate the bulky translational components such as eIF3, eIF4G plus eIF4A and the 40S ribosomal subunit.
The requirements of the RhPV IRES for canonical translation initiation factors are also distinct, both quantitatively and qualitatively, from other viral IRESs whose factor requirements have been determined. Like most other viral IRESs (except that within the cricket paralysis virus intercistronic region), the RhPV IRES from within the 5′-UTR cannot function without initiation factors eIF2 and eIF3. However, unlike well-studied IRESs from the cardio- and aphthoviruses (picornaviruses), hepatitis C virus (flaviviruses) and pestiviruses, on which ribosomes are not presumably involved in extensive scanning, the RhPV IRES-directed translation initiation is absolutely dependent on the presence of eIF1.
Thus, after the primary binding of a 43S preinitiation complex to a nucleotide sequence within the long unstructured region of the 5′-UTR of RhPV RNA (presumably assisted by eIF4G plus eIF4A, see below), the 48S complex appears to scan along the RhPV IRES to locate the initiation codon. eIF4G plus eIF4A plus ATP greatly stimulate this process, whereas eIF4B has no effect. The absence of any effect of eIF4B on RhPV IRES activity lends additional support to our conclusion that there is no significant nucleotide base-pairing in the region between the 40S ribosome entry site(s) and the initiation AUG codon, since the main functional role of eIF4B is to assist eIF4A in opening base-paired regions within the 5′-UTRs of mRNAs. As we have recently shown, its presence and functional state are especially important for 5′-UTRs that have some base-pairing in their sequences and is dispensable for unstructured 5′ leaders (6).
The initiation factor eIF4G appears to be a good candidate for the primary recruitment of the RhPV IRES onto the 40S ribosomal subunit through the interactions of eIF4G, eIF3 and the 40S subunit (19). The complex mRNA-binding site of mammalian eIF4G can bind specific structural elements in picornavirus IRESs as exemplified by the J-K domains of the encephalomyocarditis virus and foot-and-mouth disease virus IRESs (16, 22). These specific elements are not recognized by eIF4G of plant origin (28). On the other hand, in the case of 5′-end-dependent translation initiation, eIF4G seems to interact with 5′-UTRs of these mRNAs via its general mRNA-binding site (20, 36).
The amino acids constituting this nonspecific mRNA binding site within the eIF4G molecule have not been determined precisely as yet, though they are thought to be located close to the eIF4E binding site (20, 36). It is logical to suggest that this nonspecific mRNA-binding site, whatever the origin of the eIF4G, is responsible for binding to the RhPV IRES, thereby accounting, at least in part, for its “cross-kingdom” properties. However, as shown by Lomakin et al. (20), the affinity of the nonspecific interaction of eIF4G with RNA is two orders of magnitude lower than that of its specific binding to the J-K domain of the encephalomyocarditis virus IRES. Therefore, to ensure a sufficiently stable binding of the translational apparatus to the RhPV IRES, other translation initiation components should also participate in this binding.
Our studies provide convincing evidence that despite complete omission of eIF4G and eIF4A and the presence of AMPPNP, some formation of the 48S initiation complex on the RhPV IRES still occurs. Earlier data suggested that RNA possessing reduced secondary structure may bind 40S subunits in the presence of eIF3 independently of Met-tRNAi and ATP (40). More recently, Kolupaeva et al. (17) reported that U-rich sequences are capable of forming a ternary complex with 40S and eIF3. This might be the case for the RhPV IRES, since its functional part is single stranded and U-rich. Indeed, the full-length RhPV IRES but not the deletion mutant which lacks 300 nucleotides from the 3′ region is able to form a ternary complex with eIF3 and 40S in the absence of eIF2, Met-tRNA, or any of the eIF4 group proteins (Fig. 6). This binding may be the key feature of the RhPV IRES function.
These considerations imply that the mammalian 40S ribosomal subunit (assisted by eIF3) can accommodate the single-stranded RhPV IRES in its mRNA-binding cleft, even in the absence of the initiation factors of group 4 and ATP hydrolysis. This primary accommodation of the IRES by the 40S subunit is presumably followed by scanning of the downstream sequence promoted by the scanning factor eIF1 (29, 32) until the initiation AUG codon is located. Such a situation is reminiscent of the prokaryotic mode of translation initiation where the initiation factor IF3 performs the role of eIF1 (21, 32). It is plausible to suggest that the single-stranded nature of the RhPV IRES accounts for its strong but less selective potential to bind key mRNA recruiting components of the translation initiation apparatus of different origin. One may speculate that the simultaneous interaction of the unstructured RhPV IRES with the mRNA-binding cleft of the 40S subunit and initiation factors eIF3 and eIF4G may provide (additively or synergistically) sufficient stability of the primary complex of the translational initiation machinery with the 5′-UTR of RhPV RNA required for the efficient progression of the translation initiation process.
The formation of the 48S complex in the absence of group 4 initiation factors and ATP hydrolysis has been recently described for an artificial RNA construct (32) whose 5′-UTR was represented by the 100% single-stranded (CAA)n sequence (43). However, this 5′ leader was not able to function as an IRES since the insertion of a stable stem-loop structure at its 5′ end completely blocked formation of the 48S complex. One of the possible explanations for this result is that the single-stranded region separating the 5′-terminal stem-loop and the initiation codon in this construct was not long enough (only about 40 nucleotides) to accommodate the translation apparatus and hence to direct translation initiation in a 5′-end-independent way. The probability that the nucleotide composition of single-stranded RNA regions can affect their IRES activity may not be excluded, as “cross-kingdom” properties of IRES elements represented by long purine-rich sequences have been reported (5).
Our data do not imply that any long single-stranded sequence will work as an IRES in any cell of any origin. However, they clearly show that, in principle, there is no intrinsic block to the eukaryotic ribosome entering an internal RNA region, even if this region does not comprise a structural element with a sophisticated organization that specifically binds the translation initiation apparatus. This contrasts with the strict rules of the scanning hypothesis (18) which imply that the selection of the 5′ termini of mRNAs by eukaryotic ribosomes during translation initiation is an intrinsic property of the eukaryotic translation machinery. Thus, these data contribute to a growing body of evidence which suggests that there are many more similarities between prokaryotic and eukaryotic ribosomes than has previously been thought. The simplified mode of internal ribosome entry onto the 5′-UTR of RhPV RNA may confer on RhPV a greater versatility to survive and replicate under conditions of competition with cellular mRNAs of host cells.
It should be noted, however, that a low GC content of the 5′-UTR of RhPV RNA is not a common feature of members of the Dicistroviridae (cricket paralysis virus, Drosophila C virus, shrimp paralysis virus, Plautia stali intestinal virus, and others), which more likely have “conventional” IRESs due to their high GC content and multiple upstream AUG codons, some of which are in optimal nucleotide context. Remarkably, the cricket paralysis virus 5′ IRES does not support cross-kingdom activity, as it does not function in the wheat germ extract system (45). Further experiments should demonstrate what molecular mechanisms of translation initiation are employed by other members of this family.
Acknowledgments
We thank Yan Dunaevsky for help in FPLC purification of native and recombinant translation initiation factors, Petr Sergiev for providing us with the expression clone for E. coli MetRSase, and Andrey Pisarev for some participation at the initial step of the work.
This work was supported in part by a grant from INTAS (project number 01-0293) to G.J.B. and I.N.S., by grant 02-04-48798 from the Russian Foundation for Basic Research to I.N.S., and by a BBSRC award to L.O.R. and G.J.B.
REFERENCES
- 1.Ali, N., and A. Siddiqui. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 94:2249-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA. p. 869-900. In N. Sonenberg, Hershey J.W.B., and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Collier, A. J., J. Gallego, R. Klinck, P. T. Cole, S. J. Harris, G. P. Harrison, Aboul- F. Ela, G. Varani, and S. Walker. 2002. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 9:375-380. [DOI] [PubMed] [Google Scholar]
- 4.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dmitriev, S. E., A. V. Pisarev, M. P. Rubtsova, Y. E. Dunaevsky, and I. N. Shatsky. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 533:99-104. [DOI] [PubMed] [Google Scholar]
- 6.Dmitriev, S. E., Y. M. Terenin, Y. E. Dunaevsky, W. C. Merrick, and I. N. Shatsky. 2003. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 23:8925-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorokhov, Y. L., M. V. Skulachev, P. A. Ivanov, S. D. Zvereva, L. G. Tjulkina, A. Merits, Y. Y. Gleba, T. Hohn, and J. G. Atabekov. 2002. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA 99:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Miragall, O., and E. Salas. 2003. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9:1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez, J., I. Yaman, C. Huang, et al. 2005. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol. Cell 17:405-416. [DOI] [PubMed] [Google Scholar]
- 10.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 12.Hershey, J. W. B., and R. J. Jackson. 2000. The pathway and mechanism of initiation of protein synthesis., p. 33-88. In N. Sonenberg, M. B. Mathews, and Hershey J.W.B. (ed.), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, CSHL, New York.
- 13.Jan, E., T. G. Kinzy, and P. Sarnow. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. USA 100:15410-15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolupaeva, V. G., C. Hellen, and I. Shatsky. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199-1212. [PMC free article] [PubMed] [Google Scholar]
- 16.Kolupaeva, V. G., T. V. Pestova, C. U. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditic virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 17.Kolupaeva, V. G., A. Unbehaun, I. B. Lomakin, C. U. Hellen, and T. V. Pestova. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11:470-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translation initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 20.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical initiation of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G with to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomakin, I. B., V. G. Kolupaeva, A. Marintchev, G. Wagner, and T. V. Pestova. 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17:2786-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez deq Uinto, S., E. Lafuente, and E. Salas. 2001. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7:1213-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrick, W. C. 1979. Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 60:101-108. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell, S. A., K. A. Spriggs, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11:757-771. [DOI] [PubMed] [Google Scholar]
- 25.Moon, J. S., L. L. Domier, N. K. McCoppin, C. J. D'Arcy, and H. Jin. 1998. Nucleotide sequence analysis shows that Rhopalosiphum padi virus is a member of a novel group of insect-infecting RNA viruses. Virology 243:54-65. [DOI] [PubMed] [Google Scholar]
- 26.Pause, A., N. Methot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF4A define a crucial role for eIF4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova, T. V., I. N. Shatsky, and C. U. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova, T. V., S. I. Borukhov, and C. U. Hellen. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854-859. [DOI] [PubMed] [Google Scholar]
- 30.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova, T. V., and C. U. Hellen. 2001. Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA 7:1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., and V. G. Kolupaeva. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickering, B. M., S. A. Mitchell, K. A. Spriggs, M. Stoneley, and A. E. Willis. 2004. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 24:5595-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 35.Pisarev, A. V., M. A. Skabkin, A. A. Thomas, W. C. Merrick, L. P. Ovchinnikov, and I. N. Shatsky. 2002. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40S ribosomal subunits to the initiation codon of beta-globin mRNA. J. Biol. Chem. 277:15445-15451. [DOI] [PubMed] [Google Scholar]
- 36.Prevot, D., D. Decimo, C. H. Herbreteau, Roux., F J. Garin, J. L. Darlix, and T. Ohlmann. 2003. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 22:1909-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson, M. E. M., R. A. Seamons, and G. J. Belsham. 1999. A selection system for functional internal ribosome entry site (IRES) elements: Analysis of the requirement for a concerved GNRA tatraloop in the encephalomyocarditis virus IRES. RNA 5:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Royall, E., K. E. Woolaway, J. Schacherl, S. Kubick, G. J. Belsham, and L. O. Roberts. 2004. The Rhopalosiphum padi virus 5′ internal ribosome entry site is functional in Spodoptera frugiperda 21 cells and in their cell-free lysates: implications for the baculovirus expression system. J. Gen. Virol. 85:1565-1569. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki, J., and N. Nakashima. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA 97:1512-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seal, S. N., A. Schmidt, and A. Marcus. 1989. Ribosome binding to inosine-substituted mRNAs in the absence of ATP and mRNA factors. J. Biol. Chem. 264:7363-7368. [PubMed] [Google Scholar]
- 41.Shimizu, Y., A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa, and T. Ueda. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19:751-755. [DOI] [PubMed] [Google Scholar]
- 42.Sizova, D. V., V. G. Kolupaeva, T. V. Pestova, I. N. Shatsky, and C. U. Hellen. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzareva, N. V., V. I. Makhno, and I. V. Boni. 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337:189-194. [DOI] [PubMed] [Google Scholar]
- 44.Vagner, S., B. Galy, and S. Pyronnet. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, J. E., M. J. Powell, S. E. Hoover, and P. Sarnow. 2000. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20:4990-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, J. E., T. V. Pestova, C. U. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511-520. [DOI] [PubMed] [Google Scholar]
- 47.Woolaway, K. E., K. Lazardis, G. J. Belsham, M. J. Carter, and L. Roberts. 2001. The 5′ untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. J. Virol. 75:10244-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaman, I., J. Fernandez, H. Liu, et al. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113:519-531. [DOI] [PubMed] [Google Scholar]