Abstract
The transcription factor RelB is required for proper development and function of dendritic cells (DCs), and its expression is upregulated early during differentiation from a variety of progenitors. We explored this mechanism of upregulation in the KG1 cell line model of a DC progenitor and in the differentiation-resistant KG1a subline. RelB expression is relatively higher in untreated KG1a cells but is upregulated only during differentiation of KG1 by an early enhancement of transcriptional elongation, followed by an increase in transcription initiation. Restoration of protein kinase CβII (PKCβII) expression in KG1a cells allows them to differentiate into DCs. We show that PKCβII also downregulated constitutive expression of NF-κB in KG1a-transfected cells and restores the upregulation of RelB during differentiation by increased transcriptional initiation and elongation. The two mechanisms are independent and sensitive to PKC signaling levels. Conversely, RelB upregulation was inhibited in primary human monocytes where PKCβII expression was knocked down by small interfering RNA targeting. Altogether, the data show that RelB expression during DC differentiation is controlled by PKCβII-mediated regulation of transcriptional initiation and elongation.
Dendritic cells (DCs) constitute a heterogeneous population of professional antigen-presenting cells with a unique role in the activation of naive T lymphocytes and the establishment of immunological tolerance and memory (2, 3). Their central role as regulators of the immune response has led to the examination of the underlying intracellular signaling pathways and gene expression that may regulate the differentiation of precursors into DCs (21, 60), most notably, the Rel/NF-κB family of transcription factors (41, 61).
NF-κB exists in mammals as a homodimer or heterodimer of the Rel proteins NFκB1 (p50/p105), NFκB2 (p52/p100), c-Rel, RelA (p65), and RelB. The dimer can be sequestered in the cytoplasm by the inhibitory IκBs, which in the canonical activation pathway can be phosphorylated, ubiquitinated, and degraded by the proteasome to allow the nuclear translocation of NF-κB and activate transcription (63). On the other hand, the NF-κB2 precursor form p100 can inhibit directly the nuclear translocation of heterodimers due to their IκB-like ankyrin repeats and is activated by an alternative pathway that requires the NF-κB-inducing kinase (NIK) and IKKα (62). In this pathway, the bound precursor p100 is processed in a regulated fashion to the p52 form to allow translocation to the nucleus. The p105 subunit also contains IκB-like ankyrin repeats that could sequester NF-κB dimers in the cytoplasm. However, its processing to the p50 form is constitutive (22, 30).
Overexpression of IκB in mature DCs downregulates major histocompatibility complex (MHC) class II; the costimulatory molecules CD80, CD86, and CD40; and the proinflammatory cytokine tumor necrosis factor alpha (TNF-α), indicating that antigen presentation is dependent on NF-κB function (64). Likewise, the inhibitory p100 precursor can act as a negative regulator of DC function, and DCs of NF-κB2-deficient mice show increased expression of MHC class II and costimulatory molecules (51). The effects of individual NF-κB proteins in DC development and function have been assessed in chimeric and knockout mice. While these studies have shown the generation of functional DCs in mice lacking individual p50, p52, RelA, and c-Rel proteins, the combined deficiency of p50 and RelA results in a severe defect in DC development, suggesting redundant functions for NF-κB subunits (41). RelB, on the other hand, is the transcription factor that has been associated most directly with DC differentiation and function (9, 39, 40, 57-59, 61). RelB protein can be detected in human and mouse DCs, with high expression in interdigitating DCs of the thymic medulla and the deep cortex of lymph nodes (6). In addition, its nuclear expression (as a p50/RelB heterodimer) is one of the hallmarks of DC differentiation and correlates with the degree of maturation, including the activation of the antigen-presenting capacity (9, 40, 43). As a result, RelB-deficient mice show an impaired antigen-presenting cell function and cellular immunity, with a profound decrease in thymus and spleen DCs (61). In addition, antigen-primed DCs with inhibited RelB function were shown to lack typical costimulatory molecules and generated antigen-specific regulatory T cells in vivo (36). Recent studies have also shown that RelB promotes differentiation into DCs, and its inhibition impairs monocyte-derived DC development with no effect on other myeloid differentiation pathways (44).
In contrast to other family members, RelB does not homodimerize and forms heterodimers almost exclusively with the p100, p52, and p50 proteins (14, 46). Furthermore, RelB complexes are not bound by IκB, so RelB can only be inhibited in the cytoplasm by p100 (50) and RelB/p50 and RelB/p52 dimers are constitutively found in the nucleus. Therefore, while NF-κB activity is typically regulated by posttranscriptional events, RelB activity levels in cells expressing the constitutive p50 and/or p52 proteins parallel increases in RelB transcription (16, 25, 28, 55). The role of RelB in DC differentiation and the fact that its upregulation is a very early event in this process (39, 52) suggest that the mechanisms controlling RelB transcription may play an integral role in the differentiation from DC progenitors and in the determination of the functional characteristics of the generated DCs.
Functional studies of the RelB enhancer-promoter region in the HeLa, Jurkat, and BJAB cell lines have shown that the gene is regulated at the level of transcription by two proximal NF-κB binding sites. While a p50 homodimer binds to one of the sites, the other is bound preferentially by heterodimers of p50 and RelA or RelB to induce transcription (5). In addition, potential vitamin D response elements have been identified in the promoter region that may be responsible for a transcriptional inhibition caused by active vitamin D3 analogs (15). While these findings have characterized aspects of the regulation of RelB expression, the regulatory mechanisms in the context of DC differentiation remain largely undefined.
The intracellular signal transduction pathways that initiate DC differentiation are only beginning to be characterized. Primary human CD34+ hematopoietic progenitor cells (HPCs) differentiate into DCs after treatment with the cytokines granulocyte-macrophage colony-stimulating factor (GM-CSF) and TNF-α (7). The GM-CSF receptor can associate through its common beta chain with the receptor for activated C kinase 1 (RACK1) and recruit the activated beta II isoform of protein kinase C (PKCβII) upon treatment with cytokines or the PKC agonist phorbol 12-myristate 13-acetate (PMA) (17). Direct activation of PKC by PMA treatment induces differentiation of human CD34+ HPCs exclusively into mature and functional DCs, while PKC inhibitors can block the cytokine-driven differentiation (52), further suggesting the involvement of PKC signaling in the generation of DCs from primary progenitors. We have previously established the human leukemic cell line KG1 as an in vitro model of DC differentiation from CD34+ progenitors (13, 52). KG1 is a CD34+ myeloblastic cell line that expresses CD86 and MHC class II and, like CD34+ precursor cells, can differentiate into DCs after treatment with GM-CSF plus TNF-α or PMA alone. This model has been used successfully by several groups to analyze features unique to DCs such as antigen cross-presentation (1), DC differentiation (20, 53), and maturation (29), as well as the regulation of DC-specific molecules (49). A key aspect of the model is the existence of the KG1a cell line originally isolated from KG1 that, despite being closely related to its parent cell line (37), fails to differentiate into DCs under any known stimuli (52). This contrasting outcome allows probing of the molecular mechanisms driving DC differentiation.
We have used the KG1/KG1a models to investigate the mechanisms that regulate relB gene expression very early during DC differentiation. We have found that basal NF-κB protein expression is downregulated by PKC in resting cells, but RelB levels are upregulated during differentiation from KG1 cells due to increased transcription initiation and elongation. We have recently shown that the restoration of PKCβII expression in KG1a cells allows their differentiation into DCs (P. J. Cejas et al., submitted for publication). Further analysis of these PKCβII-transfected KG1a cells shows the restoration of increased RelB transcription initiation and elongation during differentiation and indicates that both mechanisms are regulated by PKCβII, can act in an independent fashion, and are sensitive to the levels of PKC activity.
MATERIALS AND METHODS
Cell culture.
KG1 and KG1a cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in Iscove's modified Dulbecco's medium-20% fetal bovine serum-100 mM l-glutamine-100 mM penicillin-streptomycin at 5 × 105 cells/ml as previously described (52). KG1a stable clones βII-E9, -E11, -C10, and -D8 expressing different levels of PKCβII-enhanced green fluorescent protein (EGFP) fusion protein and KG1a-neo control cells (unpublished data) were cultured under the same conditions as KG1 and KG1a cells plus Geneticin at 300 μg/ml. Cells were stimulated with PMA (10 ng/ml; Sigma, St. Louis, MO) for the indicated times.
Confocal microscopy.
KG1a cells were left untreated or treated with PMA (10 ng/ml) for 1 h, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in 1% bovine serum albumin, and stained with an anti-PKCβII antibody (c-18; Santa Cruz Biotechnology, Santa Cruz, CA). Cells were then stained with fluorescein isothiocyanate-conjugated donkey anti-rabbit secondary antibody (Jackson Immunoresearch, West Grove, PA). βII-E9 cells expressing the PKCβII-EGFP fusion protein were treated or not with PMA (10 ng/ml) for 1 h. Both KG1a and βII-E9 cells were imaged by confocal microscopy (Zeiss LSM-510) for the fluorescent tag.
Northern blotting analyses.
RNA was isolated from cells using the RNABee method (Tel-Test, Friendswood, TX) to carry out Northern blotting experiments as previously described (52). Briefly, 20 μg of total RNA/lane from unstimulated and PMA-stimulated cells was separated in a formaldehyde-agarose gel, transferred to a nylon membrane (Hybond-N; Amersham, Aylesbury, United Kingdom) by capillary action, and hybridized to a radiolabeled probe corresponding to exons 1 to 5 of RelB, generated by restriction digestion with AccI and EcoRI of the full-length human RelB cDNA (kind gift from Ulrich Siebenlist, National Institute of Allergy and Infectious Diseases, National Institutes of Health). Blots were washed two times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 56°C before exposure to a phosphorimaging screen (Imaging Screen-K; Bio-Rad, Hercules, CA) and analysis with the Personal Molecular Imager FX System (Bio-Rad). RNA integrity and equal loading were assessed after reprobing the blots with a human full-length glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA radiolabeled probe.
mRNA stability.
Actinomycin D (10 μg/ml; Calbiochem, San Diego, CA) was added to KG1 and KG1a cells that had been left untreated or treated with PMA for 2 h. Aliquots were taken at different time intervals, and the levels of RelB mRNA relative to GAPDH content at each time point were determined by Northern blot analysis.
Western blot analyses.
Protein lysates were generated after disrupting the cells with RIPA buffer (0.1% SDS, 1.0% Triton X-100, 1.0% deoxycholate, 5 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 10 mM benzamidine, 2 μg/ml leupeptin, 100 μM sodium orthovanadate, 10 mM p-nitrophenylphosphate, 10 mM Tris, pH 7.2). Ten micrograms of protein/lane was separated on an 8% SDS-polyacrylamide gel and transferred to nitrocellulose membranes. Blots were probed with antibodies against human RelB, p50, p52, or RelA (Santa Cruz Biotechnology) and visualized by chemiluminescence detection (ECL; Amersham, Aylesbury, United Kingdom). Equal loading was confirmed after stripping the membranes with 0.5 M NaOH and probing with anti-actin antibody (Sigma).
Cloning of the RelB promoter-enhancer reporter constructs.
A 4.6-kb DNA fragment containing the RelB promoter and extending from −4,646 bp to −46 bp from the start of translation was isolated from the LambdaFix human genomic library (Stratagene, La Jolla, CA) using a probe that corresponds to exons 1 to 3 of the human RelB cDNA. The fragment was cloned in forward and reverse orientations into the pGL2-basic vector (Promega, Madison, WI) to generate the pGRelBF and pGRelBR constructs, respectively.
Functional analysis of reporter constructs.
Before electroporation with the Nucleofector system (Amaxa Biosystems, Cologne, Germany), all cells were incubated for 24 h at a concentration of 0.5 × 106 cells/ml, with the exception of βII-E9 cells, which were kept at 0.25 × 106 cells/ml. A total of 10 × 106 cells were resuspended in 100 μl of reagent V and mixed with 10 μg of the reporter construct pGRelBF or pGRelBR plus 100 ng of the pRL-CMV plasmid (Promega) encoding Renilla luciferase to normalize for transfection efficiency. Cells were pulsed at the T-01 setting, and after overnight incubation each transfection experiment was split into two cultures of equal volumes. One of the cultures from each set was left untreated, while the other was treated with PMA for an additional 5 h (KG1 and KG1a cells) or 8 h (βII-E9 and KG1a-neo cells). Cells were harvested, lysed, and assayed for luciferase activity using the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions. The expression levels of Renilla luciferase were normalized between cell lines by analysis of cells transfected with the pDsRed2-C1 plasmid (BD Biosciences, Palo Alto, CA) plus pRL-CMV.
Gene microarray analyses.
Total RNA was isolated from KG1 and KG1a cells left untreated or treated with PMA for 2 h using the RNeasy kit (QIAGEN, Valencia, CA). Double-stranded cDNA was synthesized from total RNA by incubation with First Strand Synthesis kit reagents (Invitrogen, Carlsbad, CA) and an oligo T7-(dT)24 primer, followed by incubation with Second Strand Synthesis kit reagents (Invitrogen). Biotin-labeled cRNA was synthesized using the BioArray HighYield RNA Transcript Labeling kit (T7) (Enzo Diagnostics, Farmingdale, NY) and purified using the RNeasy kit (QIAGEN). The cRNAs were fragmented prior to hybridization to HG-U133 Plus 2.0 chips (Affymetrix, Santa Clara, CA), washed, antibody amplified, and stained as indicated in the Affymetrix Technical Manual. Chip fluorescence was normalized prior to comparison using the Affymetrix GeneChip Operating System (Affymetrix) by scaling each array to a common (trimmed) mean intensity of 500, and signal values were clipped to a threshold value of 100. All probe set signal values for CLPTM1, RelB, and SFRS16 were called present by the GeneChip Operating System program.
Generation of RelB genomic probe constructs.
All RelB genomic clone inserts were obtained from restriction digestion of the bacterial artificial chromosome clone RP11-91A12 (Children's Hospital Oakland-BACPAC Resources, Oakland, CA) as follows: probe A (5′ end), EcoRI plus HpaI, 2,397 bp; probe B (exon 4), SpeI plus PvuII, 1,922 bp; probe C (exon 5), SacI plus XmaI, 2,706 bp; probe D (3′ end), ApoI plus PstI, 2,467 bp. Each fragment, together with a 290-bp β-actin probe generated by PCR, was cloned into the pBluescript SK (+) and pBluescript SK II (−) vectors (Stratagene). The identities of all constructs were confirmed after restriction enzyme digestion and DNA sequencing.
To generate the single-stranded DNA (ssDNA) probes for nuclear runoff assays of KG1 and KG1a cells, all genomic clones were transformed into XL1-Blue MRF′ bacteria (Stratagene). Bacterial cultures were incubated with helper phage R408 (Stratagene) at 5 × 107 PFU/ml before isolation of ssDNA using the QIAprep Spin M13 Kit (QIAGEN). PKCβII is expressed in βII-E9 cells from the pPKCβ-EGFP vector, which has long regions of DNA that align with the pBluescript vector and results in unusually high background levels in the nuclear runoff assays. To avoid this problem, we used purified double-stranded DNA (dsDNA) clone inserts as hybridization probes in runoff assays from βII-E9 and KG1a-neo cells. As a negative control, a short region of pBluescript that does not align with the pPKCβ-EGFP vector was employed. Probes were generated by restriction digestion of the original constructs in pBluescript SK (+) as follows: probe A, BamHI plus SalI, 1,771 bp; probe B, XbaI plus HindIII, 1,923 bp; probe C, NarI plus SacI, 1,650 bp; probe D, NcoI, 1,710 bp. Each insert was gel purified before spotting into nylon membranes.
PKC enzyme activity assay.
PKC enzyme activity levels were determined using the SignaTECT Assay System (Promega) according to the manufacturer's instructions. Briefly, 7 × 106 cells each of KG1, KG1a, KG1a-neo, βII-E9, and βII-E11 cells were homogenized in 25 mM Tris buffer (pH 7.4) containing 0.5 mM EDTA, 0.5 mM EGTA, 10 mM β-mercaptoethanol, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 50 μg/ml phenylmethylsulfonyl fluoride and purified through a DEAE column. Same-volume aliquots of each sample were incubated in a buffer containing calcium, peptide target, and [α-32P]ATP, and the reaction was stopped with 7.5 M guanidine-HCl buffer. Each reaction was spotted onto binding paper and quantified by scintillation counting. Signal values were normalized to protein content and expressed relative to the activity of the undiluted (1/1) KG1a-neo cell lysate. Enzyme activity levels in the cell membrane was detected in a similar manner after isolating cell membrane fractions as previously described.
Nuclear transcriptional runoff assays.
Nuclear runoff experiments were carried out following the standard protocols (31, 32). Briefly, nuclei isolated from untreated and treated cells (50 × 106 each) were incubated at 30°C for 30 min in the presence of ATP, CTP, and GTP (0.25 mM each, Promega) and 300 μCi of [32P]UTP (>3,000 Ci/mol; Perkin-Elmer, Boston, MA). After additional successive incubations with RQ1 DNase (Promega) and proteinase K (Sigma), nuclear extracts were washed with phenol-chloroform, followed by precipitation and purification of the RNA through a Sephadex G-50 column (Sigma). After an additional precipitation, equal counts per minute of RNA were hybridized to probes A, B, C, and D (10 μg/dsDNA probe, 5 μg/ssDNA probe) previously spotted onto nylon membranes representing the 5′end, exon 4, exon 5, and the 3′ end of the relB gene, respectively. Probes for β-actin and pBluescript were including as loading and background controls. Signal intensities were detected by densitometry after exposing the blots to phosphorimaging screens for 24 h.
siRNA transfection of primary human monocytes.
Peripheral blood mononuclear cells were obtained from normal volunteers under University of Miami Institutional Review Board-approved protocols and purified by discontinuous-density centrifugation. Monocytes were freshly enriched by negative selection using Miltenyi Biotec monocyte isolation kit II (Miltenyi Biotec). After purification, CD14-positive cells were identified by staining with anti-CD14PE antibody (Immunotech). Enriched monocytes (10 × 106) were left untransfected or nucleofected with 5 μg of either pEGFP-siEmpty or pEGFP-siPKCβ plasmid using a Human Monocyte Nucleofector Kit (Amaxa). The PKCβ small interfering RNA (siRNA) is targeted against GGAAGCTGTGGCCATCTGC in PKCβ and is cloned into the pFRT-HIP plasmid, in which the EGFP-encoding gene is coexpressed. The empty construct (siEmpty) is the same vector without the PKCβ siRNA. Two days after transfection (to allow knockdown of PKCβ expression), monocytes were differentiated with 1,000 U/ml human grade GM-CSF and 1,000 U/ml interleukin-4 (IL-4; R&D Systems) and total protein lysates were made in RIPA buffer at 2 h post cytokine addition. Proteins were separated on an 8% gel and immunoblotted with rabbit anti-RelB, mouse anti-PKCβII (both from Santa Cruz), or rabbit anti-actin (Sigma) antibodies.
RESULTS
Regulation of RelB expression is different in KG1 and KG1a cells.
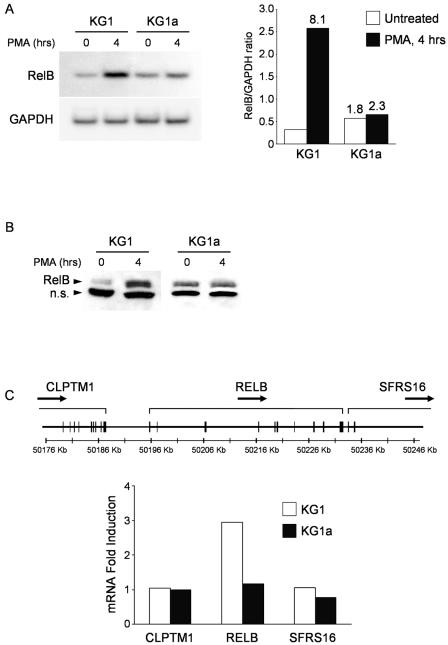
We first analyzed if the different abilities of KG1 and KG1a cells to generate DCs were reflected in a different regulation of RelB expression in the immediate-early period of PMA-induced differentiation. While RelB mRNA is readily detected in both cell lines under unstimulated conditions, we consistently observed a twofold greater amount of message in KG1a than in KG1 cells by Northern blotting (Fig. 1A) and gene array experiments (data not shown). RelB mRNA levels are upregulated during PMA-induced differentiation of KG1 cells, reaching an eightfold maximal expression increase after 4 h of treatment (Fig. 1A, right side) and subsequently decrease to a near-constant level (threefold over untreated) until the end of the 5 days of differentiation (data not shown). In contrast, RelB levels in KG1a cells are only marginally altered by PMA treatment (less than 1.5-fold increase) and remain constant throughout the 5 days. RelB protein expression follows the early mRNA upregulation induced by PMA in KG1 cells, suggesting that the primary mechanism of regulation is transcriptional (Fig. 1B).
FIG. 1.
RelB is upregulated early in the differentiation of DC progenitor KG1 cells. (A) RelB mRNA content is lower in KG1 cells than in KG1a cells, but it is soon upregulated after PMA treatment. KG1 and KG1a cells were stimulated with PMA (10 ng/ml), and RelB mRNA expression levels were analyzed in a Northern blotting assay. The membrane was subsequently hybridized to a GAPDH cDNA probe to confirm equal loading. Hybridization signals were quantified by phosphorimaging before normalizing the RelB mRNA content in each lane to the respective GAPDH signal (right side). Values at the top of the bars indicate n-fold expression over untreated KG1 cells. Shown is one representative result from at least three independent experiments. (B) RelB protein expression follows the regulation observed at the mRNA level. Western blot analyses were performed on lysates of KG1 and KG1a cells stimulated with PMA (10 ng/ml) using an anti-RelB polyclonal antibody. Shown is one representative result from at least three independent experiments. n.s. = nonspecific band. (C) Expression levels of RelB flanking genes are not affected during PMA stimulation. The relB gene expands over ∼37 kb of DNA, and it is flanked by the CLPTM1 and SFRS16 genes (CLPTM1 and SFRS16 are not represented in their entirety). Total RNA was isolated from KG1 and KG1a cells left untreated or stimulated with PMA for 2 h for a global expression analysis using the Affymetrix HG-U133 Plus 2.0 array. All probe set signal values for the CLPTM1, relB, and SFRS16 genes were called present by the GeneChip algorithm employed in the analysis. n-Fold induction in each cell line was determined by dividing the signal values of each gene at 2 h by the respective values at 0 h.
We next determined if the increase in RelB mRNA was the result of a regulatory mechanism affecting the whole genomic locus or if this effect was restricted to RelB. In humans, the relB gene is located in chromosome 19 flanked by the CLPTM1 gene in its 5′ upstream region (with less than 10 kb of intergenic DNA) and by SFRS16 in its immediate 3′ end (Fig. 1C). The relative expression values for CLPTM1, RelB, and SFRS16 were extracted from microarray data of KG1 and KG1a cells both in unstimulated conditions and after stimulation with PMA for 2 h. As shown in Fig. 1C, the mRNA levels of the flanking genes CLPTM1 and SFRS16 remained unaltered after PKC activation in KG1 cells while RelB displayed a threefold increase, suggesting that the PMA-induced upregulation is specific for RelB and is not the result of a locus regulatory mechanism. In KG1a, the expression levels of the three genes remain largely unaltered after PMA treatment.
The differences in RelB regulation cannot be explained solely on the basis of differences in promoter activity.
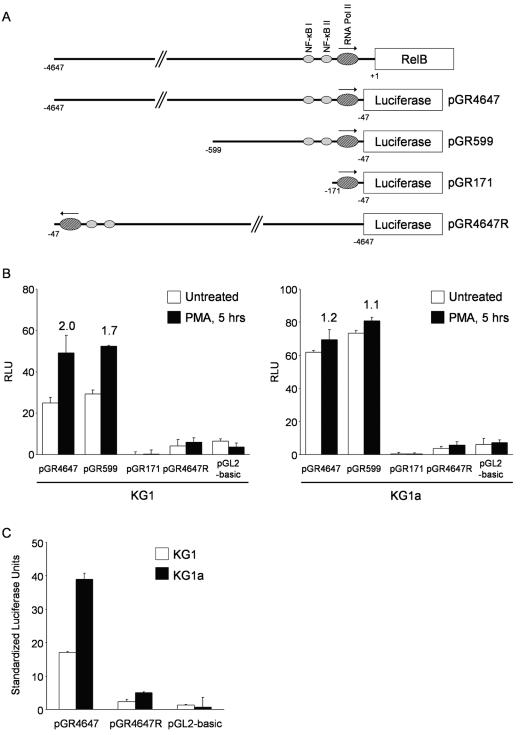
Previous studies have shown that RelB is transcriptionally regulated in the HeLa, Jurkat, and BJAB cell lines by a TATA-less promoter containing two NF-κB sites that positively regulate RelB expression. RelA/p50 and RelB/p50 heterodimers bind to these sites and enhance transcription initiation (5). To assess whether RelB mRNA expression in DC differentiation is controlled primarily by the regulation of promoter activity, we generated three reporter constructs containing different-size fragments of the 5′ upstream region of the relB gene (Fig. 2A). The pGR4647 and pGR599 constructs contain, respectively, 4.6 and 0.6 kb of DNA and include the two NF-κB-binding cis-acting sites previously described. The pGR171 construct contains less than 200 bp and lacks both NF-κB-binding sites while retaining the RNA polymerase II core promoter. The 4.6-kb DNA fragment was also cloned in reverse orientation for use as a negative control. Each reporter construct was transfected into KG1 and KG1a cells to determine the luciferase content in cells left untreated or after 5 h of PMA treatment, using a Renilla luciferase-expressing vector to normalize for transfection efficiency. As shown in Fig. 2B, the pGR4647 and pGR599 constructs have similar transcriptional activities in untreated KG1 cells (left side, open bars) while no activity was detected for pGR171. After PMA stimulation, activity levels increase approximately the same for the first two constructs and remain undetected for pGR171 (closed bars). Altogether, the data indicate that the cis-acting elements regulating transcription from the reporter constructs in unstimulated and PMA-treated cells are contained in the region located between 47 and 599 bp upstream of the RelB translation start site. Interestingly, the twofold increase in luciferase activity levels induced by PMA in pGR4647 and pGR599 is significantly lower than the eightfold upregulation previously seen in Northern blotting experiments. Incubation with PMA for shorter (2 to 4 h) or longer (8 to 24 h) times resulted in relatively lower luciferase activities. Any possible interference in the assay from the transfection itself was ruled out by Northern blotting experiments that indicated no differences in the levels and kinetics of RelB upregulation between transfected and untransfected cells (data not shown). One possible explanation for the discrepancy between promoter activity and steady-state levels of RelB mRNA is the existence of other mechanisms of gene regulation controlling RelB expression.
FIG. 2.
Changes in RelB promoter activity do not correlate fully with the regulation of RelB in KG1 cells. (A) Cloning of the promoter-containing RelB 5′ genomic region into the pGL2-basic vector. A 4.6-kb DNA fragment containing the RelB promoter-enhancer region was cloned into the pGL2-basic vector to drive the expression of luciferase (pGR4647). The pGR599 and pGR171 deletion constructs were generated by restriction digestion of the original clone. The pGR4647R construct containing the 4.6-kb DNA fragment in reverse orientation was cloned as a negative control. NF-κBI and -II indicate the two NF-κB-binding regulatory sites previously identified in the RelB promoter-enhancer region. Pol, polymerase. (B) PMA treatment induces only twofold upregulation of the RelB promoter activity in KG1 cells. KG1 and KG1a cells were transiently transfected with the reporter constructs containing the promoter-enhancer region of RelB. Renilla luciferase-encoding plasmid pRL-CMV was cotransfected to allow normalization of transfection efficiency. After overnight incubation, cells were treated with PMA for 5 h before assaying for luciferase activity. Relative light units (RLU) were determined by dividing the firefly luciferase activity by the respective Renilla luciferase activity values. Values at the top of the bars indicate n-fold induction over untreated samples. The data represent the mean ± the standard deviation of triplicate measurements from a representative experiment of three. (C) RelB promoter activity correlates with thesteady-state mRNA levels in untreated KG1 and KG1a cells. Standardized light units were determined by normalizing the relative light units from the reporter constructs in untreated KG1 and KG1a cells to the relative cytomegalovirus promoter activity in both cell lines. The relative cytomegalovirus promoter activities were determined by adjusting the Renilla luciferase activity values from the pRL-CMV plasmid to the transfection efficiency as determined by flow cytometry after cotransfection with the pDsRed2-C1 vector. Data represent the mean ± the standard deviation of triplicate measurements from a representative experiment of three.
As in KG1 cells, we found no functional differences between the pGR4647 and pGR599 constructs in untreated KG1a cells, while the smaller pGR171 plasmid showed no transcriptional activity (Fig. 2B, right side, open bars). In correlation with the Northern blotting experiments, no significant luciferase activity increase was observed after PMA treatment (closed bars).
To compare the activities of the reporter constructs between cell lines, KG1 and KG1a cells were cotransfected with the Renilla luciferase-expressing vector and a plasmid encoding red fluorescent protein. The Renilla luciferase activity was determined for each cell line and normalized to the respective transfection efficiency as determined by flow cytometry. As shown in Fig. 2C, the functional activity of pGR4647 is twofold higher in KG1a than in KG1 untreated cells, matching the relative steady-state levels of RelB mRNA found previously by Northern blotting assays.
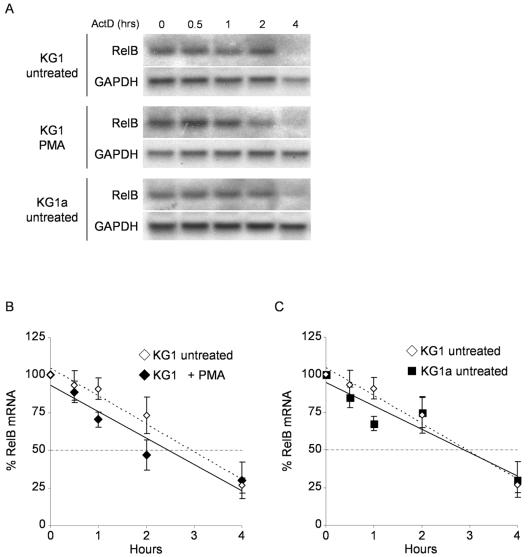
RelB expression is not regulated by alterations in mRNA stability.
To examine possible differences in RelB mRNA stability, we used actinomycin D to arrest transcription before and after PMA stimulation of KG1 and KG1a cells. Culture aliquots were taken at different intervals, and the levels of RelB and GAPDH mRNAs were determined in Northern blotting experiments (Fig. 3A). As shown in Fig. 3B, these experiments yielded similar RelB mRNA half-life values for PMA-treated (2.5 h, solid line) and untreated (3.0 h, dashed line) KG1 cells. In addition, the RelB mRNA half-life was the same in untreated KG1 and KG1a cells (Fig. 3C). These results indicate that mRNA stabilization does not contribute to the upregulation of RelB during differentiation of KG1 cells or to the relatively higher RelB expression in KG1a over KG1 untreated cells.
FIG. 3.
RelB expression is not regulated by changes in mRNA stability. (A) Determination of RelB mRNA half-life in KG1, KG1a, and PMA-treated KG1 cells. KG1 and KG1a cells left unstimulated or stimulated with PMA for 2 h were treated with actinomycin D to arrest transcription before isolation of total RNA at the indicated time points. RelB and GAPDH transcripts were detected in Northern blotting assays as previously described. (B) PMA treatment does not increase the RelB mRNA half-life in KG1 cells. Hybridization signals for the RelB and GAPDH mRNAs were quantified by phosphorimaging in KG1 cells left untreated or treated with PMA for 2 h. Each lane was normalized for equal loading by dividing the RelB content by the respective GAPDH signal and expressed relative to RelB content at 0 h for each condition. The mRNA half-life was calculated by linear regression analysis to be 3.0 h and 2.5 h for untreated and PMA-treated KG1 cells, respectively. The dashed line parallel to the x axis represents 50% mRNA decay. The data represent the mean ± the standard error from two independent experiments. (C) RelB mRNA half-life is the same in KG1 and KG1a untreated cells. A RelB mRNA half-life of 3.0 h was determined for untreated KG1a cells as described for panel B. The data represent the mean ± the standard error from two independent experiments.
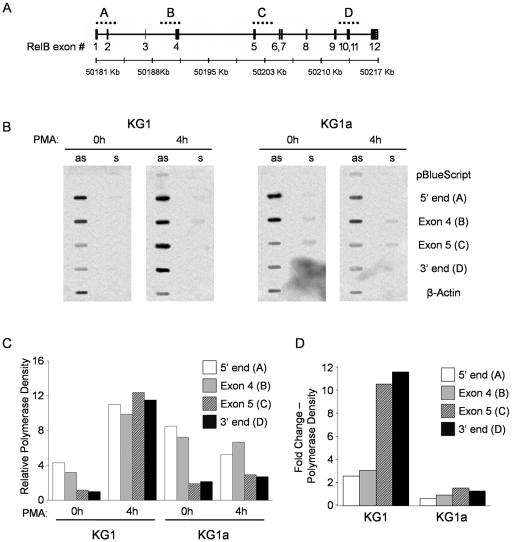
RelB transcriptional elongation is enhanced only in PMA-treated KG1 cells.
While not described for a member of the NF-κB family, regulation of transcriptional elongation has been established as an important regulatory mechanism for a variety of mammalian genes such as c-myc (27), that for heat shock protein hsp70 (11), and that for apolipoprotein A-I (33). The human relB gene contains 12 exons within 36 kb of DNA, with its longest intron extending over 3 kb between exons 4 and 5. To determine if RelB expression is additionally regulated by changes in elongation efficiency, we performed nuclear transcriptional runoff studies with KG1 and KG1a cells using four ssDNA hybridization probes that expand over the 5′ end, exon 4, exon 5, and the 3′ end of the gene, respectively (Fig. 4A). Radiolabeled nascent transcripts were isolated at 0 and 4 h after PMA treatment of KG1 and KG1a cells and hybridized to the membranes containing the probes. As shown in Fig. 4B, we detected practically no hybridization signals with the sense probes, indicating that no transcription was occurring in the antisense direction. Results were also negative for both sense and antisense probes of pBluescript, indicating no significant contribution of the vector backbone to the hybridization signal of the probes. Signal intensities for each probe were corrected for thymidine composition and normalized to the β-actin content in the respective membrane. As quantified in Fig. 4C, transcription rates (determined as polymerase density) in untreated KG1 cells dropped significantly from the 5′ end of the gene (probe A) to the 3′ end (probe D), indicating a decrease in elongation efficiency along the relB gene. No decrease was detected, however, after stimulation of KG1 cells with PMA, as all probes showed similar corrected signal intensities. Furthermore, and in agreement with the promoter studies, transcription initiation (determined by 5′-end probe values) had a twofold average increase relative to β-actin following PMA stimulation (Fig. 4D). Overall, there was an 11-fold increase in the transcription rate of full-length RelB (determined by 3′-end probe values) in KG1 cells stimulated with PMA, consistent with the results from Northern blotting experiments. In correlation also with the promoter studies, transcription initiation in untreated KG1a cells was twofold higher than in KG1 cells and showed the same decrease in transcription efficiency along the gene. However, the higher transcription initiation rate resulted in higher levels of full-length RelB mRNA (Fig. 4C). In contrast to KG1, however, transcript elongation efficiency was not relieved in KG1a cells after PMA stimulation and, as expected, no increase was seen in transcription initiation (Fig. 4D).
FIG. 4.
RelB upregulation in KG1 cells results from an increase in transcription initiation and elongation. (A) Cloning of RelB genomic probes for nuclear transcriptional runoff assays. The relB gene contains 12 exons extended over 37 kb of DNA sequence. Exons 4 and 5 are separated in the relB gene by 9.8 kb of intronic sequence. Hybridization probes A, B, C, and D were obtained by restriction digestion of bacterial artificial chromosome clone RP11-91A12 and expand over the 5′ end, exon 4, exon 5, and the 3′ end of the relB gene, respectively. (B) Nuclear transcriptional runoff assays. Nuclei were isolated from KG1 and KG1a cells left untreated or treated with PMA for 4 h. Nascent transcripts were labeled in vitro with [α-32P]UTP and hybridized to the indicated sense (s) and antisense (as) ssDNA probes. (C) RelB transcriptional elongation is enhanced exclusively in PMA-treated KG1 cells. Hybridization signals for each probe were quantified by phosphorimaging and corrected for thymidine content. The resulting values were normalized against the respective actin intensities and expressed relative to the 3′-end probe value in untreated KG1 cells. The graph shows representative results from one of three independent experiments. (D) PMA stimulation induces an 11-fold average upregulation in full RelB transcripts. Changes in n-fold expression were determined by dividing the signal intensity values of each probe at 4 h by the corresponding values in untreated samples. The graph shows representative results from one of three independent experiments.
These results provide, to our knowledge, the first evidence of regulation of transcriptional elongation as a controlling mechanism of gene expression for a member of the NF-κB family. Additional nuclear runoff experiments indicated that the increase in elongation efficiency in KG1 cells is an early event, being detected as early as 2 h after PMA treatment, when almost no increase in transcription initiation is observed (data not shown). The differential abilities of KG1 and KG1a cells to regulate transcript elongation early after PMA stimulation represent an additional mechanism responsible for the observed upregulation of RelB in KG1 cells.
Inhibition of PKCβII expression inhibits RelB upregulation in primary human monocytes.
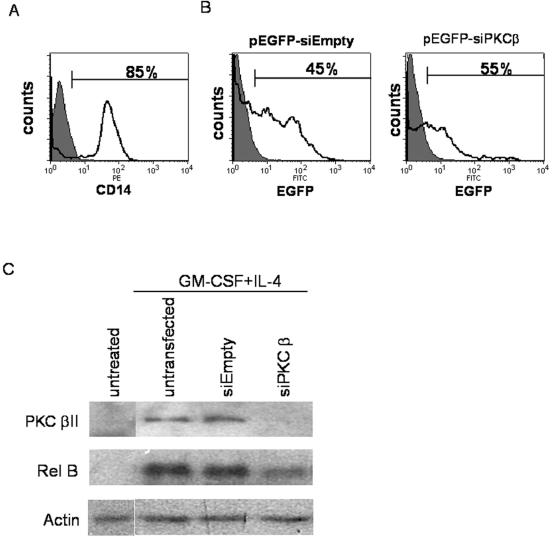
We have previously shown that PMA treatment of primary human CD34+ HPCs results in DC differentiation with induction of RelB (13), suggesting a role for PKC in the regulation of RelB expression. Similarly, the inability of KG1a cells to upregulate RelB expression after PMA stimulation could indicate a quantitative and/or qualitative defect in PKC expression, the major target of phorbol esters. In fact, in contrast to KG1 cells, KG1a cells do not express the βII isoform of PKC (19) and we have found that PKCβII-transfected KG1a cells regain the ability to differentiate into DCs (Cejas et al., submitted). In this same study, we have also found that PKCβII is the only cPKC isoform that is clearly activated by DC differentiation stimuli in both primary human DC progenitors (CD34+ HPCs, monocytes) and transformed myeloid cell lines. To assess whether PKCβII is specifically involved in RelB upregulation during DC differentiation, we first targeted PKCβII by siRNA in primary human monocytes stimulated with GM-CSF plus IL-4 (at an immediate-early time point). As shown in Fig. 5, the purity of the starting monocyte population was 85% (Fig. 5A), and the 45 to 55% transfection efficiency as measured by fluorescence-activated cell sorter (Fig. 5B) is likely an underestimation based on inspection by fluorescent microscopy. Untreated monocytes have low PKCβII expression (detectable on longer exposures of the Western blots), while untransfected or empty-vector-transfected monocytes cultured in GM-CSF plus IL-4 have increased PKCβII expression (Fig. 5C). However, PKCβII expression is considerably less in the monocytes transfected with siRNA against PKCβ. This knockdown is mirrored by a similar decrease in RelB induction in the siPKCβ-transfected monocytes under the cytokine culture conditions compared to the untransfected or empty-vector-transfected cells. These findings indicate that PKCβII plays an important role in RelB upregulation during DC differentiation in primary human monocytes.
FIG. 5.
Inhibition of PKCβII expression in primary human monocytes inhibits RelB upregulation in response to cytokine-induced DC differentiation. (A) Monocytes were enriched from PBMC as described in Materials and Methods. CD14-positive cells are shown as an open histogram and are overlaid with the isotype control (filled histogram). Percent CD14-positive cells is indicated. PE, phycoerythrin. (B) Enriched monocytes were left untransfected or transfected with pEGFP-siEmpty or pEGFP-siPKCβ constructs and analyzed for EGFP expression 2 days after transfection by fluorescence-activated cell sorter. The data are illustrated as transfected monocytes (open histogram) overlaid with untransfected monocyte controls (filled histogram). The percentage of cells expressing EGFP is indicated. FITC, fluorescein isothiocyanate. (C) Total cell lysates were made from enriched monocytes prior to transfection, untransfected or transfected monocytes incubated for 2 days in medium alone, and untransfected or transfected monocytes stimulated with GM-CSF plus IL-4 for 2 h, 2 days posttransfection. Proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed for expression of PKCβII, RelB, and actin. The data shown are representative results from one of two independent experiments.
Restoration of PKCβII expression in KG1a cells induces a pattern of RelB regulation similar to KG1 cells.
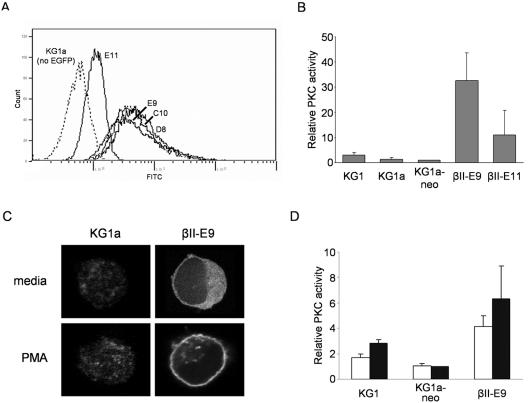
We next examined the regulation of RelB expression in PKC-βII-transfected KG1a (as noted above, we have found that PKCβII-transfected KG1a cells regain the ability to differentiate into DCs) to more carefully examine the mechanisms by which PKCβII modulates RelB expression. We analyzed the four different single cell clones of KG1a-transfected cells, βII-E11, -E9, -C10, and -D8, expressing various levels of the transfected PKCβII-EGFP fusion protein as assessed by EGFP expression (Fig. 6A). Overexpression of PKCβII in the transfectants results in a higher level of enzyme activity under unstimulated conditions. As shown in Fig. 6B, unstimulated KG1 had more kinase activity (approximately twofold higher) than KG1a or KG1a-neo cells but at much lower levels than transfected KG1a clones βII-E9 and βII-E11. In correlation with the respective amounts of PKCβII-EGFP expression detected by flow cytometry, βII-E9 cells show a higher level of enzyme activity than βII-E11 cells. Full activation of PKCβII, however, occurs only after translocation to the cell membrane and stabilization of the activated form of the protein by binding to diacylglycerol or diacylglycerol analogs like PMA. As shown in Fig. 6C, the majority of PKCβII-EGFP is localized in the cytosol of unstimulated βII-E9 cells, and full activation of the enzyme with PMA results in localization of almost all PKCβII-EGFP to the cell membrane. Correspondingly, PKC activity at the membrane after PMA stimulation increased for KG1 and βII-E9 cells, with no apparent change for KG1a-neo control cells (Fig. 6D).
FIG. 6.
Generation of single cell clones of PKCβII-transfected KG1a cells expressing different levels of kinase expression. (A) Cell clones βII-E11, -E9, -C10, and -D8 express different levels of PKCβII protein. Flow cytometry analysis of KG1a cell clones expressing different levels of PKCβII-EGFP fusion protein. FITC, fluorescein isothiocyanate. (B) Untreated PKCβII-expressing KG1a cells have significant levels of PKC enzyme activity. Total PKC activities were determined in KG1, KG1a, KG1a-neo, βII-E9, and βII-E11 untreated cell lysates by phosphorylation of a PKC-specific peptide substrate. PKC values are normalized to total protein content and are expressed relative to the PKC activity in KG1a-neo cells. The data represent the mean ± the standard deviation from three independent experiments. (C) The PKCβII-EGFP fusion protein translocates to the cell membrane after PMA stimulation. KG1a cells and βII-E9 cells were left untreated or treated with PMA (10 ng/ml) for 30 min. KG1a cells were stained for PKCβII using a fluorescein isothiocyanate-conjugated secondary antibody before visualization by confocal microscopy. βII-E9 cells were visualized directly for the EGFP tag. (D) PKC enzyme activity at the cell membrane increases after PMA stimulation. KG1, KG1a-neo, and βII-E9 cells were left unstimulated (open bars) or stimulated (closed bars) with PMA (10 ng/ml) for 45 min. Cell membrane lysates were generated for all samples before determining PKC activity levels as explained for panel B.
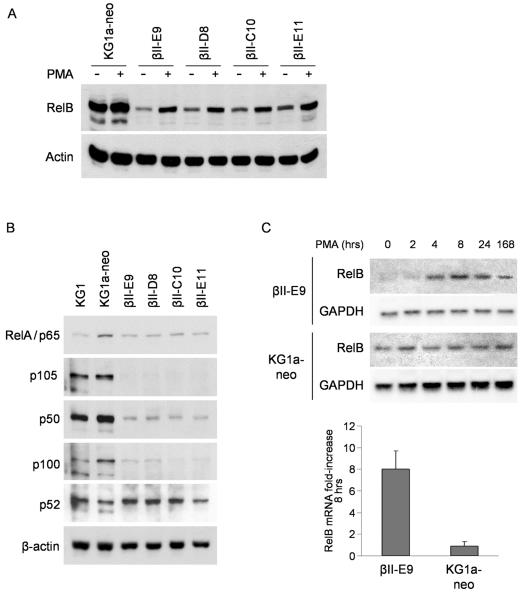
Western blot analysis show that, similar to KG1 cells, untreated PKCβII-transfected KG1a cells have lower levels of RelB protein compared to KG1a-neo cells, with an early increase after PMA treatment (Fig. 7A). These results suggest that the different regulation of RelB expression in KG1 and KG1a cells is a result of differences in the expression of PKC isoforms between the two cell lines. Furthermore, the data reveal a complex role for PKC in the control of RelB: the PKCβII activity in transfected KG1a cells under unstimulated conditions results in downregulation of RelB mRNA expression compared to cells with no PKCβII. However, full activation of PKC by PMA treatment leads to significant upregulation of RelB levels.
FIG. 7.
Forced expression of PKCβII in KG1a cells results in a mode of RelB regulation similar to that of KG1 cells. (A) PKCβII-transfected KG1a cells regulate RelB expression like KG1 cells. Protein lysates were generated for KG1a-neo, βII-E9, -D8, -C10, and -E11 cells stimulated or not with PMA (10 ng/ml) for 4 h. RelB and actin protein expression was analyzed in Western blot assays as previously described. (B) NF-κB protein expression is downregulated in PKCβII-transfected KG1a cells. Protein lysates were generated for KG1, KG1a-neo, βII-E9, -D8, -C10, and -E11 untreated cells. Expression of the RelA, p105/p50, p100/p52, and actin proteins was analyzed in Western blot assays. (C) RelB mRNA expression has a maximal ninefold upregulation in βII-E9 cells after PMA treatment. KG1a-neo and βII-E9 cells were stimulated with PMA (10 ng/ml) for the indicated times. RelB and GAPDH mRNA levels were determined in Northern blotting assays as previously described. The n-fold increase was calculated after normalizing the RelB mRNA hybridization signals to the respective GAPDH content and dividing by the value obtained at 0 h (right side). Only the maximal n-fold increase (8 h) is shown. The data represent the mean ± the standard deviation from three independent experiments.
RelB transcription is positively regulated by heterodimers of p50 and the subunit RelA or RelB, so it is possible that the low levels of RelB mRNA found in unstimulated PKCβII-transfected cells is the result of downregulation of these transcription factors. Consistent with this, Western blot analysis shows that all PKCβII-transfected KG1a cell clones have lower expression levels of RelA and p50 proteins compared to KG1a-neo control cells (Fig. 7B). The downregulation is especially dramatic for p50 and the longer isoform p105, as the expression levels are even lower than those found in KG1 cells. Both KG1 and KG1a cells contain low levels of p100 and p52 proteins. While p100 protein levels are even lower in PKCβII-transfected cells, we did not detect downregulation of the expression of p52 protein.
PKC activation upregulates RelB expression in KG1 cells via increased transcription initiation and elongation. To find out if these mechanisms were restored in PKCβII-transfected KG1a cells, we analyzed the molecular events leading to the upregulation of RelB in the βII-E9 clone. Unstimulated βII-E9 cells show a level of RelB protein expression even lower than those found in KG1 cells. However, Northern blotting analysis showed the same maximal increase in RelB mRNA expression after PMA treatment in βII-E9 cells that was previously observed for KG1 cells (eightfold), with only a slight delay in the kinetics (4 h for KG1 versus 8 h for βII-E9) (Fig. 7C).
RelB transcriptional elongation efficiency is high in unstimulated βII-E9 cells; transcriptional initiation is upregulated after PMA stimulation.
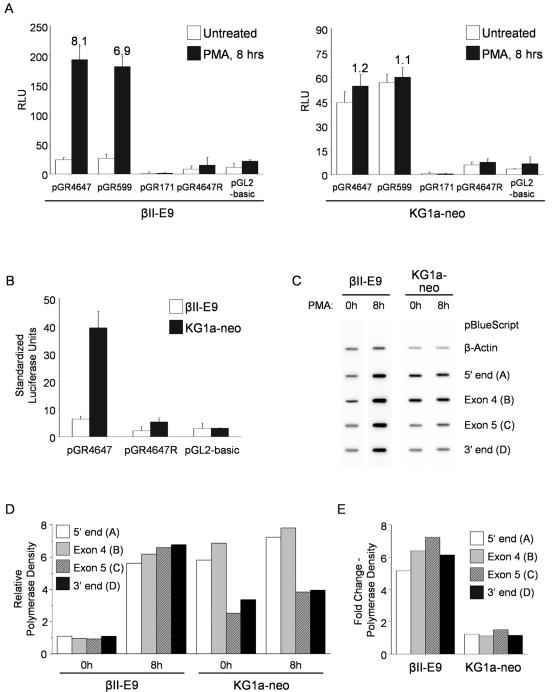
To determine if the increase in RelB expression in βII-E9 cells correlates with higher promoter activity, we transfected the reporter constructs (described in Fig. 2A) and assayed for luciferase activity in cells both under unstimulated conditions and after 8 h of PMA treatment. As shown for KG1 cells, the luciferase activity was similar for the larger pGR4647 and pGR599 constructs in unstimulated βII-E9 cells, while no activity was detected for pGR171 (Fig. 8A, left side, open bars). However, PMA treatment results in a sevenfold average increase in promoter activity in βII-E9 cells (closed bars), which is in agreement with the increase observed in mRNA at that same time point. We detected no additional increase in luciferase activity at earlier or later time points after PMA treatment. The results obtained with KG1a-neo cells conform to those previously seen for KG1a cells (Fig. 8A, right side). To determine if the lower expression of the transcription factors p50 and RelA in unstimulated βII-E9 cells correlated with lower RelB promoter activity, we normalized the Renilla luciferase activity levels as described for KG1 and KG1a cells. As shown in Fig. 8B, expression of PKCβII in βII-E9 cells results in a fivefold lower promoter activity from the pGR4647 construct compared to KG1a-neo control cells.
FIG. 8.
RelB transcription initiation in βII-E9 cells increases after PMA treatment and is not attenuated. (A) PMA treatment induces a sevenfold upregulation of the RelB promoter activity in βII-E9 cells. βII-E9 and KG1a-neo cells were transiently transfected with reporter constructs containing the promoter-enhancer region of RelB. Renilla luciferase-encoding plasmid pRL-CMV was cotransfected to allow normalization of transfection efficiency. After overnight incubation, cells were treated with PMA for 8 h before assaying for luciferase activity. Relative light units (RLU) were determined by dividing the firefly luciferase activity by the respective Renilla luciferase activity values. Values at the top of the bars indicate n-fold induction over untreated samples. The graphs show representative results from one of three independent experiments. (B) PKCβII expression results in downregulation of the RelB promoter activity in untreated βII-E9 cells. Standardized light units were determined by normalizing the relative light units from the reporter constructs in untreated KG1a-neo and βII-E9 cells to the relative activity of the cytomegalovirus promoter in both cell lines. The relative cytomegalovirus promoter activities were determined by adjusting the Renilla luciferase activity values from the pRL-CMV plasmid to the transfection efficiency as determined by flow cytometry after cotransfection with the pDsRed2-C1 vector. (C) Nuclear transcriptional runoff assays. Nuclei were isolated from βII-E9 and KG1a-neo cells left untreated or treated with PMA for 8 h. Nascent transcripts were labeled in vitro with [α-32P]UTP and hybridized to the indicated dsDNA probe inserts. (D) Untreated βII-E9 cells show low transcription initiation rates and no decrease in RelB transcript elongation. Hybridization signals for each probe were quantified by phosphorimaging and corrected for thymidine content. The resulting values were normalized against the respective actin intensities and expressed relative to the average probe value in untreated βII-E9 cells. The graph shows representative results from two independent experiments. (E) PMA stimulation induces a sixfold average upregulation in RelB transcription in βII-E9 cells. Changes (n-fold) in expression for each probe were determined by dividing the signal intensity values at 8 h by the corresponding values in untreated samples. The graph shows representative results from two independent experiments.
The results suggest that the levels of RelB expression in unstimulated and PMA-stimulated βII-E9 cells can be explained solely on the basis of promoter activity, with no indirect evidence for changes in elongation efficiency previously seen in untreated KG1 cells. We next carried out nuclear transcriptional runoff assays to directly assess the elongation rates in βII-E9 cells and also to verify the relative levels of transcription initiation suggested by the promoter activity assays.
Radiolabeled nascent transcripts were isolated at 0 and 8 h after PMA treatment of βII-E9 and KG1a-neo cells and hybridized to membranes containing probes A through D (described in Fig. 4A). As shown in Fig. 8C, probes A through D had a uniform level of hybridization signal in untreated βII-E9, indicating no decrease in elongation efficiency. On the other hand, unstimulated KG1a-neo control cells showed a stronger signal for the 5′ end of the gene (probe A) than for the 3′ end (probe D), demonstrating the same decrease in transcript elongation efficiency seen in untransfected KG1a cells. Signal intensities increased for all probes in βII-E9 cells after 8 h of PMA treatment, with no change for KG1a-neo cells. As quantified in Fig. 8D, RelB transcription initiation (measured as polymerase density of the 5′-end probe) is fourfold lower in βII-E9 cells than in KG1a-neo untreated cells, confirming the results found in promoter activity experiments. On the other hand, the decrease in transcript elongation observed in KG1a and untreated KG1 cells does not occur in βII-E9 cells, as all probes show similar corrected hybridization signal intensities at 0 h. We have found that PKCβII overexpression in transfected KG1a cells results in a partially differentiated DC phenotype characterized by the expression of surface markers typical of mature DCs such as CD80, CD86, CD40, and CD83 while still requiring phorbol ester stimulation to acquire DC morphology and induce allogeneic T-cell proliferation (Cejas et al., submitted). This semiactivated phenotype is reflected also in the mechanism of RelB regulation, as βII-E9 cells show high elongation efficiency but, at the same time, low levels of transcript initiation. The transcription rate for probes A through D increases by a sixfold average after PMA treatment (Fig. 8E), very similar to the levels observed previously by Northern blotting and promoter activity experiments. As in untransfected KG1a cells, elongation efficiency decreases along the RelB locus in KG1a-neo cells and is unaffected by PMA treatment.
DISCUSSION
A considerable number of different DC progenitors have been identified in humans and mice, as well as a variety of stimuli capable of driving their differentiation (48). However, relatively little is known about the early intracellular signaling events that trigger the cascade of genetic events that underlies DC differentiation. RelB is the transcription factor that has been associated most directly with DC differentiation and maturation, where it undergoes a very early but sustained upregulation of its expression (39). New studies indicate that RelB promotes the differentiation of DCs by enhancing monopoiesis (44). However, RelB-expressing DC progenitors still require treatment with cytokines like GM-CSF and TNF-α to generate DCs, indicating the need for additional signaling events that cooperate with RelB to induce DC differentiation. Different from other NF-κB proteins, the p50/RelB dimer is not retained in the cytosol; thus, its total expression level directly correlates with the levels in the cell nucleus and therefore with its transcriptional and biological activities (25, 28, 46). How relB gene expression is regulated, however, is largely unknown. To further understand the mechanisms controlling RelB expression in the setting of DC differentiation, we compared how RelB is regulated in the KG1 cell line model of a DC progenitor and in KG1a, a subline that fails to undergo DC differentiation.
KG1 and KG1a were first recognized as two different sublines by their different responses to PMA treatment (26). More specifically, they differ in their PKC activity levels, translocation, and substrate phosphorylation, which can be traced, at least partly, to a selective loss of PKCβII isoform expression in KG1a cells (19). We have recently shown that restoration of the enzyme expression in KG1a cells allows their differentiation into DCs (Cejas et al., submitted), so we extended our analysis to these PKCβII-transfected cells to analyze if their new ability to differentiate into DCs was reflected in a change in the mechanism regulating relB gene expression.
The data presented here show that RelB expression is controlled by the combination of two mechanisms acting at the transcriptional level: first, an NF-κB-mediated regulation of promoter activity (both positive and negative) and second, the regulation of transcript elongation along the RelB locus. We found that while KG1 and KG1a cells under unstimulated conditions have similar levels of transcript elongation efficiency, the transcription initiation rate (determined by nuclear runoff and promoter activity assays) is greater in KG1a cells, resulting in larger amounts of full-length RelB mRNA and increased RelB protein levels. When PKCβII expression is restored in KG1a cells (as in βII-E9 cells), the transcription initiation rate under unstimulated conditions drops, indicating a negative regulatory effect of intermediate levels of PKC activity on RelB promoter function and suggesting that the lack of this specific isoform is the cause of the higher transcription initiation observed in KG1a cells. The downregulation of RelB expression in the DC progenitor could be a requirement for the differentiation process to occur. A similar phenomenon has been described in chicken bone marrow cells expressing a conditional v-Rel estrogen receptor fusion protein (4). These studies found that, despite the requirement for c-Rel expression for proper DC differentiation, inactivation of the Rel fusion protein was required first for the cells to differentiate into DCs. The negative regulation of RelB promoter activity observed in untreated cells is no longer seen after full activation of PKC. In fact, RelB transcription initiation increases in PMA-stimulated KG1 and βII-E9 cells.
One possible explanation for these dual opposing effects on transcription initiation could be the existence of negative and positive regulatory elements in the relB promoter that are directly and selectively activated in unstimulated versus stimulated PKCβII-expressing cells. A similar mechanism has been described for NF-κB2, which is autoregulated positively in PMA-stimulated cells and negatively in unstimulated cells (34). These studies revealed at least four κB elements in the promoter that positively regulate promoter activity after PMA activation and other negative regulatory elements acting in untreated cells that overlap with a subset of the κB sites. In addition, a nuclear complex was identified with a putative inhibitory or repressive effect that binds to this subset of κB elements in unstimulated cells and does not contain any NF-κB1, NF-κB2, RelA, or c-Rel subunits. In the case of RelB, vitamin D response elements have been identified in the promoter that negatively regulate its activity after stimulation with the steroid 1α,25-dihydroxyvitamin D3. The inhibition is independent of the positive effect of NF-κB activity on RelB transcription, and downregulation of this activity by glucocorticoid treatment caused an additional decrease in the RelB promoter function. The repression of basal gene transcription in an inactivated state would allow the cells to maintain tight control of NF-κB activity.
An alternative explanation is that intermediate levels of PKCβII enzyme activity downregulate positive regulatory elements driving transcription from the relB promoter in untreated cells, followed by an increase in their activity after PMA treatment. Consistent with this, we found a lower level of expression of p50 and RelA proteins (which have been shown to positively regulate RelB transcription) in KG1 than in KG1a cells. More importantly, all the PKCβII-transfected KG1a clones also demonstrated a decrease in the expression of both NF-κB subunits. We have found no previous report of PKC-mediated downregulation of NF-κB protein levels. Transcription from the NF-κB1, NF-κB2, and relB promoters is positively regulated by NF-κB activity (5, 10, 35) while RelA transcripts levels are regulated primarily through SP1-binding sites in the promoter (56). Therefore, PKC-mediated downregulation of p50 and RelA expression would lead to a decrease in p50/p105, p52/p100, and RelB transcripts and further diminish their steady-state expression levels. In effect, the downregulation of p50/p105 and RelB protein levels in untreated PKCβII-transfected KG1a cells is more dramatic than the drop in RelA protein expression. While the basal levels of p52 protein are already low in KG1 and KG1a cells and are basically unaffected in transfected KG1a cells, we detected a downregulation of the levels of the precursor form p100, suggesting also a decrease in transcription.
The ability of full PKC activation to induce NF-κB nuclear activity is well established, as studies using PKCβ-deficient mice have shown that the enzyme has an essential role in the phosphorylation and activation of IKKα (probably through PKC-associated kinase PKK/RIP4) (38) and the immediate activation of NF-κB (47). The translocation of NF-κB to the nucleus subsequently induces RelB transcription in KG1 and transfected KG1a cells to upregulate the expression levels responsible for long-term NF-κB activity.
We have found that RelB transcriptional elongation is enhanced in PMA-stimulated KG1 cells and, together with an increase in transcription initiation, results in strong upregulation of RelB mRNA and protein during early DC differentiation. Full transcript elongation is an early event detected in KG1 cells 2 h before the increase in transcription initiation. The control of transcriptional elongation has not been described before as a regulatory mechanism of NF-κB expression, although it has been characterized for several other mammalian genes, such as the proto-oncogenes c-myc (27) and c-myb (42, 54), heat shock protein hsp70 (11), and apolipoprotein A-I (33). The exact mechanism regulating transcriptional elongation can vary. Transcript attenuation (the regulated elongation block at a specific site in the locus) has been described in more detail close to the transcription start site (<100 bp), and it is thought to be linked to promoter activity (27). There are some examples of transcriptional attenuation occurring at longer distances (even >1 kb), but they have not been studied yet in greater detail (33, 42, 54). Finally, transcription activators can act by enhancing elongation efficiency rather than initiation (18, 23).
Intrinsic elements in the DNA sequence can induce destabilizing secondary conformations in the nascent RNA and result in polymerase II pause, arrest, or termination without other intervening factors (24). In other studies, DNase hypersensitivity sites have been mapped to the attenuation site, together with DNA-binding proteins that presumably hinder transcript elongation (12, 42), and the relief of attenuation correlates with loss of hypersensitivity sites. Analysis of c-myb regulation in murine erythroleukemia cells has shown that its transcriptional attenuation is prevented by the expression of functional RelB/p50 dimers and correlates with binding of the complex to κB sites flanking the transcriptional pause site (42, 54). However, untreated KG1a cells contain nuclear p50/RelB dimers, suggesting that RelB transcript attenuation is not autoregulated. PKC activity has also been implicated in the regulation of transcriptional attenuation. One example is the increased elongation of c-myc transcription mediated by PKCɛ in erythroid cells after erythropoietin stimulation (8). We have not ruled out either transcriptional attenuation or elongation as the additional mechanism regulating RelB expression. Our results show a strong decrease in transcript elongation between exons 4 and 5 of the relB gene, but more experiments are required to determine if transcripts are blocked at a specific site in the intron (attenuation) or if there is a gradual decline in nascent transcripts throughout the locus. The presence of more potent polymerase pause sites within exons 4 and 5 could explain why the decrease in elongation seems more dramatic at this site.
To our surprise, βII-E9 cells show a semiactivated phenotype at the molecular level, and no decrease in transcript elongation is detected under unstimulated conditions. A possible explanation is that the molecular events increasing the efficiency of RNA polymerase elongation are triggered at intermediate levels of PKC activity, like those found in PKCβII-overexpressing βII-E9 cells. Further evidence of the semiactivated phenotype of resting βII-E9 cells (in addition to upregulation of DC surface markers) is the expression of several genes that are selectively transcribed during DC differentiation of KG1 while remaining absent in PMA-stimulated KG1a cells (Cejas et al., submitted).
Our initial findings using siRNA-mediated knockdown also indicate that PKCβ is important for RelB upregulation in primary human monocytes, under culture conditions where the DC differentiation stimuli are cytokines (GM-CSF and IL-4). Although the siRNA used in our studies targets both the βI and βII mRNAs (βI is the shorter, alternatively spliced mRNA of the gene for PKCβ), we have found in both primary human monocytes and CD34+ HPCs that only βII (and not βI) is activated during DC differentiation stimulated by either PMA or cytokines (GM-CSF, TNF-α, IL-4) (Cejas et al., submitted). The PKCβ knockdown data are also consistent with our findings that PKC activation is a downstream signaling component of cytokine-induced DC differentiation from both primary human CD34+ HPCs (52) and monocytes (Cejas et al., submitted) and that restoration of PKCβII expression reconstitutes the ability of KG1a to differentiate to DC and with previous studies demonstrating recruitment of PKCβII activation by the activated GM-CSF receptor (17). Although our characterization of PKCβII in DC differentiation is ongoing, we hypothesize that regulation of PKCβII expression in DC precursors by exogenous factors may be one mechanism that determines the immunologic function of the differentiated DC, in part due to the downstream effects on RelB expression. From a pathogenesis standpoint, the dual observations that certain intracellular pathogens express molecules that inhibit PKC activation (e.g., lipophosphoglycan from Leishmania in monocytes/macrophages) and that inhibition of RelB upregulation generates DC that induce antigen-specific Treg (36) suggest the possibility that such pathogens might blunt the adaptive immune response by infecting myeloid DC precursors to generate immunoregulatory DC.
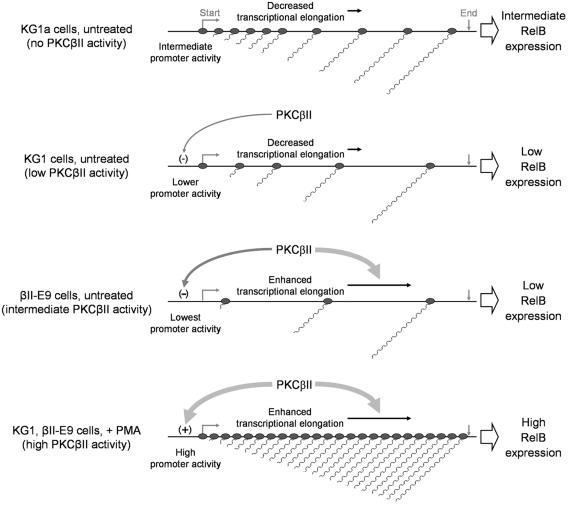
Based on our data, we present a model for RelB regulation during early DC differentiation (Fig. 9). KG1a cells with no detectable PKCβII expression actively initiate transcription from the relB promoter, but only a fraction of the nascent transcripts can generate full-length RelB mRNA. Transcription initiation in KG1 cells, which have low (basal) levels of PKCβII activity, is lower due to a decrease in p50/RelA protein expression. In addition, the relatively inefficient transcript elongation in untreated KG1 cells results in further downregulation of RelB expression. In contrast, the higher PKC activity in PKCβII-overexpressing βII-E9 cells allows the full elongation of all generated transcripts to the 3′ end of the gene. However, the higher PKC activity also results in decreased transcription initiation, which is ultimately responsible for the observed low levels of RelB mRNA expression. When PKC is fully activated (as in PMA-treated cells), transcriptional elongation occurs uninterrupted but now combines with an increase in transcription initiation (due to enhanced levels and nuclear translocation of p50/RelA protein) to elevate the amounts of RelB mRNA and protein.
FIG. 9.
PKCβII controls RelB expression by regulation of transcriptional initiation and elongation. KG1a cells actively initiate transcription from the RelB promoter, but elongation is relatively inefficient. Basal PKCβII activity in untreated KG1 cells downregulates the promoter activity with no increase in elongation efficiency, resulting in lower levels of full-length RelB transcripts. Increased expression (and enzyme activity) of PKCβII in untreated βII-E9 cells enhances transcript elongation, but the stronger downregulation in transcriptional initiation results in even lower levels of RelB mRNA. PMA-mediated activation of PKCβII results in an increase in promoter activity that combines with enhanced elongation to upregulate RelB mRNA levels.
The proposed model implies the existence of distinct signaling pathways that are selectively triggered at different levels of PKC signaling. Such a model is supported by studies that found different thresholds of PKC activity required for lineage commitment from a hematopoietic precursor cell line model (45). It is also possible that the increases in transcript elongation and initiation are not triggered by different thresholds of PKCβII activity but rather by different locations of activated enzyme. As previously shown, transfected KG1a cells express PKCβII protein at high levels, so the concentration of activated kinase in untreated cells (defined by Kact), although small, is much higher than the basal levels found in untreated KG1 cells and is not necessarily localized in the cell membrane. This level of PKC signaling is sufficient to trigger molecular events that result in the semiactivated phenotype of PKCβII-transfected KG1a cells. However, full PKC activation by PMA treatment is required for the increase in RelB transcription initiation and differentiation, leading to higher PKC activity levels and localization of the enzyme to the membrane.
The data presented here indicate that RelB expression, and therefore its activity, is regulated in the context of DC differentiation by transcriptional initiation and elongation. We also provide evidence that the two mechanisms can act independently, are regulated by PKCβII, and are sensitive to the levels of kinase signaling. The action of two independent pathways suggests how RelB expression can be finely and tightly regulated during DC differentiation.
Acknowledgments
We thank Edward W. Harhaj and John R. Bethea for critical reading of the manuscript.
This work was supported by NIH CA85208 and CA95829.
REFERENCES
- 1.Ackerman, A. L., and P. Cresswell. 2003. Regulation of MHC class I transport in human dendritic cells and the dendritic-like cell line KG-1. J. Immunol. 170:4178-4188. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245-252. [DOI] [PubMed] [Google Scholar]
- 4.Boehmelt, G., J. Madruga, P. Dorfler, K. Briegel, H. Schwarz, P. J. Enrietto, and M. Zenke. 1995. Dendritic cell progenitor is transformed by a conditional v-Rel estrogen receptor fusion protein v-RelER. Cell 80:341-352. [DOI] [PubMed] [Google Scholar]
- 5.Bren, G. D., N. J. Solan, H. Miyoshi, K. N. Pennington, L. J. Pobst, and C. V. Paya. 2001. Transcription of the RelB gene is regulated by NF-κB. Oncogene 20:7722-7733. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco, D., R. P. Ryseck, and R. Bravo. 1993. Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development 118:1221-1231. [DOI] [PubMed] [Google Scholar]
- 7.Caux, C., B. Vanbervliet, C. Massacrier, C. Dezutter-Dambuyant, B. de Saint-Vis, C. Jacquet, K. Yoneda, S. Imamura, D. Schmitt, and J. Banchereau. 1996. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to GM-CSF + TNFα. J. Exp. Med. 184:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C., and A. J. Sytkowski. 2001. Erythropoietin activates two distinct signaling pathways required for the initiation and the elongation of c-myc. J. Biol. Chem. 276:38518-38526. [DOI] [PubMed] [Google Scholar]
- 9.Clark, G. J., S. Gunningham, A. Troy, S. Vuckovic, and D. N. Hart. 1999. Expression of the RelB transcription factor correlates with the activation of human dendritic cells. Immunology 98:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogswell, P. C., R. I. Scheinman, and A. S. Baldwin, Jr. 1993. Promoter of the human NF-κB p50/p105 gene. Regulation by NF-κB subunits and by c-REL. J. Immunol. 150:2794-2804. [PubMed] [Google Scholar]
- 11.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 17:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallinger, G., H. Oberkofler, C. Seelos, and W. Patsch. 1999. Transcriptional elongation of the rat apolipoprotein A-I gene: identification and mapping of two arrest sites and their signals. J. Lipid Res. 40:1229-1239. [PubMed] [Google Scholar]
- 13.Davis, T. A., A. A. Saini, P. J. Blair, B. L. Levine, N. Craighead, D. M. Harlan, C. H. June, and K. P. Lee. 1998. Phorbol esters induce differentiation of human CD34+ hemopoietic progenitors to dendritic cells: evidence for protein kinase C-mediated signaling. J. Immunol. 160:3689-3697. [PubMed] [Google Scholar]
- 14.Dobrzanski, P., R. P. Ryseck, and R. Bravo. 1995. Specific inhibition of RelB/p52 transcriptional activity by the C-terminal domain of p100. Oncogene 10:1003-1007. [PubMed] [Google Scholar]
- 15.Dong, X., T. Craig, N. Xing, L. A. Bachman, C. V. Paya, F. Weih, D. J. McKean, R. Kumar, and M. D. Griffin. 2003. Direct transcriptional regulation of RelB by 1α,25-dihydroxyvitamin D3 and its analogs: physiologic and therapeutic implications for dendritic cell function. J. Biol. Chem. 278:49378-49385. [DOI] [PubMed] [Google Scholar]
- 16.Francis, D. A., R. Sen, N. Rice, and T. L. Rothstein. 1998. Receptor-specific induction of NF-κB components in primary B cells. Int. Immunol. 10:285-293. [DOI] [PubMed] [Google Scholar]
- 17.Geijsen, N., M. Spaargaren, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 1999. Association of RACK1 and PKCβ with the common beta-chain of the IL-5/IL-3/GM-CSF receptor. Oncogene 18:5126-5130. [DOI] [PubMed] [Google Scholar]
- 18.Guo, S., Y. Yamaguchi, S. Schilbach, T. Wada, J. Lee, A. Goddard, D. French, H. Handa, and A. Rosenthal. 2000. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408:366-369. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, W. C., R. T. Abraham, C. L. Ashendel, and G. E. Woloschak. 1989. Differential responsiveness to phorbol esters correlates with differential expression of protein kinase C in KG-1 and KG-1a human myeloid leukemia cells. Biochim. Biophys. Acta 1013:47-54. [DOI] [PubMed] [Google Scholar]
- 20.Hulette, B. C., G. Rowden, C. A. Ryan, C. M. Lawson, S. M. Dawes, G. M. Ridder, and G. F. Gerberick. 2001. Cytokine induction of a human acute myelogenous leukemia cell line (KG-1) to a CD1a+ dendritic cell phenotype. Arch. Dermatol. Res. 293:147-158. [DOI] [PubMed] [Google Scholar]
- 21.Ju, X. S., C. Hacker, J. Madruga, S. M. Kurz, S. Knespel, G. Blendinger, S. Rose-John, and Z. Martin. 2003. Towards determining the differentiation program of antigen-presenting dendritic cells by transcriptional profiling. Eur. J. Cell Biol. 82:75-86. [DOI] [PubMed] [Google Scholar]
- 22.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 23.Keegan, B. R., J. L. Feldman, D. H. Lee, D. S. Koos, R. K. Ho, D. Y. Stainier, and D. Yelon. 2002. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development 129:1623-1632. [DOI] [PubMed] [Google Scholar]
- 24.Keene, R. G., A. Mueller, R. Landick, and L. London. 1999. Transcriptional pause, arrest and termination sites for RNA polymerase II in mammalian N- and c-myc genes. Nucleic Acids Res. 27:3173-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kistler, B., A. Rolink, R. Marienfeld, M. Neumann, and T. Wirth. 1998. Induction of nuclear factor-κB during primary B cell differentiation. J. Immunol. 160:2308-2317. [PubMed] [Google Scholar]
- 26.Koeffler, H. P., R. Billing, A. J. Lusis, R. Sparkes, and D. W. Golde. 1980. An undifferentiated variant derived from the human acute myelogenous leukemia cell line (KG-1). Blood 56:265-273. [PubMed] [Google Scholar]
- 27.Krumm, A., T. Meulia, M. Brunvand, and M. Groudine. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6:2201-2213. [DOI] [PubMed] [Google Scholar]
- 28.Lernbecher, T., B. Kistler, and T. Wirth. 1994. Two distinct mechanisms contribute to the constitutive activation of RelB in lymphoid cells. EMBO J. 13:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J., M. L. Mbow, L. Sun, L. Li, G. Yang, D. E. Griswold, A. Schantz, D. J. Shealy, T. J. Goletz, J. Wan, and D. Peritt. 2004. Induction of dendritic cell maturation by IL-18. Cell. Immunol. 227:103-108. [DOI] [PubMed] [Google Scholar]
- 30.Lin, L., G. N. DeMartino, and W. C. Greene. 1998. Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell 92:819-828. [DOI] [PubMed] [Google Scholar]
- 31.Linial, M., and M. Groudine. 1985. Transcription of three c-myc exons is enhanced in chicken bursal lymphoma cell lines. Proc. Natl. Acad. Sci. USA 82:53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linial, M., N. Gunderson, and M. Groudine. 1985. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science 230:1126-1132. [DOI] [PubMed] [Google Scholar]
- 33.Lin-Lee, Y. C., S. M. Soyal, A. Surguchov, S. Sanders, W. Strobl, and W. Patsch. 1995. Thyroid hormone influences conditional transcript elongation of the apolipoprotein A-I gene in rat liver. J. Lipid Res. 36:1586-1594. [PubMed] [Google Scholar]
- 34.Liptay, S., R. M. Schmid, E. G. Nabel, and G. J. Nabel. 1994. Transcriptional regulation of NF-κB2: evidence for κB-mediated positive and negative autoregulation. Mol. Cell. Biol. 14:7695-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombardi, L., P. Ciana, C. Cappellini, D. Trecca, L. Guerrini, A. Migliazza, A. T. Maiolo, and A. Neri. 1995. Structural and functional characterization of the promoter regions of the NFκB2 gene. Nucleic Acids Res. 23:2328-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, E., B. O'Sullivan, P. Low, and R. Thomas. 2003. Antigen-specific suppression of a primed immune response by dendritic cells mediated by regulatory T cells secreting interleukin-10. Immunity 18:155-167. [DOI] [PubMed] [Google Scholar]
- 37.Mrozek, K., S. M. Tanner, K. Heinonen, and C. D. Bloomfield. 2003. Molecular cytogenetic characterization of the KG-1 and KG-1a acute myeloid leukemia cell lines by use of spectral karyotyping and fluorescence in situ hybridization. Genes Chromosomes Cancer 38:249-252. [DOI] [PubMed] [Google Scholar]
- 38.Muto, A., J. Ruland, L. M. McAllister-Lucas, P. C. Lucas, S. Yamaoka, F. F. Chen, A. Lin, T. W. Mak, G. Nunez, and N. Inohara. 2002. Protein kinase C-associated kinase (PKK) mediates Bcl10-independent NF-κB activation induced by phorbol ester. J. Biol. Chem. 277:31871-31876. [DOI] [PubMed] [Google Scholar]
- 39.Neumann, M., H. Fries, C. Scheicher, P. Keikavoussi, A. Kolb-Maurer, E. Brocker, E. Serfling, and E. Kampgen. 2000. Differential expression of Rel/NF-κB and octamer factors is a hallmark of the generation and maturation of dendritic cells. Blood 95:277-285. [PubMed] [Google Scholar]
- 40.O'Sullivan, B. J., K. P. MacDonald, A. R. Pettit, and R. Thomas. 2000. RelB nuclear translocation regulates B cell MHC molecule, CD40 expression, and antigen-presenting cell function. Proc. Natl. Acad. Sci. USA 97:11421-11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouaaz, F., J. Arron, Y. Zheng, Y. Choi, and A. A. Beg. 2002. Dendritic cell development and survival require distinct NF-κB subunits. Immunity 16:257-270. [DOI] [PubMed] [Google Scholar]
- 42.Perkel, J. M., M. C. Simon, and A. Rao. 2002. Identification of a c-myb attenuator-binding factor. Leuk. Res. 26:179-190. [DOI] [PubMed] [Google Scholar]
- 43.Pettit, A. R., K. P. MacDonald, B. O'Sullivan, and R. Thomas. 2000. Differentiated dendritic cells expressing nuclear RelB are predominantly located in rheumatoid synovial tissue perivascular mononuclear cell aggregates. Arthritis Rheum. 43:791-800. [DOI] [PubMed] [Google Scholar]
- 44.Platzer, B., A. Jorgl, S. Taschner, B. Hocher, and H. Strobl. 2004. RelB regulates human dendritic cell subset development by promoting monocyte intermediates. Blood 104:3655-3663. [DOI] [PubMed] [Google Scholar]
- 45.Rossi, F., M. McNagny, G. Smith, J. Frampton, and T. Graf. 1996. Lineage commitment of transformed haematopoietic progenitors is determined by the level of PKC activity. EMBO J. 15:1894-1901. [PMC free article] [PubMed] [Google Scholar]
- 46.Ryseck, R. P., P. Bull, M. Takamiya, V. Bours, U. Siebenlist, P. Dobrzanski, and R. Bravo. 1992. RelB, a new Rel family transcription activator that can interact with p50-NF-κB. Mol. Cell. Biol. 12:674-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saijo, K., I. Mecklenbrauker, A. Santana, M. Leitger, C. Schmedt, and A. Tarakhovsky. 2002. Protein kinase C beta controls nuclear factor κB activation in B cells through selective regulation of the IκB kinase alpha. J. Exp. Med. 195:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shortman, K., and Y. J. Liu. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151-161. [DOI] [PubMed] [Google Scholar]
- 49.Soilleux, E. J., R. Barten, and J. Trowsdale. 2000. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 165:2937-2942. [DOI] [PubMed] [Google Scholar]
- 50.Solan, N. J., H. Miyoshi, E. M. Carmona, G. D. Bren, and C. V. Paya. 2002. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 277:1405-1418. [DOI] [PubMed] [Google Scholar]
- 51.Speirs, K., L. Lieberman, J. Caamano, C. A. Hunter, and P. Scott. 2004. Cutting edge: NF-κB2 is a negative regulator of dendritic cell function. J. Immunol. 172:752-756. [DOI] [PubMed] [Google Scholar]
- 52.St Louis, D. C., J. B. Woodcock, G. Fransozo, P. J. Blair, L. M. Carlson, M. Murillo, M. R. Wells, A. J. Williams, D. S. Smoot, S. Kaushal, J. L. Grimes, D. M. Harlan, J. P. Chute, C. H. June, U. Siebenlist, and K. P. Lee. 1999. Evidence for distinct intracellular signaling pathways in CD34+ progenitor to dendritic cell differentiation from a human cell line model. J. Immunol. 162:3237-3248. [PubMed] [Google Scholar]
- 53.Suciu-Foca Cortesini, N., F. Piazza, E. Ho, R. Ciubotariu, J. LeMaoult, R. Dalla-Favera, and R. Cortesini. 2001. Distinct mRNA microarray profiles of tolerogenic dendritic cells. Hum. Immunol. 62:1065-1072. [DOI] [PubMed] [Google Scholar]
- 54.Suhasini, M., and R. B. Pilz. 1999. Transcriptional elongation of c-myb is regulated by NF-κB (p50/RelB). Oncogene 18:7360-7369. [DOI] [PubMed] [Google Scholar]
- 55.Suhasini, M., C. D. Reddy, E. P. Reddy, J. A. DiDonato, and R. B. Pilz. 1997. cAMP-induced NF-κB (p50/relB) binding to a c-myb intronic enhancer correlates with c-myb up-regulation and inhibition of erythroleukemia cell differentiation. Oncogene 15:1859-1870. [DOI] [PubMed] [Google Scholar]
- 56.Ueberla, K., Y. Lu, E. Chung, and W. A. Haseltine. 1993. The NF-κB p65 promoter. J. Acquir. Immune Defic. Syndr. 6:227-230. [PubMed] [Google Scholar]
- 57.Valero, R., M. L. Baron, S. Guerin, S. Beliard, H. Lelouard, B. Kahn-Perles, B. Vialettes, C. Nguyen, J. Imbert, and P. Naquet. 2002. A defective NF-κB/RelB pathway in autoimmune-prone New Zealand black mice is associated with inefficient expansion of thymocyte and dendritic cells. J. Immunol. 169:185-192. [DOI] [PubMed] [Google Scholar]
- 58.Weih, D. S., Z. B. Yilmaz, and F. Weih. 2001. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 167:1909-1919. [DOI] [PubMed] [Google Scholar]
- 59.Weih, F., G. Warr, H. Yang, and R. Bravo. 1997. Multifocal defects in immune responses in RelB-deficient mice. J. Immunol. 158:5211-5218. [PubMed] [Google Scholar]
- 60.Wilson, H. L., and H. C. O'Neill. 2003. Identification of differentially expressed genes representing dendritic cell precursors and their progeny. Blood 102:1661-1669. [DOI] [PubMed] [Google Scholar]
- 61.Wu, L., A. D'Amico, K. D. Winkel, M. Suter, D. Lo, and K. Shortman. 1998. RelB is essential for the development of myeloid-related CD8α− dendritic cells but not of lymphoid-related CD8α+ dendritic cells. Immunity 9:839-847. [DOI] [PubMed] [Google Scholar]
- 62.Xiao, G., E. W. Harhaj, and S. C. Sun. 2001. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7:401-409. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto, Y., and R. B. Gaynor. 2004. IκB kinases: key regulators of the NF-κB pathway. Trends Biochem. Sci. 29:72-79. [DOI] [PubMed] [Google Scholar]
- 64.Yoshimura, S., J. Bondeson, B. M. Foxwell, F. M. Brennan, and M. Feldmann. 2001. Effective antigen presentation by dendritic cells is NF-κB dependent: coordinate regulation of MHC, costimulatory molecules and cytokines. Int. Immunol. 13:675-683. [DOI] [PubMed] [Google Scholar]