Abstract
LINE-1 (L1) retrotransposons comprise ∼17% of human DNA, yet little is known about L1 integration. Here, we characterized 100 retrotransposition events in HeLa cells and show that distinct DNA repair pathways can resolve L1 cDNA retrotransposition intermediates. L1 cDNA resolution can lead to various forms of genetic instability including the generation of chimeric L1s, intrachromosomal deletions, intrachromosomal duplications, and intra-L1 rearrangements as well as a possible interchromosomal translocation. The L1 retrotransposition machinery also can mobilize U6 snRNA to new genomic locations, increasing the repertoire of noncoding RNAs that are mobilized by L1s. Finally, we have determined that the L1 reverse transcriptase can faithfully replicate its own transcript and has a base misincorporation error rate of ∼1/7,000 bases. These data indicate that L1 retrotransposition in transformed human cells can lead to a variety of genomic rearrangements and suggest that host processes act to restrict L1 integration in cultured human cells. Indeed, the initial steps in L1 retrotransposition may define a host/parasite battleground that serves to limit the number of active L1s in the genome.
Long interspersed element 1 (LINE-1 or L1) is an abundant retrotransposon that comprises ∼17% of human DNA (43, 69). Most L1s are retrotransposition defective because they are 5′ truncated, contain internal rearrangements, or harbor mutations within their open reading frames (25, 43). However, the average human genome is estimated to contain ∼80 to 100 retrotransposition-competent L1s (RC-L1s), and approximately 10% of these elements are classified as highly active or “hot” (6, 63).
Human RC-L1s are ∼6.0 kb and contain a 5′ untranslated region (UTR), two nonoverlapping open reading frames (ORF1 and ORF2), and a 3′ UTR that ends in a poly(A) tail (Fig. 1A) (13, 53, 66). ORF1 encodes a 40-kDa nucleic acid binding protein (30, 31, 33), whereas ORF2 has the potential to encode a 150-kDa protein with demonstrated endonuclease (L1 EN) and reverse transcriptase (L1 RT) activities (15, 19, 22, 51). ORF2p also contains a cysteine-rich domain (CX3CX7HX4C) of unknown function (17, 54). Both proteins are required for retrotransposition in cis (54), which most probably occurs by a mechanism termed “target site primed reverse transcription” (TPRT) (19, 47, 54, 72). However, how L1 integration is completed remains a mystery.
FIG. 1.
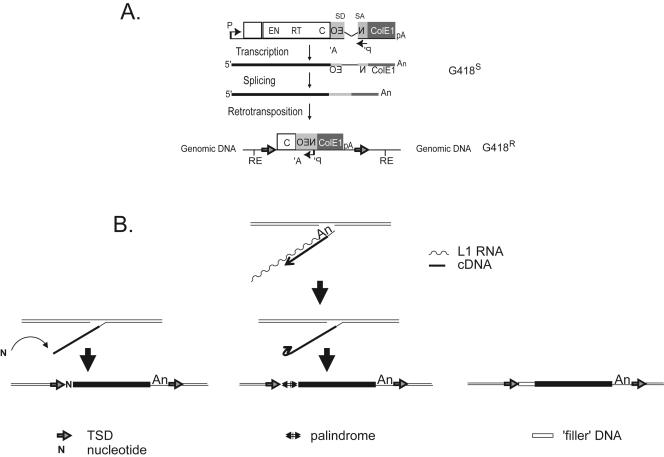
Simple sequence alterations at the 5′ genomic DNA/L1 junction. A. Rationale of the assay. The 3′ UTR of a human RC-L1 was tagged with a reporter cassette designed to detect retrotransposition events. Open rectangles indicate L1 ORF1 and L1 ORF2, respectively. The relative positions of the endonuclease (EN), RT, and cysteine-rich domains (C) are indicated. The position of the L1 promoter (P) and the simian virus 40 late polyadenylation signal (pA) needed for L1 expression also are indicated. The mneoI gene and ColE1 bacterial origin of replication are indicated by light and dark gray rectangles, respectively. The relative positions of the prokaryotic/eukaryotic promoter (P′) and the thymidine kinase polyadenylation signal (A′) required for reporter gene expression also are shown. The mneoI gene is interrupted by an intron (γ-globin intron 2) in the same transcriptional orientation as the L1. SD and SA indicate the splice donor and splice acceptor sites, respectively. This arrangement ensures that a functional NEO transcript will be translated only following L1 retrotransposition. The putative structure of a resultant retrotransposition event that confers G418 resistance (G418r) to HeLa cells is shown at the bottom. Horizontal arrows flanking the resultant insertion indicate the TSD. Black lines flanking the insertion indicate HeLa genomic DNA, and RE represents the cleavage site for the restriction enzyme present in flanking genomic DNA. B. Extra sequences at the 5′ genomic DNA/L1 junction. The top center panel indicates a Y-branched L1 retrotransposition intermediate following the initial stages of TPRT. A variety of sequence alterations were found at the 5′ genomic DNA/L1 junction including the addition of single “untemplated” nucleotides (indicated by the “N” in the bottom left panel), the formation of a palindromic repeat (indicated by the arrows in the bottom middle panel), and the addition of “filler” DNA (indicated by the open rectangle in the bottom right panel).
We recently developed a plasmid-based rescue system that allows the recovery of L1 insertions in cultured human HeLa cells with minimal influence from selective pressures that occur during genome evolution. We found that L1 retrotransposition is associated with various forms of genetic instability and that the nascent L1 cDNA can undergo recombination with endogenous L1 elements, resulting in the formation of chimeric L1s. Consistent findings by Symer et al., using a colon cell line (HCT116) with an essentially normal karyotype, have led to the hypothesis that L1 retrotransposition can lead to various types of genomic instability (21, 72).
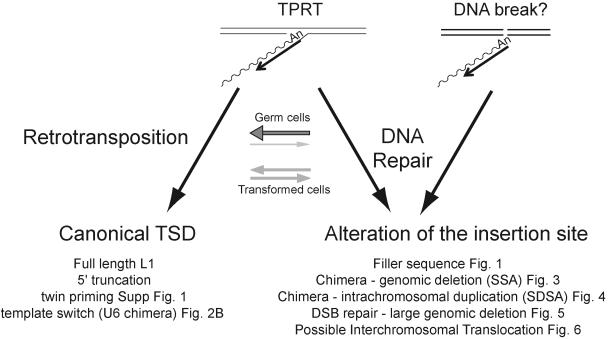
Here, we describe the analysis of 100 L1 retrotransposition events in HeLa cells derived from four previously characterized RC-L1s (L1.2A, LRE-2, L1.3, and L1RP). Consistent with previous studies, we have found that retrotransposition is associated with the generation of intrachromosomal deletions, the creation of chimeric L1 elements, and the addition of non-L1 nucleotides at the 5′ insertion junction (21, 56, 72). In addition, we have observed novel rearrangements, including the mobilization of U6 small uracil-rich nuclear RNA (U6 snRNA) to a new genomic location, the formation of intrachromosomal duplications, intra-L1 rearrangements, and the generation of a possible interchromosomal translocation. Finally, we have determined that the L1 RT can faithfully replicate its own transcript and has a base misincorporation error rate of ∼1/7,000 bases. Together, these data indicate that the resolution of L1 retrotransposition intermediates in transformed human cell lines can lead to a variety of genomic rearrangements and lead us to propose that host processes act to restrict L1 retrotransposition during integration, limiting the number of full-length L1s in the genome.
MATERIALS AND METHODS
Oligonucleotides and DNA sequences.
Oligonucleotide sequences are available upon request or can be accessed at www.med.umich.edu/hg/RESEARCH/FACULTY/Moran/moranweb.htm.
All insertion sequences are available upon request or can be accessed at www.med.umich.edu/hg/RESEARCH/FACULTY/Moran/moranweb.htm.
Plasmids.
pJM101, pK7/L1mneoI/ColE1, and pCEP4/L1mneoI/ColE1 plasmid DNAs were previously described in the work of Moran et al. (54) and Gilbert et al. (21).
(i) pJM140/L1.3/delta2K7.
The mneoI-ColE1 fragment from pK7/L1mneoI/ColE1 was amplified by PCR using the NotIBamNeo and ColE1SmaBam primers. The PCR product was cloned into pBS-KS− digested with BamHI, to generate pBSNeoColE1. The 3,106-bp mneoI-ColE1 fragment from pBSNeoColE1 was then digested with BamHI, and the ends were made blunt with T4 DNA polymerase. The blunt restriction fragment was introduced into an engineered SmaI site at position 5980 of the L1.3 3′ UTR present in pJCC5/L1.3. Swapping the 9,204-bp NotI-BamHI fragment containing the engineered L1 into pJM140/L1.3 ΔCMVΔSV40 poly(A)+ (52) created pJM140/L1.3/delta2K7.
(ii) pJM140/L1.2/delta2K7, pJM140/LRE2/delta2K7, and pJM140/RP/delta2K7.
Swapping the 5,973-bp NotI-Bstz17i fragment from pJM101/L1.2, pJM104, and RPcmv with the corresponding fragment from pJM140/L1.3/delta2K7 created pJM140/L1.2/delta2K7, pJM140/LRE2/delta2K7, and pJM140/RP/delta2K7, respectively.
(iii) pBSSP140/L1.3K7.
A PmeI site was created in the L1.3 3′ UTR (at position 5826) using the QuickChange site-directed mutagenesis kit (Stratagene) and primers PmeIfwd and PmeIrev on a plasmid containing a 621-bp SpeI-BamHI fragment of the L1 3′ UTR originating from pJCC5/L1.3. Swapping the 382-bp NcoI-BamHI fragment containing the PmeI site with the corresponding fragment from pJCC5/L1.3 created pBSL1.3PmeI. The 3,106-bp mneoI-ColE1-blunted BamHI fragment from the pBSNeoColE1 plasmid was then introduced into the PmeI site to create pBSSP140/L1.3K7.
(iv) pSP140/L1.3K7.
Swapping the 9,195-bp NotI-BamHI fragment of pBSSP140/L1.3K7 into pJM140/L1.3 ΔCMVΔSV40 poly(A)+ (52) created pSP140/L1.3K7.
(v) pSP101/L1.3K7.
Construct pSP101/L1.3K7 contains a modification of the L1 poly(A) tail compared to pSP140/L1.3K7. The sequence is TAT(A)2T(A)3 instead of TAT(A)2T(A)23G(A)3.
(vi) pSP140/L1.3K7ΔL1pA.
The L1 pA signal of pSP140/L1.3K7 was deleted, creating the A-rich tail TAT(A)28G(A)3.
DNA preparation.
Plasmid DNAs were purified on QIAGEN Midi Prep columns (QIAGEN). Rescued plasmids were purified using Wizard S/V Mini Prep kits (Promega). HeLa genomic DNA was isolated using either the blood and cell Midi Prep kit (QIAGEN) or the cell and tissue DNA isolation kit (Puregene; Gentra).
L1 retrotransposition assay.
HeLa cells were grown at 37°C in an atmosphere containing 7% carbon dioxide and 100% humidity in Dulbecco's modified Eagle's medium lacking pyruvate (Gibco BRL). Dulbecco's modified Eagle's medium was supplemented with 10% fetal bovine calf serum and 1× penicillin-streptomycin-l-glutamine (a 100× stock is sold by Gibco BRL). Cell passage and cloning (by either limiting dilution or colony lifting) were performed using standard techniques. Retrotransposition was monitored using the transient retrotransposition assay (76).
Rescue procedure.
To clone the retrotransposition events, genomic DNA was extracted from cell lines derived from single or small pools (10 to 250) of G418r colonies. Ten micrograms of DNA was digested overnight with HindIII, BglII, BclI, or BamHI (New England Biolabs). The enzyme was either heat inactivated or removed using the Wizard DNA cleanup kit (Promega). The resultant fragments were ligated overnight at 14°C in a 500-μl volume with T4 DNA ligase (2,400 U; New England Biolabs). Ligations were concentrated by centrifugation through a Microcon-100 concentrator at 500 × g for 14 min, and the entire concentrated ligation was transformed into 1 ml of XL1-Blue MRF′ cells (Stratagene), which were made to competencies of >1 × 108 transformants/μg (35, 73). One to 15 transformants were visible after overnight growth at 37°C on LB agar supplemented with 50 μg/ml kanamycin. The sizes of the rescued plasmids varied from ∼3 kb to >12 kb in size.
Sequence analysis.
Sequences flanking each L1 insertion were used as probes in BLAT searches to identify the preintegration site in the human genome working draft sequence (HGWD) using either the 04/03 or the 07/03 freezes (http://genome.cse.ucsc.edu) (38). Some rescued plasmids contained L1s that were truncated at a restriction site used in their recovery. To obtain the remaining L1 sequence, we used the HGWD to design an oligonucleotide primer from sequences presumed to flank the 5′ end of the retrotransposed L1. The 5′ primer then was used in conjunction with 173NEOas to PCR amplify the 5′ flanking sequence from genomic DNA of cell lines harboring the relevant insertions. BigDye terminator cycle sequencing (ABI PRISM) was performed on an Applied Biosystems DNA sequencer (model ABI377 or ABI3700) at the University of Michigan Core facilities. For chimeric L1s, the nucleotide-nucleotide BLAST interface (blastn; http://www.ncbi.nlm.nih.gov/BLAST/) (1) or BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) (38) was used to identify an endogenous L1 in the HGWD with 100% identity to that present in the chimeric L1. In each case, the endogenous L1s and their associated flanking sequences were 100% identical to the sequences present in the recovered plasmids.
PCR amplification.
Amplification of pre- and postintegration sequences was conducted following the protocol provided with the Expand Long Template PCR System (Roche). Each PCR amplification was carried out in a 50-μl volume containing, 500 ng of genomic DNA, 300 nM of each primer, 350 μM of each deoxynucleoside triphosphate, and 1 μl of enzyme mix. PCRs were conducted using the following cycling conditions: one cycle of 94°C for 2 min; 30 cycles of 94°C for 10 s, 60°C for 30 s, and 68°C for 1 min per kb; and one cycle of 68°C for 10 min. To amplify products larger than 6 kb, the final extension time at 68°C was extended to 30 min. Amplification was conducted on genomic DNA isolated from either clonal cell lines or polyclonal cell lines derived from 10 to 250 G418-resistant foci. For each PCR, HeLa DNA was used as a control to demonstrate that the observed genomic rearrangement was not present in naïve cells. For the results shown in Fig. 7, all nucleotide misincorporations were verified by sequencing PCR products isolated from their respective cell lines.
FIG. 7.
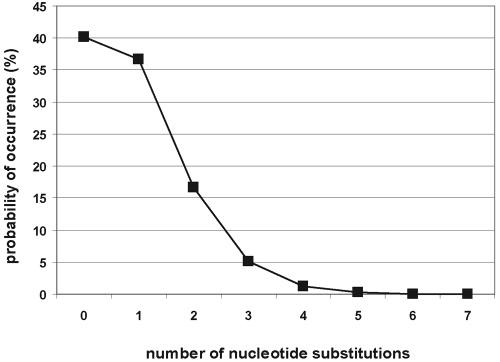
Misincorporation error rate of the L1 RT. The binomial distribution was used to calculate the percentage of full-length L1s that are faithful copies of their respective progenitor element. The calculations are based on a base misincorporation rate of 1 error/6,584 bp (15 errors in 98,758 bases of L1 sequenced).
RESULTS
A cohort of 100 L1 retrotransposition events.
We previously have used various strategies to recover 41 L1 retrotransposition events from cultured human HeLa cells (21, 54). Here, we developed a second-generation recovery vector (see Materials and Methods) and describe the characterization of 59 additional L1 retrotransposition events in HeLa cells that are derived from four previously characterized RC-L1s (L1.2A, LRE-2, L1.3, and L1RP) (13, 14, 32, 39, 54). Each RC-L1 is a member of the Ta (transcribed active) subfamily (5, 68) and exhibits different retrotransposition efficiencies in cultured cells. L1RP displays the highest retrotransposition efficiency (39), whereas L1.2A has the lowest retrotransposition efficiency because of two amino acid changes downstream of the conserved cysteine-rich motif (48). This cohort of 100 L1 retrotransposition events (Table 1) combined with the improved assembly of the HGWD provides an opportunity to comprehensively assess L1 retrotransposition in a transformed human cell line.
TABLE 1.
Sequence characteristics of 100 L1s in cultured cellsa
| Clone no. | Typeb | Extrac | Length (bp)d | Cleavagee | TSDf (bp) | Del. or dupl. |
|---|---|---|---|---|---|---|
| 1 | Full length | 5′ untemplated (G) | 6,090 | TCTT/A | 13 | |
| 2 | Full length | 5′ untemplated (G) | 6,090 | TTTC/A | 10 | |
| 3 | Full length | 5,984 | TTTT/A | 13 | ||
| 4 | Full length | 5,428 | TCTT/A | 15 | ||
| 5 | Full length | 5,986 | TTTT/A | 17 | ||
| 6 | Full length | 3,123 | TTTA/A | 15 | ||
| 7 | 5′ truncation | 5′ duplication (6/11 nt) | 472 | TTTT/A | 67 | |
| 8 | 5′ truncation | 5′ templated seq. palindrome | 27 | TTTT/G | Del. 3 | |
| 9 | 5′ truncation | 5′ untemplated (A) | 281 | TTTT/C | 6,319 | |
| 10 | 5′ truncation | 5′ untemplated (A) | 687 | TTTT/G | 116 | |
| 11 | 5′ truncation | 5′ untemplated (G) | 27 | TTCT/G | 5 | |
| 12 | 5′ truncation | 5′ untemplated (T) | 415 | TTTT/A | 39 | |
| 13 | 5′ truncation | 5′ untemplated nt (ATAA) | 120 | TTTT/A | 15 | |
| 14 | 5′ truncation | 5′ untemplated nt (TTTATTAT) | 306 | TTTT/A | 45 | |
| 15 | 5′ truncation | 5′ untemplated nt (TTGAAGATTC) | 2,685 | TTTC/A | 162 | |
| 16 | 5′ truncation | 5′ untemplated nt (GAAGACATATTTG) | 744 | ATTT/G | 13 | |
| 17 | 5′ truncation | 5′ untemplated nt (AATGGTTTTTAAGGGTTTT) | 646 | TTTT/A | 75 | |
| 18 | 5′ truncation | 5′ untemplated nt (TATGTTTTATGTTTTAAATA) | 752 | TTTT/A | Blunt | |
| 19 | 5′ truncation | Inverted hnRNP on 5′ (TTTC/A) | 306 | TTTT/G | Del. 14101 | |
| 20 | 5′ truncation | Template switch U6 | 101 | TTTT/A | 17 | |
| 21 | 5′ truncation | Templated seq. (81 bp) | 3,948 | TTTT/A | 125 | |
| 22 | 5′ truncation | Templated seq. (55 bp) | 300 | AGGT/G | Del. 7567 | |
| 23 | 5′ truncation | Templated seq. and translocation | 242 | TCTT/G | NA | |
| 24 | 5′ truncation | 1,308 | TTTT/A | 2 | ||
| 25 | 5′ truncation | — | TTTT/A | 217 | ||
| 26 | 5′ truncation | — | TTTT/A | Del. 5 | ||
| 27 | 5′ truncation | 549 | TCTT/G | 34 | ||
| 28 | 5′ truncation | 53 | TTCT/A | 17 | ||
| 29 | 5′ truncation | 270 | TTTT/A | 98 | ||
| 30 | 5′ truncation | 600 | TTTA/A | 323 | ||
| 31 | 5′ truncation | 420 | TTTT/C | Del. 20 | ||
| 32 | 5′ truncation | 155 | TTTT/G | 18 | ||
| 33 | 5′ truncation | 5,428 | TTTT/G | 13 | ||
| 34 | 5′ truncation | 1,052 | TATT/A | 15 | ||
| 35 | 5′ truncation | 135 | TTTT/A | 17 | ||
| 36 | 5′ truncation | 138 | TTTT/G | Del. 2 | ||
| 37 | 5′ truncation | 307 | TTTC/A | 55 | ||
| 38 | 5′ truncation | 3 | TTCT/A | 14 | ||
| 39 | 5′ truncation | 172 | TTTT/G | 127 | ||
| 40 | 5′ truncation | 1,661 | TATT/G | Del. 17 | ||
| 41 | 5′ truncation | 433 | TTTT/A | 13 | ||
| 42 | 5′ truncation | 582 | TCTT/G | Blunt | ||
| 43 | 5′ truncation | 501 | TTTT/A | Del. 47 | ||
| 44 | 5′ truncation | 376 | TTCT/G | 16 | ||
| 45 | 5′ truncation | 288 | TTTT/G | 49 | ||
| 46 | 5′ truncation | 9 | TTTT/A | 135 | ||
| 47 | 5′ truncation | 536 | TCTT/G | 11 | ||
| 48 | 5′ truncation | 221 | TTCT/G | 76 | ||
| 49 | 5′ truncation | 691 | TTTT/G | 12 | ||
| 50 | 5′ truncation | 60 | TTTT/G | 15 | ||
| 51 | 5′ truncation | 118 | ACTT/A | 15 | ||
| 52 | 5′ truncation | 18 | TTTT/A | 28 | ||
| 53 | 5′ truncation | 10 | ATTT/A | 7 | ||
| 54 | 5′ truncation | 120 | TTCT/A | Blunt | ||
| 55 | 5′ truncation | 1,173 | CTTT/C | 27 | ||
| 56 | 5′ truncation | 33 | TTTT/G | Del. 31 | ||
| 57 | 5′ truncation | 150 | TCTT/A | 7 | ||
| 58 | 5′ truncation | 245 | TTTT/A | 14 | ||
| 59 | 5′ truncation | 29 | ATTT/A | 15 | ||
| 60 | Chimera | 3′ duplication (9/15 and 267/270) | 1,190 | TTTT/A | Del. 2985/3014 | |
| 61 | Chimera | 514 | TTTT/A | Del. 503/545 | ||
| 62 | Chimera | 577 | TCTT/A | Del. 193/238 | ||
| 63 | Chimera | 1,090 | TTTA/A | NA | ||
| 64 | Endo. indep. and chimera | 3′ untemplated (GTG) | 775 | GTGA/G | Del. 1296 | |
| 65 | Endo. indep. and chimera | 5′ flank duplication and 3′ truncation | 332 | 3 pos. | Del 131076 | |
| 66 | Inv./del. and chimera | Intrachromosomal dupl. | 1,879 | TTTT/G | 15 | 15- to 38-bp del. |
| 67 | Inv./del. and chimera | Intrachromosomal dupl. and 5′ untemplated (T) | 1,617 | TTTT/A | 14 | 4,467- to 4,469-bp del. |
| 68 | Inversion/deletion | Internal deletion (246-nt del.) | 3,817 | TTTT/A | 15 | 7- to 9-bp del. |
| 69 | Inversion/deletion | Untemplated nt (ATATATAACAGAGC) | 2,731 | TTCT/A | 14 | 175- to 179-bp del. |
| 70 | Inversion/deletion | 211 | TCTT/A | 13 | 12- to 22-bp del. | |
| 71 | Inversion/deletion | 327 | TTTA/A | 15 | 13- to 16-bp del. | |
| 72 | Inversion/deletion | 514 | TTTT/G | 11 | 563- to 568-bp del. | |
| 73 | Inversion/deletion | 2,233 | TTAT/A | 16 | 569- to 574-bp del. | |
| 74 | Inversion/deletion | 5,512 | TCTT/A | 16 | 2- to 4-bp del. | |
| 75 | Inversion/deletion | 1,370 | TTCT/A | 14 | 536- to 538-bp del. | |
| 76 | Inversion/deletion | 132 | TTTC/A | 11 | 246- to 253-bp del. | |
| 77 | Inversion/deletion | 272 | TCTT/A | 16 | 9- to 11-bp del. | |
| 78 | Inversion/deletion | 275 | TTTA/A | 16 | 3- to 5-bp del. | |
| 79 | Inversion/deletion | 240 | TCTT/A | 14 | 33- to 35-bp del. | |
| 80 | Inversion/deletion | 1,891 | TTCT/A | 16 | 4- to 10-bp del. | |
| 81 | Inversion/deletion | 2,029 | TTCT/A | NA | NA | |
| 82 | Inversion/duplication | 552 | TTTT/G | 15 | 5- to 6-bp dupl. | |
| 83 | Inversion/duplication | 6,074 | TTTT/A | 15 | 8- to 11-bp dupl. | |
| 84 | 5′ truncation | 5′ untemplated (G) | 353 | TTTA/A | Del. 2 | |
| 85 | 5′ truncation | 5′ untemplated (G) | 153 | ATTA/G | 55 | |
| 86 | 5′ truncation | 5′ untemplated nt (ACAAGTTAT) | — | TTTA/A | 186 | |
| 87 | 5′ truncation | 349 | TTTA/A | 20 | ||
| 88 | 5′ truncation | 2,005 | TTTT/A | 235 | ||
| 89 | 5′ truncation | 399 | TCTT/G | 108 | ||
| 90 | 5′ truncation | 20 | TTTT/A | 14 | ||
| 91 | 5′ truncation | 496 | TTTT/A | 113 | ||
| 92 | 5′ truncation | 226 | TTTT/G | 10 | ||
| 93 | 5′ truncation | 117 | TTCT/A | Blunt | ||
| 94 | 5′ truncation | — | TATT/A | Blunt | ||
| 95 | 5′ truncation | 1,127 | TGTT/A | 43 | ||
| 96 | 5′ truncation | 399 | TCTT/G | 12 | ||
| 97 | 5′ truncation | 194 | TGTT/G | 153 | ||
| 98 | 5′ truncation | 51 | TTTT/G | Del. 2 | ||
| 99 | Chimera | 239 | TTTT/A | 159 | ||
| 100 | Inversion/deletion | 28 | TTTA/A | 16 | 22- to 23-bp del. |
An expanded version of Table 1 is present in the supplemental material. Abbreviations: del., deletion; dupl., duplication; nt, nucleotide(s); seq., sequence; inv., inversion; endo. indep., endonuclease independent; NA, not applicable; pos., position(s).
Structural feature(s) associated with the insertion. —, no L1 segment was reverse transcribed.
Sequence alteration(s) associated with the insertion.
Length of L1 insertion, not including the size of the retrotransposition indicator cassette (2.3 or 2.7 kb, respectively).
Sequence of the “bottom-strand” target site cleavage site.
Length of target site duplication.
Engineered L1 elements readily insert into genes.
Consistent with previous analyses, our data indicate that new L1 insertions occur throughout the genome and that engineered L1 elements readily can insert into genes (21, 52, 54, 72). Thirty insertions occurred into introns of known (17) or hypothetical (13) genes (Table 1). Another 46 L1s inserted into “predicted” genes; however, it remains to be determined whether these events represent insertions into actual genes. Two insertions could not be mapped definitively. The first inserted into a sequence that is present at least 20 times in the HGWD (insertion 31, Table 1), whereas the second (insertion 53) occurred into a sequence absent from the HGWD.
Most insertions occur at consensus L1 EN cleavage sites and structurally resemble endogenous L1s.
Ninety-seven events preferentially inserted into sequences that resemble L1 endonuclease consensus cleavage sites (e.g., 5′ TTTT/A and related sequences) (10, 19, 21, 56, 72). Thus, most L1 retrotransposition events seem to be initiated by conventional endonuclease-dependent TPRT. Interestingly, in 14/97 events the endonucleolytic cleavage did not occur at the phosphodiester bond between a T/A but instead occurred between either a C/A (four events) or an A/A (10 events). Thus, if TPRT involves base pairing between the L1 RNA poly(A) tail and single-stranded DNA at the target site, our data indicate that the L1 RT can extend terminal RNA/DNA base mismatches. Three events (insertions 22, 64, and 65, Table 1) occurred into sequences that are not preferential L1 endonuclease cleavage sites and may have used genomic nicks or breaks to initiate TPRT (see below) (21, 56).
Consistent with previous analyses, the recovered L1 retrotransposition events generally resemble endogenous L1s in structure (21, 54, 72). Six L1s are full-length insertions (i.e., they contain a full-length copy of the RC-L1 and the ∼2.4-kb spliced mneoI/ColE1 reporter cassette). Ninety-four L1s are variably 5′ truncated. Of those, 19 contain inversion/deletion or inversion/duplication structures, which commonly are generated upon retrotransposition (21, 34, 58, 72). Each class of insertion is described in greater detail below.
Full-length L1 insertions.
Six L1s (insertions 1 to 6, Table 1) represent full-length insertions into a preferred L1 endonuclease cleavage site, and each is flanked by a perfect target site duplication (TSD) that ranges in size from 10 to 17 bp. Five L1s (insertions 21, 33, 68, 74, and 83, Table 1) have retrotransposed greater than 6 kb of sequence. Thus, 11/100 events have retrotransposed a genomic equivalent of L1 RNA. However, because we cannot recover retrotransposition events containing less than 2.4 kb in this assay, our results may overestimate the actual percentage of full-length L1 retrotransposition events. Alternatively, it remains possible that the recovery procedure allows the more efficient recovery of 5′-truncated L1s, leading to an underrepresentation of full-length L1s in our data set.
Two full-length L1s (insertions 1 and 2, Table 1) contain an extra guanosine residue at their respective 5′ ends, which may be derived from reverse transcription of the 7-methyl guanosine cap presumed to be present on L1 RNA (2, 4, 54). Interestingly, insertion 4 contains a short patch of four nucleotide changes in its 5′ UTR, whereas insertion 6 contains seven nucleotide changes (one in the 5′ UTR, two in ORF1, and four at the beginning of ORF2). One of the changes in insertion 6 results in a missense mutation (H335Y) in ORF1p. Thus, the changes present in insertions 4 and 6 could result from L1 RT errors. Alternatively, these insertions may represent chimeric L1s that could, in principle, be formed by either nonallelic cDNA-mediated recombination with an endogenous L1 or RNA-mediated template switching during TPRT (see below and the Discussion for possible mechanisms).
5′-truncated insertions. (i) Target site alterations.
Seventy-two endonuclease-dependent L1 insertions contained simple variable 5′ truncations. The majority of elements (52/72; ∼72%) were flanked by variably sized TSDs (see Table S1 in the supplemental material). The sizes of the TSDs were binned as follows: (i) short TSDs that range from 2 to 49 bp (32 instances), (ii) unconventional long TSDs that range from 50 to 323 bp (19 instances), and (iii) a very long TSD that was 6,319 bp in length (21). Consistent with previous studies, we sometimes observed short regions of microcomplementarity at the 5′ genomic DNA/L1 junction; however, the degree of microhomology is not as pronounced as that found in inversion/deletion L1s (see Fig. S1C in the supplemental material) (49, 50, 56, 65, 72, 74).
Eighteen of 72 endonuclease-dependent L1 insertions (∼26%) either were associated with target site deletions (insertions 8, 19, 26, 31, 36, 40, 43, 56, 60 to 62, 84, and 98, Table 1) or did not contain a target site alteration (insertions 18, 42, 54, 93, and 94, Table 1). The sizes of the deletions were binned as follows: (i) short deletions that range from 2 to 47 bp (nine instances) and (ii) large deletions that range from 193 bp to >14 kb (four instances).
We could not unambiguously determine the target site alterations associated with two insertions. The first (insertion 23, Table 1) represents a possible interchromosomal translocation. The second (insertion 63, Table 1) represents a chimeric L1. Both events are discussed in greater detail below.
(ii) Nucleotide additions at the genomic DNA/5′ L1 junction.
Several 5′-truncated L1 elements have nucleotide additions at the genomic DNA/5′ L1 junction. In six instances (insertions 9 to 12, 84, and 85, Table 1) we observed single-nucleotide additions, which represent “untemplated” nucleotides that may result from terminal transferase activity associated with the L1 RT (Fig. 1B, left panel). However, unlike the situation described above for the two full-length L1 insertions, the additional nucleotides appear to be random. Two (insertions 9 and 10) contained an extra adenosine residue, three (insertions 11, 83, and 84) contained an extra guanosine residue, and the last (insertion 12) contained an extra thymidine residue.
In seven instances (insertions 13 to 18 and 86, Table 1), we observed short nucleotide additions at the genomic DNA/5′ L1 junction that range in size from 4 to 20 bp (Fig. 1B, right panel). These DNA segments generally are A/T rich, and we determined that segments longer than 10 bp were not complementary to sequences in L1 RNA. Thus, they are not part of an inversion/deletion that is formed via “twin priming” (see below). Instead, these short DNA segments most probably represent “filler DNAs” that sometimes are found in eukaryotic cells at insertion, deletion, and translocation breakpoints (23, 24, 40, 44-46, 61, 62). In one instance (insertion 21 on chromosome 12, Table 1), we observed an 81-bp DNA fragment at the genomic DNA/5′ L1 junction that seemingly is derived from chromosome 10. PCR confirmed that this sequence is not present at the preintegration site. Thus, it was either “captured” at the genomic DNA/5′ L1 junction during TPRT or copied from another chromosomal template.
In another instance (insertion 8, Table 1), four nucleotides (5′-TAGT-3′) were added to the genomic DNA/5′ L1 junction. Interestingly, the 22 nucleotides at the genomic DNA/5′ L1 junction of that insertion (which includes the 5′-TAGT-3′ nucleotides plus two nucleotides from flanking genomic DNA) can form a perfect palindrome. Thus, addition of the 5′-TAGT-3′ could result if the first-strand cDNA formed a fold-back structure, allowing the use of the L1 cDNA as a template in second-strand cDNA synthesis (Fig. 1B, middle panel). An event similar to this previously was observed in Chinese hamster ovary cells (56).
Inversions/deletions or inversions/duplications.
We identified 19 internally rearranged L1s. Sixteen (insertions 66 to 80 and 100, Table 1) contained an inversion/deletion of L1 sequences, with the size of the L1 deletion varying from 2 to 4,469 bp. Two (insertions 82 and 83, Table 1) contained an internal inversion/duplication of L1 sequences, with the size of the L1 duplication varying from 5 to 11 bp (see Fig. S1 in the supplemental material). We were unable to unambiguously map one event (insertion 81, Table 1) because we could not PCR amplify at the 5′ end of the insertion using primers based on the HGWD.
Unlike the situation observed for the 5′-truncated L1s, all the inversion/deletion or inversion/duplication L1 retrotransposition events are flanked by canonical-length TSDs that range in size from 11 to 16 bp. Interestingly, in six events (insertions 70, 71, 78, 79, 82, and 100, Table 1) the inversion breakpoint is located between either the mneoI gene and ColE1 or between the ColE1 and L1 polyadenylation site. Thus, we likely will miss some retrotransposition events that confer G418r on HeLa cells but contain rearrangements inside the ColE1origin of replication because they cannot be recovered as autonomously replicating plasmids in Escherichia coli.
Microcomplementarity at the genomic DNA/5′ inversion and the L1/L1 inversion junctions.
The “twin priming” model has been proposed to explain the formation of inversion/deletion and inversion/duplication L1 structures (58). In accord with this model, we have observed sequence microcomplementarity at the genomic DNA/5′ inversion junction between L1 RNA and the target site that ranges from 2 to 8 nucleotides (14 are ≥4 bases; see Fig. S1B in the supplemental material). We also have observed imperfect regions of microcomplementarity (e.g., three of six bases in insertion 82, four of eight bases in insertion 79, and four of seven bases in insertions 77 and 100; see Fig. S1B in the supplemental material). In five cases (insertions 74, 79, 82, 83, and 100; see Fig. S1B in the supplemental material), this imperfect microcomplementarity results in a terminal mismatch between the 5′ target sequence and L1 RNA. Thus, these data lend further support to the notion that the L1 RT does not require terminal base pairing between the primer and template to initiate reverse transcription.
We also observed sequence microcomplementarity at the L1/L1 inversion junction that ranges from 1 to 6 bp (see Fig. S1B in the supplemental material). As above, we occasionally observed imperfect regions of microcomplementarity (e.g., three of five bases on insertion 68 and six of eight bases in insertion 73, Table 1; see also Fig. S1B in the supplemental material). In one instance we did not observe microcomplementarity between the two opposing cDNAs (e.g., insertion 68; Table 1; see also Fig. S1B in the supplemental material). Thus, as for the case in conventional TPRT, we propose that terminal base pairing is not strictly required for second-strand cDNA synthesis.
Finally, in two instances (insertions 67 and 69, Table 1), added bases appear at the L1/L1 inversion junction. Insertion 67 contains a single base addition (thymidine), which may result from terminal transferase activity associated with the L1 RT. Insertion 69 contains a 14-bp insertion, which likely was either “captured” at the L1/L1 junction or copied from another template in chromosomal DNA after the initiation of twin priming.
Non-L1 cDNAs at the genomic DNA/5′ L1 junction.
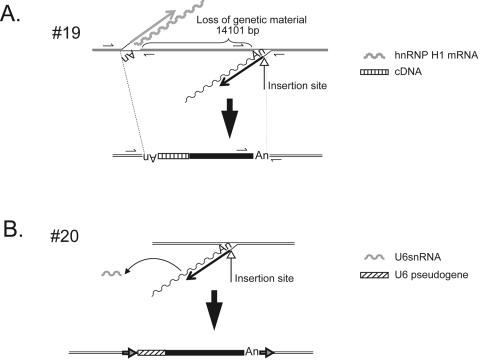
Two insertions contained non-L1 cDNAs at the genomic DNA/5′ L1 junction and represent processed pseudogene/L1 chimeras. The first (insertion 19, Table 1) is associated with an hnRNP H1 processed pseudogene that is present in the opposite orientation of newly integrated L1 sequence, and its formation resulted in a ∼14-kb deletion of genomic DNA (Fig. 2A). Both the L1 and the hnRNP H1 pseudogene sequences end in a poly(A) tail, and each inserted at a preferred L1 endonuclease cleavage site (5′-TTTT/G-3′ and 5′-TTTC/A-3′, respectively). Thus, it is likely that retrotransposition of the L1 and hnRNP H1 processed pseudogene occurred contemporaneously at a similar genomic location. Microhomology-mediated recombination between the two minus-strand cDNAs then led to the formation of the processed pseudogene/L1 chimera and the concomitant deletion of target site nucleotides. Consistent with this model, we observed three nucleotides of microhomology at the cDNA/cDNA junction.
FIG. 2.
cDNA additions at the 5′ genomic DNA/L1 junction. A. A processed pseudogene/L1 chimera. Insertion 19 was accompanied by the addition of a 5′-truncated cDNA copy of hnRNP H1 mRNA, which is located in the opposite transcriptional orientation of the L1. The undulating and straight gray lines represent hnRNP H1 mRNA and hnRNP H1 minus-strand cDNA, respectively. The undulating and straight black lines represent L1 mRNA and minus-strand L1 cDNA, respectively. Recombination between the resultant cDNAs resulted in a genomic deletion of approximately 14.1 kb. The structure of the hnRNP H1/L1 chimera is shown at the bottom of the figure. The striped and black rectangles indicate the integrated hnRNP H1 and L1 cDNAs, respectively. Double black lines indicate flanking genomic DNA. “An” indicates the poly(A) tail at the ends of the hnRNP H1 and L1 cDNAs. Small arrows indicate the positions of PCR primers used to characterize the insertion. B. A U6/L1 chimera. Insertion 20 was accompanied by the addition of a full-length U6 cDNA copy in the same transcriptional orientation as the L1. The undulating and straight black lines represent L1 mRNA and minus-strand L1 cDNA, respectively. The gray undulating line indicates U6 snRNA. The structure of the resultant chimera is shown at the bottom of the figure. The striped and black rectangles indicate the integrated U6 and L1 cDNAs, respectively. Double black lines indicate flanking genomic DNA. “An” indicates the poly(A) tail at the end of the L1 cDNA. Horizontal arrows indicate TSDs that flank the chimera.
The second processed pseudogene/L1 chimera (insertion 20, Table 1) contains 107 bp of U6 snRNA cDNA at the genomic DNA/5′ L1 junction. The U6 sequence is in the same transcriptional orientation as the L1, and the chimera integrated into a preferred L1 endonuclease cleavage site (5′-TTTT/A-3′) and is flanked by a 17-bp TSD (Fig. 2B). This structure most probably was generated by template switching of the L1 RT from L1 RNA to U6 snRNA during TPRT. Interestingly, recent in silico analyses have identified similar chimeric pseudogenes in the human genome draft sequence (7, 8). Moreover, we have determined that U6/L1 chimeras can be generated readily in HeLa cells (N. Gilbert, A. Doucet, and J. V. Moran, unpublished data). Thus, these data provide yet another example of how the cultured cell retrotransposition system can recapitulate processes that have occurred during the course of human genome evolution.
Chimeric L1s can be created by a variety of mechanisms. (i) Chimeric L1s associated with deletions.
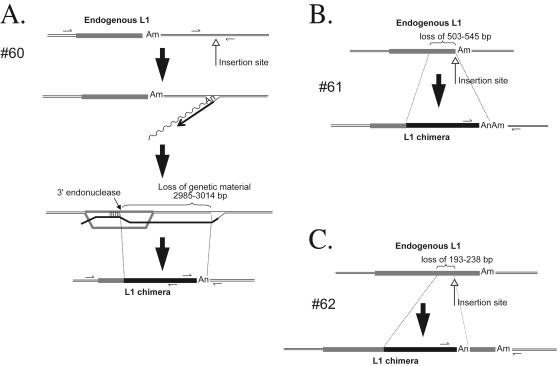
We identified three chimeric L1 sequences (insertions 60 to 62 [Table 1; Fig. 3]) that contain the 5′ end of an endogenous L1 fused to the corresponding position in our engineered L1 element. In each case, the insertion occurred into an L1 EN consensus cleavage site and generation of the chimera resulted in target site deletions that range in length from 193 bp to ∼3.1 kb. The chimeras probably formed during TPRT, when the nascent L1 cDNA underwent homeologous recombination with an endogenous L1 element located upstream of the insertion site. Indeed, this mechanism would be analogous to single-strand annealing (SSA) and would explain the concomitant deletion of target site nucleotides (Fig. 3A to C) (21, 71).
FIG. 3.
Chimeric L1 insertions associated with genomic deletions. Insertions 60, 61, and 62 (A, B, and C, respectively) resulted in the formation of chimeric L1s and the concomitant deletion of target site nucleotides. Each insertion initiated by TPRT then likely was joined to target site DNA by single-strand annealing (the process is depicted in full in panel A). The undulating and straight black lines represent L1 mRNA and minus-strand L1 cDNA, respectively. The gray rectangles represent endogenous L1s, and “Am” represents the position of their poly(A) tails. The upward-pointing open-headed arrow indicates the L1 integration site. The gray/black-shaded rectangle indicates the resultant chimeric L1. “An” indicates the poly(A) tail at the end of the newly integrated L1. Small horizontal arrows indicate the positions of PCR primers used to characterize the insertion. The sizes of the target site deletions (in base pairs) are indicated in each panel.
(ii) Chimeric L1s associated with intrachromosomal duplications.
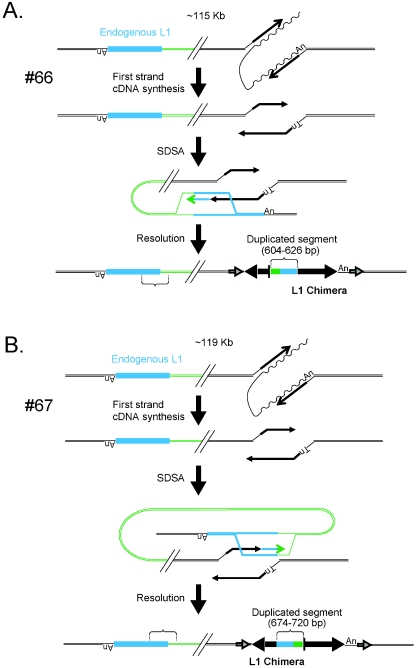
We identified a new category of insertions, which we have termed intrachromosomal duplications. The first (insertion 66 [Table 1; Fig. 4A]), integrated into a preferred L1 endonuclease cleavage site (5′-TTTT/G-3′), is flanked by a 15-bp TSD and contains a 604- to 626-bp insertion at the L1/L1 inversion junction. The sequence at the L1/L1 inversion junction most probably was copied from a chromosomal template located in the inverse orientation 115 kb upstream of the L1 integration site. The process is analogous to synthesis-dependent strand annealing (SDSA) and results in the capture of both non-L1 and endogenous L1 DNA at the L1/L1 inversion junction, the creation of a chimeric L1 element, and the inversion of the original chromosomal template sequence (Fig. 4A) (57).
FIG. 4.
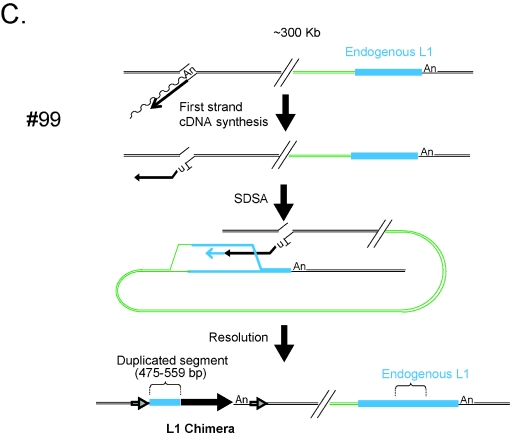
Chimeric L1 insertions associated with genomic duplications. Insertions 66, 67, and 99 resulted in the formation of chimeric L1s and the duplication of an intrachromosomal segment of DNA. Each insertion initiated by TPRT and likely was repaired by synthesis-dependent strand annealing. The undulating and straight black lines represent L1 mRNA and minus-strand L1 cDNA, respectively. In event 66 (A), twin priming resulted in cDNA synthesis using the 3′-OH present at the top and bottom strands of the target site. The bottom-strand cDNA then used an endogenous L1 located ∼115 kb upstream of the insertion site (light blue rectangle) as a template for SDSA. As a result the newly integrated L1 was joined to the endogenous L1 as well as its flanking DNA (green line). Resolution of the intermediate resulted in the inversion/duplication of a 604- to 626-bp segment of DNA and the formation of a chimeric L1. The entire insertion is flanked by a 15-bp TSD (indicated as in Fig. 1). In event 67 (B), twin priming resulted in two L1 cDNAs using the 3′-OH present at the top and bottom strands of the target site. The top-strand cDNA then used an endogenous L1 located ∼119 kb upstream of the insertion site (light blue rectangle) as a template for SDSA. As a result the newly integrated L1 cDNA was covalently joined to the endogenous L1 as well as its flanking DNA (green line). Resolution of the intermediate resulted in the duplication of a 674- to 720-bp segment of DNA and the formation of a chimeric L1. The entire insertion is flanked by a 14-bp TSD (indicated as in Fig. 1). In event 99 (C), TPRT resulted in the initiation of L1 cDNA synthesis. The resultant cDNA then used an endogenous L1 located ∼300 kb downstream of the insertion site (light blue rectangle) as a template for SDSA. As a result the newly integrated L1 cDNA was covalently joined to the endogenous L1 (green line). Resolution of the intermediate resulted in the duplication of a 475- to 559-bp segment of L1 DNA and the formation of a chimeric L1. The entire insertion is flanked by a 159-bp TSD (indicated as in Fig. 1). The large horizontal black arrows indicate the transcriptional orientation of new inserted L1 fragments. The vertical bar is a schematic of the junction of the inverted fragments. In the three cases, the presence of discriminating single nucleotide polymorphisms between the engineered L1 and the endogenous L1 was used to determine the size ranges of the duplicated L1 fragments.
The second (insertion 67, Table 1) originally was defined as a deletion (21). Additional characterization of the flanking sequence from subsequent PCR products proved that it instead represents an intrachromosomal duplication. In this instance, the L1 integrated into a preferred L1 endonuclease cleavage site (5′-TTTT/A-3′), is flanked by a 14-bp TSD, and contains a 674- to 720-bp insertion at the L1/L1 inversion junction. The sequence at the L1/L1 inversion junction most probably was copied from a chromosomal template located in an inverse orientation 119 kb upstream of the L1 integration site. Again, this process is analogous to SDSA and results in the creation of a chimeric L1 element with the capture of both endogenous L1 and non-L1 DNA at the L1/L1 inversion junction (Fig. 4B).
The third (insertion 99, Table 1) is a 5′-truncated chimera. In this instance, the L1 integrated into a preferred L1 endonuclease cleavage site (5′-TTTT/A-3′), is flanked by a 16-bp TSD, and contains a 475- to 559-bp insertion at the 5′ genomic DNA/L1 junction (Fig. 4C). The sequence at the 5′ genomic DNA/L1 junction most probably was copied from a chromosomal template located ∼300 kb downstream of the L1 integration site. Similarly to the cases observed above, SDSA results in the creation of a chimeric L1 element and the capture of endogenous L1 DNA at the 5′ genomic DNA/L1 junction (Fig. 4C).
Despite extensive efforts, we could not completely characterize insertion 63 (Table 1). The L1 clearly is a chimera because there are 42 sequence changes from L1.3 between nucleotide positions 3658 and 4900. Analysis of the HGWD did not reveal an endogenous L1 sequence with sequence identity to the segment of L1 DNA present in the chimera. Thus, we could not determine unambiguously whether the formation of the chimera was accompanied by a genomic rearrangement. However, the endogenous L1 sequence with the greatest identity to that in the chimera (>97% identical over ∼1,300 bp) is present on the same chromosome as insertion 63 and is located more than 366 kb upstream of the insertion site. Therefore, it remains possible that the chimera was formed by an SDSA-like mechanism.
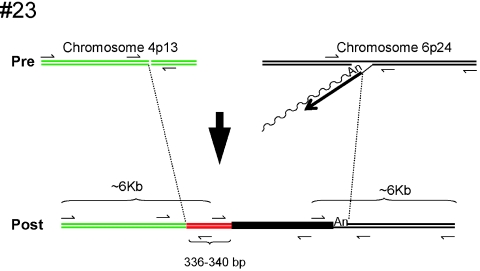
A possible interchromosomal translocation.
One event (insertion 23 [Table 1; Fig. 5]) could not be attributed to any of the groups described above. The insertion occurred at a preferred L1 EN consensus cleavage site (5′-TCTT/G-3′) located on chromosome 6. The genomic DNA/5′ L1 junction segment contains a ∼340-bp segment of DNA derived from chromosome 3. That sequence then is flanked by a sequence derived from chromosome 4. PCR analysis confirmed that the chromosome 6/chromosome 3, chromosome 3/chromosome 4, and chromosome 4/chromosome 6 junctions are not present in naïve HeLa cells. Additional characterization revealed that the 5′ end of the insertion is flanked by at least 6 kb of sequence derived from chromosome 4 and that its 3′ end is flanked by at least 6 kb of sequence derived from chromosome 6. Thus, these data suggest that an interchromosomal translocation occurred upon L1 retrotransposition and that a fragment of chromosome 3 somehow was captured as “filler DNA” during the process (Fig. 5; see reference 59 for possible mechanistic similarities).
FIG. 5.
A possible interchromosomal translocation generated upon L1 retrotransposition. Green and black lines indicate the structures of the preintegration sites on chromosomes 4 and 6, respectively. The insertion initiated at an integration site that map to chromosome 6 by endonuclease-dependent TPRT. The undulating and straight black lines represent L1 mRNA and minus-strand L1 cDNA, respectively. The resultant insertion is shown at the bottom of the figure. The filled black rectangle indicates the newly integrated L1. The red line indicates a 336- to 340-bp segment of DNA derived from chromosome 3. Small arrows indicate the positions of PCR primers used to characterize the insertion. PCR demonstrated that the respective junction sequences were not present in naïve HeLa cells and that the insertion is flanked by at least 6 kb of genomic DNA that maps to chromosomes 4 and 6, respectively.
Insertions at atypical L1 endonuclease cleavage sites.
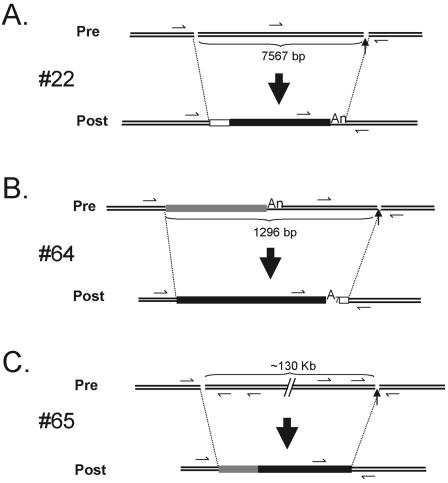
Three of 100 events integrated into sequences that are atypical L1 endonuclease cleavage sites and may represent insertions that initiated reverse transcription from nicks or double-stranded breaks in genomic DNA (56). Interestingly, each insertion apparently was associated with a genomic deletion that ranges in size from ∼1.3 kb to potentially >130 kb (Fig. 6).
FIG. 6.
Insertions at atypical integration sites. Insertions 22, 64, and 65 (A, B, and C, respectively) occurred at sequences that do not resemble an L1 EN consensus cleavage site. Each event resulted in the deletion of target site nucleotides, and the sizes of the deletions are indicated below the respective preintegration sites. A 55- to 59-bp MaLR sequence is located at the 5′ genomic DNA/L1 junction in insertion 22 (depicted by a white box). An endogenous L1 (gray rectangle) was completely replaced by the newly integrated L1 in insertion 64, and three untemplated base pairs are present at the L1/3′ genomic DNA junction. A 292- to 527-bp segment of endogenous L1 (gray rectangle) is located at the 5′ genomic DNA/L1 junction in insertion 65. The origin of this DNA segment remains unknown. Small arrows indicate the positions of PCR primers used to characterize the insertions.
The first (insertion 22, Table 1), integrated into an atypical sequence (5′-AGGT/G-3′), contains a 55- to 59-bp insertion at the genomic DNA/5′ L1 junction that is derived from the long terminal repeat (LTR) of an ancient MaLR retrotransposon and is associated with a 7,567-bp deletion of genomic DNA (Fig. 6A). PCR experiments proved that the MaLR sequence was not present at the empty site prior to insertion. The genomic origin of the MaLR sequence with 100% identity is from chromosome 17 (HGWD, assembly May 2004) and was either “captured” at the genomic DNA/5′ L1 junction during TPRT or copied from another chromosomal template.
The second (insertion 64, Table 1) integrated into an atypical sequence (5′-GTGA/G-3′) and contains a short poly(A) tail of 7 nucleotides as well as a 3-nucleotide addition (5′-GTG-3′) at the 3′ L1/genomic DNA junction. The insertion resulted in the formation of a chimeric L1 and is associated with a 1,296-bp deletion of genomic DNA. Interestingly, the chimeric L1 most probably was formed by the previously mentioned SSA-type mechanism (21, 71). The only difference is that the new L1 sequence has totally replaced the endogenous L1 sequence present at the preintegration site (Fig. 6B).
The last event (insertion 65, Table 1), integrated at an atypical sequence (5′-GAGC/T-3′, 5′-AGCT/G-3′, or 5′-GCTG/C-3′), lacks a poly(A) tail, contains a 292- to 527-bp insertion of an endogenous L1 at the genomic DNA/5′ L1 junction, and is associated with a genomic deletion that may be >131 kb in length (Fig. 6C). Moreover, the genomic DNA at the 5′-end junction of the insertion presents a short duplication of 16 nucleotides. PCR demonstrated that the 292- to 527-bp endogenous L1 is not present at the preintegration site. BLAT revealed that this L1 segment could originate from an intronic region of the Rabconnectin-3 gene (accession no. Q8TDJ6), which is located on chromosome 15 (position: chr15, 49662615 to 49664907). The exact mechanism for how this insertion was formed requires further study.
Intra-L1 rearrangements.
In three instances, we observed intra-L1 rearrangements that resulted in a small deletion or duplication of L1 sequence (see Fig. S2 in the supplemental material). In every case, PCR confirmed that the deletion or duplication was present in HeLa chromosomal DNA at the postintegration site and that it did not arise during the recovery process (data not shown). The first (insertion 68, Table 1) contains a 246-bp deletion between positions 5019 and 5266 of L1 ORF2. The deletion occurred within a 5-bp direct repeat (5′-CAAAA-3′; see Fig. S2B in the supplemental material). Interestingly, similar internal deletions can be found for a cohort of L1s in the HGWD; however, it remained unclear whether these structures were formed during retrotransposition. Our data strongly suggest that internal deletions can occur during retrotransposition. The second (insertion 7, Table 1) contains a 6- to 11-bp duplication at the 5′ end of L1 (see Fig. S2C in the supplemental material). The third instance (insertion 60, Table 1) contains two duplications near the 3′ L1/genomic DNA junction. The first is 267 to 270 bp in size, whereas the second is 9 to 15 bp in size (see Fig. S2D in the supplemental material). As is the case for insertion 68, there is microhomology at each duplication junction (four nucleotides for case 7, AATG, and three nucleotides for both junctions of case 60, CAA or TAA). Those regions of homology might have been involved in the rearrangement during reverse transcription.
L1 RT accuracy.
We did not find any significant structural differences between the retrotransposition events generated from the four RC-L1s. The average size of the recovered insertions is ∼2,950 bp for L1.2A, ∼4,200 bp for LRE-2, ∼3,575 bp for L1.3, and ∼3,165 bp for L1RP. Thus, retrotransposition efficiency does not seem to be strictly correlated with L1 RT processivity. To determine the L1 RT misincorporation rate, we compared 98,758 bp of L1 sequence derived from the recovered insertion to the L1 progenitor sequence. Since the ColE1 and Neo sequences are subject to selective pressure, they were excluded from the analysis. We also excluded regions of retrotransposed L1s that contain “clustered” nucleotide changes, as they may represent “short patch” gene conversion tracts that do not result from base misincorporation. We detected 15 single-base changes between the retrotransposed L1 and its progenitor, allowing us to estimate the L1 RT misincorporation error rate as 1 error/6,584 bp (i.e., 15/98,758 bp). Using the binomial distribution, we estimate that ∼40% of full-length retrotransposed L1s will be faithful copies of the progenitor element, whereas ∼37% will contain one base change (Fig. 7). Unlike the case for retroviruses, we did not observe any base preferences in mutation (27).
DISCUSSION
In sum, we have used a high-throughput system to characterize de novo L1 retrotransposition events in transformed human cells before they are blurred by selective pressures that occur during evolution. These data build on previous analyses (21, 56, 72) and provide the most comprehensive data set to date for studying the fate of retrotransposition intermediates in cultured cells. As a result of this study, we have determined that retrotransposition can result in the alteration of L1 structure and occasionally is associated with various forms of genomic instability.
Why is L1 retrotransposition often associated with genetic instability? We propose that there are at least two non-mutually exclusive mechanisms to resolve L1 retrotransposition intermediates in HeLa cells. The first pathway, which we term conventional retrotransposition, involves the initiation of TPRT at a consensus L1 endonuclease cleavage site. After the onset of minus-strand L1 cDNA synthesis, top-strand cleavage provides a primer for the initiation of plus-strand L1 cDNA synthesis. Repair DNA polymerases then can repair the resultant “gap,” generating a newly integrated L1 containing the hallmarks of TPRT (see reference 21).
Conventional retrotransposition is analogous to the mechanism of R2Bm retrotransposition and can account for the structural features associated with full-length L1s, inversion/deletion L1s, and the cohort of 5′-truncated L1s that have canonical structures (47). For full-length L1s, second-strand cleavage may occur after the completion of minus-strand L1 cDNA synthesis. For conventional inversion/deletion and simple 5′-truncated L1s, second-strand cleavage may occur before the completion of minus-strand cDNA synthesis, providing an opportunity for the minus-strand L1 cDNA to anneal to complementary nucleotides at the 3′ overhang present at the target site. The cDNA/target site pairing may enable premature initiation of plus-strand L1 cDNA synthesis, leading to incomplete L1 replication. This represents a refinement of previous models and could account for microhomology observed at the 5′ genomic DNA/L1 junctions of both inversion/deletion and some 5′-truncated L1s (21, 49, 65, 72, 74).
The second pathway, which we term abortive retrotransposition, likely involves the premature termination of minus-strand L1 cDNA synthesis, leading to a Y-branched intermediate containing an incompletely replicated L1 cDNA joined to target site DNA. Premature termination of minus-strand L1 cDNA synthesis could occur if the L1 RT prematurely disassociates from its RNA template. Consistently, Farley et al. have provided evidence that the reduced retrotransposition efficiency of L1.2A in vitro may be due to mutations that decrease L1 RT processivity (18). However, despite exhibiting relatively low retrotransposition efficiency in vitro, we found that LRE-2 was able to generate full-length retrotransposition events at an efficiency comparable to those of both L1.3 and L1RP and to present a similar average size of de novo inserts. These data argue that the reduced retrotransposition efficiency observed for LRE-2 is not due simply to a decrease in L1 RT processivity. Instead, although it is possible that other mechanisms also act to cause the 5′ truncations associated with L1 insertions, we propose that a host repair process (or processes) may act to disassociate the L1 RT from its nascent cDNA. Thus, L1 integration may represent a host/parasite battleground, where L1 is in a “race” to complete cDNA synthesis before being acted upon by host defense mechanisms that counteract retrotransposition. Consistent with this scenario, Eickbush and colleagues have found that the Bombyx mori R2Bm retrotransposon has evolved a highly processive reverse transcriptase activity (3).
What is the fate of the proposed Y-branched intermediate generated by abortive L1 retrotransposition? The major outcome likely is 5′ truncation, which could occur if the minus-strand L1 cDNA undergoes microhomology-mediated DNA repair with complementary sequences at the target site. Moreover, we have provided evidence that the truncated minus-strand L1 cDNA can complete integration using other DNA repair pathways. For example, the formation of chimeric L1s and the concomitant deletion of target site nucleotides (Fig. 3) could occur if the minus-strand L1 cDNA undergoes repair by single-strand annealing (21, 71). The formation of chimeric L1s also could occur if minus- or plus-strand L1 cDNA undergoes repair by a synthesis-dependent strand annealing mechanism using an intrachromosomal (or perhaps interchromosomal) nonallelic L1 as a repair template (Fig. 4 and 5) (57). Interestingly, repair through an SDSA-like mechanism can result in intrachromosomal duplications and perhaps interchromosomal translocations and illustrates yet another means by which an L1 retrotransposition-associated process can shuffle DNA to new genomic locations. Indeed, this mechanism could explain the evolution of new mouse L1 subfamilies as well as the “swapping” of promoters that occurs for non-LTR retrotransposons in certain avian species (26, 29, 64). In principle, chimeric L1s also can be formed by RNA-mediated recombination events; however, our studies show that template switching of the L1 RT between heterologous L1 RNAs is relatively rare (Gilbert, Doucet, and Moran, unpublished data).
We also characterized three L1 insertions that integrated into an atypical L1 endonuclease cleavage site. These insertions were structurally similar to EN-independent retrotransposition events observed in Chinese hamster ovary cell lines, and each event was accompanied by a deletion of target site nucleotides that ranged in size from 1.3 kb to possibly >130 kb. Thus, it is tempting to speculate that these events initiated TPRT from an existing genomic DNA lesion by an EN-independent mechanism (56). If so, the chimeric L1 observed in insertion 64, which was likely resolved by SSA, suggests that EN-dependent and EN-independent L1 retrotransposition events can complete integration via similar repair pathways.
How second (i.e., top)-strand cleavage of target site DNA occurs merits further study. Although 97 retrotransposition events initiated at an L1 EN consensus cleavage site (5′-TTTT/A), the analysis of L1s flanked by canonical TSDs did not allow the definition of a strict consensus second-strand cleavage site. When the analysis was restricted to the 19 inversion/deletion and inversion/duplication L1s, we did observe a weak preference for the sequence 5′-TYTN/R, which is consistent with previous studies conducted by Jurka (36). Thus, there may be some specificity for top-strand cleavage when retrotransposition events are formed by “twin priming.” That being said, the following possibilities remain: (i) L1 encodes an unconventional endonuclease that exhibits site-specific bottom-strand cleavage activity but has a relaxed (or perhaps) no sequence specificity for top-strand cleavage, (ii) L1 encodes a second nuclease activity that is required for top-strand cleavage (11), or (iii) host factors act to cleave top strand. Finally, it is formally possible that the retrotransposition machinery initially recognizes a nicked DNA substrate and that L1 EN actually mediates second-strand cleavage at a consensus cleavage site.
How do L1 retrotransposition events generated in cultured cells compare with those represented in the human genome working draft sequence? Besides the many above-mentioned similarities, we also observed the formation of chimeric U6/L1 retrotransposons in HeLa cells, increasing the repertoire of noncoding RNAs that still are being mobilized by the L1 retrotransposition machinery. U6/L1 chimeric elements formed many independent times over the course of genome evolution (7, 8). Thus, these data once again show that the cultured cell retrotransposition assay can recapitulate events that occurred during the course of human genome evolution.
There also are notable differences between retrotransposition events generated in cultured cells and those present in the HGWD. First, many L1 retrotransposition events generated in cultured cells were flanked by unusually long TSDs. Although it remains possible that recombination between the long TSDs will render these events unstable during genome evolution, it is more likely that peculiarities associated with HeLa cells (e.g., a less compact chromatin structure at certain chromosomal loci) facilitate their formation. Second, the frequency of L1-associated genomic rearrangements appears to be more prevalent in cultured cells. Again differences in the intracellular environment of HeLa cells (e.g., the lack of p53 function or occult mutations in DNA repair proteins) may provide a permissive milieu for their formation. However, it is possible that some large deletions and the formation of possible interchromosomal translocations may be subject to negative selective pressure over evolutionary time and thus are present at reduced frequency in the HGWD if at all. Finally, it is noteworthy that the unusual structural features associated with chimeric L1s and EN-independent insertions present difficult assembly problems, which may exclude their presence in the HGWD.
Are L1-associated genomic rearrangements limited to transformed cells? Although this scenario remains formally possible, genomic rearrangements can accompany L1, Alu, and simple poly(A) insertions in vivo (9, 20, 28, 37, 41, 42, 60, 67, 70, 75). Thus, it is unlikely that these arrangements are peculiar to HeLa cells. Instead, we propose that differences in the cellular environments between transformed and germ cells may cause a shift in balance between conventional and abortive L1 retrotransposition (Fig. 8). For example, the lack of p53 function increase in transformed cells may allow the Y-branched intermediates formed during abortive retrotransposition to utilize alternative DNA pathways to complete integration, leading to the observed genomic rearrangements. Alternatively, it is possible that abortive retrotransposition intermediates are not well tolerated in “normal” cells and may be subject to repair pathways, which remove the L1 cDNA from target site DNA.
FIG. 8.
Competing pathways for L1 retrotransposition in cultured human cells. L1 retrotransposition can be initiated in two ways. The major pathway uses L1 EN to initiate TPRT (left panel). A secondary pathway could use a DNA lesion to initiate reverse transcription (right panel) (56). We propose that the resultant cDNA intermediate can be resolved using distinct DNA repair pathways (see Discussion). “Canonical” retrotransposition (left panel) results in the formation of L1 insertions with standard hallmarks of TPRT. “Abortive” retrotransposition (right panel) can lead to a variety of structures, depending on the DNA repair pathway that is utilized to resolve the L1 cDNA intermediates. The balance between “conventional” and “abortive” retrotransposition likely depends on the cellular milieu. Conventional retrotransposition may be more apparent in germ cells (depicted by a large horizontal gray arrow over a thin light gray arrow), whereas “abortive” retrotransposition may be more apparent in transformed cultured cells (depicted with equally sized gray horizontal arrows).
In closing, it is noteworthy that the ability to repair a TPRT-initiated cDNA intermediate by DNA-based recombination is not peculiar to L1s. For example, a similar mechanism is responsible for the asymmetric coconversion patterns observed during group II intron mobility and an intramolecular SDSA-like gene conversion mechanism probably is utilized during abortive group II intron retrohoming (12, 16, 55). Moreover, recent data indicate group II introns also have the ability to initiate retrotransposition from preexisting genomic lesions in vivo (77). Thus, our findings further highlight mechanistic similarities that are shared among this diverse family of retroelements.
Supplementary Material
Acknowledgments
We thank members of the University of Michigan DNA Sequencing Core for help with sequencing and José Luis Garcia Perez, Deanna Kulpa, and Amy Hulme for critically evaluating the manuscript.
This work was supported in part by grants to J.V.M. from the W. M. Keck Foundation and the National Institutes of Health (GM60518). The University of Michigan Cancer Center helped defray some of the DNA sequencing costs.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Athanikar, J. N., R. M. Badge, and J. V. Moran. 2004. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32:3846-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibillo, A., and T. H. Eickbush. 2002. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J. Biol. Chem. 277:34836-34845. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D. 2003. The unusual phylogenetic distribution of retrotransposons: a hypothesis. Genome Res. 13:1975-1983. [DOI] [PubMed] [Google Scholar]
- 5.Boissinot, S., P. Chevret, and A. V. Furano. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17:915-928. [DOI] [PubMed] [Google Scholar]
- 6.Brouha, B., J. Schustak, R. M. Badge, S. Lutz-Prigge, A. H. Farley, J. V. Moran, and H. H. Kazazian, Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100:5280-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzdin, A., E. Gogvadze, E. Kovalskaya, P. Volchkov, S. Ustyugova, A. Illarionova, A. Fushan, T. Vinogradova, and E. Sverdlov. 2003. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzdin, A., S. Ustyugova, E. Gogvadze, T. Vinogradova, Y. Lebedev, and E. Sverdlov. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics 80:402-406. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, M. L., A. M. Roy-Engel, S. V. Nguyen, A. H. Salem, E. Vogel, B. Vincent, J. Myers, Z. Ahmad, L. Nguyen, M. Sammarco, W. S. Watkins, J. Henke, W. Makalowski, L. B. Jorde, P. L. Deininger, and M. A. Batzer. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311:17-40. [DOI] [PubMed] [Google Scholar]
- 10.Cost, G. J., and J. D. Boeke. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37:18081-18093. [DOI] [PubMed] [Google Scholar]
- 11.Cost, G. J., Q. Feng, A. Jacquier, and J. D. Boeke. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson, L., S. Connell, H. R. Huang, R. M. Henke, L. Liu, and P. S. Perlman. 2004. Abortive transposition by a group II intron in yeast mitochondria. Genetics 168:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dombroski, B. A., S. L. Mathias, E. Nanthakumar, A. F. Scott, and H. H. Kazazian, Jr. 1991. Isolation of an active human transposable element. Science 254:1805-1808. [DOI] [PubMed] [Google Scholar]
- 14.Dombroski, B. A., A. F. Scott, and H. H. Kazazian, Jr. 1993. Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl. Acad. Sci. USA 90:6513-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ergun, S., C. Buschmann, J. Heukeshoven, K. Dammann, F. Schnieders, H. Lauke, F. Chalajour, N. Kilic, W. H. Stratling, and G. G. Schumann. 2004. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 279:27753-27763. [DOI] [PubMed] [Google Scholar]
- 16.Eskes, R., L. Liu, H. Ma, M. Y. Chao, L. Dickson, A. M. Lambowitz, and P. S. Perlman. 2000. Multiple homing pathways used by yeast mitochondrial group II introns. Mol. Cell. Biol. 20:8432-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fanning, T., and M. Singer. 1987. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 15:2251-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farley, A. H., E. T. Luning Prak, and H. H. Kazazian, Jr. 2004. More active human L1 retrotransposons produce longer insertions. Nucleic Acids Res. 32:502-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, Q., J. V. Moran, H. H. Kazazian, Jr., and J. D. Boeke. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905-916. [DOI] [PubMed] [Google Scholar]
- 20.Garvey, S. M., C. Rajan, A. P. Lerner, W. N. Frankel, and G. A. Cox. 2002. The muscular dystrophy with myositis (mdm) mouse mutation disrupts a skeletal muscle-specific domain of titin. Genomics 79:146-149. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, N., S. Lutz-Prigge, and J. V. Moran. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315-325. [DOI] [PubMed] [Google Scholar]
- 22.Goodier, J. L., E. M. Ostertag, K. A. Engleka, M. C. Seleme, and H. H. Kazazian, Jr. 2004. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 13:1041-1048. [DOI] [PubMed] [Google Scholar]
- 23.Gorbunova, V., and A. A. Levy. 1997. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25:4650-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorbunova, V. V., and A. A. Levy. 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4:263-269. [DOI] [PubMed] [Google Scholar]
- 25.Grimaldi, G., J. Skowronski, and M. F. Singer. 1984. Defining the beginning and end of KpnI family segments. EMBO J. 3:1753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas, N. B., J. M. Grabowski, J. North, J. V. Moran, H. H. Kazazian, and J. B. Burch. 2001. Subfamilies of CR1 non-LTR retrotransposons have different 5′UTR sequences but are otherwise conserved. Gene 265:175-183. [DOI] [PubMed] [Google Scholar]
- 27.Harris, R. S., A. M. Sheehy, H. M. Craig, M. H. Malim, and M. S. Neuberger. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immunol. 4:641-643. [DOI] [PubMed] [Google Scholar]
- 28.Hayakawa, T., Y. Satta, P. Gagneux, A. Varki, and N. Takahata. 2001. Alu-mediated inactivation of the human CMP- N-acetylneuraminic acid hydroxylase gene. Proc. Natl. Acad. Sci. USA 98:11399-11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayward, B. E., M. Zavanelli, and A. V. Furano. 1997. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics 146:641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hohjoh, H., and M. F. Singer. 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15:630-639. [PMC free article] [PubMed] [Google Scholar]
- 31.Hohjoh, H., and M. F. Singer. 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16:6034-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes, S. E., B. A. Dombroski, C. M. Krebs, C. D. Boehm, and H. H. Kazazian, Jr. 1994. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat. Genet. 7:143-148. [DOI] [PubMed] [Google Scholar]
- 33.Holmes, S. E., M. F. Singer, and G. D. Swergold. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267:19765-19768. [PubMed] [Google Scholar]
- 34.Hutchison, C. A., S. C. Hardies, D. D. Loeb, W. R. Shehee, and M. H. Edgell. 1989. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome, p. 593-617. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. ASM Press, Washington, D.C.
- 35.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 36.Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 94:1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kass, D. H., M. A. Batzer, and P. L. Deininger. 1995. Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol. Cell. Biol. 15:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimberland, M. L., V. Divoky, J. Prchal, U. Schwahn, W. Berger, and H. H. Kazazian, Jr. 1999. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 8:1557-1560. [DOI] [PubMed] [Google Scholar]
- 40.Kirik, A., S. Salomon, and H. Puchta. 2000. Species-specific double-strand break repair and genome evolution in plants. EMBO J. 19:5562-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima, T., K. Nakajima, and K. Mikoshiba. 2000. The disabled 1 gene is disrupted by a replacement with L1 fragment in yotari mice. Brain Res. Mol. Brain Res. 75:121-127. [DOI] [PubMed] [Google Scholar]
- 42.Kutsche, K., B. Ressler, H. G. Katzera, U. Orth, G. Gillessen-Kaesbach, S. Morlot, E. Schwinger, and A. Gal. 2002. Characterization of breakpoint sequences of five rearrangements in L1CAM and ABCD1 (ALD) genes. Hum. Mutat. 19:526-535. [DOI] [PubMed] [Google Scholar]
- 43.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 44.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, Y., and A. S. Waldman. 2001. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics 158:1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, Y., and A. S. Waldman. 2001. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 29:3975-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 48.Lutz, S. M., B. J. Vincent, H. H. Kazazian, Jr., M. A. Batzer, and J. V. Moran. 2003. Allelic heterogeneity in LINE-1 retrotransposition activity. Am. J. Hum. Genet. 73:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin, S. L., and F. D. Bushman. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin, S. L., W.-L. P. Li, A. V. Furano, and S. Boissinot. The structures of mouse and human L1 elements reflect their insertion mechanism. Mol. Genet. Genomics, in press. [DOI] [PubMed]
- 51.Mathias, S. L., A. F. Scott, H. H. Kazazian, Jr., J. D. Boeke, and A. Gabriel. 1991. Reverse transcriptase encoded by a human transposable element. Science 254:1808-1810. [DOI] [PubMed] [Google Scholar]
- 52.Moran, J. V., R. J. DeBerardinis, and H. H. Kazazian, Jr. 1999. Exon shuffling by L1 retrotransposition. Science 283:1530-1534. [DOI] [PubMed] [Google Scholar]
- 53.Moran, J. V., and N. Gilbert. 2002. Mammalian LINE-1 retrotransposons and related elements, p. 836-869. In A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 54.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 55.Moran, J. V., S. Zimmerly, R. Eskes, J. C. Kennell, A. M. Lambowitz, R. A. Butow, and P. S. Perlman. 1995. Mobile group II introns of yeast mitochondrial DNA are novel site-specific retroelements. Mol. Cell. Biol. 15:2828-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrish, T. A., N. Gilbert, J. S. Myers, B. J. Vincent, T. D. Stamato, G. E. Taccioli, M. A. Batzer, and J. V. Moran. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31:159-165. [DOI] [PubMed] [Google Scholar]
- 57.Nassif, N., J. Penney, S. Pal, W. R. Engels, and G. B. Gloor. 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14:1613-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ostertag, E. M., and H. H. Kazazian, Jr. 2001. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 11:2059-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paques, F., W. Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18:2045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salem, A. H., G. E. Kilroy, W. S. Watkins, L. B. Jorde, and M. A. Batzer. 2003. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 20:1349-1361. [DOI] [PubMed] [Google Scholar]
- 61.Sargent, R. G., M. A. Brenneman, and J. H. Wilson. 1997. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 17:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sargent, R. G., J. L. Meservy, B. D. Perkins, A. E. Kilburn, Z. Intody, G. M. Adair, R. S. Nairn, and J. H. Wilson. 2000. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 28:3771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sassaman, D. M., B. A. Dombroski, J. V. Moran, M. L. Kimberland, T. P. Naas, R. J. DeBerardinis, A. Gabriel, G. D. Swergold, and H. H. Kazazian, Jr. 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16:37-43. [DOI] [PubMed] [Google Scholar]
- 64.Saxton, J. A., and S. L. Martin. 1998. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J. Mol. Biol. 280:611-622. [DOI] [PubMed] [Google Scholar]
- 65.Schwarz-Sommer, Z., L. Leclerq, E. Goebel, and H. Saedler. 1987. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 6:3873-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott, A. F., B. J. Schmeckpeper, M. Abdelrazik, C. T. Comey, B. O'Hara, J. P. Rossiter, T. Cooley, P. Heath, K. D. Smith, and L. Margolet. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segal, Y., B. Peissel, A. Renieri, M. de Marchi, A. Ballabio, Y. Pei, and J. Zhou. 1999. LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am. J. Hum. Genet. 64:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skowronski, J., T. G. Fanning, and M. F. Singer. 1988. Unit-length Line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 8:1385-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smit, A. F. 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6:743-748. [DOI] [PubMed] [Google Scholar]
- 70.Su, L. K., G. Steinbach, J. C. Sawyer, M. Hindi, P. A. Ward, and P. M. Lynch. 2000. Genomic rearrangements of the APC tumor-suppressor gene in familial adenomatous polyposis. Hum. Genet. 106:101-107. [DOI] [PubMed] [Google Scholar]
- 71.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Symer, D. E., C. Connelly, S. T. Szak, E. M. Caputo, G. J. Cost, G. Parmigiani, and J. D. Boeke. 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110:327-338. [DOI] [PubMed] [Google Scholar]
- 73.Tang, X., Y. Nakata, H. O. Li, M. Zhang, H. Gao, A. Fujita, O. Sakatsume, T. Ohta, and K. Yokoyama. 1994. The optimization of preparations of competent cells for transformation of E. coli. Nucleic Acids Res. 22:2857-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voliva, C. F., S. L. Martin, C. A. Hutchison III, and M. H. Edgell. 1984. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J. Mol. Biol. 178:795-813. [DOI] [PubMed] [Google Scholar]
- 75.Wang, T., I. Lerer, Z. Gueta, M. Sagi, L. Kadouri, T. Peretz, and D. Abeliovich. 2001. A deletion/insertion mutation in the BRCA2 gene in a breast cancer family: a possible role of the Alu-polyA tail in the evolution of the deletion. Genes Chromosomes Cancer 31:91-95. [DOI] [PubMed] [Google Scholar]
- 76.Wei, W., T. A. Morrish, R. S. Alisch, and J. V. Moran. 2000. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 284:435-438. [DOI] [PubMed] [Google Scholar]
- 77.Zhong, J., and A. M. Lambowitz. 2003. Group II intron mobility using nascent strands at DNA replication forks to prime reverse transcription. EMBO J. 22:4555-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.